Our study reveals Drg1 as a new binding partner of Dishevelled. The Drg1–Dishevelled association regulates Daam1 and RhoA interactions and activity, leading to polymerization and stability of the actin cytoskeleton, a process that is essential for proper multiciliation.
Abstract
Cilia are critical for proper embryonic development and maintaining homeostasis. Although extensively studied, there are still significant gaps regarding the proteins involved in regulating ciliogenesis. Using the Xenopus laevis embryo, we show that Dishevelled (Dvl), a key Wnt signaling scaffold that is critical to proper ciliogenesis, interacts with Drg1 (developmentally regulated GTP-binding protein 1). The loss of Drg1 or disruption of the interaction with Dvl reduces the length and number of cilia and displays defects in basal body migration and docking to the apical surface of multiciliated cells (MCCs). Moreover, Drg1 morphants display abnormal rotational polarity of basal bodies and a decrease in apical actin and RhoA activity that can be attributed to disruption of the protein complex between Dvl and Daam1, as well as between Daam1 and RhoA. These results support the concept that the Drg1–Dvl interaction regulates apical actin polymerization and stability in MCCs. Thus, Drg1 is a newly identified partner of Dvl in regulating ciliogenesis.
Introduction
Ciliogenesis is an essential developmental process for organogenesis and tissue homeostasis in vertebrates. Cilia are microtubule-based structures generated from the basal body, a modified centriole, and protrude from the cell membrane. Most cells in vertebrates possess nonmotile monocilia (primary cilia) that contribute to signal transduction (e.g., shh) and the sensing of environmental stimuli during development (Goetz and Anderson, 2010). Multiciliated cells (MCCs) are represented in the brain ventricles, respiratory track, and reproductive track in mammals and the mucociliary embryonic epidermis of Xenopus laevis. Multicilia are critical for regulating extracellular molecules and fluid flow (Brooks and Wallingford, 2014; Meunier and Azimzadeh, 2016; Spassky and Meunier, 2017). To generate multicilia, basal bodies, a specialized form of centriole, migrate collectively to the apex of MCCs and dock at the MCC plasma membrane (Dawe et al., 2007; Zhang and Mitchell, 2016). Basal body docking requires the formation of an apical actin meshwork that is regulated by actin regulators, such as RhoA (Pan et al., 2007; Werner et al., 2011).
As motile cilia develop in MCCs, the planar cell polarity (PCP) components play a critical role in transducing polarization information and refining metachronal ciliary beating (Park et al., 2006, 2008; Gray et al., 2009a; Mitchell et al., 2009; Yasunaga et al., 2011). The core PCP component Dishevelled (Dvl; referred to as Dsh elsewhere) plays a variety of roles in both normal development and disease states (Gao and Chen, 2010). The developmental roles of Dvl are primarily governed by dynamic changes in location and stability upon the interaction with different binding partners (Wong et al., 2003; Lee et al., 2018). In part, it is the particular Wnt ligands and receptors that ultimately determine Dvl-mediated signaling. Canonical Wnt signaling will lead to β-catenin stabilization and induction of Wnt target genes through TCF/LEF (T-cell factor/lymphoid enhancer-binding factor) transcription factors (MacDonald et al., 2009), and the canonical Wnt signal has been implicated in vertebrate ciliogenesis (Shi et al., 2014; Zhao et al., 2016). Noncanonical Wnt signaling leads to the activation of RhoA through an interaction with Daam1 and promotes JNK signaling (Schlessinger et al., 2009). Another noncanonical route of signaling involves the transduction of signals through G proteins to phospholipase C and inositol triphosphate, resulting in the release of calcium (Komiya and Habas, 2008). To fulfill this variety of roles, Dvl collaborates with numerous binding partners largely through three conserved domains (DIX, PDZ, and DEP), along with two regions (the basic region and the proline-rich region) that reside between the domains (Gao and Chen, 2010). The DIX and PDZ domains are required for activation of the canonical Wnt signal by interacting with Frizzled receptors and recruiting the Axin–GSK3β complex to the plasma membrane, leading to β-catenin stabilization and localization in the nucleus (Cliffe et al., 2003; Wong et al., 2003). The C-terminal DEP and PDZ domains are essential for Wnt/PCP signaling (Liu et al., 2008) and control a variety of PCP components that direct polarized cell behavior, including ciliogenesis (Pan et al., 2004; Yu et al., 2007; Ganner et al., 2009). There are three Dvl genes in vertebrates (Dvl1, Dvl2, and Dvl3), and they display divergent expression patterns and distinct developmental roles among different species (Gray et al., 2009b).
As a core PCP component, Dvl regulates multiciliation by modulating the formation of the apical actin meshwork and basal body docking in the epidermis of the Xenopus embryo (Park et al., 2008), and loss of Dvl disrupts the function of MCCs in adult mouse ependymal cells (Ohata et al., 2014). Although Dvl is also known to contribute to ciliogenesis in MCCs through an interaction with ERK7 or serving as a substrate for PTEN (Miyatake et al., 2015; Shnitsar et al., 2015), it remains unclear how Dvl modulates ciliogenesis in cooperation with its binding partners during MCC development. Daam1, a member of the Formin family of actin nucleation factors (Kühn and Geyer, 2014) and PCP effectors, associates with Dvl (Habas et al., 2001). Daam1 has a critical role in morphogenetic processes during vertebrate embryonic development (Li et al., 2011; Miller et al., 2011; Ossipova et al., 2018), and a recent report demonstrated that knockdown of Daam1 also causes ciliogenesis defects in MCCs (Yasunaga et al., 2015), but whether a Dvl and Daam1 interaction is necessary to control multicilia formation in MCCs was not addressed.
Developmentally regulated GTP-binding protein 1 (Drg1) is a highly conserved Obg family GTPase among different species (Li and Trueb, 2000), and it displays a broad range of expression in both adult tissues and vertebrate embryos (Sazuka et al., 1992; Kumar et al., 1993; Li and Trueb, 2000). There are two DRG family members, Drg1 and Drg2 (Li and Trueb, 2000). These proteins contain a GTP-binding domain, followed by a ThrRS, GTPase, and SpoT (TGS) domain toward the C-terminus (Pérez-Arellano et al., 2013). The generalized role of the TGS domain relates to protein–protein interactions, such as ligand binding, or a regulatory role in the context of certain enzymes (e.g., GTPases; Wolf et al., 1999). While Drg1 and Drg2 have 55% identity at the amino acid level, they demonstrate a similar broad spatial expression pattern by whole-mount in situ hybridization (Ishikawa et al., 2009). It was previously reported that Drg1 stimulates the transforming activity of c-myc and ras by an interaction with TAL1 (Mahajan et al., 1996) and that Drg1 modulates protein translation in complex with Dfrp1 in mammalian cells (Ishikawa et al., 2005, 2009). A recent study also indicates that Drg1 locates to mitotic spindle checkpoint proteins and promotes tumorigenesis through an effect on cell cycle (Lu et al., 2016). In Xenopus, Drg1 expression has been reported in several regions of the brain, eye, otic vesicle, branchial arches, somites, and all along the neural tube (Sazuka et al., 1992; Kumar et al., 1993), while broad expression was found in mouse and human tissues. The physiological role of Drg1 during vertebrate development is still unclear, with no null vertebrate models reported. In our study, we identify Drg1 as a novel binding partner of Dvl that is required for the proper Dvl interaction with Daam1. These interactions play a crucial role in modulating the formation of the apical actin meshwork and basal body docking in the MCCs.
Results
Drg1 associates with Dvl2 and Dvl3 in Xenopus embryos and human cell lines
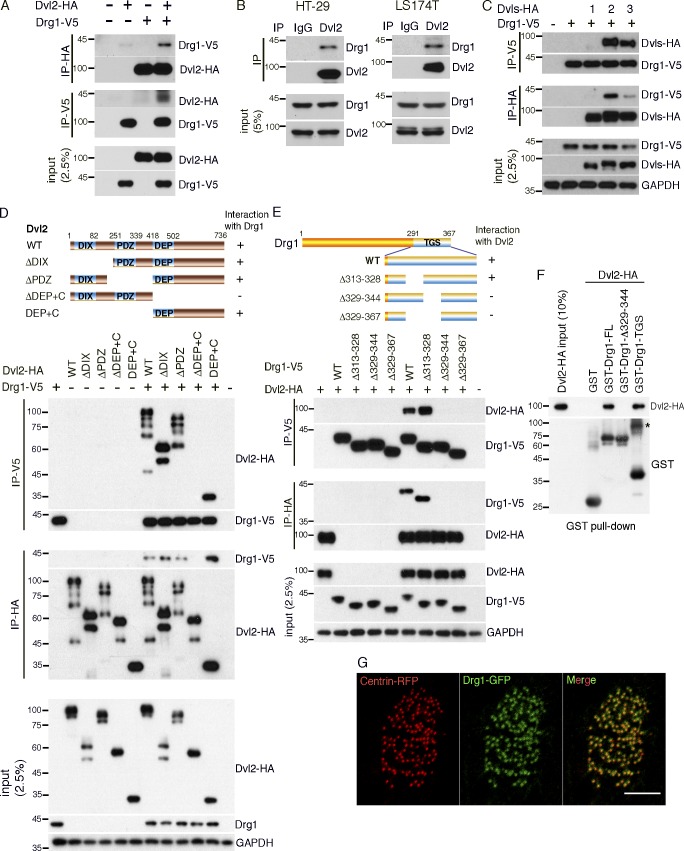
Dvl have several interacting proteins that mediate or regulate their morphogenetic functions during development (Sharma et al., 2018). To identify novel binding partners of Dvl in Xenopus embryos, we overexpressed a Dvl2 protein lacking the DIX domain that is associated with canonical Wnt signaling and performed immunoprecipitation (IP) and mass spectrometry analysis. This analysis revealed Drg1 as a candidate Dvl2 binding partner in Xenopus embryos (Table S1). To validate the Drg1–Dvl2 interaction, we performed co-IP using embryos exogenously expressing tagged versions of both proteins. Drg1-V5 was detected in Dvl2-HA immune complexes, and Dvl2-HA was detected in Drg1-V5 immune complexes (Fig. 1 A). Co-IP analysis from two human colon cancer cell lines, HT29 and LS174T, demonstrated endogenous Drg1 in Dvl2 immune complexes (Fig. 1 B), confirming that Dvl2 and Drg1 associate in vivo and validating the IP/mass spectrometry data. Since Dvl consists of three family members (Dvl1, Dvl2, and Dvl3; Gray et al., 2009b), we determined which members associate with Drg1 by coexpressing each one along with Drg1 in embryos. Interestingly, reciprocal co-IP analysis shows that Drg1 shows the most affinity with Dvl2, followed by Dvl3, but displays little interaction with Dvl1 (Fig. 1 C).
Figure 1.
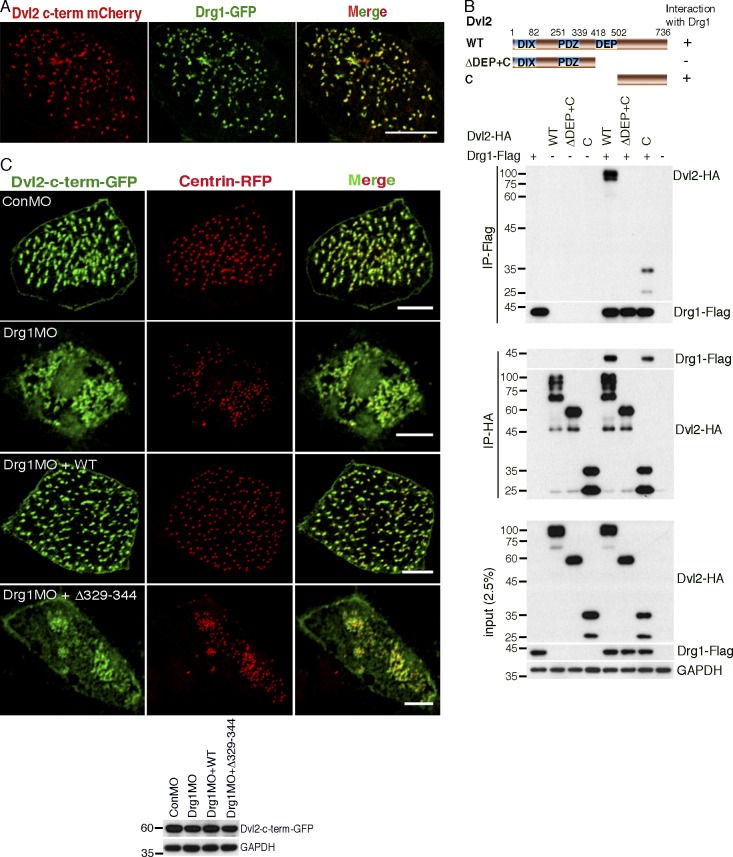
Drg1 associates with Dvl. (A) Exogenous Drg1 interacts with Dvl2. Dvl2-HA and Drg1-V5 mRNAs were injected in Xenopus embryos, and reciprocal co-IPs were performed. 2.5% of input was loaded. (B) Endogenous Drg1 associates with Dvl2. Using lysates from HT-29 and LS174T colon cancer cells, endogenous co-IPs were performed by precipitating endogenous Dvl2. 2.5% of input was loaded. IgG pull-down is a negative control. (C) Drg1 interacts with Dvl2 and Dvl3, but not Dvl1. The indicated Dvl-HA and Drg1-V5 mRNA were coinjected, and co-IPs were conducted by pull-down in both directions. 2.5% of input was loaded. (D) Drg1 interaction domain mapping of Dvl2. The DEP+C region of Dvl2 is necessary and sufficient to interact with Drg1. The indicated WT and deletion mutants of Dvl2-HA and Drg1-V5 mRNAs were expressed in Xenopus embryos, and co-IPs were performed in both directions. 2.5% of input was loaded. (E) Dvl2 interaction domain mapping of Drg1. WT and the indicated deletion mutants of Drg1-V5 and Dvl2-HA were expressed in Xenopus embryos, and co-IPs were conducted in both directions. Amino acids between 329 and 344 of Drg1 are required for the association with Dvl2. 2.5% of input was loaded. (F) Dvl2-HA associates with Drg1-GST in vitro. 1 µg of the purified recombinant proteins was used for the in vitro binding assay. The asterisk marks a dimerized form of isolated TGS domain. 10% of input was loaded. (G) The subcellular localization of Drg1 in an MCC. Drg1-GFP (green) and centrin-RFP (red, basal body marker) mRNAs were coinjected into the marginal region of both ventral blastomeres at the four-cell stage. The embryos were observed at stage 25. Images were generated by maximum intensity projection from serial z-stack images. Scale bar, 5 µm.
The DEP domain and the contiguous C-terminal region (DEP+C) domain of Dvl and the TGS domain of Drg1 are required for Dvl2–Drg1 interactions
Dvl have a host of interacting partners that bind via the various domains within the protein (Gao and Chen, 2010). To locate the region within Dvl2 necessary for an association with Drg1, deletion mutants of Dvl2 described in a previous study (Lee et al., 2006) were used (Fig. 1 D). Co-IP analysis indicated that the Dvl2 mutant lacking the DEP+C domain significantly decreased the interaction between Dvl2 and Drg1. Moreover, a protein consisting of the DEP+C region was sufficient to interact with Drg1 (Fig. 1 D). Consistent with these results, the DEP+C domain of Dvl3 is also required for an interaction with Drg1 (Fig. S1 A).
Protein domain mapping of Drg1 was performed to determine the region necessary for an interaction with Dvl2 (Fig. S1 B). Four deletion mutants of Drg1 (Δ1–65 [ΔN], Δ66–150 [ΔM1], Δ151–290 [ΔM2], and Δ291–367 [ΔTGS]) were generated and expressed along with Dvl2 in embryos. Co-IP analysis showed that the TGS domain is required for a Drg1–Dvl2 interaction (Fig. S1 B). Further testing was performed with several internal deletion mutants within the TGS domain (Fig. 1 E), and we found that amino acids 329–344 of Drg1 is necessary for a robust association with Dvl2. In contrast, a construct with the adjacent amino acids 313–328 deleted maintained the interaction with Dvl2 (Figs. 1 E and S1 C), suggesting that amino acids 329–344 in the TGS domain are required for the Drg1–Dvl2 interaction. To address whether Drg1 has a direct interaction with Dvl2, we performed an in vitro binding assay using purified recombinant proteins and observed that the amino acids 329–344 are necessary to associate with Dvl2 and that the TGS domain of Drg1 is sufficient for the Drg1–Dvl2 interaction (Fig. 1 F), consistent with the Drg1 domain mapping data (Fig. 1 E).
Drg1 localizes to the basal body area in MCCs
In Xenopus embryos, Drg1 transcripts are abundant in several developing tissues and organs such as head, developing eye, neural crest, pronephros, and notochord at the early tailbud stages (Ishikawa et al., 2003). Since the development of MCCs begins around the neurula stage (Zhang and Mitchell, 2016), we examined the localization of Drg1 transcripts in the embryos at these stages using whole-mount in situ hybridization. Drg1 transcripts are expressed in the ectoderm and neural plate (Fig. S2 A). Although Drg1 protein localizes to both the cytoplasm and nucleus in cultured cells (Ishikawa et al., 2005; Lu et al., 2016), proteomic studies suggest that Drg1 may be a potential component of cilia (Liu et al., 2007; Ishikawa et al., 2012). Thus, we expressed a Drg1-GFP fusion protein along with centrin-RFP (as a basal body marker) in embryos. In the MCCs, Drg1 localized to the basal body region (Fig. 1 G), implicating a potential role of Drg1 in ciliogenesis.
Drg1 is required for ciliogenesis in MCCs
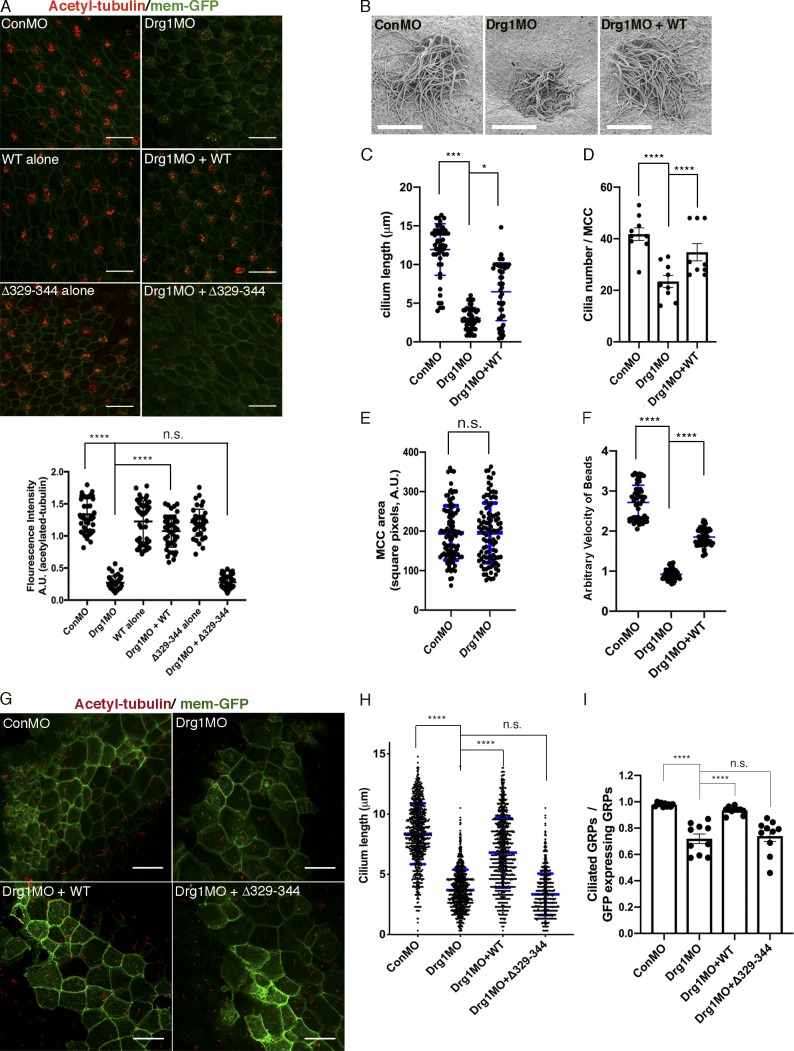
Several lines of evidence indicate a potential role for Drg1 in ciliogenesis, including studies identifying Drg1 as part of the cilia proteome (Liu et al., 2007; Ishikawa et al., 2012), the demonstration that Dvl plays a critical role in ciliogenesis (Park et al., 2008), and the preceding experiments showing the basal body localization of Drg1 (Fig. 1 G). To test whether Drg1 participates in ciliogenesis in MCCs, morpholino oligonucleotides (MOs) against Drg1 were used. To target morpholinos to the epidermis of embryonic MCCs, they were microinjected into the marginal regions of both ventral blastomeres of four-cell-stage embryos. Immunostaining showed acetylated tubulin was significantly decreased in the MCCs of Drg1 morphants (Figs. 2 A and S2 D), which is similar to phenotypes observed with MOs against Dvl2 and Dvl3 in the MCCs (Fig. S2 E). The reduced acetylated tubulin staining observed upon Drg1 knockdown was rescued by MO-resistant WT Drg1 expression (Fig. 2 A). The Drg1 MO–mediated ciliogenesis defects and the rescue by WT Drg1 were also confirmed by scanning EM of MCCs from the Drg1 morphants (Fig. 2 B). This analysis shows both the length and number of cilia in Drg1 morphant MCCs decrease, and MO-resistant WT Drg1 expression partially but significantly rescues the length and population of MCC cilia (Fig. 2, C and D). However, when compared with control, the Drg1 knockdown did not affect the surface area of MCCs on the epidermis (Fig. 2 E). Also, we observed that when compared with control embryos, Drg1 morphants displayed a significant decrease in the fluid flow across the epidermis (Fig. 2 F and Videos 1, 2, and 3), suggesting that the Drg1 knockdown compromises the function of cilia in the MCCs. The specificity of MOs and the efficient expression of MO-resistant Drg1 mRNAs were affirmed using immunoblots (Fig. S2, B and C). As an experimental control, exogenous expression of WT or Drg1 mutants alone showed no effect on MCCs. Intriguingly, a mutant Drg1 lacking amino acids 329–344 (required for an interaction with Dvl) failed to rescue acetylated tubulin staining in the Drg1 morphants (Fig. 2 A), suggesting Drg1 may modulate ciliogenesis in cooperation with Dvl in the MCCs.
Figure 2.
The Drg1–Dvl interaction is important for ciliogenesis in MCCs and the GRP. (A) Drg1 knockdown causes a significant reduction of acetylated tubulin on the epidermis of tadpoles, and the expression of WT Drg1, but not the Δ329–344 mutant, rescues the knockdown phenotype. A cocktail of synthetic mRNAs and morpholinos (7.5 ng each) was injected into both ventral blastomeres at the four-cell stage of embryos. The injected embryos were fixed at stage 27. For immunostaining, anti-acetylated tubulin antibody was used to visualize multicilia (red), and membrane-GFP (green) was used as a tracer. Relative acetylated tubulin intensity is quantified (image n = 45 from 15 embryos for each condition). ****, P < 0.0001, one-way ANOVA; scale bars, 100 µm. Error bars indicate ± SD. (B) Scanning EM of embryonic epidermis confirms Drg1 knockdown causes ciliogenesis defects. 7.5 ng of control and Drg1 morpholinos was injected into both marginal ventral blastomeres at the four-cell stage, and embryos were fixed at stage 31. Scale bars, 12 µm. (C) Quantification of cilia length in MCCs. Data are plotted as the mean, with error bars representing SD. Measured cilia, n > 100; embryos per group, n = 3; ***, P < 0.001; *, P = 0.012, one-way ANOVA. (D) Quantification of cilia number per MCC, n = 10; embryos per group, n = 3; ****, P < 0.0001, one-way ANOVA. Error bars indicate ± SD. (E) The area of epidermal MCCs of stage 27 embryos; MCCs, n = 100; embryos per group, n = 10; two-tailed unpaired t test. Error bars indicate ± SD. (F) Quantification of the fluid flow with green fluorescent beads (Videos 1, 2, and 3). Velocity of five beads along the body axis was manually measured. Measured bead velocity per embryo, n = 5; embryos (stage 27–28) per group, n = 10; stage 27 embryos; ****, P < 0.0001, one-way ANOVA. Error bars indicate ± SD. (G) Drg1 knockdown decreases the length of cilia in the GRP. The indicated mRNAs and MOs were injected to both marginal dorsal blastomeres at the four-cell stage, and embryos were harvested at stage 18. Dissected GRPs were fixed, followed by immunostaining using acetylated tubulin antibody (red) and anti-GFP antibody (green). Mem-GFP is used as a tracer. Scale bars, 30 µm. (H) Quantification of cilia length in the GRP, measured cilia per group; n > 900, dissected GRPs per group; n = 10, ****, P < 0.0001, one-way ANOVA. Error bars indicate ± SD. (I) Quantification of cilia number on GRP; the ratio of the number of ciliated cells expressing GFP to total GRP cells expressing GFP in experimental groups is normalized to the control and expressed as a relative value to the control (set as 1). GRPs per group; n = 10, ****, P < 0.0001, one-way ANOVA. Error bars indicate ± SD. a.u., arbitrary units; n.s., not significant.
Drg2 has >50% amino acid identity with Drg1 (Li and Trueb, 2000). Thus, we tested whether Drg2 is capable of associating with Dvl, and although exogenously expressed Flag-tagged Drg2 formed an immune complex with HA-Dvl2 and HA-Dvl3 (Fig. S2 F), the endogenous Drg2 failed to interact with either Dvl2 or Dvl3 in HT29 cells (Fig. S2 G). Moreover, unlike Drg1 morphants, Drg2 morphants did not show any alterations in acetylated tubulin staining (Fig. S2 H). These data clearly suggest that despite the similarity in amino acid sequence between Drg1 and Drg2, only Drg1 appreciably contributes to ciliogenesis in MCCs.
In addition to multicilia in MCCs, there are motile monocilia in the gastrocoel roof plate (GRP) of neurula-stage Xenopus embryos, which contribute to the determination of left–right asymmetry (Schweickert et al., 2007). We tested whether Drg1 is required for motile monocilia formation in the GRP. To target the GRP cilia, a mixture of MOs and mRNAs were injected to both dorsal marginal regions of four-cell-stage embryos. Drg1 knockdown caused a reduction in the number and length of cilia in the GRP, and the shortened length and the reduced number of cilia in GRP cells was partially restored by MO-resistant WT Drg1 expression, but not by the Δ329–344 mutant that fails to interact with Dvl (Fig. 2, G–I). These data suggest that Drg1 affects ciliogenesis in both MCCs and the GRP and that the interaction between Drg1 and Dvl may be required for ciliogenesis.
Drg1 is required for planar polarization and apical docking of basal bodies in MCCs
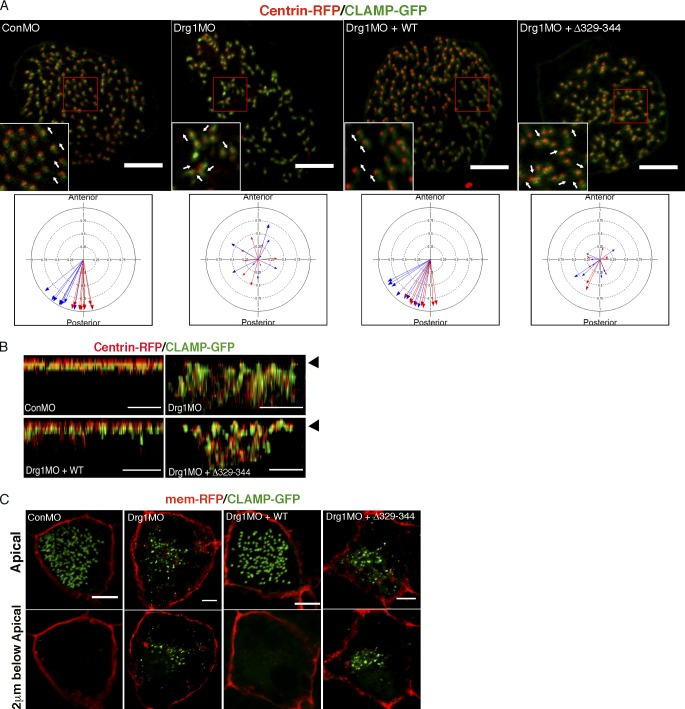
Since the planar polarization of basal bodies is essential to multicilia function in the MCCs (Mitchell et al., 2007; Park et al., 2008), we assessed the orientation of basal bodies in Drg1 morphants using centrin-RFP (a basal body marker) and CLAMP-GFP (a rootlet marker) as previously described (Park et al., 2008). To determine the direction of basal body/rootlets, we measured the mean orientation and the circular SD of the rootlets within a single MCC relative to the anterior–posterior axis of an embryo. The mean direction of basal body/rootlets in the MCCs from control embryos at stage 27 oriented toward the posterior direction as expected (Mitchell et al., 2007); however, the Drg1 morphants displayed marked directional misorientation of basal body/rootlets (Fig. 3 A). Although WT Drg1 expression rescued the misorientation of the rootlets in Drg1 morphants, the Drg1 Δ329–344 mutant failed to rescue the posterior directionality, resulting in a random orientation of basal body/rootlets (Fig. 3 A).
Figure 3.
Drg1 is required for planar polarization and apical docking of basal bodies in MCCs. (A) Planar polarization defect in Drg1 morphants. CLAMP-GFP (rootlet marker) and centrin-RFP (basal body marker) were coinjected with the mixture of the indicated MOs and mRNAs. Areas enclosed by the red box are magnified in the insets, and arrows indicate the orientation of basal body/rootlets in MCCs. The circular plots depict polarity of basal body/rootlets in MCCs. Each arrow indicates the mean polar direction of basal body/rootlets in a single MCC. Arrow length depicts 1 minus circular variance of basal body/rootlet orientation around the mean. Different colored arrows represent data from different embryos. Scale bars, 5 µm. (B) Basal body docking defect at the apical regions of MCCs upon Drg1 knockdown. CLAMP-GFP and centrin-RFP mRNAs were coinjected with the cocktail of the indicated MOs and mRNAs. Serial z-stack confocal images were projected in the y-z plane. Arrowheads indicate the position of MCC apical surface. Scale bars, 3 µm. (C) Basal body docking defect at the MCC apical region upon Drg1 knockdown. CLAMP-GFP (green) and membrane-RFP (red) mRNAs were injected with the indicated morpholinos and mRNAs. Top panels indicate apical region of MCCs and bottom panels 2 µm below the apical region. Scale bars, 5 µm.
During multicilia formation, basal bodies migrate and dock to the apical region of the MCCs (Boisvieux-Ulrich et al., 1990; Werner and Mitchell, 2012; Zhang and Mitchell, 2016). We expressed CLAMP-GFP and centrin-RFP in embryos as markers of rootlets and basal bodies, respectively (Park et al., 2008; Werner et al., 2014). CLAMP is enriched at the rootlets associated with motile cilia and regulates PCP signaling as well as asymmetric microtubule accumulation in MCCs (Kim et al., 2018). To test whether Drg1 affects this process, embryos expressing CLAMP-GFP and centrin-RFP were also injected with Drg1 MO alone or along with WT or Drg1 Δ329–344 mutant RNAs. Comparing lateral views of MCCs (via z-stacks), the apical location of CLAMP-GFP and centrin-RFP was significantly disrupted upon Drg1 knockdown when compared with control MO–injected embryos (Fig. 3 B). Even when examining the focal plane 2 µm below the apical region, apical localization of CLAMP-GFP was disrupted upon Drg1 knockdown, resulting in CLAMP-GFP also localizing below the apical surface (Fig. 3 C). In the Drg1 morphants, expression of MO-resistant WT Drg1 restored the apical location of CLAMP-GFP in the MCCs, as shown by the lack of CLAMP-GFP signal below the apical plane (Fig. 3 C). In contrast, Drg1 Δ329–344 (Dvl-interaction mutant) failed to rescue this defect (Fig. 3 B and C).
One plausible explanation for the apparent defect in basal body docking upon Drg1 knockdown might be failure of the MCCs to migrate to the epidermal surface. To determine whether this was the case, we performed immunofluorescence (IF) using an α-tubulin antibody to mark MCC progenitors in the epidermal epidermis. MCC progenitors maintain a high level of α-tubulin expression (Chevalier et al., 2015). When compared with the control morphants, Drg1 morphants did not display any significant migration defects of the MCC progenitors (Fig. S3 A). Consistent with this finding, examination of the surface area of MCCs in earlier-stage embryos (stage 22) showed no significant difference in the MCC surface area between control and Drg1 morphants (Fig. S3 B). This result is similar to that observed in later stage (stage 27) embryos (Fig. 2 E). These data are also supported by the scanning electron micrographs showing defective cilia at the epidermal surface (Fig. 2 B). Taken together, these data indicate that Drg1 regulates the behavior of basal bodies in the MCCs through an interaction with Dvl.
An interaction between Drg1 and Dvl is required for apical actin meshwork formation
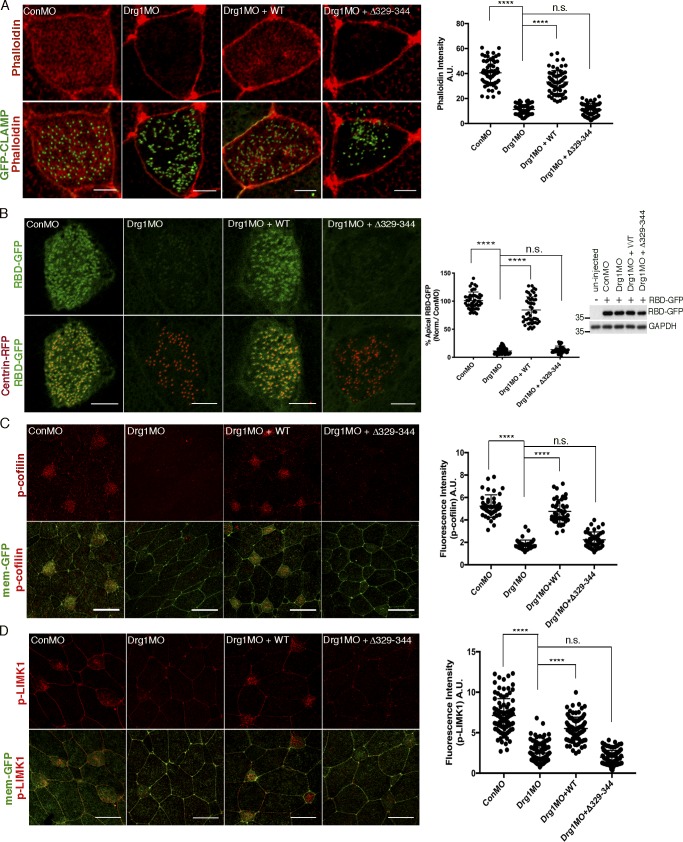
The apical and subapical actin meshwork is required for the basal bodies to dock within the apical region of the MCCs (Boisvieux-Ulrich et al., 1990; Pan et al., 2007). Using phalloidin staining to examine the actin layer in MCCs, we found that Drg1 morphants demonstrate significantly decreased phalloidin staining in the apical region (Figs. 4 A and S3 C). Restoration of phalloidin staining was observed when WT Drg1 was expressed in the presence of the Drg1 MO. Unlike WT Drg1, no rescue was observed by expressing the Drg1 Δ329–344 mutant (Figs. 4 A and S3 C), suggesting that the interaction between Drg1 and Dvl contributes to the formation of the actin meshwork in the apical region of MCCs.
Figure 4.
Drg1–Dvl interaction modulates apical actin enrichment in MCCs through Rho activation. (A) Apical actin meshwork decreases upon Drg1 knockdown. The loss of apical actin signal is rescued by expression of Drg1 WT, but not by the Drg1 Δ329–344 mutant. The embryos were injected with the indicated MOs and mRNAs, fixed at stage 25, and stained with phalloidin (red) to visualize cortical F-actin in apical region of MCCs. GFP-CLAMP (green) marks rootlets. ****, P < 0.0001, one-way ANOVA; n = 65. Scale bars, 5 µm. Error bars indicate ± SD. (B) Drg1 knockdown decreases Rho activity. Synthetic mRNAs of RBD-GFP (green) and centrin-RFP (red) were coinjected with the indicated MOs and mRNAs, and embryos were fixed at stage 25. Fluorescence intensity of experimental groups is normalized to the control MCCs and expressed as a percentage relative to control. Expression level of RBD-GFP was tested by Western blot. Images are generated by maximum intensity projection of serial z-stack confocal images from apical (0 µm) to subapical region (−2.5 µm). ****, P < 0.0001, one-way ANOVA; n = 49. Scale bars, 5 µm. Error bars indicate ± SD. (C and D) Phosphorylation of Rho downstream targets is decreased by Drg1 knockdown. The embryos injected with the indicated MOs and mRNAs were stained for phospho-cofilin (C) and phospho-LIMK1 (D). Statistical analysis by one-way ANOVA with post hoc Turkey’s multiple range tests. All results are derived from three independent experiments. ****, P < 0.0001, one-way ANOVA; n = 42 (C), 40 (D). Error bars represent mean ± SD. Scale bars, 30 µm. a.u., arbitrary units; n.s., not significant.
Small GTPases such as RhoA regulate actin cytoskeleton organization and remodeling (Hall, 1998; Sit and Manser, 2011; Spiering and Hodgson, 2011), and Dvl-mediated regulation of Rho activity is required for ciliogenesis in MCCs (Park et al., 2008). Thus, we hypothesized that the Drg1–Dvl interaction has an impact on Rho activity, thereby affecting formation of the apical actin meshwork. To test this hypothesis, the Rho-binding domain of Rhotekin-GFP (RBD-GFP) was used to detect endogenous active Rho, and centrin-RFP was used as a basal body marker. Drg1 knockdown in embryos led to a marked reduction of RBD-GFP signal in the MCCs when compared with the abundant RBD-GFP signal accumulated at the basal bodies in the control MCCs (Fig. 4 B). This decrease in RBD-GFP signal was rescued upon Drg1 WT expression, but not upon expression of the Drg1 Δ329–344 mutant expression (Fig. 4 B). The Western blot for RBD-GFP showed that Drg1 knockdown did not affect the amount of RBD-GFP protein (Fig. 4 B). Collectively, these data suggest that the association between Drg1 and Dvl is critical to forming the apical actin meshwork in the MCCs through Rho activation.
One prediction from this concept is that important downstream targets of Rho GTPases such as LIM kinase (LIMK) and cofilin, which are consequential to remodeling of the actin cytoskeleton (Etienne-Manneville and Hall, 2002), would also be affected by Drg1 knockdown. LIMK directly phosphorylates cofilin, leading to actin fiber stabilization and polymerization (Bernstein and Bamburg, 2010). Thus, we assessed whether Drg1 knockdown affects the activity of Rho downstream targets in the MCCs by performing IF with phospho-LIMK1 (phospho-Thr508) and phospho-cofilin (phospho-Ser3) antibodies. The phospho-LIMK1 and phospho-cofilin expression in the Drg1 morphant MCCs was substantially decreased when compared with the control MO–injected embryos (Fig. 4, C and D). Expression of WT Drg1, but not the Δ329–344 mutant, rescued phosphorylation of the Rho downstream targets in the Drg1 MO bearing MCCs (Fig. 4, C and D). Since the available antibodies were unable to detect endogenous total cofilin or LIMK as references, we exogenously expressed a minimally detectable (by IF and immunoblotting [IB]) amount of HA-tagged cofilin and HA-tagged LIMK1 protein and found that the expression of these proteins was unaltered in the absence of Drg1 (Fig. S3 D). These data suggest that the decrease in phospho-cofilin and phospho-LIMK1 found in Drg1 morphants is not likely due to total protein reduction. These data confirm that the association between Drg1 and Dvl is required for Rho activation in the MCCs, resulting in downstream targets modulating the formation of the apical actin meshwork.
Drg1 GTPase activity is dispensable for ciliogenesis in MCCs
It remains unclear what physiological role is imparted by the GTPase activity of Drg1 in vertebrate development. Therefore, we assessed whether this activity may be required for ciliogenesis in the MCCs. We generated a mutant Drg1 harboring two mutations (S78N-T100D) that have been reported to greatly diminish the GTPase activity (Francis et al., 2012; Pérez-Arellano et al., 2013; Schellhaus et al., 2017). Similar to WT Drg1, the expression of the S78N-T100D mutant restored the acetylated tubulin expression in the Drg1 morphants (Fig. S4 A). MO-resistant WT and mutant Drg1 protein expression was confirmed by Western blot (Fig. S4, B and C). Of note, the interaction between Drg1 and Dvl2, as evidenced by co-IP analysis, was unchanged by the mutation (Fig. S4 D). Although cilia morphology was restored by the GTPase mutant, we tested whether ciliary function was also restored by assaying fluid flow. While the Drg1 morphants displayed a significant decrease in the fluid flow across the epidermis, the exogenous expression of the S78N-T100D mutant or the WT control rescued fluid flow (Fig. S4 E). Collectively, these data indicate that the GTPase activity of Drg1 is not required for proper ciliogenesis in the MCCs.
Drg1 knockdown has a mild effect on Dvl localization to the basal body region in MCCs
Dvl2 is known to localize to basal body/rootlet areas in MCCs (Park et al., 2008). Since Drg1 also localized to the basal body region (Fig. 1 G), it was possible that Dvl2 may colocalize with Drg1 in the MCCs. To assess this possibility, we carefully titrated a minimal amount of RNA encoding the Dvl2 C-terminal fragment fused to mCherry (Dvl2-C-term-mCherry) that would allow detection of basal body/rootlet regions without causing phenotypic effects in cilia (Park et al., 2008). Dvl2-C-term-mCherry was coexpressed with Drg1-GFP, and a small region of overlap is observed with the two proteins in the MCCs (Fig. 5 A), which was confirmed by co-IP, indicating that Drg1 associates with the Dvl2 C-terminal fragment (Fig. 5 B). We then tested whether knockdown of Drg1 affects Dvl2 localization in the MCCs. The indicated mRNAs and MOs were injected into two ventral blastomeres at the four-cell stage, and immunostaining was performed. In the control MO–injected MCC controls, the C-terminal fragment of Dvl2 localized adjacent to centrin-RFP. Upon Drg1 knockdown, the localization of the C-terminal fragment of Dvl2 was not as restricted to the centrin-RFP and spread more easily within the cytosolic region, but a considerable amount of C-terminal fragments were still colocalized with the centrin-RFP (Fig. 5 C). WT Drg1 expression in the Drg1 morphants led to restricted localization of the Dvl2 C-terminal fragments to the basal body areas, but the Drg1 Δ329–344 Dvl2-interaction mutant failed to suppress the Drg1 knockdown effect (Fig. 5 C). Notably, Drg1 localization to basal bodies was not affected by the knockdown of Dvl (Fig. S5 A); however, as expected with Dvl morphants, the spacing and distribution of the basal bodies was disrupted (Fig. S5 A). Collectively, these data indicate that Drg1 is not a primary contributor to the regulation of Dvl location to the basal body/rootlet region and that Dvl is not required for Drg1 localization to the basal body area in MCCs.
Figure 5.
Drg1 knockdown has a mild effect on Dvl localization to the basal body region in MCCs. (A) Drg1 colocalizes with the Dvl2 C terminus in MCCs. The mRNAs of the Dvl2 C-terminal fragment tagged with mCherry and Drg1-GFP were coinjected into both marginal ventral blastomeres of four-cell-stage embryos and fixed at stage 25. Images are generated by maximum intensity projection of serial z-stack confocal images. Scale bar, 10 µm. (B) The C-terminus of Dvl2 associates with Drg1. The indicated HA-tagged Dvl2 mutants were coexpressed with Drg1-Flag, and co-IP was performed. (C) Drg1 knockdown diminishes Dvl2 C-terminal localization to the basal body area. mRNAs of Dvl2-c-term-GFP and centrin-RFP (basal body marker) were coinjected with the indicated MOs and mRNAs. The expression level of Dvl2-c-term-GFP was tested by Western blot. Embryos were fixed at stage 25–27. Serial z-stack images were generated. A maximum intensity projection image is shown. Scale bars, 5 µm.
Drg1 associates with Daam1 through Dvl
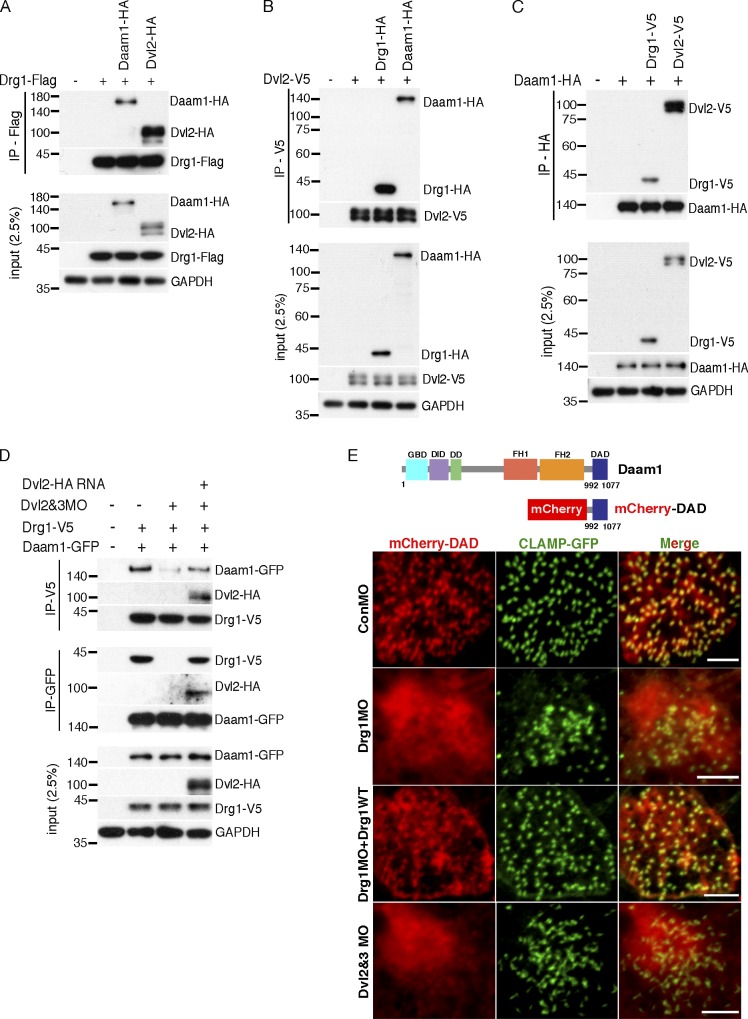
Having established that Drg1 has only a limited effect on Dvl localization, we examined how a Dvl-interacting partner, Daam1, might be impacted. Daam1 has been shown to directly interact with Dvl (Habas et al., 2001) and play a critical role in ciliogenesis in the MCCs (Yasunaga et al., 2015). Since the preceding experiments established that Drg1 interacts with Dvl and affects ciliogenesis, we posited the question of whether Daam1 may be a component of the Drg1–Dvl complex. To examine this concept, we tested whether Drg1 associates with Daam1 in embryos exogenously expressing Drg1 and Daam1-GFP. Co-IP analysis showed that Daam1 interacted with WT Drg1, but not the Drg1 Δ329–344 mutant (Fig. S5 B), revealing that amino acids 329–344 of Drg1, which are required for an interaction with Dvl, are also required for an association with Daam1. One clear possibility is that Dvl may mediate the interaction between Drg1 and Daam1 or that Daam1 mediates the Drg1–Dvl interaction. Since antibodies to detect endogenous Drg1, Dvl2, and Daam1 proteins in Xenopus are not available, the extent of overexpression of these proteins relative to the endogenous proteins cannot be quantified. To determine whether there was a marked difference in the affinity of protein interactions among Drg1, Dvl2, and Daam1, we adjusted the expression levels of two proteins tagged with the same epitope to comparable levels while expressing the other binding partner tagged with a different epitope for IP. With the same expression levels of Dvl2 and Daam1, Drg1 tended to interact with Dvl2 somewhat more robustly (Fig. 6 A). Reciprocally, Dvl2 preferred associating with Drg1 relative to Daam1 when Drg1 and Daam1 were similarly expressed (Fig. 6 B). Likewise, when Drg1 and Dvl2 were expressed at similar levels, the affinity between Daam1 and Dvl2 was slightly more robust than that between Drg1 and Daam1 (Fig. 6 C). Overall, these data did not indicate a dramatic difference in affinity of partner interactions among Drg1, Dvl2, and Daam1 but showed that the Drg1–Dvl2 association was stronger than the Drg1–Daam1 association. While the data collectively suggest that Dvl may mediate the interaction between Drg1 and Daam, it was still unclear. Thus, we explored whether removing one of the endogenous proteins in the co-IP analysis would more definitively determine which protein may be the critical link of the complex. As both Dvl2 and Dvl3 interact with Drg1 (Figs. 1 C and S2 G) and both are present in embryos, we tested whether the association between Drg1 and Daam1 could be detected in the absence of Dvl2 and Dvl3. Embryos were injected with RNA encoding Drg1 and Daam1-GFP along with MOs against Dvl2 and Dvl3. Co-IP analysis showed that the Drg1–Daam1 interaction decreased upon the knockdown of Dvl2 and Dvl3 and that MO-resistant Dvl2 expression was sufficient to restore the association between the two proteins (Fig. 6 D). Interestingly, knockdown of Daam1 did not affect the Drg1–Dvl2 interaction (Fig. S5 C), supporting the concept that Dvl functions as a central scaffold to form a complex that includes Drg1 and Daam1.
Figure 6.
Drg1 is required for Dvl–Daam1 signaling in MCCs. (A–C) The affinity of protein interactions among Drg1, Dvl2, and Daam1 were tested by IP with the adjusted expression levels of two proteins tagged with the same epitope while expressing the other binding partner tagged with a different epitope. The indicated mRNAs were injected at the one-cell stage, and co-IPs were performed using embryos at stage 12. (D) The Drg1–Daam1 interaction is disrupted by Dvl knockdown. Drg1-V5 and Daam1-GFP mRNAs were coinjected with Dvl2 and Dvl3 MOs and/or Dvl2-Flag (MO resistant) mRNA, and co-IPs were performed with embryos harvested at stage 20. The reduced interaction between Drg1 and Daam1 is restored by Dvl2-Flag expression. (E) mCherry-DAD and GFP-CLAMP mRNAs were coinjected with the indicated mRNAs and MOs, and MCCs of embryos at stage 25 were observed. The close location of mCherry-DAD to CLAMP-GFP was disrupted by Drg1 or Dvl knockdown in MCCs. Images were generated by maximum intensity projection of serial z-stack confocal images from surface to subapical regions (up to −2 µm). Scale bars, 5 µm. DD, dimerization domain; DID, diaphanous inhibitory domain; FH1, formin homology 1; FH2, formin homology 2; GBD, GTPase-binding domain.
Drg1 is required for proper basal body and Daam1 localization in MCCs
Since our data indicate that Dvl mediates the interaction between Drg1 and Daam1, we tested Daam1 localization within MCCs. Using IF microscopy in embryos exogenously expressing Daam1, Daam1 localized to the apical and subapical area of MCCs and was detected in a diffuse manner both proximal and distal to the basal bodies (Fig. S5 D). This locale is consistent with a previous report showing Daam1 localization in the proximal and/or distal side of basal bodies as well as decorating basal bodies (Yasunaga et al., 2015). Exogenous expression of full-length Daam1 was present in a broader pattern than Dvl, which may not be unexpected, since Daam1 contains a Formin-homology domain that associates with actin filaments. To test whether Daam1 localization within MCCs is affected by the presence of Drg1, we expressed the Daam1-GFP protein in the presence or absence of the Drg1 MO. In contrast to the Daam1 localization in control embryos, the usual even spatial distribution of Daam1-GFP around the basal bodies at the apical and subapical regions of MCCs was severely disrupted in embryos injected with the Drg1 MO (Fig. S5 E). Expression of WT Drg1 restored the spatial distribution pattern, but the Dvl-binding mutant (Drg1 Δ329–344) failed to restore the normal distribution of Daam1-GFP in Drg1 morphant MCCs (Fig. S5 E).
Due to the more diffuse localization of full-length Daam1, it was difficult to assess whether the Daam1 that is associated with Dvl at the basal body area was affected by the loss of Drg1.
Therefore, we tested whether the diaphanous autoregulatory domain (DAD) that interacts with Dvl may be responsible for localizing Daam1 with Dvl in MCCs. We generated mCherry fusion proteins, one with the N-terminal fragment (as a control) and another with the DAD of Daam1. As expected, the DAD-mCherry fusion protein colocalized with GFP-CLAMP in MCCs (Fig. S5 F), whereas Dvl mostly localizes in MCCs (Park et al., 2008). Unlike the N-terminal fragment of Daam1, mCherry-DAD colocalized with GFP-Drg1 (Fig. S5 F), suggesting that the DAD of Daam1 may play an important role in localizing Daam1 with the Drg1–Dvl complex in MCCs.
Having observed that the DAD of Daam1 is responsible for Daam1 localization in MCCs (Fig. S5 F), we tested whether Drg1 knockdown has an impact on the DAD localization in MCCs. A minimal amount of DAD-mCherry RNA that was observed to have no effect on ciliogenesis was coinjected with the CLAMP-GFP RNA and the indicated MOs. Drg1 knockdown markedly disrupted the localization of DAD-mCherry to the basal body/rootlet regions, as indicated by the lack of colocalization with CLAMP-GFP (Fig. 6 E). The introduction of an MO-resistant Drg1 RNA partially but substantively rescued the localization of DAD-mCherry. As expected, the knockdown of both Dvl2 and Dvl3 phenocopied the DAD-mCherry localization observed in the Drg1 morphants (Fig. 6 E). Despite the disruption of basal body spacing and distribution in Daam1 morphant MCCs, Daam1 knockdown did not affect Drg1 localization to basal bodies (Fig. S5 G). Together, these data suggest that the Drg1–Dvl association plays a substantive role in localizing Daam1 in MCCs.
Active Daam1 expression is sufficient to suppress Drg1 knockdown phenotypes in MCCs
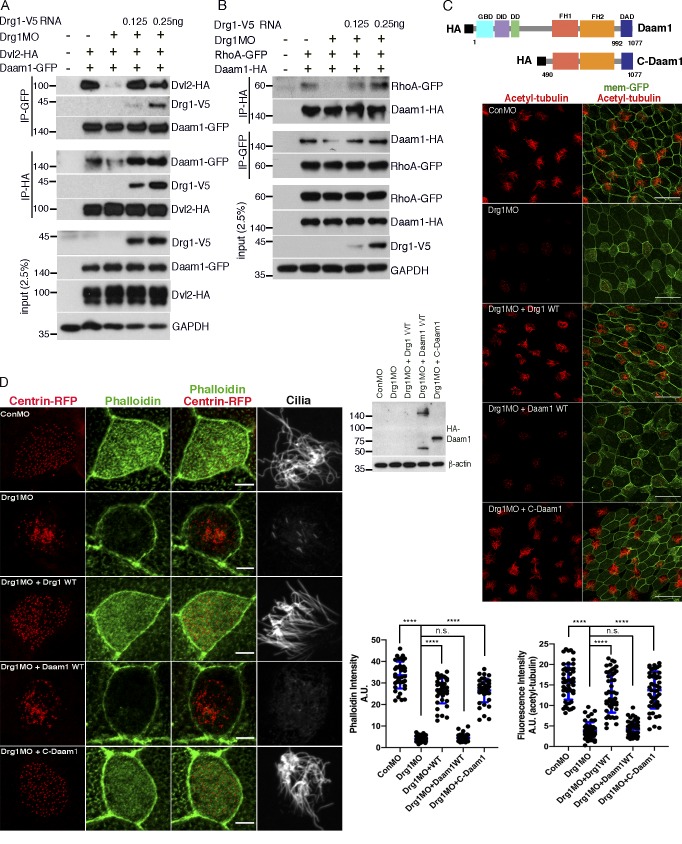
It has been shown previously that the interaction between Daam1 and Dvl is critical for Daam1 activation via release from an autoinhibited state, which allows Daam1 to have enhanced interactions with Rho-GTP (Liu et al., 2008). Our data suggest that Drg1 plays a role in proper Daam1 localization with Dvl as well as RhoA activation. Although we have shown that Daam1 knockdown does not affect the Drg1–Dvl interaction (Fig. S5 C), our data showing that DAD-mCherry mislocalizes away from the basal body/rootlets in the Drg1 morphants (Fig. 6 E) suggest that Drg1 may affect the association between Daam1 and Dvl. Moreover, it was also important to determine whether Drg1 has a role in the known Daam1 interaction with RhoA (Habas et al., 2001). To address these issues, we performed co-IP analyses of embryonic lysates from embryos exogenously expressing Daam1 and Dvl2 in the presence of Drg1 MO to block endogenous Drg1 expression. Drg1 knockdown decreased the Daam1–Dvl2 interaction, which was restored by expressing an MO-resistant WT Drg1 (Fig. 7 A). Likewise, we performed co-IP analysis of lysates from embryos expressing HA-tagged Daam1 and RhoA-GFP in the presence of the Drg1 MO. The interaction between Daam1 and RhoA was also reduced by Drg1 knockdown and restored by re-expression of WT Drg1 (Fig. 7 B). These data suggest that Drg1 plays a substantive role in the Dvl–Daam1 interaction, which is an important determinant of RhoA activity and the formation of the apical actin network.
Figure 7.
Drg1 modulation of Daam1 activity is critical for multiciliation. (A) Drg1 knockdown reduces the Dvl2–Daam1 interaction. The cocktail of the indicated mRNAs and MOs was injected into embryos, and co-IPs were conducted with embryos harvested at stage 20. MO-resistant Drg1-V5 expression rescues decreased the Dvl2–Daam1 interaction upon Drg1 knockdown. (B) Drg1 knockdown decreases the Daam1–RhoA interaction. The indicated mRNAs and morpholinos were coinjected into embryos, which were collected at stage 20 for co-IP analysis. (C) C-Daam1 expression rescues reduced acetylated tubulin staining upon Drg1 knockdown. The indicated mRNAs and MOs were injected to two ventral blastomeres at the four-cell stage. For C-Daam1 expression, 2.5 pg of HA-tagged C-Daam1 encoding plasmid DNAs were injected. The embryos fixed at stage 27 were used for immunostaining. Expression levels of Daam1-WT and C-Daam1 were tested by Western blot. Relative acetylated tubulin intensity is quantified (image n = 40 from 15 embryos for each condition). ****, P < 0.0001, one-way ANOVA. Scale bars, 50 µm. (D) C-Daam1 expression restores the formation of the apical actin meshwork that was decreased upon Drg1 knockdown. The indicated mRNAs and MOs were coinjected into two ventral blastomeres of four-cell-stage embryos and fixed at stage 27 for phalloidin and acetylated tubulin staining. ****, P < 0.0001, one-way ANOVA; n = 40. Scale bars, 5 µm. Error bars indicate ± SD. n.s., not significant.
One important test of the Drg1–Dvl–Daam1 pathway model is whether Daam1 activation is sufficient to suppress the Drg1 knockdown effects on ciliogenesis. We employed a constitutively active form of Daam1 consisting of the Daam1 C-terminus (C-Daam1; Liu et al., 2008). The C-Daam1 encoding plasmid was coinjected with the indicated MOs and RNAs, and acetylated tubulin staining was performed. We titrated the C-Daam1 encoding DNA and injected the minimum amount required to restore the acetylated tubulin staining of the cilia that had been markedly inhibited in the Drg1 morphants (Fig. 7 C). As expected, a similar amount of nonactivated WT Daam1 protein expression failed to rescue the cilia-associated acetylated tubulin staining in the Drg1 morphants. Moreover, phalloidin staining clearly showed that the active Daam1 rescued the ciliogenesis defects in the Drg1 morphants through restoration of the apical actin meshwork in MCCs (Fig. 7 D). Thus, these data suggest that upon Drg1 knockdown, Daam1 has decreased interactions with RhoA and may compromise actin polymerization mediated by these molecules and their downstream effectors (Liu et al., 2008). Taken together, the Drg1–Dvl interaction maintains the proper interactions between Dvl and Daam1 in the MCCs and regulates the actin nucleation activity of Daam1.
Discussion
In the current study, we used overexpression of N-terminally truncated Dvl2 in Xenopus embryos along with IP and mass spectrometry to identify a Dvl-interacting protein, Drg1. Since Dvl is required for both apical docking and planar polarization of basal bodies in vertebrate mucociliary epithelial development (Park et al., 2008), we examined the role of the Drg1–Dvl interaction in multicilia formation. As multicilia are specialized structures required for tissue homeostasis and organ development and functions, multiciliogenesis consists of several critical steps, including basal body docking and apical actin meshwork formation, that require fine-tuned choreography among a variety of proteins. One example is the PCP proteins governing multicilia formation (Brooks and Wallingford, 2014; Meunier and Azimzadeh, 2016). In MCCs, the core PCP component Dvl modulates basal body polarization, docking, and Rho activity (Park et al., 2008) in cooperation with the PCP effector Inturned (Park et al., 2006). Although Daam1, another PCP effector, is a known downstream target of Dvl, it remains unclear whether Dvl–Daam1 interactions contribute to multicilia formation. Instead, Daam1’s association with NPHP4, an actin nucleating factor, is known to be required for subapical actin formation in MCCs (Yasunaga et al., 2015). Although it is not a PCP component, ERK7, an atypical mitogen-activated protein kinase, also associates with Dvl through the DEP+C domain, leading to phosphorylation of CapZIP, a regulator of actin and ciliogenesis (Miyatake et al., 2015). Since interaction with this region of the protein is shared by other molecules, it may be required to generate a signaling complex to modulate ciliogenesis.
Since the basal body is a microtubule organizing center, the basal body localization of Drg1 (Fig. 1 G) is consistent with previous reports demonstrating that Drg1 may play a role in microtubule dynamics (Lu et al., 2016; Schellhaus et al., 2017). We hypothesized that the novel Dvl binding partner Drg1 modulates ciliogenesis through the interaction with Dvl in the MCCs. This concept was based on the following compelling initial evidence: (1) Drg1 localized to basal body regions (Fig. 1 G), (2) Drg1 colocalized with the C terminus of Dvl in MCCs (Fig. 5 A), and (3) proteomic studies indicated that Drg1 may be a ciliary protein (Liu et al., 2007; Ishikawa et al., 2012).
Within the region inhabited by basal bodies, the Drg1–Dvl complex controls apical actin enrichment by two different but related mechanisms (Fig. S5 H). One mechanism is that the Drg1–Dvl complex stabilizes the apical actin cytoskeleton through RhoA- and GTPase-associated kinases that target LIMK-1 (Fig. 4 D), which in turn phosphorylates cofilin on serine (Fig. 4 C) and compromises cofilin severing activity, leading to actin fiber stabilization (Bernstein and Bamburg, 2010; Fig. 4 A). In support of such a process, cofilin-1 phosphorylation increases during MCC differentiation in human airway epithelial cells (Chevalier et al., 2015). Using another mechanism, the Drg1–Dvl complex can interact with Daam1 (Fig. 6, A–D; and Fig. S5 B), and relieve its autoinhibition, thus activating Daam1’s ability to polymerize actin (Liu et al., 2008; Fig. 4 A). Consistent with both mechanisms, knockdown of Drg1 caused a significant decrease in the interaction between Dvl and Daam1 (Fig. 7 A) as well as the association of RhoA with Daam1 (Fig. 7 B). Drg1 MO–mediated disruption of the Drg1–Dvl–Daam1 complex led to a loss of active Rho in the MCCs (Fig. 4 B). This mechanism is supported by our data showing that the constitutively active Daam1 (C-Daam1) expression is sufficient to bypass the requirement for Drg1 and suppress Drg1 knockdown effects by restoring the apical actin meshwork in MCCs (Fig. 7, C and D). Together, these data indicate that Drg1 scaffold function is critical for Daam1 activation, resulting in apical actin enrichment.
Although Drg1 belongs to the subfamily of Obg GTPases, the physiological role of Drg1 as a GTPase is largely unknown. Expression of the Drg1 mutant (S78N-T100D) rescued the acetylated tubulin levels (Fig. S4 A) as well as the velocity of fluid flow on MCCs that were diminished upon Drg1 knockdown, and these mutants maintained the ability to associate with Dvl2 (Fig. S4, D and E). Thus, the role of Drg1 and its interaction with Dvl is independent of GTPase activity and may function as a scaffold to recruit Dvl and other binding partners to the basal body region.
It is of interest that our data indicate that the other Drg subfamily member, Drg2, may not play a role in ciliogenesis in Xenopus MCCs. The association between Drg2 and Dvl occurred only when overexpressed (Fig. S2, F and G). A similar differential interaction of Drg1 and Drg2 with respect to other proteins was observed in a previous study (Ishikawa et al., 2009) that showed Drg1 interacts with Dfrp2 only when ectopically overexpressed, while Drg2 endogenously interacts with Dfrp2 (Ishikawa et al., 2009). Furthermore, contrary to an expected functional redundancy between Drg1 and Drg2 based upon the structural similarities, they do not share physiological functions. For example, the Drg1–Dfrp1 complex is found in ribosomal subunits, but Drg2–Dfrp2 was not detected in ribosomal fractions. This differential subcellular distribution pattern between Drg1 and Drg2 may contribute to the exclusive endogenous interaction of Drg1 with Dvl. In addition, the Drg1 amino acid sequence (329–344) that is required for the Dvl interaction is highly homologous but not identical in the Drg2 counterpart. The amino acid differences may impact the interaction with Dvl. For example, threonine 327 and serine 328 of Drg2 are not shared by Drg1 (isoleucine 330 and lysine 331) and may represent potential phosphorylation sites leading to protein structural changes upon phosphorylation. We cannot comprehensively preclude the possibility that Drg2 may have involvement in ciliogenesis in a different context (e.g., nonmotile primary cilia), since Drg2 has been reported to modulate Rab5 activity in human cells (Mani et al., 2016). Rab5 is implicated in ciliary endomembrane processes at or near the ciliary base (Blacque et al., 2018). Also, Drg2 appears to regulate membrane tubulin stability cooperatively with Rac1 in mammalian cells (Mani et al., 2017). However, our Drg2 MO results (Fig. S2 G), along with the lack of an interaction between endogenous Drg2 and Dvl (Fig. S2 H), strongly suggest that the ciliary role for Drg1 is not shared with Drg2.
Taken together, the Drg1–Dvl interaction plays a critical role in ciliogenesis in MCCs by regulating the localization of Daam1 and commensurate RhoA activity, contributing to basal body docking and the formation of the apical actin meshwork. In addition to its function in multicilia, Drg1 might contribute to other types of cilia such as monocilia (motile or nonmotile). This assertion is supported by our evidence showing that Drg1 knockdown in the GRP reduced the length of motile monocilia (Fig. 2, G–I). Additionally, Drg1 morphants in which the morpholinos were targeted to dorsal embryonic regions show a reduced head size, decreased Vax1 expression, and diminished separation of the two eye fields. These phenotypes are quite similar to the cranial defects observed in hedgehog signaling mutants (Fig. S5 I). Since mutant proteins required for cilia function are implicated in impaired hedgehog signaling (Murdoch and Copp, 2010), the comparable phenotypes of Drg1 morphants may also suggest a role of Drg1 in the formation of nonmotile monocilia. Consistent with this concept, other studies identify Drg1 is a potential component of the mouse photoreceptor sensory cilium complex (Liu et al., 2007) as well as primary cilia from mouse kidney cells (Ishikawa et al., 2012). Thus, Drg1 may play a similar Dvl-dependent scaffold role during the development of a variety of cilia.
Materials and methods
Xenopus embryo microinjection and manipulation
To obtain eggs, female frogs were injected with human chorionic gonadotropin (SG-10; Sigma-Aldrich) at 600 U a day before microinjection. The next day, collected eggs were in vitro fertilized with sperm for 30 min before dejellying embryos. SP6 mMESSAGE mMACHINE kit (AM1340; Life Technologies) was used to prepare synthetic mRNAs for microinjection. Drg1 morpholinos (Drg1-MO1: 5′-CTAGCCAGTGTCCCGCTCATTGTTC-3′, and Drg1-MO2: 5′-CCGGTGTAGGTCGCTTCACAGCAAA-3′), Dvl2 morpholino (Dvl2-MO: 5′-TCACTTTAGTCTCCGCCATTCTGCG-3′), Daam1 morpholino (Daam1 MO: 5′-GCCGCAGGTCTGTCAGTTGCTTCTA-3′), Dvl3 morpholino (Dvl3-MO: 5′-GGTAGATGACCTTGGTCTCCCCCAT-3′), and control MOs were purchased from Gene Tools and prepared according to the manufacturer’s instructions. Embryos were microinjected with RNAs and/or MOs at the animal pole at the one-cell stage for co-IP experiments. To target MCCs on the epidermis, morpholinos and mRNAs were injected to both marginal ventral blastomeres of embryos at the four-cell stage. To target cilia on GRP, morpholinos and mRNAs were injected to both marginal dorsal blastomeres of embryos at the four-cell stage.
Immunoblots
Embryos were lysed using TNSG buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 10% glycerol, and protease inhibitors). Protein lysates were loaded on SDS–polyacrylamide gels (8–12%) and transferred to polyvinylidene difluoride membrane. After blocking in TBST (TBS buffer with 0.1% Tween20) containing 5% nonfat dry milk, antibodies were incubated as per the manufacturer’s instructions.
Co-IP
For co-IPs of exogenous protein interactions, the injected embryos were lysed with TNSG buffer including protease inhibitors. Tagged proteins were precipitated with monoclonal anti-HA agarose beads (A2095; Sigma-Aldrich), anti-V5 agarose beads (A7345; Sigma-Aldrich), or anti-Flag M2 affinity gel (A2220; Sigma-Aldrich). For GFP-tagged protein pull-down, anti-GFP antibody was used. The resultant immunocomplexes were formed for 8 h at 4°C on a nutator and washed with lysis buffer four times. For co-IPs of endogenous protein interactions, HT29 and LS174T cells were lysed with TNSG containing 0.5% NP-40, and 10 µl of anti-Dvl2 antibody was incubated with 2 mg of total lysates at 4°C overnight, followed by an additional incubation with protein A/G plus agarose beads (sc-2003; Santa Cruz Biotechnology) for 2 h. Bead immunocomplexes were washed three times with the same lysis buffer. To examine coprecipitation of Drg1 with Dvl2, IB with Drg1 antibody was followed.
Recombinant protein purification and in vitro binding assay
The pGEX4T-2 plasmids encoding WT, Δ329–344, and the TGS domain of Drg1 or Dvl2-HA were transformed into Rosetta (DE3)-competent cells (70954; Sigma-Aldrich). The production of GST fusion proteins was induced by isopropyl-β-D-thiogalactopyranoside at 0.1 mM. GST-Drg1 (WT, Δ329–344, and the TGS domain) proteins were precipitated by glutathione sepharose 4B resin and eluted by incubating with glutathione at 184 mM. For the purification of Dvl2-HA protein, the complex of GST-Dvl2-HA and glutathione resin was incubated with thrombin. For the in vitro binding assay, 1–2 µg of the purified recombinant proteins was used, and the proteins were incubated in in vitro binding buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40, and PMSF) for 1–2 h before washing with the in vitro binding buffer.
IF and confocal microscopy
Embryos were fixed with MEMFA (4% formaldehyde in 1× MEM salt) for 2 h at RT or 4°C overnight and permeabilized with 0.2% Triton X-100 in PBS. For immunostaining to detect phospho-proteins, embryos were fixed with 2% TCA (T6399; Sigma-Aldrich) for 20–30 min at RT and permeabilized with 0.3% Triton X-100 in PBS for 30 min, followed by blocking embryos with PBS containing 2% BSA. Then, embryos were incubated with primary antibody in blocking solution at 4°C overnight. For secondary antibody incubation, Alexa Fluor 488 goat anti-rabbit IgG (dilution 1:500, A11034; Invitrogen), Alexa Fluor 594 goat anti-mouse IgG (dilution 1:500, A11005; Invitrogen), and Alexa Fluor 647 goat anti-mouse IgG (dilution 1:500, A28181; Invitrogen) were used. Confocal microscopy was performed with an LSM 780 microscope equipped with a Plan-Apochromat 63×/1.4 oil differential interference contrast objective. Serial z-stack images for a single MCC and multiple MCCs (low magnification) were taken with 0.2 µm and 0.8 µm distance, respectively.
Quantification of PCP defects in MCCs
The images of CLAMP-GFP and centrin-RFP were taken using Plan-Apochromat 63×/1.4 oil differential interference contrast objective. The planar polarization defects were quantified as previously described (Werner and Mitchell, 2013). The relative angle composed of a reference anterior-posterior axis and the line connecting basal body and rootlet was measured using Fiji and analyzed using Oriana 4.0 (Kovach Computing Services) circular statistics software.
Antibodies
The following antibodies were used for IB, IF, and IP experiments: anti-Drg1 antibody (dilution 1:1,000 for IB, PAB20044; Abnova), anti-Drg2 antibody (1:1,000 for IB, NBP2-16227; Novus Biological), anti-acetylated tubulin antibody (1:1,000 for IF, T7451; Sigma-Aldrich), monoclonal anti–α-tubulin antibody (1:500 for IF, T9026; Sigma-Aldrich), anti-Dvl2 antibody (1:1000 for IB, 1:200 for IP, 3216S; Cell Signaling Technology), anti-Dvl3 antibody (1:2,000 for IB, 2 µg for IP, 13444–1-AP; Proteintech), anti-phospho-cofilin (Ser3) polyclonal antibody (1:500 for IF, 44-1072G; Thermo Fisher Scientific), anti-phospho LIMK1 (phospho T508) antibody (1:500 for IF, ab38508; Abcam), anti-GAPDH antibody (1:5,000 for IB, FL-335; Santa Cruz Biotechnology), anti-GFP antibody (1 µl/tube for IP, 1:500 for IF, NB600-308; Novus Biological), anti-GST antibody (1:2,000 for IB, ab19256; Abcam), HA-tag (C29F4) antibody (1:1,000 for IF, 3724; Cell Signaling Technology), anti-V5-peroxidase antibody (dilution 1:10,000 for IB, V2260; Sigma-Aldrich), and monoclonal anti-Flag-M2 HRP antibody (dilution 1:5,000 for IB, A8592; Sigma-Aldrich).
Molecular cloning and plasmids
Xenopus Drg1 and Xenopus Drg2 constructs were kindly provided by Dr. Jun-ichiro Inoue (The University of Tokyo, Tokyo, Japan). The PCR-amplified coding regions were cloned into pCS2. To produce proteins with tags (e.g., HA, Flag, and V5), tag coding sequences were added to reverse primers. The point mutants of Drg1 (S78N-T100D) were generated by a site-directed mutagenesis method. The deletion mutants of Xenopus of Dvl2 were previously described (Lee et al., 2006; Yoon et al., 2018). Full-length Xenopus Dvl1 and Dvl3 were PCR amplified from cDNA pools generated from mRNAs of stage 20 embryos and subcloned into pCS2 vector. The deletion mutants of Drg1 (Δ313–328 and Δ329–344) and Dvl3 (ΔPDZ) were generated by site-directed mutagenesis. For the other deletion mutants of Drg1 and Dvl3, inserts were PCR amplified and subcloned into pCS2 vector. Daam1-HA plasmid is a gift from Dr. Sergei Sokol (Icahn School of Medicine at Mount Sinai, New York, NY). To generate mutants N-Daam1, C-Daam1, and DAD (Liu et al., 2008; Ju et al., 2010), the DNA sequences were PCR amplified and subcloned into pCS2 or pCS2-mCherry (34936; Addgene). HA-tag encoding DNA sequence was added to a forward primer. CLAMP-GFP and Centrin4-RFP constructs were kindly provided by Dr. John Wallingford (The University of Texas at Austin, Austin, TX). The sequences of all subcloned plasmids were verified by DNA sequencing.
Scanning EM
The embryos at stage 31 were fixed in a cocktail of 4% formaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer and post-fixed using a 1% osmium tetroxide solution, followed by dehydration in a series of graded alcohols ranging from 35% to 100% with a final dehydration course of tetramethylsilane and air dried. The samples were subsequently mounted on stubs, sputter coated with a thin layer of iridium, and imaged at 2.0 kV using a Hitachi S-4500 field emission scanning electron microscope.
Assessment of fluid flow
The experimental setup and materials followed the previous publication (Werner and Mitchell, 2013). To visualize fluid flow, a dissecting scope equipped with a digital camera (Canon Powershop G10) was used. Fluorescent beads (F8836; Invitrogen) were diluted 1:20 with 0.1× Modified Barth’s Saline (MBS) and washed two or three times by centrifuging at 5,000 rpm for 1 min. Bead pellets were resuspended with 0.1× MBS including 1.5% glycerol and dropped onto anesthetized embryos at stage 27 before imaging started. Videos were taken with 10 embryos per group, and velocity of fluid flow was manually measured by tracking individual beads along the body axis on full zoom with a 27-inch monitor using the 2× reverse playing mode of QuickTime player.
Mass spectrometry
Embryos were microinjected with mRNAs encoding Xenopus V5-tagged Dvl2 lacking the DIX domain, and 600 embryos per group were harvested, followed by IP. Gel running was conducted using a 4–12% Bis-Tris gradient gel (NP0321PK2; Thermo Fisher Scientific). After staining with SimplyBlue SafeStain (LC6060; Invitrogen), gel bands were excised and digested with trypsin. Digested peptides were analyzed by an Easy-nLC 1000 instrument with a LTQ Velos Pro mass spectrometer (Thermo Fisher Scientific), as previously described (Ding et al., 2017). Acquired tandem mass spectrometry spectra UniProt Xenopus protein database with SEQUEST interfaced with BioWorks 3.3 (Thermo Fisher Scientific) was used to identify the acquired tandem mass spectrometry spectra.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as previously described (Harland, 1991). Briefly, we used MEMFA (100 mM MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde) as a fixative. Fixed embryos were dehydrated in methanol at −20°C overnight and rehydrated in PBST (PBS + 0.1% Tween20), followed by incubation in 100 mM triethanolamine. After incubation in 100 mM triethanolamine containing 0.25% acetic anhydride, embryos were refixed in 4% paraformaldehyde and washed with PBST. RNA probes were generated from Xenopus Drg1 cDNA (nucleotide from +1 to +1,104; Ishikawa et al., 2003) using MEGAshortscript T7 kit (AM1354; Invitrogen), and embryos were hybridized with RNA probes at 60°C overnight. Embryos were washed with 2× SSC and 0.2× SSC solutions at 60°C and treated with RNases (RNaseA and RNaseT). Following blocking, embryos were incubated with anti–Digoxigennin-AP antibody (11266027; Roche). BM purple alkaline phosphatase substrate (19457200; Roche) was used for chromogenic reaction.
Statistics
The one-way ANOVA with Turkey’s multiple comparisons test and the unpaired two-tailed t test were performed using Prism8.
Online supplemental material
Fig. S1 shows that Dvl3 interacts with Drg1 through DEP+C and that the TGS domain of Drg1 is required for the Dvl2–Drg1 association. Fig. S2 shows that Drg1 knockdown with MO2 having another target sequence also causes decreased acetylated tubulin staining and that coknockdown of Dvl2 and Dvl3 phenocopies the Drg1 knockdown; however, Drg2 knockdown does not induce ciliogenesis defects. Fig. S3 shows that the migration of MCC progenitors to the surface of the epidermis is not delayed upon Drg1 knockdown and that Drg1 knockdown does not affect total amount of exogenous cofilin and LIMK1 proteins. Fig. S4 demonstrates that Drg1 knockdown phenotypes are independent of the GTPase activity. Fig. S5 shows the DAD of Daam1 colocalizes with Drg1 in MCCs and that Drg1 knockdown causes abnormal localization of Daam1 in MCCs. Table S1 shows a list of proteins detected in the mass spectrometric analysis of Dvl2 IPs from Xenopus embryos expressing Dvl2. Videos 1, 2, and 3 show that Drg1 knockdown causes defects in ciliary fluid flow across the epidermis of embryos at stage 27–28. Videos 4, 5, 6, and 7 show the GTPase activity of Drg1 is not required for ciliary function in the MCCs.
Supplementary Material
Acknowledgments
We are grateful to P. McCrea, T. Yamaguchi, and C. Westlake for critically reading the manuscript and providing helpful comments. We thank L. Jenkins for mass spectrometry tests, K. Peifley for technical support for microscopy, and all members of the Cancer and Developmental Biology Laboratory for discussion and comments.
This research was supported by the National Cancer Institute, National Institutes of Health, Intramural Research Program, and was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The authors declare no competing financial interests.
Author contributions: M. Lee designed and carried out the experiments with the help of Y. Hwang, J. Yoon, and J. Sun. A. Harned and K. Nagashima assisted with scanning EM. M. Lee and I.O. Daar wrote the manuscript. I.O. Daar supervised the project. All of the authors discussed the results and commented on the manuscript.
References
- Bernstein B.W., and Bamburg J.R.. 2010. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20:187–195. 10.1016/j.tcb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Scheidel N., and Kuhns S.. 2018. Rab GTPases in cilium formation and function. Small GTPases. 9:76–94. 10.1080/21541248.2017.1353847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E., Lainé M.C., and Sandoz D.. 1990. Cytochalasin D inhibits basal body migration and ciliary elongation in quail oviduct epithelium. Cell Tissue Res. 259:443–454. 10.1007/BF01740770 [DOI] [PubMed] [Google Scholar]
- Brooks E.R., and Wallingford J.B.. 2014. Multiciliated cells. Curr. Biol. 24:R973–R982. 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B., Adamiok A., Mercey O., Revinski D.R., Zaragosi L.E., Pasini A., Kodjabachian L., Barbry P., and Marcet B.. 2015. miR-34/449 control apical actin network formation during multiciliogenesis through small GTPase pathways. Nat. Commun. 6:8386 10.1038/ncomms9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe A., Hamada F., and Bienz M.. 2003. A role of Dishevelled in relocating Axin to the plasma membrane during wingless signaling. Curr. Biol. 13:960–966. 10.1016/S0960-9822(03)00370-1 [DOI] [PubMed] [Google Scholar]
- Dawe H.R., Farr H., and Gull K.. 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120:7–15. 10.1242/jcs.03305 [DOI] [PubMed] [Google Scholar]
- Ding X., Philip S., Martin B.K., Pang Y., Burkett S., Swing D.A., Pamala C., Ritt D.A., Zhou M., Morrison D.K., et al. . 2017. Survival of BRCA2-Deficient Cells Is Promoted by GIPC3, a Novel Genetic Interactor of BRCA2. Genetics. 207:1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2002. Rho GTPases in cell biology. Nature. 420:629–635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Francis S.M., Gas M.E., Daugeron M.C., Bravo J., and Séraphin B.. 2012. Rbg1-Tma46 dimer structure reveals new functional domains and their role in polysome recruitment. Nucleic Acids Res. 40:11100–11114. 10.1093/nar/gks867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganner A., Lienkamp S., Schäfer T., Romaker D., Wegierski T., Park T.J., Spreitzer S., Simons M., Gloy J., Kim E., et al. . 2009. Regulation of ciliary polarity by the APC/C. Proc. Natl. Acad. Sci. USA. 106:17799–17804. 10.1073/pnas.0909465106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., and Chen Y.G.. 2010. Dishevelled: The hub of Wnt signaling. Cell. Signal. 22:717–727. 10.1016/j.cellsig.2009.11.021 [DOI] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R.S., Abitua P.B., Wlodarczyk B.J., Szabo-Rogers H.L., Blanchard O., Lee I., Weiss G.S., Liu K.J., Marcotte E.M., Wallingford J.B., and Finnell R.H.. 2009a The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat. Cell Biol. 11:1225–1232. 10.1038/ncb1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R.S., Bayly R.D., Green S.A., Agarwala S., Lowe C.J., and Wallingford J.B.. 2009b Diversification of the expression patterns and developmental functions of the dishevelled gene family during chordate evolution. Dev. Dyn. 238:2044–2057. 10.1002/dvdy.22028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R., Kato Y., and He X.. 2001. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 107:843–854. 10.1016/S0092-8674(01)00614-6 [DOI] [PubMed] [Google Scholar]
- Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514. 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- Harland R.M. 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36:685–695. 10.1016/S0091-679X(08)60307-6 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Thompson J., Yates J.R. III, and Marshall W.F.. 2012. Proteomic analysis of mammalian primary cilia. Curr. Biol. 22:414–419. 10.1016/j.cub.2012.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Azuma S., Ikawa S., Morishita Y., Gohda J., Akiyama T., Semba K., and Inoue J.. 2003. Cloning and characterization of Xenopus laevis drg2, a member of the developmentally regulated GTP-binding protein subfamily. Gene. 322:105–112. 10.1016/j.gene.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Azuma S., Ikawa S., Semba K., and Inoue J.. 2005. Identification of DRG family regulatory proteins (DFRPs): specific regulation of DRG1 and DRG2. Genes Cells. 10:139–150. 10.1111/j.1365-2443.2005.00825.x [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Akiyama T., Ito K., Semba K., and Inoue J.. 2009. Independent stabilizations of polysomal Drg1/Dfrp1 complex and non-polysomal Drg2/Dfrp2 complex in mammalian cells. Biochem. Biophys. Res. Commun. 390:552–556. 10.1016/j.bbrc.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Ju R., Cirone P., Lin S., Griesbach H., Slusarski D.C., and Crews C.M.. 2010. Activation of the planar cell polarity formin DAAM1 leads to inhibition of endothelial cell proliferation, migration, and angiogenesis. Proc. Natl. Acad. Sci. USA. 107:6906–6911. 10.1073/pnas.1001075107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Zhang S., Werner M.E., Brotslaw E.J., Mitchell J.W., Altabbaa M.M., and Mitchell B.J.. 2018. CLAMP/Spef1 regulates planar cell polarity signaling and asymmetric microtubule accumulation in the Xenopus ciliated epithelia. J. Cell Biol. 217:1633–1641. 10.1083/jcb.201706058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y., and Habas R.. 2008. Wnt signal transduction pathways. Organogenesis. 4:68–75. 10.4161/org.4.2.5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., and Geyer M.. 2014. Formins as effector proteins of Rho GTPases. Small GTPases. 5:e29513 10.4161/sgtp.29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Iwao M., Yamagishi T., Noda M., and Asashima M.. 1993. Expression of GTP-binding protein gene drg during Xenopus laevis development. Int. J. Dev. Biol. 37:539–546. [PubMed] [Google Scholar]
- Lee H.S., Bong Y.S., Moore K.B., Soria K., Moody S.A., and Daar I.O.. 2006. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat. Cell Biol. 8:55–63. 10.1038/ncb1344 [DOI] [PubMed] [Google Scholar]
- Lee H., Cheong S.M., Han W., Koo Y., Jo S.B., Cho G.S., Yang J.S., Kim S., and Han J.K.. 2018. Head formation requires Dishevelled degradation that is mediated by March2 in concert with Dapper1. Development. 145:145 10.1242/dev.143107 [DOI] [PubMed] [Google Scholar]
- Li B., and Trueb B.. 2000. DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta. 1491:196–204. 10.1016/S0167-4781(00)00025-7 [DOI] [PubMed] [Google Scholar]
- Li D., Hallett M.A., Zhu W., Rubart M., Liu Y., Yang Z., Chen H., Haneline L.S., Chan R.J., Schwartz R.J., et al. . 2011. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 138:303–315. 10.1242/dev.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Tan G., Levenkova N., Li T., Pugh E.N. Jr., Rux J.J., Speicher D.W., and Pierce E.A.. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics. 6:1299–1317. 10.1074/mcp.M700054-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L.W., and Habas R.. 2008. Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. USA. 105:210–215. 10.1073/pnas.0707277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Lv Y., Dong J., Hu S., and Peng R.. 2016. DRG1 is a potential oncogene in lung adenocarcinoma and promotes tumor progression via spindle checkpoint signaling regulation. Oncotarget. 7:72795–72806. 10.18632/oncotarget.11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K., and He X.. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 17:9–26. 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M.A., Park S.T., and Sun X.H.. 1996. Association of a novel GTP binding protein, DRG, with TAL oncogenic proteins. Oncogene. 12:2343–2350. [PubMed] [Google Scholar]
- Mani M., Lee U.H., Yoon N.A., Kim H.J., Ko M.S., Seol W., Joe Y., Chung H.T., Lee B.J., Moon C.H., et al. . 2016. Developmentally regulated GTP-binding protein 2 coordinates Rab5 activity and transferrin recycling. Mol. Biol. Cell. 27:334–348. 10.1091/mbc.e15-08-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M., Lee U.H., Yoon N.A., Yoon E.H., Lee B.J., Cho W.J., and Park J.W.. 2017. Developmentally regulated GTP-binding protein 2 is required for stabilization of Rac1-positive membrane tubules. Biochem. Biophys. Res. Commun. 493:758–764. 10.1016/j.bbrc.2017.08.110 [DOI] [PubMed] [Google Scholar]
- Meunier A., and Azimzadeh J.. 2016. Multiciliated Cells in Animals. Cold Spring Harb. Perspect. Biol. 8:a028233 10.1101/cshperspect.a028233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Canny S.G., Jang C.W., Cho K., Ji H., Wagner D.S., Jones E.A., Habas R., and McCrea P.D.. 2011. Pronephric tubulogenesis requires Daam1-mediated planar cell polarity signaling. J. Am. Soc. Nephrol. 22:1654–1664. 10.1681/ASN.2010101086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B., Jacobs R., Li J., Chien S., and Kintner C.. 2007. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 447:97–101. 10.1038/nature05771 [DOI] [PubMed] [Google Scholar]
- Mitchell B., Stubbs J.L., Huisman F., Taborek P., Yu C., and Kintner C.. 2009. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr. Biol. 19:924–929. 10.1016/j.cub.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake K., Kusakabe M., Takahashi C., and Nishida E.. 2015. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat. Commun. 6:6666 10.1038/ncomms7666 [DOI] [PubMed] [Google Scholar]
- Murdoch J.N., and Copp A.J.. 2010. The relationship between sonic Hedgehog signaling, cilia, and neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 88:633–652. 10.1002/bdra.20686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata S., Nakatani J., Herranz-Pérez V., Cheng J., Belinson H., Inubushi T., Snider W.D., García-Verdugo J.M., Wynshaw-Boris A., and Alvarez-Buylla A.. 2014. Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron. 83:558–571. 10.1016/j.neuron.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O., Kerney R., Saint-Jeannet J.P., and Sokol S.Y.. 2018. Regulation of neural crest development by the formin family protein Daam1. Genesis. 56:e23108 10.1002/dvg.23108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., You Y., Huang T., and Brody S.L.. 2007. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 120:1868–1876. 10.1242/jcs.005306 [DOI] [PubMed] [Google Scholar]
- Pan W.J., Pang S.Z., Huang T., Guo H.Y., Wu D., and Li L.. 2004. Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 14:324–330. 10.1038/sj.cr.7290232 [DOI] [PubMed] [Google Scholar]
- Park T.J., Haigo S.L., and Wallingford J.B.. 2006. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat. Genet. 38:303–311. 10.1038/ng1753 [DOI] [PubMed] [Google Scholar]
- Park T.J., Mitchell B.J., Abitua P.B., Kintner C., and Wallingford J.B.. 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40:871–879. 10.1038/ng.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Arellano I., Spínola-Amilibia M., and Bravo J.. 2013. Human Drg1 is a potassium-dependent GTPase enhanced by Lerepo4. FEBS J. 280:3647–3657. 10.1111/febs.12356 [DOI] [PubMed] [Google Scholar]
- Sazuka T., Kinoshita M., Tomooka Y., Ikawa Y., Noda M., and Kumar S.. 1992. Expression of DRG during murine embryonic development. Biochem. Biophys. Res. Commun. 189:371–377. 10.1016/0006-291X(92)91568-B [DOI] [PubMed] [Google Scholar]
- Schellhaus A.K., Moreno-Andrés D., Chugh M., Yokoyama H., Moschopoulou A., De S., Bono F., Hipp K., Schäffer E., and Antonin W.. 2017. Developmentally Regulated GTP binding protein 1 (DRG1) controls microtubule dynamics. Sci. Rep. 7:9996 10.1038/s41598-017-10088-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger K., Hall A., and Tolwinski N.. 2009. Wnt signaling pathways meet Rho GTPases. Genes Dev. 23:265–277. 10.1101/gad.1760809 [DOI] [PubMed] [Google Scholar]
- Schweickert A., Weber T., Beyer T., Vick P., Bogusch S., Feistel K., and Blum M.. 2007. Cilia-driven leftward flow determines laterality in Xenopus. Curr. Biol. 17:60–66. 10.1016/j.cub.2006.10.067 [DOI] [PubMed] [Google Scholar]
- Sharma M., Castro-Piedras I., Simmons G.E. Jr., and Pruitt K.. 2018. Dishevelled: A masterful conductor of complex Wnt signals. Cell. Signal. 47:52–64. 10.1016/j.cellsig.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Galati D., Winey M., and Klymkowsky M.W.. 2014. Chibby functions in Xenopus ciliary assembly, embryonic development, and the regulation of gene expression. Dev. Biol. 395:287–298. 10.1016/j.ydbio.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnitsar I., Bashkurov M., Masson G.R., Ogunjimi A.A., Mosessian S., Cabeza E.A., Hirsch C.L., Trcka D., Gish G., Jiao J., et al. . 2015. PTEN regulates cilia through Dishevelled. Nat. Commun. 6:8388 10.1038/ncomms9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit S.T., and Manser E.. 2011. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 124:679–683. 10.1242/jcs.064964 [DOI] [PubMed] [Google Scholar]
- Spassky N., and Meunier A.. 2017. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18:423–436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Spiering D., and Hodgson L.. 2011. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adhes. Migr. 5:170–180. 10.4161/cam.5.2.14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.E., and Mitchell B.J.. 2012. Understanding ciliated epithelia: the power of Xenopus. Genesis. 50:176–185. 10.1002/dvg.20824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.E., and Mitchell B.J.. 2013. Using Xenopus skin to study cilia development and function. Methods Enzymol. 525:191–217. 10.1016/B978-0-12-397944-5.00010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.E., Hwang P., Huisman F., Taborek P., Yu C.C., and Mitchell B.J.. 2011. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 195:19–26. 10.1083/jcb.201106110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.E., Mitchell J.W., Putzbach W., Bacon E., Kim S.K., and Mitchell B.J.. 2014. Radial intercalation is regulated by the Par complex and the microtubule-stabilizing protein CLAMP/Spef1. J. Cell Biol. 206:367–376. 10.1083/jcb.201312045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y.I., Aravind L., Grishin N.V., and Koonin E.V.. 1999. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9:689–710. [PubMed] [Google Scholar]
- Wong H.C., Bourdelas A., Krauss A., Lee H.J., Shao Y., Wu D., Mlodzik M., Shi D.L., and Zheng J.. 2003. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell. 12:1251–1260. 10.1016/S1097-2765(03)00427-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga T., Itoh K., and Sokol S.Y.. 2011. Regulation of basal body and ciliary functions by Diversin. Mech. Dev. 128:376–386. 10.1016/j.mod.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga T., Hoff S., Schell C., Helmstädter M., Kretz O., Kuechlin S., Yakulov T.A., Engel C., Müller B., Bensch R., et al. . 2015. The polarity protein Inturned links NPHP4 to Daam1 to control the subapical actin network in multiciliated cells. J. Cell Biol. 211:963–973. 10.1083/jcb.201502043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Hwang Y.S., Lee M., Sun J., Cho H.J., Knapik L., and Daar I.O.. 2018. TBC1d24-ephrinB2 interaction regulates contact inhibition of locomotion in neural crest cell migration. Nat. Commun. 9:3491 10.1038/s41467-018-05924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Rual J.F., Tamai K., Harada Y., Vidal M., He X., and Kirchhausen T.. 2007. Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev. Cell. 12:129–141. 10.1016/j.devcel.2006.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., and Mitchell B.J.. 2016. Basal bodies in Xenopus. Cilia. 5:2 10.1186/s13630-016-0024-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Shi J., Winey M., and Klymkowsky M.W.. 2016. Identifying domains of EFHC1 involved in ciliary localization, ciliogenesis, and the regulation of Wnt signaling. Dev. Biol. 411:257–265. 10.1016/j.ydbio.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.