Microtubule bundles in the spindle midzone have been reported to either promote or hinder chromosome movement. Pamula et al. examine the assembly dynamics of midzone microtubule bundles during anaphase and how chromosome segregation is impacted by aberrant bundle assembly.
Abstract
In the spindle midzone, microtubules from opposite half-spindles form bundles between segregating chromosomes. Microtubule bundles can either push or restrict chromosome movement during anaphase in different cellular contexts, but how these activities are achieved remains poorly understood. Here, we use high-resolution live-cell imaging to analyze individual microtubule bundles, growing filaments, and chromosome movement in dividing human cells. Within bundles, filament overlap length marked by the cross-linking protein PRC1 decreases during anaphase as chromosome segregation slows. Filament ends within microtubule bundles appear capped despite dynamic PRC1 turnover and submicrometer proximity to growing microtubules. Chromosome segregation distance and rate are increased in two human cell lines when microtubule bundle assembly is prevented via PRC1 knockdown. Upon expressing a mutant PRC1 with reduced microtubule affinity, bundles assemble but chromosome hypersegregation is still observed. We propose that microtubule overlap length reduction, typically linked to pushing forces generated within filament bundles, is needed to properly restrict spindle elongation and position chromosomes within daughter cells.
Introduction
Specialized microtubule arrays perform critical functions in diverse cellular contexts. During cell division, kinetochore microtubules assemble from stabilized filaments and, in anaphase, drive chromosome-to-pole movement (Asbury, 2017). During anaphase, an array of overlapping microtubules called the spindle midzone or central spindle assembles between segregating sister chromosomes (Eggert et al., 2006). Within this structure, microtubules from opposite half-spindles interdigitate at their plus ends, forming bundles of antiparallel microtubules (Euteneuer and McIntosh, 1980; Heidemann and McIntosh, 1980). Microtubule bundles assemble during anaphase in diverse eukaryotes including yeast, worms, and humans, suggesting a conserved function (Ding et al., 1993; Mastronarde et al., 1993; Oegema and Hyman, 2006).
The organization of midzone microtubules has been examined using electron microscopy, which has revealed that microtubules in the midzone overlap extensively and likely undergo relative sliding as anaphase progresses (McIntosh et al., 1975a,b). Live-cell imaging has revealed that these bundled microtubules coexist alongside polymerizing microtubules in the spindle midzone and undergo two kinds of dynamics (Shelden and Wadsworth, 1990; Mastronarde et al., 1993; Yamashita et al., 2015). First, compared with unbundled growing microtubules, which have a half-life of tens of seconds, a subset of midzone microtubules are stabilized ∼10-fold (Salmon et al., 1984; Saxton et al., 1984). The plus ends of these filaments, revealed by driving monopolar spindles into anaphase, are capped in a Kif4-dependent manner, an activity that likely suppresses plus end dynamics in bipolar spindle midzones as well (Hu et al., 2011). Second, the length of filament overlap within microtubule bundles decreases with increasing chromosome segregation distance, proposed to be a result of relative microtubule sliding (Saxton and McIntosh, 1987; Mastronarde et al., 1993). However, due in part to challenges in imaging microtubule bundles within the full volume of the spindle midzone at high temporal frequency and with isotropic resolution, we do not know how the 3D organization of microtubule bundles changes during the fast dynamics of anaphase in human cells.
Early laser-cutting experiments in fungi support a model in which midzone bundles function to limit the separation rate of chromosomes during anaphase (Aist and Berns, 1981). Similar results were observed in the first mitotic division of Caenorhabditis elegans embryos (Grill et al., 2001), where molecular dissection has revealed a role for kinesin-5 motors in restricting relative filament sliding and pole separation during anaphase (Saunders et al., 2007). In contrast, laser-cutting experiments in diatoms, fission yeast, human cells, and C. elegans oocytes support a model in which microtubule bundles in the spindle midzone function to drive chromosome separation and spindle elongation (Leslie and Pickett-Heaps, 1983; Khodjakov et al., 2004; Laband et al., 2017; Vukušić et al., 2017). In human cells, a specialized array of overlapping microtubules termed “bridging fibers” have been described that link kinetochore fibers on sister chromatids during metaphase (Kajtez et al., 2016). Severing both kinetochore microtubules and bridging fibers during early anaphase in human U2OS cells prevents the segregation of sister chromatids (Vukušić et al., 2017). In contrast to these data, a study using another vertebrate species (porcine kidney cells) has shown that the spindle midzone restricts chromosome segregation in a kinesin-5–dependent manner (Collins et al., 2014). We note that the use of fast infrared laser in recent studies has partially overcome the potential toxicity of UV radiation (Brugués and Needleman, 2014; Vukušić et al., 2017); however, it is still difficult to exclude potential indirect effects due to local protein damage, and accounting for the conflicting results remains challenging.
Targeted protein knockdown has revealed the requirement for several key proteins in the assembly of the spindle midzone (Glotzer, 2009). Among these is PRC1, a member of the conserved Ase1/PRC1/MAP65 family of microtubule cross-linking proteins. Knockdown of PRC1 or its homologues prevents midzone microtubule bundle assembly in diverse eukaryotes without preventing metaphase bipolar spindle formation or activating the spindle assembly checkpoint (Jiang et al., 1998; Verbrugghe and White, 2004; Vernì et al., 2004). This approach has been used extensively in human cells to examine the consequence on metaphase microtubule bundle assembly, anaphase midzone assembly, and cytokinesis (Jiang et al., 1998; Mollinari et al., 2002, 2005; Kurasawa et al., 2004; Polak et al., 2017). However, the consequence on chromosome segregation rates in human cells has not been studied. Further, it is unclear how select biochemical properties of PRC1, such as microtubule binding affinity, are tuned to allow microtubule bundles in the midzone to accomplish species-specific functions.
Here, we combine high-resolution live-cell imaging with protein knockdown and structure-guided mutagenesis to investigate how microtubule bundle assembly in the midzone impacts chromosome segregation dynamics in human cells. We use lattice light sheet microscopy (LLSM) to relate anaphase chromosome movement to the dynamics of bundled microtubules and growing filaments in different stages of anaphase. We examine time-dependent changes in bundle overlap marked by PRC1 in cells expressing wild-type or mutant GFP-tagged protein. Our data suggest that the proper functioning of the midzone brake depends not only on the assembly of microtubule bundles but also on the timely reduction in overlap length within microtubule bundles during chromosome segregation.
Results
3D analysis of microtubule bundles and chromosomes during anaphase
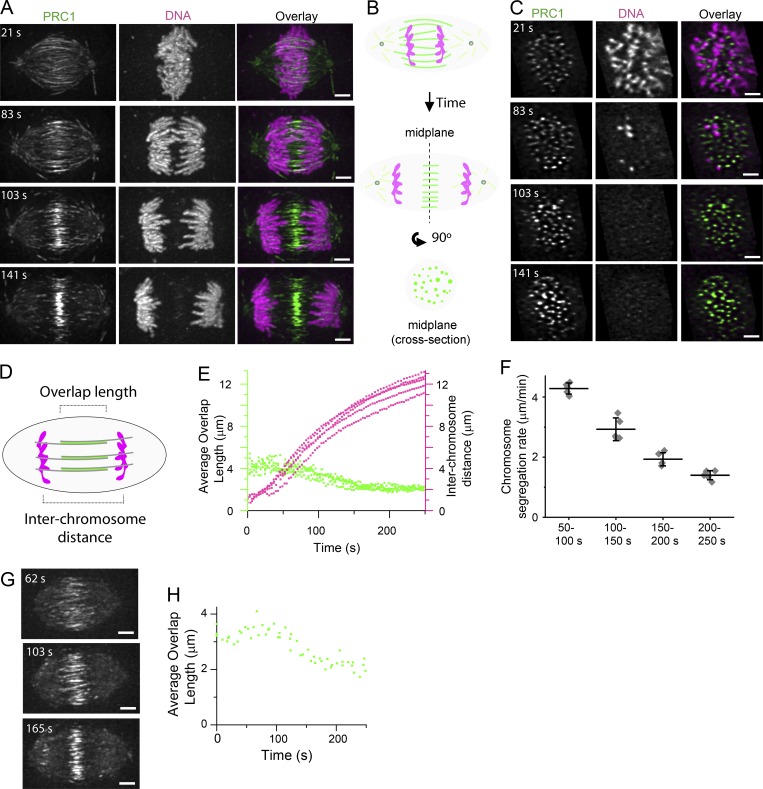
To relate the dynamics of microtubule bundles to chromosome segregation, we imaged dividing hTERT-RPE1 cells stably expressing GFP-tagged PRC1 (hereafter referred to as GFP-PRC1) and treated with a DNA dye (SiR-DNA). In vitro studies have shown that PRC1 selectively binds antiparallel microtubule overlaps (Bieling et al., 2010; Subramanian et al., 2010). Therefore, we reasoned that GFP-PRC1 localization in dividing cells could be used as a readout of antiparallel overlap within microtubule bundles. We used LLSM to image whole cell volumes (101 frames at 300-nm step size) in two colors at rates up to 20 cell volumes/min (see Materials and methods; Chen et al., 2014). Consistent with our immunofluorescence staining of hTERT-RPE1 cells (Fig. S1, A and B), GFP-PRC1 signal was detected in between the segregating chromosomes and between poles and the cell cortex (Fig. 1 A and Video 1) likely corresponding to end tags, micrometer-sized zones at the plus ends of microtubules that accumulate PRC1 and Kif4 (Subramanian et al., 2013). We also note that the localization pattern of PRC1 is reminiscent of the “stem bodies” observed in the midzone of anaphase spindles by electron microscopy (Brinkley and Cartwright, 1971; McIntosh and Landis, 1971). We examined select cross-sectional planes perpendicular to the spindle pole-to-pole axis and selected the plane equidistant from the two spindle poles (hereafter, the spindle midplane) for further examination (Fig. 1 B). In the spindle midplane, GFP-PRC1 appeared as spots of signal intensity throughout anaphase, typically <1 µm in diameter, revealing individual bundle cross sections and spatial distributions (Fig. 1 C). As the majority of GFP-PRC1 signal was localized to microtubule bundles located in the midzone (Fig. S1, C and D), we focused on this region for further examination.
Figure 1.
3D analysis of microtubule bundles and chromosomes in dividing cells. (A and C) Near-simultaneous two-color LLSM was used to image GFP-PRC1 and chromosomes during anaphase in hTERT-RPE1 cells. Cell volumes (101 images in each channel at 300-nm step size) were captured at 3-s intervals. T = 0 was assigned to the frame immediately before that with detectable chromatid separation. (A) Single-channel images (maximum-intensity projections) and overlays from select time points show GFP-PRC1 (green) and chromosomes (magenta). Time-lapse recording is provided in Video 1. Scale bar, 3 µm. (B) Schematic of an anaphase spindle shows PRC1 (green) and chromosomes (magenta) at a plane incident with the pole-to-pole axis and at the spindle midplane, a cross-sectional plane orthogonal to the pole-to-pole axis and equidistant between the two poles. (C) Single-channel images (single image planes) and overlays show the spindle midplane of the cell shown in A. Scale bar, 3 µm. (D) Schematic of an anaphase spindle shows region of microtubule overlap marked by PRC1 (green) and chromosomes (magenta). The length of microtubule overlap and interchromosome distance is indicated. (E) Plots of average microtubule overlap length (green dots) and interchromosome distance (magenta dots) versus time in hTERT-RPE1 cells. Average microtubule overlap length was determined for each cell at each frame (n = 5 cells). (F) Average chromosome segregation rates following anaphase onset. Data were binned in 50-s time intervals. Error bars are SD. (G) LLSM was used to image Halo-PRC1 during anaphase in HeLa cells. Cell volumes (101 images at 300-nm step size) were captured at 9.5–20.5-s intervals. Single-channel images (maximum-intensity projections) show select time points from an example cell imaged at 20.5-s intervals. T = 0 was assigned to the frame immediately before that with detectable spindle elongation. Scale bar, 3 µm. (H) Plot of average microtubule overlap length versus time in HeLa cells. Average microtubule overlap length was determined for each cell at each frame (n = 3 cells).
To identify the localization of GFP-PRC1, we processed the LLSM image data using a multistep approach that included 3D watershed and segmentation analysis (see Materials and methods). We examined the length of GFP-PRC1 decoration on microtubule bundles in the spindle midzone and tracked chromosome segregation distance (Fig. 1 D). At T = 0, the frame immediately before that with detectable chromatid separation (anaphase onset), the average length of GFP-PRC1 decoration was 4.0 ± 0.4 µm (mean ± SD, n = 5 cells), spanning ∼40% of the total spindle length (Fig. 1 E, green points). The length of GFP-PRC1 decoration began to decrease after an ∼50-s delay, reaching 2.1 ± 0.3 µm at T = 200 s. During this time window, chromosome separation distance increased from 3.4 ± 0.6 µm (T = 50 s) to 11.1 ± 0.8 µm (T = 200 s; Fig. 1 E, magenta points). After this time, a substantial change in the length of GFP-PRC1 decoration or chromosome separation distance was not observed. We determined the rate of chromosome separation in different time intervals (50-s bin size) and found that the fastest rate of chromosome movement occurred between 50 and 100 s (4.3 ± 0.2 µm/min, n = 5 cells; Fig. 1 F), approximately four times faster than the rate of GFP-PRC1 length shortening (∼1 µm/min) during this time. We repeated these analyses in an independent cell line (HeLa cells expressing Halo-tagged PRC1 [hereafter, Halo-PRC1]) and found similar results (Fig. 1 G). In HeLa cells, the length of Halo-PRC1 decoration was 3.4 ± 0.2 µm at T = 0 s and decreased to 2.4 ± 0.2 µm by T = 200 s (n = 3 cells; Fig. 1 H). It is likely that the change in GFP-PRC1 and Halo-PRC1 decoration reflects a change in the overlap length of microtubules in bundles. Together, these data suggest that in the absence of filament growth within microtubule bundles, changes in the length of filament overlap are unlikely to contribute substantially to chromosome separation distance during anaphase in human cells.
Dynamics of microtubule bundles and FRAP analysis of PRC1
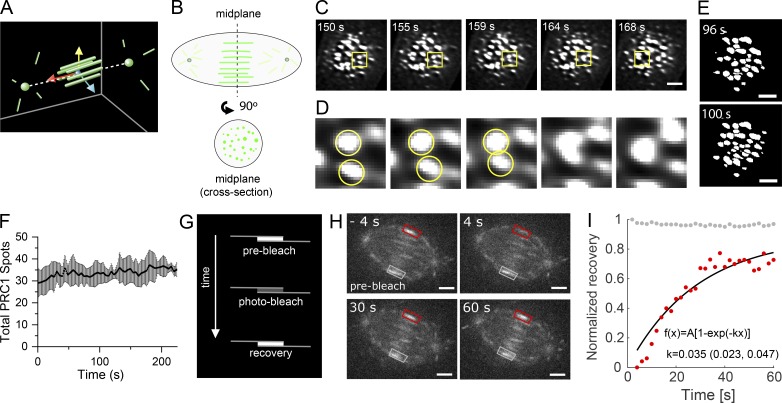
To determine how chromosome segregation is temporally correlated with changes in the organization of microtubule bundles within the spindle midzone, we examined GFP-PRC1 localization in the spindle midplane (Fig. 2, A and B). We found that GFP-PRC1 spots frequently appeared to change shape between sequential frames (Fig. 2 C). Occasionally, we observed spots approaching one another in consecutive image frames and fusing, forming a single spot with higher signal density (Fig. 2 D) that rarely split back into two spots. Such bundle fusion events could be detected throughout anaphase before furrow ingression and typically could be detected once every few frames (∼10 s).
Figure 2.
Dynamics of microtubule bundles and FRAP analysis of PRC1. (A) Schematic indicating position of PRC1-tagged microtubules (green bars) and position of the spindle midplane, the plane formed by passing through yellow and blue vectors, orthogonal to the red vector, equidistant to the two spindle poles (green spheres). (B) Schematic of an anaphase spindle shows GFP-PRC1 (green) localization in a plane incident with the pole-to-pole axis and in the spindle midplane. (C–E) LLSM was used to image GFP-PRC1 during anaphase in hTERT-RPE1 cells. T = 0 s was assigned to the frame immediately before detectable pole separation. Cells were imaged at 4.4-s intervals. (C) Single-channel images (single cross-sectional plane) at select time points. Scale bar, 3 µm. (D) Inset from yellow boxes in C, magnified 5.3×. Two spots of GFP-PRC1 intensity in consecutive frames are indicated (yellow circles). (E) Single-channel images (single cross-sectional plane) after watershed processing. Images from two consecutive frames are shown. Scale bar, 3 µm. (F) Plot of the number of spots versus time detected in watershed-processed images (mean ± SD). (G) Schematic of FRAP experiment. Single microtubule bundles were targeted for photobleaching and the fluorescence recovery monitored over time. (H) Analysis of GFP-PRC1 FRAP in hTERT-RPE1 cells imaged using spinning disk confocal microscopy. Time = 0 is time of photobleach laser pulse. Overlay indicates region targeted for photobleaching (red box) and an eqivalent unphotobleached control area (gray box). Scale bar, 3 µm. (I) Plot of normalized recovery for photobleached GFP-PRC1 signal (red dots) and unbleached GFP-PRC1 control signal (gray dots) for the cell shown in H. Data were fitted to the following equation: f(x) = A[1 − exp(−kx)]; k = 0.035 (95% confidence bounds 0.023 to 0.047).
In metaphase, the number of PRC1-tagged microtubule bundles has been proposed to be determined by the number of chromosome pairs (Polak et al., 2017) and likely corresponds to the number of bundles at the start of anaphase. In hTERT-RPE1 cells, which have 46 chromosomes, as few as five fusion events could decrease the number of bundles by 10%. To investigate this possibility, we performed 2D watershed analysis on segmented LLSM images of GFP-PRC1 in the spindle midplane (Fig. 2 E). We counted the number of spots in each frame and found that the total number of spots fluctuated in consecutive image frames and overall exhibited a modest increase (Fig. 2 F), suggesting that fusion between bundles is supplemented by the assembly of additional microtubule bundles. This observation is consistent with centrosome-independent microtubule formation pathways being active during anaphase and the observation that midzone bundles can assemble in the absence of preanaphase microtubules (Canman et al., 2000; Goshima et al., 2008).
To examine the dynamics of PRC1 cross-linkers within microtubule bundles during early to midanaphase when the length of microtubule overlap within bundles is actively decreasing, we performed FRAP experiments in hTERT-RPE1 cells expressing GFP-PRC1 (Fig. 2 G). In the absence of chromosome markers, we estimated the time in anaphase based on the length of microtubule overlap marked by GFP-PRC1. We selected cells that had microtubule bundle overlap lengths of 2–3.5 µm, indicating that cells were in the first ∼2 min of anaphase (Fig. 2 H). We found that the rate of recovery of GFP-PRC1 within microtubule bundles had a t1/2 of 36 ± 9 s (n = 5 cells; mean ± SD; Fig. 2 I). Previous studies have shown that microtubules in the midzone are stabilized during anaphase, with turnover times of several minutes (Saxton and McIntosh, 1987; Hu et al., 2011). Our FRAP data suggest that the PRC1 molecules turn over faster than the microtubules to which they bind. Further, these data suggest that unlike PRC1-tagged microtubule bundles found in metaphase spindles (Polak et al., 2017), the position of anaphase microtubule bundles do not appear to be constrained by the position of kinetochore pairs. Overall, these results uncover unappreciated midzone bundle dynamics that occur as chromosomes separate.
Examining PRC1 and EB1 localization in cross-sectional planes of dividing cells
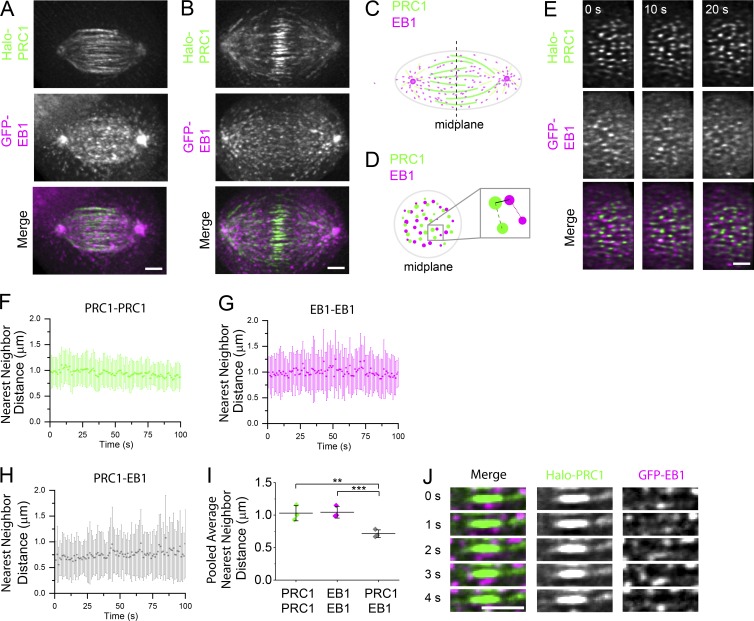
In the spindle midzone, PRC1-tagged microtubule bundles are interspersed within a dense network of EB1-tagged dynamic filaments. To analyze the dyanmics of these filaments we generated stable hTERT-RPE1 cells expressing both GFP-tagged EB1 (hereafter, GFP-EB1), a plus end tip-tracking protein, and Halo-PRC1. We imaged dividing cells using two-color LLSM after treating with a Halo-reactant dye (Fig. 3, A and B; and Video 2; see Materials and methods). We examined cells after spindle elongation began, which is coordinated with anaphase onset (sister chromatid separation) in these cells (Su et al., 2016). Halo-PRC1 signal intensity was highest near the spindle midplane (Fig. 3, A and B), similar to endogenous PRC1 (Fig. S1, A and B). We performed 3D tracking of EB1 comets (Fig. S2, A and B) and found that the average track velocity (0.33 ± 0.08 µm/s; mean ± SD) was similar to that reported in a previous study (Yamashita et al., 2015).
Figure 3.
Examining PRC1 and EB1 localization in cross-sectional planes of dividing cells. (A and B) Near-simultaneous two-color LLSM was used to image Halo-PRC1 and GFP-EB1 during anaphase in hTERT-RPE1 cells. Cell volumes (58 images in each channel at 350-nm step size) were captured at 1-s intervals. A select time point from an early anaphase (A) and a late anaphase (B) spindle from two different cells is shown. Single-channel images (maximum-intensity projections) and overlays show Halo-PRC1 (green) and GFP-EB1 (magenta). Time-lapse recording is provided in Video 2. Scale bar, 3 µm. (C and D) Schematics of anaphase spindles show PRC1 and EB1 decoration in a plane incident with the pole-to-pole axis (C) or in the spindle midplane, the plane equidistant from the two spindle poles and perpendicular to the spindle long axis (D). (C) The position of the spindle midplane is indicated (dashed line). (D) Inset shows schematic of nearest neighbor distances measured between two PRC1 spots (dashed line), two EB1 spots (dotted line), or one PRC1 and one EB1 spot (solid line). (E) Midplane of the cell shown in B. T = 0 indicates the start of movie. Time-lapse recording is provided in Video 3. (F–H) Nearest-neighbor distances versus time show mean ± SD for each frame of the video for the cell shown in B and E. T = 0 indicates the start of movie. Measurements between pairs of PRC1 spots (F), pairs of EB1 spots (G), and PRC1 and EB1 spots (H) are shown. (I) Average nearest-neighbor distance measurements (pooled from the first 30 frames of each movie, n = 3 cells; PRC1–PRC1: 1.03 ± 0.12 µm; EB1–EB1: 1.04 ± 0.09 µm; PRC1–EB1: 0.71 ± 0.06 µm; **, P < 0.02; ***, P < 0.004). Error bars are SD. (J) Time series of a select region (single plane) in the midzone from the cell shown in B, indicating relative position of a single microtubule bundle and GFP-EB1 signal. Single-channel images and overlays show Halo-PRC1 (green) and GFP-EB1 (magenta) in consecutive frames. Scale bar, 3 µm.
We next examined Halo-PRC1 and EB1-GFP at the spindle midplane (Fig. 3, C and D). Halo-PRC1 and GFP-EB1 signal intensity both appear as spots typically <1 µm in diameter (Fig. 3 E). Two-color overlays showed that PRC1-tagged microtubules and dynamic EB1-tagged microtubules coexist in a dense network at the spindle midplane (Fig. 3 E and Video 3). We performed 2D localization analysis on GFP-EB1 and Halo-PRC1 spots and measured the nearest-neighbor distance between spots (Fig. 3 D). The average nearest-neighbor distance between Halo-PRC1 and other Halo-PRC1 spots was ∼1 µm and did not change over the course of the video (100 frames at 1 frame/s; Fig. 3 F). We observed a similar trend for the nearest-neighbor distance between GFP-EB1 and other GFP-EB1 spots (Fig. 3 G). The average nearest-neighbor distance between PRC1 and EB1 spots was smaller, ∼0.6–0.7 µm (Fig. 3 H). We pooled the data from the first 30 frames of each video (n = 3 cells; n = 44 ± 10 EB1-EB1 pairs per frame; 39 ± 12 PRC1-PRC1 pairs per frame; Fig. 3 I) and found this difference to be significant (P < 0.02; two-tailed t test). Importantly, each of these measurements was similar for cells at different stages of anaphase, regardless of the length of microtubule overlap.
To address whether the observed changes in microtubule overlap length were directly related to the extent of microtubule sliding, we considered the possibility that microtubules in bundles could be growing. We inspected individual Halo-PRC1–tagged microtubule bundles, focusing on the ends of microtubule overlap regions. The region of overlap marked by Halo-PRC1 appeared spatially separated from GFP-EB1 signal (Fig. 3 J). In one rare example, we could observe GFP-EB1 colocalizing with one end of a microtubule overlap region, but we did not detect an increase in overlap length (Fig. S2 C). These data suggest that the ends of microtubules in overlap regions are nondynamic and support a model in which the change in overlap length within microtubule bundles during anaphase is directly related to the extent of microtubule sliding. Earlier work has shown that tubulin subunits can incorporate into midzone filaments (Saxton et al., 1984). However, this occurs over the course of several minutes in late anaphase and telophase cells, time points that we did not examine. Therefore, we do not believe these data are contradictory. Together, these data show that the spacing of PRC1 decorated microtubule bundles and growing EB1-tagged microtubules are largely independent of each other and of chromosome position along the spindle’s long axis during anaphase.
Selective disruption of microtubule bundling results in increased chromosome segregation rates and pole elongation rates
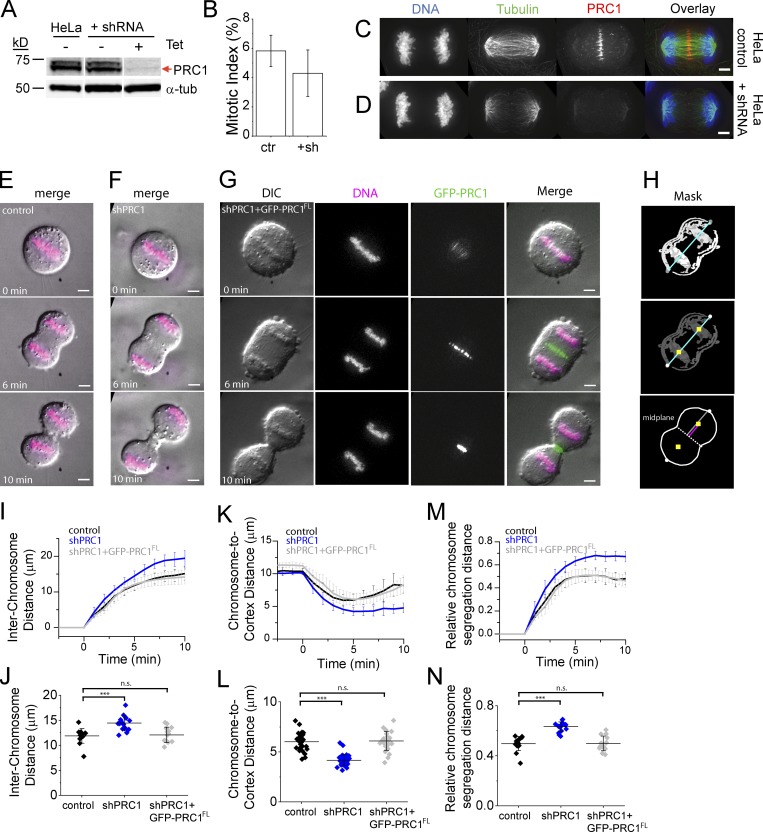
To examine how selective disruption of microtubule bundle assembly impacts chromosome segregation dynamics, we knocked down PRC1. We adopted an inducible shRNA-based approach, as genetic deletion leads to cumulative defects that result in senescence (McKinley and Cheeseman, 2017). Western blots of metaphase-arrested cell lysates showed that the efficiency of PRC1 knockdown was 95 ± 1% (n = 3 blots) in HeLa cells ∼72 h after shRNA induction (hereafter, shPRC1; Figs. 4 A and S3 A). The mitotic index in these cells (4.3 ± 1.6%, n = 1,225 cells) was not significantly different from that in HeLa controls (HeLa lacking the shRNA construct; 5.8 ± 2.0%, n = 2,944 cells; P > 0.2; Fig. 4 B). Consistent with PRC1’s role in completion of cell division, a substantial fraction of cells in this population appeared to have more than one nucleus, indicating at least one cell division failure (Fig. S3 B). In HeLa control cells, endogenous PRC1 localizes to microtubules between segregating chromosomes (Fig. 4 C), similar to what we observed in hTERT-RPE1 cells (Fig. S1, A and B). PRC1 signal was detected on 100% of anaphase spindles in control cells (n = 77 cells), indicated by a peak in signal intensity at the center of the spindle in linescans drawn across the pole-to-pole axis. In contrast, PRC1 signal was not detected over background in >90% of shPRC1 cells (Fig. 4 D). In shPRC1 cells where endogenous PRC1 signal was detected, the intensity was relatively low and appeared to associate with only a few (<10) microtubule bundles. Anaphase cells were detected at a similar frequency (∼10%) in both shPRC1 and control cells, indicating that the loss of PRC1 does not alter metaphase-to-anaphase progression, consistent with earlier work (Jiang et al., 1998; Mollinari et al., 2002, 2005).
Figure 4.
PRC1 knockdown results in increased chromosome segregation rate and distance. (A) Western blot analysis of cell lysates of HeLa control cells and HeLa cells containing shRNA to PRC1 before (− tet) and after (+ tet) tetracycline induction of shRNA construct. Antibodies against α-tubulin (α-tub) and PRC1 are indicated. Expected position of PRC1 protein is indicated (red arrow). Full blots are provided in Figs. S3 and S5. (B) Analysis of mitotic index from control cells (ctr) and those expressing shRNA to PRC1 (+sh) cells. (C and D) Immunofluorescence of HeLa cells. Single-channel images (maximum-intensity projections) and overlays show DNA (blue), tubulin (green), and PRC1 (red) in HeLa control cells (C) and cells expressing shRNA to PRC1 (D). Scale bar, 3 µm. (E–G) Live-cell imaging of HeLa cells. Single-channel (single z slice) and overlay images show differential interference contrast (DIC) images (gray), chromosomes (magenta), and GFP-PRC1 (green) in HeLa control cells (E), HeLa cells expressing shRNA to PRC1 (shPRC1; F), and HeLa cells expressing shRNA to PRC1 and shRNA-resistant GFP-tagged full-length PRC1 (shPRC1+GFP-PRC1FL; G). T = 0 was assigned to the frame immediately before that with detectable chromatid separation. Time-lapse recordings are provided in Videos 4 and 5. Scale bar, 3 µm. (H) Segmented binary image showing outline of cortex and chromosome mask. The position of chromosome centroid (yellow squares), the axis of chromosome segregation (blue line), the position of the midplane (dotted line), the distance from the midplane to the cortex (gray line), and the distance from chromosome to cortex (pink line) are indicated. (I–N) Analysis of chromosome and cortical position in HeLa control (black; n = 15), shRNA (blue; n = 19), and shPRC1+GFP-PRC1 (gray; n = 14) cells. Mean ± SD. (I and J) Interchromosome distance over time (I) and at T = 5 min (J; control: 11.9 ± 1.5 µm; shPRC1: 14.5 ± 1.4 µm; shPRC1+GFP-PRC1FL: 12.1 ± 1.5 µm; P < 0.003). (K and L) Chromosome-to-cortex distance over time (K) and at T = 5 min (L; control: 6.0 ± 0.9 µm; shPRC1: 4.2 ± 0.5 µm; shPRC1+GFP-PRC1FL: 5.9 ± 0.7 µm; P << 0.0001). (M and N) Relative chromosome distance to cortex over time (M) and at T = 5 min (N; control: 0.50 ± 0.06; shPRC1: 0.63 ± 0.04; shPRC1+GFP-PRC1FL: 0.50 ± 0.06; P < 0.0001). n.s., not significant.
We also generated “addback” cells expressing both shRNA to endogenous PRC1 and shRNA-resistant GFP-tagged full-length PRC1 (hereafter, shPRC1+GFP-PRC1FL; see Materials and methods). We imaged all three cell lines live after treating with a DNA dye (SiR-DNA; Fig. 4, E–G; and Videos 4 and 5). In each cell line, furrow ingression initiated at a similar time following anaphase onset, and cell length appeared similar in all three cell lines (Fig. S3, C and D), suggesting that cell elongation is not impacted. We tracked chromosome position, taken as the centroid of the chromosome mask (Fig. 4 H, yellow squares), to monitor interchromosome distance. Shortly after anaphase onset, we observed that the distance between chromosomes was consistently larger in shPRC1 cells (n = 19; Fig. 4 I, blue trace) compared with control (n = 15; Fig. 4 I, black trace) or shPRC1+GFP-PRC1FL cells (n = 14; Fig. 4 I, gray trace). At T = 5 min, a time that precedes furrow ingression in >90% of cells but is after microtubule bundle overlap reaches a steady-state length, we could observe distinct differences between cell lines. Chromosomes in shPRC1 cells had segregated 20% further (14.5 ± 1.4 µm; Fig. 4 J) than control cells by this time (11.9 ± 1.5 µm; Fig. 4 J). This chromosome “hypersegregation” defect in shPRC1 cells was statistically significant (P < 0.003, two-tailed t test). Expression of GFP-PRC1FL rescued this defect (12.1 ± 1.5 µm; Fig. 4 J, gray).
We examined the absolute distance between chromosomes and the cell cortex (Fig. 4 K). Control and GFP-PRC1 cells showed a characteristic decrease in chromosome-to-cortex distance from anaphase onset until ∼5 min, followed by an increase (Fig. 4 K, black and gray traces, respectively). This increase after ∼5 min was not observed in shPRC1 cells (Fig. 4 K, blue trace). Compared with control cells, chromosomes in shPRC1 cells were positioned closer to the cortex by 5 min into anaphase (4.2 ± 0.5 µm in shPRC1 cells and 6.0 ± 0.9 µm in control cells) and this difference was significant (±SD; P << 0.0001; Fig. 4 L). The expression of GFP-PRC1FL rescued this defect (Fig. 4 L, gray). We also determined the relative chromosome distance to the cortex, defined as the distance between the chromosomes and the cortex (Fig. 4 H, magenta line) divided by the distance between the midplane and the cortex (Fig. 4 H, gray line). In control and shPRC1+GFP-PRC1FL cells, chromosomes segregated until reaching ∼50% of the distance between the midplane and the distal cell cortex, while those in shPRC1 cells moved a larger relative distance (Fig. 4, M and N).
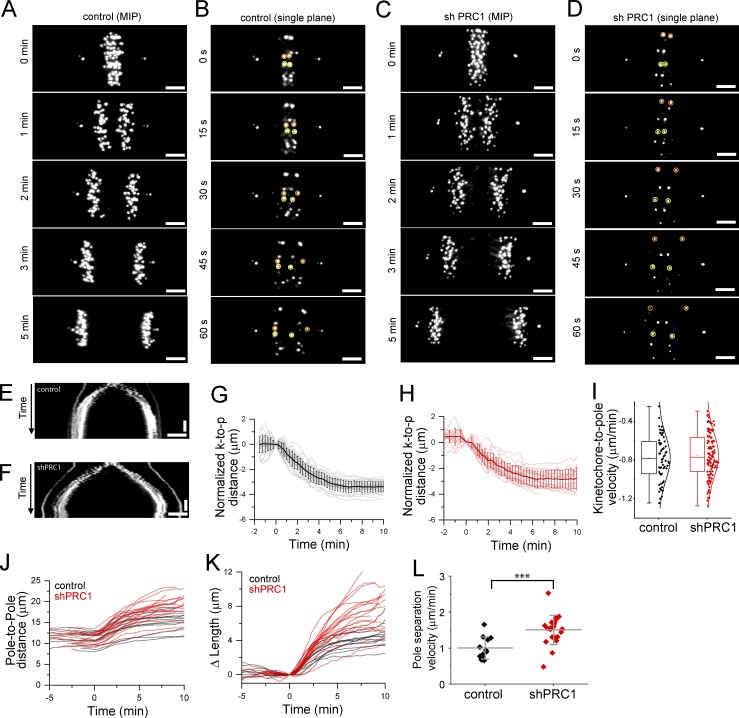
Chromosome segregation is a combination of chromosome-to-pole movement and spindle elongation. To determine how loss of microtubule bundle formation in the spindle midzone impacts these rates, we imaged hTERT-RPE1 cells expressing GFP-Centrin and GFP-CENP-A (hereafter, control cells), which mark spindle poles and kinetochores, respectively (Magidson et al., 2011; Fig. 5, A and B; and Video 6). We generated a cell line from these cells that expresses shRNA to PRC1 (hereafter, shPRC1; Fig. 5, C and D; and Video 7). The efficiency of knockdown was 80 ± 8% (Fig. S4 A), and PRC1 signal was absent or below the level of detection in 85% of anaphase cells (n = 13 cells; Fig. S4, B and C). Kymographs generated by drawing linescans across the pole-to-pole axis showed that chromosomes in shPRC1 cells segregated without an increased incidence of lagging chromosomes compared with control cells (Fig. 5, E and F), similar to what we observed following PRC1 knockdown in HeLa cells (Fig. 4 F).
Figure 5.
Defects in microtubule bundle assembly increase the rate of anaphase spindle elongation. (A–D) Live-cell imaging of centromere and spindle pole position by spinning disk confocal microscopy. Example hTERT-RPE1 cells expressing GFP-CENP-A to label the kinetochores and GFP-Centrin to label the centrosomes are shown. Select images from time series are shown. A control cell (A and B) and a cell expressing shRNA to PRC1 (shPRC1; C and D) are shown. T = 0 was assigned to the frame immediately prior to that with detectable sister kinetochore separation. (A and C) Maximum-intensity projections (MIPs) show select time points from a cell in anaphase. Time-lapse recordings are provided in Videos 6 and 7. (B and D) Single image planes show consecutive images (15-s intervals) from the cells shown in A (B) or C (D). Selected kinetochore pairs are highlighted (orange and yellow circles). (E and F) Kymograph generated from the time-lapse videos of the cell shown in A (E) or C (F). Horizontal scale bar, 3 µm. Vertical scale bar, 2 min. (G and H) Analysis of normalized kinetochore-to-pole (k-to-p) distance show individual kinetochores (thin lines) and average (bold line) traces from an example control cell (G) and an example shPRC1 cell (H). Error bars are SD. (I) Box-and-whisker plots of kinetochore-to-pole velocity. Each point represents the velocity of a single kinetochore. Velocity was determined by linear fit to kinetochore-to-pole distance versus time in the T = 0.5–3-min time window. Negative sign indicates direction of slope. Data were pooled data from all experiments (n = 59 and 111 kinetochores for control and shPRC1 cells, respectively) and fit to a Gaussian. (J) Plots of pole-to-pole distance versus time for control (n = 12 cells; black traces) and shPRC1 (n = 21 cells; red traces) cells. (K) Plot of relative pole separation distance (ΔLength), defined as the pole-to-pole distance at time t (Lt) minus the pole-to-pole distance at T = 0 (L0). (L) Pole separation velocity for control and shPRC1 cells (***, P < 0.003). Mean ± SD.
We examined kinetochore-to-pole distance over time using a 3D localization and tracking algorithm (see Materials and methods). We were able to track kinetochore position from anaphase onset through kinetochore release, identified by a synchronous increase in kinetochore-to-pole distance (Fig. S4, D and E). In control and shPRC1 cells, tracks of individual kinetochores revealed that anaphase onset occurs with a synchronous decrease in kinetochore-to-pole distance (Fig. 5, G and H). To determine the average rate of kinetochore-to-pole movement, we focused on the T = 0.5–3 min time window when this rate is reported to be fastest (Su et al., 2016). We pooled the rates for individual kinetochore tracks that had fits with R-squared values ≥0.85 (Fig. 5 I). The average rate of kinetochore-to-pole movement for control cells, −0.78 ± 0.23 µm/min (n = 59 kinetochores), was not significantly different than that for PRC1 knockdown cells, −0.75 ± 0.23 µm/min (n = 111 kinetochores; P > 0.1).
We also tracked the pole-to-pole distance as a function of time (Fig. 5 J). The pole-to-pole distance began to increase immediately after anaphase onset in control (Fig. 5 J, black traces) and PRC1 knockdown (Fig. 5 J, red traces) cells. Traces for all control and shPRC1 cells are shown (n = 12 and 21 cells, respectively). A plot of the change in anaphase spindle length (ΔLength) as a function of time t highlighted differences in the rate of pole separation (Fig. 5 K). ΔLength at each time point was calculated as the pole-to-pole distance at time t minus the pole-to-pole distance at t = 0, assigned to the frame immediately prior to that with detectable sister kinetochore separation. Maximum pole separation velocities were observed 1–3 min after anaphase onset in 94% of spindles (n = 33 cells). We therefore determined the average pole-to-pole separation rates using data from this time window (Fig. 5 L). Spindle elongation proceeded at an average rate of 0.98 ± 0.31 µm/min in control cells (n = 11 cells). Spindles in shPRC1 cells elongated ∼50% faster, at 1.51 ± 0.41 µm/min (n = 21 cells; P < 0.003).
Together, these data show that the increased rate of chromosome separation observed in two human cell lines after PRC1 knockdown is due to an increase in pole separation speed, rather than an increased rate of kinetochore-to-pole movement.
Mutations in PRC1 that reduce microtubule binding affinity can form microtubule bundles but cannot rescue chromosome hypersegregation defects
Previous structural studies of PRC1 have identified the amino acids involved in contacting microtubules (Subramanian et al., 2010; Kellogg et al., 2016). We generated two mutant constructs, one with two point mutations (K387A and K390A) in the spectrin domain (GFP-PRC1AA) and one with a truncation of the terminal 464–620 residues in the unstructured C-terminus (GFP-PRC1ΔC). Based on in vitro characterization of similar constructs, both constructs are likely to reduce microtubule binding affinity while maintaining the ability to dimerize, a prerequisite for microtubule cross-linking (Subramanian et al., 2010, 2013). Unlike the point mutations in GFP-PRC1AA, the truncation in GFP-PRC1ΔC likely disrupts interaction with some MAPs that interact with PRC1 at the C-terminus (Jiang et al., 1998; Neef et al., 2007).
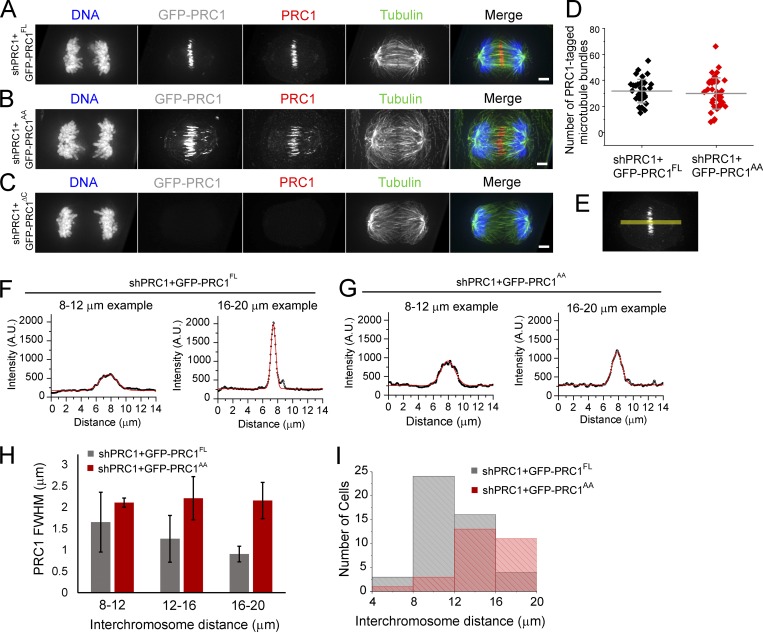
We examined three shRNA-resistant constructs—full-length GFP-PRC1 (GFP-PRC1FL) and our two mutant constructs, GFP-PRC1AA and GFP-PRC1ΔC—in the context of endogenous protein knockdown. Western blot analysis of metaphase-arrested cell lysates showed that the expression levels of the GFP constructs were comparable to those of endogenous PRC1 in control cells (GFP-PRC1FL: 69 ± 44%, n = 3; GFP-PRC1AA: 140 ± 95%, n = 4), and knockdown efficiency of endogenous PRC1 was high (shPRC1+GFP-PRC1FL: 94 ± 1%, n = 3; shPRC1+GFP-PRC1AA: 93 ± 3%, n = 4; Fig. S5 A). Knockdown efficiency in shPRC1+GFP-PRC1ΔC cells could not be measured because GFP-PRC1ΔC ran to a similar position as the endogenous PRC1 protein and could not be resolved by SDS-PAGE. However, Western blots revealed a GFP-reactant band at the expected size, indicating that the protein was expressed in cells (Fig. S5 B). Immunofluorescence analysis of fixed cells showed that the decoration of GFP-PRC1FL appeared similar to that of endogenous PRC1 in unmodified control cells (Fig. 6 A). GFP-PRC1AA also decorated microtubules in the midzone in anaphase cells (Fig. 6 B). We did not detect PRC1 signal above background on microtubules in any anaphase cells expressing GFP-PRC1ΔC in the context of endogenous protein knockdown (Figs. 6 C and S5 B). These cells looked morphologically similar to PRC1 knockdown alone (Fig. 4 D), and therefore we did not examine them further. The average number of microtubule bundles in mid- to late anaphase spindles (chromosome segregation distance 12–16 µm) was similar in both cell lines (shPRC1+GFP-PRC1FL: 32 ± 9; shPRC1+GFP-PRC1AA: 30 ± 12; P > 0.04; Fig. 6 D). These data suggest that after knockdown of endogenous PRC1, expression of either GFP-PRC1FL or GFP-PRC1AA can rescue the assembly of microtubule bundles.
Figure 6.
Mutations in PRC1 that reduce microtubule binding affinity can form microtubule bundles but cannot rescue chromosome hypersegregation defects. (A–C) Immunofluorescence analysis of HeLa cells in anaphase. Single-channel images (maximum-intensity projections) and overlays show DNA (blue), PRC1 (red), and tubulin (green). GFP fluorescence (maximum-intensity projection) is shown as reference (gray). HeLa cells coexpressing shRNA to endogenous PRC1 (shPRC1) and shRNA-resistant GFP-PRC1FL (A), GFP-PRC1AA (B), or GFP-PRC1ΔC (C). Scale bar, 3 µm. (D) Analysis of the number of PRC1-tagged microtubule bundles in cells with interchromosome distances between 12 and 16 µm. Error bars are SD. (E–G) Analysis of PRC1 signal intensity in fixed cells. (E) PRC1 channel image from A showing position of linescan (yellow) used to measure PRC1 signal intensity along the pole-to-pole axis. (F and G) Plots show example traces from cells expressing shRNA to PRC1 (shPRC1) and shRNA-resistant GFP-PRC1FL (F) or GFP-PRC1AA (G). Signal intensity data were fit to a Gaussian (red traces) to determine the FWHM. Gaussian fits with R2 values >0.90 were retained for analysis. (H) Bar chart of average FWHM, binned by interchromosome distances. Error bars are SD. (I) Histogram of the number of anaphase cells, binned by interchromosome distance. n = 53 (shPRC1+GFP-PRC1FL) and 28 (shPRC1+GFP-PRC1AA) cells.
When comparing cells with equivalent chromosome segregation distances, we observed that the decoration of GFP-PRC1AA tended to be wider than that of GFP-PRC1FL on microtubules in the midzone. To quantify this difference, we determined the full width at half-maximum (FWHM) signal in maximum-intensity images of fixed cells by drawing linescans across the pole-to-pole axis (see Materials and methods; Fig. 6 E). In each cell, we also measured the interchromosome distance and used it to separate the data into 4-µm bins (Fig. 6, F and G). The average FWHM of PRC1 signal in shPRC1+GFP-PRC1FL cells decreased with increasing interchromosome distance, indicating a reduction in the length of microtubule overlap (Fig. 6 H, gray). This trend was not observed in cells expressing GFP-PRC1AA. Instead, the FWHM measurement was similar for cells with different interchromosome distances (Fig. 6 H, red), suggesting that microtubule overlap length did not decrease as chromosomes separated. Based on the location of the mutations within the microtubule-binding spectrin domain in the PRC1 homodimer (Subramanian et al., 2010, 2013), we favor the possibility that the mutations do not impact the specificity of PRC1 antiparallel cross-linking and that the decoration of GFP-PRC1AA does indeed track the region of microtubule overlap in cells. However, further studies will be needed (e.g., correlative light and electron microscopy) to examine this.
We next examined the distribution of chromosome segregation distance in anaphase cells expressing shRNA to endogenous PRC1 and detectable GFP-PRC1FL or GFP-PRC1AA (Fig. 6 I). Only a small fraction (<9%) of anaphase cells expressing GFP-PRC1FL were found to exhibit interchromosome distances >16 µm, suggesting that this may be the upper limit to chromosome segregation distance in this context (Fig. 6 I, gray). By contrast, ∼39% of cells expressing GFP-PRC1AA were observed with interchromosome distances >16 µm, some up to 20 µm (Fig. 6 I, red). This chromosome hypersegregation defect was similar to that observed in PRC1 knockdown cells alone (Fig. 4 D). Together, these observations suggest that the reduction of microtubule overlap length within bundles may be needed for the midzone to act as a brake to properly restrict spindle elongation and position chromosomes within daughter cells.
Discussion
Our data show that microtubule bundle assembly is required to restrict chromosome segregation during anaphase. We find that the region of microtubule overlap marked by GFP-PRC1 decreases in length as chromosomes segregate, while the interbundle spacing does not. Impairing microtubule bundle assembly by knockdown of PRC1 results in increased distance and rate of chromosome segregation largely due to an increase in spindle elongation rate. Cells expressing a mutant PRC1 with reduced microtubule affinity assemble microtubule bundles that have unusually long regions of GFP-tagged mutant PRC1 and cannot rescue chromosome hypersegregation defects. Together, these data suggest an unexpected requirement for the reduction in overlap length within microtubule bundles to restrict chromosome movement and properly position chromosomes within daughter cells.
Why would the reduction in microtubule overlap length within bundles be important for the midzone to function as a brake? In vitro, PRC1 forms a complex with Kif4, and together these proteins can function in two ways: to suppress microtubule plus end dynamics and to slide microtubules apart (Bieling et al., 2010; Wijeratne and Subramanian, 2018). One study, which used microtubule minus ends anchored to the assay chamber, showed that PRC1/Kif4 can regulate microtubule overlaps through Kif4-dependent capping of filament plus ends (Bieling et al., 2010). A study that used unanchored, nondynamic microtubules showed that PRC1-Kif4 complexes can also slide antiparallel microtubules apart (Wijeratne and Subramanian, 2018). In this assay, the density of both PRC1 and Kif4 in the region of antiparallel overlap increased as microtubules slid apart. Kif4-mediated sliding stalled before the microtubules completely slid apart, likely due to molecular crowding that inhibits motor stepping (Leduc et al., 2012; Wijeratne and Subramanian, 2018). We propose that these crowded PRC1-Kif4 complexes may not just stall but also resist relative filament sliding.
At this stage, we cannot exclude the possibility that PRC1 itself acts as a brake, as has been proposed for Ase1, the budding yeast homologue. We do not favor this possibility for the following reasons. First, our FRAP experiments reveal that the turnover of PRC1 in the midzone of early anaphase spindles is relatively fast, ∼10× faster than that of GFP-Ase1 in the budding yeast midzone (Schuyler et al., 2003). In vitro experiments have shown that Ase1 accumulates in overlap regions as a direct result of the reduction in overlap length and resists further filament movement due to entropic forces (Lansky et al., 2015). This mechanism of force generation requires a long residence time of protein cross-links. Given the relatively fast rate of PRC1 turnover during early stages of anaphase, when overlap length is decreasing and chromosomes are segregating, it is unlikely that such entropic forces could be generated. However, at later stages, as PRC1 and several other proteins accumulate, the turnover may be slower and such entropic forces may become more important. Second, optical trap measurements of the force–velocity relationship show that single human PRC1 bound to a microtubule can generate frictional forces, but the relative magnitude of these forces are low and therefore not likely to be sufficient to resist motor proteins, particularly at relatively slow velocities (<0.1 pN at 1 µm/min sliding velocity; Forth et al., 2014). Third, unlike Ase1, which increases in density in overlap regions as filaments slide apart (Braun et al., 2011; Lansky et al., 2015), human PRC1 alone does not (Subramanian et al., 2010). However, the range of PRC1 concentrations that have been examined in vitro is narrow, and the possibility remains that at high soluble PRC1 concentration, different interaction modes, including those that involve PRC1–PRC1 binding, may be possible.
Other candidate proteins that can contribute to the brake-like function of the spindle midzone microtubules are Mklp1 (a kinesin-6) and kinesin-5. PRC1 interactions with centralspindlin (a 2:2 complex of Mklp1 and RacGap) are required to maintain the mechanical resilience of the central spindle in C. elegans (Lee et al., 2015). Consistent with this, knockdown of Mklp1 disrupts the stability of microtubules in the spindle midzone, and thus deciphering its role in relative filament sliding during anaphase has been challenging (Vukušić et al., 2017). In the case of kinesin-5, there are conflicting data. Experiments using porcine cells suggest that kinesin-5 activity restricts spindle elongation (Collins et al., 2014), while a more recent report using cultured human cells indicated that kinesin-5 does not impact sliding rates in early anaphase (Vukušić et al., 2017). Within the metaphase spindle, the extent of microtubule overlap near the spindle equator is likely high, as suggested by PRC1 immunofluorescence, electron microscopy studies, and our examination of GFP-PRC1 signal in live cells at the start of anaphase (McIntosh and Landis, 1971; Mastronarde et al., 1993; Jiang et al., 1998; Mollinari et al., 2002). As cells transition to anaphase, the relative sliding of the filaments associated with poleward flux (Mitchison, 1989) reduces and nearly stops (Zhai et al., 1995; Ganem et al., 2005). Our near-simultaneous GFP-EB1 and Halo-PRC1 imaging support a model in which the dynamics of microtubule plus ends within bundles are suppressed in anaphase of mammalian cells. Therefore, kinesin-5 inhibition effects may be revealed only if the initial phase of relative sliding is blocked. Photo-switchable chemical probes (Sadakane et al., 2018) or similar faster-acting inhibition strategies are needed to properly dissect the role of kinesin-5 in anaphase microtubule sliding. In vitro, ensembles of kinesin-5 can resist filament sliding, and the magnitude of this resistance scales with the number of motors and length of microtubule overlap (Shimamoto et al., 2015). Our data show that cells with longer microtubule overlaps exhibit chromosome hypersegregation, suggesting that increasing the length of microtubule overlap is not sufficient to restrict chromosome segregation speed. Therefore, at this stage, we favor the idea that kinesin-5 is not the primary molecule responsible for the midzone brake.
Our LLSM data show that microtubule bundles and dynamic microtubules coexist within the spindle midzone. Once anaphase starts, the dynamic organization of each of these two populations are largely independent of each other. Remarkably, while the length of microtubule overlap within bundles decreases as chromosomes separate, the spacing between neighboring microtubule bundles (i.e., interbundle) remains nearly constant. This is distinct from the spacing between overlapping filaments within bundles (i.e., intrabundle), which our imaging approach does not have sufficient resolution to analyze. Maintaining this interbundle spacing under compression or extension likely involves anchoring of microtubules near minus ends by microtubule cross-linking proteins (e.g., NuMA [Elting et al., 2017] or ASPM [Jiang et al., 2017]) or proteins that can bind chromatin and microtubules (e.g., chromokinesins; Wang and Adler, 1995). The chromosomes themselves may serve as physical spacers that prevent interactions between bundles, allowing only the occasional fusion events (one event/10 s) that we observe. The number of bundles in early anaphase is likely established by the chromosomes and kinetochore fibers (Polak et al., 2017), but new bundles do emerge, reflected in our data as a moderate increase in the number of PRC1-tagged microtubule bundles. The formation of these new microtubule bundles may depend on centrosome-independent pathways or the cross-linking and subsequent stabilization of dynamic microtubules, which our findings reveal are distributed between these bundles. The addition of new bundles while maintaining a constant interbundle spacing would allow the midzone to retain proximity to the cell cortex before furrow ingression and thereby contribute to the positioning of the cell division plane (Su et al., 2014). Approaches to specifically inhibit the formation of these additional bundles or modify their relative spacing will be needed to dissect the functional importance of these assembly and fusion dynamics.
Bundled microtubules in the spindle midzone can function to generate pushing forces or act as brakes (Aist and Berns, 1981; Leslie and Pickett-Heaps, 1983; Grill et al., 2001). We posit that in human cells, mechanisms responsible for these activities may coexist, and transitions between modes of function can be readily achieved. In early anaphase, microtubule bundles can contribute to the segregation of chromosomes (Vukušić et al., 2017), but as anaphase progresses, posttranslational modifications would dissociate some molecules and recruit others. Microtubule bundles are recurring motifs across diverse eukaryotes, including in the centrosome-free spindles of oocytes and in the phragmoplast of plant cells. Similar switching between pushing and braking forces may play critical roles in achieving the self-organization of these micrometer-scale structures and for their essential cellular functions.
Materials and methods
Cell lines used for LLSM imaging
Stable hTERT-RPE1 cells (RRID: CVCL_4388) expressing GFP-PRC1 were generated by retroviral transduction as described previously (Subramanian et al., 2013). Stable HeLa T-REx Flp-in cells expressing Halo-PRC1 were generated following standard vendor protocol (Invitrogen). The HaloTag sequence (Promega) was introduced at the N-terminus of human PRC1 isoform II coding sequence (NCBI accession no. NM_199413). The construct was cloned into the pCDNA5/FRT/TO (Invitrogen) expression plasmid. Expression of Halo-PRC1 was induced by adding 2 µg/ml tetracycline (Thermo Fisher Scientific) into the medium 48 h before imaging. Cells were incubated with 50 nM JF549 for 45 min before imaging. Stable hTERT-RPE1 cells expressing GFP-EB1 and Halo-PRC1 were generated by retroviral transduction following a protocol similar to that described in Subramanian et al., 2013.
Cell lines used for PRC1 knockdown and confocal imaging
To allow for tetracycline-inducible expression of the shRNA construct in hTERT-RPE1 cells, we first cloned the tetracycline repressor sequence into the pMSCVblast expression vector. We generated cells constitutively expressing the tet repressor protein via retroviral transduction. The plasmid was first transfected into Ampho-293 cells for retrovirus production. The medium was then harvested and added directly to hTERT-RPE1 cells in the presence of 4 µg/ml polybrene after filtering through a 0.45-µm filter. Blasticidin (InvivoGen) was used to select for cells that had stably incorporated the construct into the genomic DNA.
We selected the shRNA target sequence in PRC1 (5′-GTGATTGAGGCAATTCGAG-3′), which had been previously shown to suppress PRC1 in cells (Voets et al., 2015). We generated double-stranded oligomers encoding sense and antisense target sequence separated by a 9-bp hairpin sequence. The oligomers were cloned into the pSuperior.retro.puro (Oligoengine) backbone between the BglII and HindIII restriction sites. To generate HeLa and RPE1 cells with tetracycline-inducible control of PRC1 shRNA expression, we introduced the shRNA construct by retroviral transduction into the appropriate cell lines as described above. Puromycin (Sigma-Aldrich) was used to to select for cells that had stably incorporated the construct into the genomic DNA. To induce shRNA expression, 2 µg/ml tetracycline was added 72 h before imaging. The medium was replaced with fresh tetracycline once 24 h before imaging.
We generated an shRNA-resistant PRC1 construct by introducing three silent mutations into the coding sequence for human PRC1 isoform I (NCBI accession no. NM_003981.3; A888C, T891A, and A894G, noted as bold letters in the target sequence above). We introduced a GFP tag at the N-terminus of PRC1 coding sequence and cloned this construct into the pCDNA5/FRT expression plasmid (Invitrogen). To generate HeLa cells constitutively expressing shRNA-resistant GFP-PRC1, we transfected the construct into HeLa T-REx Flp-in cells that had already stably incorporated the PRC1 shRNA construct following standard vendor protocol (Invitrogen). Hygromycin B (Invitrogen) was used to select for cells that had stably incorporated the construct into the engineered Flp-in site.
Tissue culture
HeLa T-Rex Flp-in cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Sigma-Aldrich) and 2 mM l-glutamine (Gibco) and incubated in 5% CO2 at 37°C. hTERT-RPE1 cells were cultured in DMEM/F-12 GlutaMAX (Gibco) supplemented with 10% FBS (Sigma-Aldrich) and incubated in 5% CO2 at 37°C.
Immunological methods
Cells were grown on 12-mm-diameter glass coverslips for 2 d before fixation. Cells were fixed in a buffer containing formaldehyde (100 mM Pipes, 1 mM MgCl2, 10 mM EGTA, 0.2% Triton X-100, and 4% formaldehyde, pH 6.8) prewarmed to 37°C for 20 min. Cells were blocked in 1× PHEM (60 mM K-Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgSO4, pH 6.9) plus 2% BSA for 2 h at room temperature or overnight at 4°C. Coverslips were then incubated with anti-PRC1 antibody (raised against PRC1 [aa 341–466] in rabbit, affinity purified, and used at 0.5 µg/ml) overnight at 4°C. After three washes with 1× PHEM for 5 min at room temperature, the coverslips were incubated for 2 h at room temperature with FITC-conjugated mouse anti-tubulin monoclonal antibody (F2168; 1:1,000 dilution; Sigma-Aldrich). After three washes, coverslips were incubated in donkey anti-rabbit Texas red–conjugated secondary antibody (1:1,000 dilution; Jackson Immunoresearch) in 1× PHEM plus 6% donkey serum. After three washes, DNA was stained with 1 µm/ml DAPI for 10 min at room temperature, mounted (20 mM Tris-HCl, pH 8, 0.5% propyl gallate, and 90% glycerol), and sealed with nail polish.
For Western blot analysis, the anti-PRC1 antibody was used at 0.5 µg/ml, and monoclonal anti–α-tubulin antibody was used at 1:1,000 dilution (T6199; Sigma-Aldrich). Dye-conjugated secondary antibodies raised in donkey were purchased from LI-COR.
Fixed cell analysis
Images of fixed cells were acquired as z stacks with 200-nm spacing using a 100×, 1.35 NA objective on a DeltaVision Image Restoration Microscope (Applied Precision Instruments and Olympus). The immunofluorescence micrographs were processed by iterative constrained deconvolution (SoftWoRx; Applied Precision Instruments). Maximum-intensity projections were converted to tiff files, and linescans were generated using Fiji software (Schindelin et al., 2012).
Live-cell imaging using LLSM
Cells were cultured on 5-mm-diameter coverslips (64-0700; Warner Instruments). hTERT-RPE1 cells were maintained in standard culture medium (1:1 DMEM/F-12 GlutaMAX [Gibco] supplemented with 10% [vol/vol] FBS [Sigma-Aldrich]) at 37°C. HeLa cells were maintained in standard culture medium (DMEM [Gibco] supplemented with 10% [vol/vol] FBS [Signma-Aldrich] and 2 mM l-glutamine [Gibco]) at 37°C. hTERT-RPE1 cells expressing GFP-PRC1 were incubated in 500 nM SiR-DNA in standard culture medium for 30–60 min before imaging. hTERT-RPE1 cells expressing GFP-EB1 and Halo-PRC1 and HeLa T-REx Flp-In cells expressing Halo-PRC1 were incubated in 50 nM JF-646 in standard culture medium for 30–60 min before imaging. Images were collected on a custom-built instrument located at the Advanced Imaging Center at Janelia Research Campus, similar to the one described in Chen et al. (2014). The microscope was controlled by custom-made software (LabView). 3D lattice light-sheet imaging was performed using an annular mask of outer NA equal to 0.55 and inner NA equal to 0.44. Images were acquired using an sCMOS camera (Orca Flash 4.0 v2; Hamamatsu). Cells were imaged in Leibovitz’s L-15 medium (Gibco), without phenol red, supplemented with 10% FBS (Sigma-Aldrich) and 2 mM l-glutamine (Gibco) at 37°C. Dividing cells were selected, and the exposure time for each channel was adjusted to optimize the signal-to-noise ratio and ranged from 10 to 40 ms. Cells were excited sequentially using a 488-nm laser to visualize either GFP-PRC1 or GFP-EB1 and a 642-nm laser to visualize either chromosomes or Halo-PRC1. For dual GFP-PRC1 and chromosome imaging in hTERT-RPE1 cells or for Halo-PRC1 imaging in HeLa cells, 3D stacks consisting of 101 optical sections spaced 300 nm apart were captured in each channel at rates up to 20 cell volumes/min. For dual GFP-EB1 and Halo-PRC1 imaging in hTERT-RPE1 cells, 3D stacks consisting of 58 optical sections spaced 350 nm apart were captured in each channel at rates up to 60 cell volumes/min.
Live-cell imaging using spinning disk confocal
Cells were cultured on 22-mm square coverslips and mounted in a custom Rose chamber or grown on 35-mm glass-bottomed plates. Cells were imaged in Leibovitz’s L-15 medium (Gibco) supplemented with 10% FBS and 2 mM l-glutamine at 35–37°C.
Confocal GFP fluorescence micrographs of hTERT-RPE1 cells expressing GFP-Centrin/GFP-CENP-A were acquired using a Nikon TE2000 microscope (Morrell Instruments) with a 100× oil objective (PlanApo, 1.4 NA) equipped with a z piezo stage. With 0.4-µm spacing between z planes, micrographs were taken through the entire cell with a PerkinElmer Wallac UltraView confocal head and 488-nm excitation laser (Coherent). Images were acquired with an sCMOS Prime95B camera (Photometrics) using NIS-Elements software (Nikon).
Confocal fluorescence and differential interference contrast micrographs were acquired using an Inverted Axiovert 200 microscope (Zeiss/Perkin-Elmer) with a 100× oil objective (PlanApo, 1.4 NA). Images consisting of single z planes were taken with a PerkinElmer Wallac UltraView confocal head using solid-state 491-nm and/or 644-nm lasers for excitation (Spectral Applied). Images were acquired with an EMCCD iXon camera (Andor) using Metamorph software (MDS Analytical Technologies).
FRAP
FRAP experiments were performed on an inverted Axiovert 200 equipped with a spinning disk confocal head (UltraView; Perkin-Elmer). hTERT-RPE1 cells expressing GFP-PRC1 were excited with a 491-nm solid state laser (Spectral Applied) and imaged with an NA 1.49 oil immersion 100× objective lens. Images were acquired using an EMCCD camera (iXon; Andor). The regions of interest covering bleached areas were defined with a micromirror array system (Mosaic Digital Diaphragm System; Photonics). Bleaching was performed with a 2–3-s pulse, and recovered fluorescence was collected every 3 s with a 200–300-ms exposure time. Fluorescence recovery, R(t), was normalized and fitted to the following equation to determine the recovery constant k: R(t) = A × [1 − exp(−k × t)], where A is a constant. Fitting was performed in MATLAB (2018a). All acquisition parameters were controlled with MetaMorph software.
Data analysis on LLSM images
After image acquisition, several processing steps were necessary before quantitative analyses could be performed. First, due to the geometry of the excitation/emission objective pair, the raw image data were deskewed to obtain image stacks in the reference frame of the cell. Data were then deconvolved using a Richardson–Lucy algorithm and the system’s point-spread function, measured in independent experiments using 100-nm-diameter fluorescent beads. We next compensated for spindle rotation and translation within the imaging window by applying a rigid transform using MATLAB’s imregtform function (MATLAB 2018a). This transform was defined and applied between subsequent frames in the PRC1 channel and then applied to each DNA-channel frame. We then translated and rotated the data such that the long axis of the spindle was aligned to the x axis and centered in the image window.
To describe the geometry of single microtubule bundles in 3D, LLSM datasets were first denoised using a third-party bandpass function in MATLAB (Crocker and Grier, 1996). Next, a watershed routine (Hodneland et al., 2013) was applied to segment individual bundles and object geometry was extracted using MATLAB’s regionprops3 function. The midzone was defined as a region centered between spindle poles that spanned a total of 60 pixels (~6.2 µm) in length along the xy plane, with a height encompassing the full diameter of the spindle (100 pixels or 10.4 µm). Bundles with a center of mass inside the 60 × 100-pixel box defining the midzone were analyzed.
To analyze the number of GFP-PRC1–tagged microtubule bundles at the spindle midplane, datasets were processed using ImageJ’s Classic Watershed plugin (Legland et al., 2016). The minimum value was set to 0 and the maximum value to one less than the maximum value in the given frame. The total number of microtubule bundles in each frame was determined as the total number of particles identified in the watershed analysis, counted using ImageJ’s Analyze Particles command. To determine the nearest-neighbor distance between EB1-GFP and/or Halo-PRC1 spots at the spindle midplane, the position of individual spots was first identified using TrackMate (Tinevez et al., 2017). An estimated object diameter of 1 µm and a threshold of 200–2,000 was used, determined for each movie based on maximum intensity values. Typically a value between Y and Z was used. The 2D distance between each spot and all other spots was computed in MATLAB.
3D EB1 and CENP-A tracking
Aligned EB1 and CENP-A datasets were processed in three sequential steps using the Diatrack software package (Vallotton et al., 2017). First, images were background subtracted and Gaussian filtered with a sigma of 1 pixel to reduce image noise. Second, local maxima were detected and localized using a 3D Gaussian fitting routine. FWHMs of 1 and 2 pixels were used for localizing EB1 comets and CENP-A, respectively; localizations were intensity-filtered, with an intensity cutoff that depended on Eb1 and CENP-A expression levels. Typically, cutoff intensities ranged from 200 to 500 analog-to-digital units. Finally, localized peaks were tracked; peaks belonged to the same track if they appeared in sequential frames and moved within a radius of 5 and 10 pixels for Eb1 and CenpA, respectively. Search radii varied by up to three pixels per dataset, depending on the temporal resolution and localization density.
EB1 tracks were processed as previously described (Yamashita et al., 2015). Briefly, tracks longer than three frames, with a straightness factor defined as the ratio of displacement to length S > 0.6, were conserved. EB1 speeds were computed using these processed tracks, where all postprocessing analyses were performed using custom-written scripts in MATLAB (2018a). Cenp-A tracks consisting of <30 frames were discarded. Each track was paired with the nearest cell pole using MATLAB’s knnsearch function, and kinetochore-to-pole distances were computed over time.
Quantification and statistical analysis
Data presented in the text are expressed as the mean ± SD. Statistical significance was determined using two-sample unequal variance t test with two-tailed distribution. Replicates, number of quantified microtubules, and statistical results are indicated in figure legends for the respective experiments.
Online supplemental material
Fig. S1 shows analysis of PRC1 localization in hTERT-RPE1 cells. Fig. S2 shows analysis of GFP-EB1 and Halo-PRC1 in dividing cells. Figs. S3 and S4 show Western blot and immunofluorescence analysis of cells expressing shRNA to PRC1 in HeLa and hTERT-RPE1 cells, respectively. Fig. S5 shows Western blot analysis of HeLa cells expressing shRNA to PRC1 and shRNA-resistant GFP-tagged PRC1 constructs. Video 1 shows a time-lapse recording of GFP-PRC1 and chromosomes in a dividing cell. Videos 2 and 3 show time-lapse recordings of Halo-PRC1 and GFP-EB1 in a dividing cell. Videos 4 and 5 show time-lapse recordings of HeLa cells expressing shRNA to PRC1. Videos 6 and 7 show time-lapse recordings of hTERT-RPE1 cells expressing GFP-CENP-A and GFP-Centrin.
Supplementary Material
Acknowledgments
We sincerely thank all the staff at the Advanced Imaging Center, especially Dr. John Heddleston, Dr. Eric Wait, and Dr. Leong Chew for their support.
LLSM work was performed at the Advanced Imaging Center, Janelia Research Campus, jointly sponsored by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation. This work is supported by the National Institutes of Health (RO1GM65933 and R35GM130234 to T.M. Kapoor; and R35GM130298 to A. Khodjakov) and the Swiss National Science Foundation (project P2ELP3_175277 to L. Carlini).
The authors declare no competing financial interests.
Author contributions: T.M. Kapoor, M.C. Pamula, L. Carlini, and S. Forth designed experiments. M.C. Pamula, L. Carlini, S. Forth, and P. Verma collected data. M.C. Pamula, L. Carlini, and S. Forth developed software and analyzed data. A. Khodjakov provided resources. E. Betzig and W.R. Legant provided instrumentation, assisted with data curation of LLSM data, and were supported by the Howard Hughes Medical Institute. M.C. Pamula and L. Carlini generated figures. M.C. Pamula, P. Verma, and S. Suresh generated cell lines. T.M. Kapoor and M.C. Pamula wrote the manuscript with contributions from all authors. T.M. Kapoor secured funding.
References
- Aist J.R., and Berns M.W.. 1981. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J. Cell Biol. 91:446–458. 10.1083/jcb.91.2.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C.L. 2017. Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles. Biology (Basel). 6:E15 10.3390/biology6010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., and Surrey T.. 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 142:420–432. 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Braun M., Lansky Z., Fink G., Ruhnow F., Diez S., and Janson M.E.. 2011. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat. Cell Biol. 13:1259–1264. 10.1038/ncb2323 [DOI] [PubMed] [Google Scholar]
- Brinkley B.R., and Cartwright J. Jr. 1971. Ultrastructural analysis of mitotic spindle elongation in mammalian cells in vitro. Direct microtubule counts. J. Cell Biol. 50:416–431. 10.1083/jcb.50.2.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués J., and Needleman D.. 2014. Physical basis of spindle self-organization. Proc. Natl. Acad. Sci. USA. 111:18496–18500. 10.1073/pnas.1409404111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman J.C., Hoffman D.B., and Salmon E.D.. 2000. The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr. Biol. 10:611–614. 10.1016/S0960-9822(00)00490-5 [DOI] [PubMed] [Google Scholar]
- Chen B.-C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. . 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E., Mann B.J., and Wadsworth P.. 2014. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken). 71:136–144. 10.1002/cm.21158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J.C., and Grier D.G.. 1996. Materials and methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 179:298–310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Ding R., McDonald K.L., and McIntosh J.R.. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151. 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U.S., Mitchison T.J., and Field C.M.. 2006. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75:543–566. 10.1146/annurev.biochem.74.082803.133425 [DOI] [PubMed] [Google Scholar]
- Elting M.W., Prakash M., Udy D.B., and Dumont S.. 2017. Mapping Load-Bearing in the Mammalian Spindle Reveals Local Kinetochore Fiber Anchorage that Provides Mechanical Isolation and Redundancy. Curr. Biol. 27:2112–2122.e5. 10.1016/j.cub.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U., and McIntosh J.R.. 1980. Polarity of midbody and phragmoplast microtubules. J. Cell Biol. 87:509–515. 10.1083/jcb.87.2.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S., Hsia K.-C., Shimamoto Y., and Kapoor T.M.. 2014. Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell. 157:420–432. 10.1016/j.cell.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Upton K., and Compton D.A.. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. 10.1016/j.cub.2005.08.065 [DOI] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20. 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R.D.. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gönczy P., Stelzer E.H.K., and Hyman A.A.. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633. 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Heidemann S.R., and McIntosh J.R.. 1980. Visualization of the structural polarity of microtubules. Nature. 286:517–519. 10.1038/286517a0 [DOI] [PubMed] [Google Scholar]
- Hodneland E., Kögel T., Frei D.M., Gerdes H.-H., and Lundervold A.. 2013. CellSegm - a MATLAB toolbox for high-throughput 3D cell segmentation. Source Code Biol. Med. 8:16 10.1186/1751-0473-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.-K., Coughlin M., Field C.M., and Mitchison T.J.. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824. 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Jimenez G., Wells N.J., Hope T.J., Wahl G.M., Hunter T., and Fukunaga R.. 1998. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell. 2:877–885. 10.1016/S1097-2765(00)80302-0 [DOI] [PubMed] [Google Scholar]
- Kajtez J., Solomatina A., Novak M., Polak B., Vukušić K., Rüdiger J., Cojoc G., Milas A., Šumanovac Šestak I., Risteski P., et al. . 2016. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat. Commun. 7:10298 10.1038/ncomms10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.H., Howes S., Ti S.-C., Ramírez-Aportela E., Kapoor T.M., Chacón P., and Nogales E.. 2016. Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc. Natl. Acad. Sci. USA. 113:9430–9439. 10.1073/pnas.1609903113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., La Terra S., and Chang F.. 2004. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 14:1330–1340. 10.1016/j.cub.2004.07.028 [DOI] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W.C., Mochizuki Y., Dohmae N., and Todokoro K.. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23:3237–3248. 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laband K., Le Borgne R., Edwards F., Stefanutti M., Canman J.C., Verbavatz J.-M., and Dumont J.. 2017. Chromosome segregation occurs by microtubule pushing in oocytes. Nat. Commun. 8:1499 10.1038/s41467-017-01539-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansky Z., Braun M., Lüdecke A., Schlierf M., ten Wolde P.R., Janson M.E., and Diez S.. 2015. Diffusible crosslinkers generate directed forces in microtubule networks. Cell. 160:1159–1168. 10.1016/j.cell.2015.01.051 [DOI] [PubMed] [Google Scholar]
- Leduc C., Padberg-Gehle K., Varga V., Helbing D., Diez S., and Howard J.. 2012. Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc. Natl. Acad. Sci. USA. 109:6100–6105. 10.1073/pnas.1107281109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-Y., Esmaeili B., Zealley B., and Mishima M.. 2015. Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat. Commun. 6:7290 10.1038/ncomms8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legland D., Arganda-Carreras I., and Andrey P.. 2016. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 32:3532–3534. 10.1093/bioinformatics/btw413 [DOI] [PubMed] [Google Scholar]
- Leslie R.J., and Pickett-Heaps J.D.. 1983. Ultraviolet microbeam irradiations of mitotic diatoms: investigation of spindle elongation. J. Cell Biol. 96:548–561. 10.1083/jcb.96.2.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., and Khodjakov A.. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N., McDonald K.L., Ding R., and McIntosh J.R.. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489. 10.1083/jcb.123.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., and Landis S.C.. 1971. The distribution of spindle microtubules during mitosis in cultured human cells. J. Cell Biol. 49:468–497. 10.1083/jcb.49.2.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Cande W.Z., and Snyder J.A.. 1975a Structure and physiology of the mammalian mitotic spindle. Soc. Gen. Physiol. Ser. 30:31–76. [PubMed] [Google Scholar]
- McIntosh J.R., Cande Z., Snyder J., and Vanderslice K.. 1975b Studies on the mechanism of mitosis. Ann. N. Y. Acad. Sci. 253:407–427. 10.1111/j.1749-6632.1975.tb19217.x [DOI] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2017. Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53-Dependent Responses to Cell-Cycle Defects. Dev. Cell. 40:405–420.e2. 10.1016/j.devcel.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J. 1989. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109:637–652. 10.1083/jcb.109.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.-P., Jiang W., Schoehn G., Hunter T., and Margolis R.L.. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186. 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.-P., Saoudi Y., Jablonski S.A., Perard J., Yen T.J., and Margolis R.L.. 2005. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol. Biol. Cell. 16:1043–1055. 10.1091/mbc.e04-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., and Barr F.A.. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444. 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Oegema K., and Hyman A.A.. 2006. Cell division. WormBook.:1–40. 10.1895/wormbook.1.72.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak B., Risteski P., Lesjak S., and Tolić I.M.. 2017. PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 18:217–230. 10.15252/embr.201642650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakane K., Alrazi I.M.D., and Maruta S.. 2018. Highly efficient photocontrol of mitotic kinesin Eg5 ATPase activity using a novel photochromic compound composed of two azobenzene derivatives. J. Biochem. 164:295–301. 10.1093/jb/mvy051 [DOI] [PubMed] [Google Scholar]
- Salmon E.D., Leslie R.J., Saxton W.M., Karow M.L., and McIntosh J.R.. 1984. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99:2165–2174. 10.1083/jcb.99.6.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A.M., Powers J., Strome S., and Saxton W.M.. 2007. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 17:R453–R454. 10.1016/j.cub.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., and McIntosh J.R.. 1987. Interzone microtubule behavior in late anaphase and telophase spindles. J. Cell Biol. 105:875–886. 10.1083/jcb.105.2.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., Stemple D.L., Leslie R.J., Salmon E.D., Zavortink M., and McIntosh J.R.. 1984. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 99:2175–2186. 10.1083/jcb.99.6.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Materials and methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., and Pellman D.. 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160:517–528. 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden E., and Wadsworth P.. 1990. Interzonal microtubules are dynamic during spindle elongation. J. Cell Sci. 97:273–281. [DOI] [PubMed] [Google Scholar]
- Shimamoto Y., Forth S., and Kapoor T.M.. 2015. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev. Cell. 34:669–681. 10.1016/j.devcel.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K.-C., Bement W.M., Petronczki M., and von Dassow G.. 2014. An astral simulacrum of the central spindle accounts for normal, spindle-less, and anucleate cytokinesis in echinoderm embryos. Mol. Biol. Cell. 25:4049–4062. 10.1091/mbc.e14-04-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K.-C., Barry Z., Schweizer N., Maiato H., Bathe M., and Cheeseman I.M.. 2016. A Regulatory Switch Alters Chromosome Motions at the Metaphase-to-Anaphase Transition. Cell Reports. 17:1728–1738. 10.1016/j.celrep.2016.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., and Kapoor T.M.. 2010. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 142:433–443. 10.1016/j.cell.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R., Ti S.-C., Tan L., Darst S.A., and Kapoor T.M.. 2013. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 154:377–390. 10.1016/j.cell.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Vallotton P., van Oijen A.M., Whitchurch C.B., Gelfand V., Yeo L., Tsiavaliaris G., Heinrich S., Dultz E., Weis K., and Grünwald D.. 2017. Diatrack particle tracking software: Review of applications and performance evaluation. Traffic. 18:840–852. 10.1111/tra.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe K.J.C., and White J.G.. 2004. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 14:1755–1760. 10.1016/j.cub.2004.09.055 [DOI] [PubMed] [Google Scholar]
- Vernì F., Somma M.P., Gunsalus K.C., Bonaccorsi S., Belloni G., Goldberg M.L., and Gatti M.. 2004. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 14:1569–1575. 10.1016/j.cub.2004.08.054 [DOI] [PubMed] [Google Scholar]
- Voets E., Marsman J., Demmers J., Beijersbergen R., and Wolthuis R.. 2015. The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1. Sci. Rep. 5:14798 10.1038/srep14798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukušić K., Buđa R., Bosilj A., Milas A., Pavin N., and Tolić I.M.. 2017. Microtubule Sliding within the Bridging Fiber Pushes Kinetochore Fibers Apart to Segregate Chromosomes. Dev. Cell. 43:11–23.e6. 10.1016/j.devcel.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Z., and Adler R.. 1995. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J. Cell Biol. 128:761–768. 10.1083/jcb.128.5.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne S., and Subramanian R.. 2018. Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. eLife. 7:e32595 10.7554/eLife.32595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N., Morita M., Legant W.R., Chen B.-C., Betzig E., Yokota H., and Mimori-Kiyosue Y.. 2015. Three-dimensional tracking of plus-tips by lattice light-sheet microscopy permits the quantification of microtubule growth trajectories within the mitotic apparatus. J. Biomed. Opt. 20:101206 10.1117/1.JBO.20.10.101206 [DOI] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P.J., and Borisy G.G.. 1995. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 131:721–734. 10.1083/jcb.131.3.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.