Pereira and Arraiano preview work from the Wolin laboratory that uncovers an important regulatory role for the RNA exosome in stem cell development.
Abstract
In this issue, Belair et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201811148) show that, together with a complex network of transcription factors and chromatin modifiers, the RNA exosome regulates embryonic stem cell (ESC) differentiation and pluripotency.
Human embryonic stem cells (hESCs) are prototypical pluripotent cells derived from the inner cell mass of blastocyst-stage embryos, which have the capacity of self-renewal and can differentiate into all three embryonic germ layers (endoderm, mesoderm, and ectoderm; 1). The characteristics of ESCs are quite promising for clinical therapies, and therefore their differentiation process has been intensively studied (2). Their differentiation from pluripotency requires extensive rewiring of the cellular state, involving changes in cell morphology and distinct metabolic, transcriptional, and epigenetic states. During this process, there is a balanced expression of a complex network of transcription factors (TFs) such as NANOG, OCT4, and SOX2, as well as the action of epigenetic mechanisms (3). The singularity of ESCs relies on their “open” chromatin state, allowing the rapid switch of transcriptional programs upon differentiation (4). In addition, ESC chromatin has unique histone modification patterns with “bivalent” domains containing both repressive and activating histone marks, which may silence many developmentally regulated genes, keeping them poised for activation as cells enter various differentiation pathways (4, 5). The role of TFs in controlling the identity of hESCs has been well characterized, but the mechanisms by which post-transcriptional processes regulate the fate of hESCs are still largely unknown.
Convincing evidence points to the importance of RNA turnover in the regulation of the cellular levels of mRNAs that encode certain TFs with relevance in inducing pluripotency as well as in differentiating ESCs (6). Moreover, certain miRNAs and RNA-binding proteins seem to control the expression of a wide range of genes in these types of cells. It has also been demonstrated that specific cytoplasmic RNA decay pathways, namely nonsense-mediated RNA decay (NMD), have a central role in the regulation of ESC differentiation, given that NMD enhances mesoderm formation and limits definitive endoderm formation (7). In addition to NMD, other RNA decay pathways can be important in the processing of immature transcripts and removal of faulty and/or unstable RNAs (8). The eukaryotic RNA exosome is a versatile RNA-degradation machinery that cleaves RNAs from their 3′ termini, playing critical roles in the maturation, surveillance, and quality control of RNAs within the cell (9). Some studies have indicated that the exosome is involved in the regulation of the expression levels of specific mRNAs involved in cell proliferation, differentiation, and development (10, 11). However, little is known about the involvement of the RNA exosome in the regulation of hESC pluripotency as well as the post-transcriptional surveillance pathways that degrade differentiation-related RNAs. This scientific question is addressed in this issue by Belair et al., who show that the RNA exosome restrains hESC differentiation into the endoderm, mesoderm, and ectoderm (12).
To accomplish this goal, Belair et al. set up an experimental methodology based on (1) the generation of EXOSC3-depleted hESC lines (RNA exosome EXOSC3 subunit depletion destabilizes the catalytic subunits EXOSC10 and DIS3) using shRNAs; (2) the application of siRNAs directed against other sites on EXOSC3 mRNA in hESC lines (here designated by siEXOSC3-treated cells); and (3) the generation of EXOSC3-depleted hESCs differentiated into embryoid bodies (EBs), which are three-dimensional structures that contain all three germ layers. Consistent with earlier studies on EXOSC3-depleted human HeLa cells (13), Belair et al. found that EXOSC3-depleted hESC proliferation rates were unaffected, and, in addition, no changes were observed in the levels of mRNA expression encoding pluripotency markers (OCT4, NANOG, and SOX2). However, the authors observed that upon differentiation, both siEXOSC3-treated cells and EXOSC3-depleted EBs presented increased levels of mRNAs that are markers of early ectoderm (DLX5, PAX6, and ZIC1), mesoderm (MIXL1, EOMES, and TBXT), and endoderm (GATA4 and SOX17). Together, these data demonstrate that exosome is involved in hESC differentiation in vitro and help to explain why elevated exosome levels are important for maintaining pluripotency.
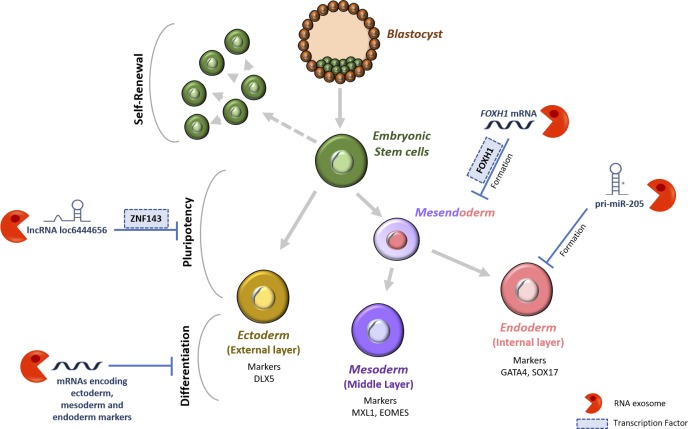
Belair et al. (12) identified which differentiation/pluripotency-related RNAs are targets of the exosome and how the exosome could contribute to differentiation and pluripotency by modulating RNA levels. Using high-throughput sequencing and immunoprecipitation, Belair et al. identified new RNAs whose expression levels are increased in EXOSC3-depleted cells when compared with the control cells. Among the identified RNAs that act as substrates of the exosome were transcripts derived from L1 LINE; pri-miR-205 involved in the differentiation of ESCs into the endoderm; long non-coding RNA (lncRNA) loc6444656 that encodes a TF (zinc finger protein 143 [ZNF143]) important for hESC pluripotency; and specific subsets of mRNAs encoding or regulating key developmental regulators (Fig. 1).
Figure 1.
Regulation of hESC differentiation and pluripotency by the RNA exosome. Belair et al. show that the RNA exosome acts by regulating the cellular levels of several RNA classes (mRNAs, lncRNAs, and pri-miRNAs) and, consequently, the levels of certain TFs with relevance in inducing pluripotency and differentiation in hESCs. Pri-miR-205, primary miRNA transcript 205.
The FOXH1 (Forkhead box protein H1) mRNA was identified by Belair et al. (12) as an important player that could account for the repression of hESC differentiation by exosomes. FOXH1 is a crucial TF for genes such as NODAL, LEFTY1, CER1, and MIXL1, which are involved in the formation of the mesendoderm (ME), the precursor to mesoderm and endoderm. As expected, Belair et al. observed that the exosome modulated ME formation by degrading FOXH1 mRNA, and the expression levels of ME markers increased in EXOSC3-depleted EBs in comparison to the control EBs.
The current study provides evidence that exosome restrains hESC differentiation by a mechanism that includes reduction of the expression of FOXH1 mRNA. Belair et al. suggest an additional role for the exosome in maintaining hESC pluripotency by selective degradation of RNAs that encode key developmental regulators (12). A pri-miRNA was identified as a target of the exosome (i.e., the EXOSC10-containing exosomes affect miR-205 levels by reducing the pri-miRNA). These findings suggest that increased levels of miR-205 could contribute to endoderm formation. The 3′ to 5′ exoribonuclease EXOSC10 (also called hRRP6) is the major catalytic subunit of the exosome in nucleoli, whereas Dis3 (also known as hRRP44) is the main catalytic subunit of the nucleoplasmic exosome, being also present in the cytoplasm (8, 9, 13). Dis3, Dis3L1, and Dis3L2 belong to the same RNB/RNase II family of exoribonucleases that has been linked to human diseases (8, 13, 14). It is interesting to point out that knockdown of cytoplasmic Dis3L2 in mouse ESCs leads to the stabilization of pre-let-7, and the major function of let-7 genes seems to be the promotion of terminal differentiation and tumor suppression (15).
In summary, the study by Belair et al. (12) supports the idea that RNA decay pathways make key contributions to regulate the fate of hESCs. The work also sheds light on an important connection between exosome, differentiation, and pluripotency. This is an important breakthrough with potential translational application since it could lead to new RNA-based clinical therapies.
Acknowledgments
This work was financially supported by project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular), which is funded by Fundo Europeu de Desenvolvimento Regional (FEDER) through COMPETE2020 (Programa Operacional Competitividade e Internacionalização), and by Fundação para a Ciência e a Tecnologia (Portugal), including grant PTDC/BIA-MIC/1399/2014 to C.M. Arraiano (which also supported a post-doctoral fellowship to P. Pereira).
The authors declare no competing financial interests.
References
- 1.Thomson J.A., et al. Science. 1998 doi: 10.1126/science.282.5391.1145. [DOI] [Google Scholar]
- 2.Hu J., and Wang J. Clin. Transplant. 2019 doi: 10.1111/ctr.13573. [DOI] [Google Scholar]
- 3.De Los Angeles A., et al. Nature. 2015 doi: 10.1038/nature15515. [DOI] [Google Scholar]
- 4.Gaspar-Maia A., et al. Nat. Rev. Mol. Cell Biol. 2011 doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein B.E., et al. Cell. 2006 doi: 10.1016/j.cell.2006.02.041. [DOI] [Google Scholar]
- 6.Neff A.T., et al. Genome Res. 2012 doi: 10.1101/gr.134312.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lou C.-H., et al. Stem Cell Rep. 2016. [DOI]
- 8.Arraiano C.M., et al. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbagrm.2013.03.009. [DOI] [Google Scholar]
- 9.Mitchell P., et al. Cell. 1997 doi: 10.1016/S0092-8674(00)80432-8. [DOI] [Google Scholar]
- 10.Mistry D.S., et al. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloret-Llinares M., et al. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky817. [DOI] [Google Scholar]
- 12.Belair C., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201811148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomecki R., et al. EMBO J. 2010 doi: 10.1038/emboj.2010.121. [DOI] [Google Scholar]
- 14.Reis F.P., et al. Wiley Interdiscip. Rev. RNA. 2013 doi: 10.1002/wrna.1180. [DOI] [PubMed] [Google Scholar]
- 15.Chang H.M., et al. Nature. 2013 doi: 10.1038/nature12119. [DOI] [Google Scholar]