Abstract
Background: Wildlife poses a significant burden for the complete eradication of bovine tuberculosis (bTB). In particular, wild boar (Sus scrofa) is one of the most important reservoirs of Mycobacterium bovis, the causal agent of bTB. Wild boar can display from mild TB lesions, usually found in head lymph nodes, to generalized TB lesions distributed in different anatomical regions; but rarely clinical signs, which complicates the diagnosis of Mycobacterium bovis infection and bTB control. Among the possibilities for this variability in lesion distribution is the influence of the host-beneficial commensal-primed immune barrier. In this respect, beneficial microbes may delay bTB dissemination as a consequence of an antagonistic competition for nutrients and phagocytes. In order to explore this possibility, we have tested whether typical commensals such as lactobacilli have the capacity to reduce the survival rate of the surrogate M. bovis strain Bacillus Calmette-Guerin (BCG); and to modulate its phagocyte intake.
Results: Three Lactobacillus species, L. casei, L. plantarum, and L. salivarius, isolated from wild boar feces displayed a pH-dependent inhibitory activity against BCG and influenced its intake by porcine blood phagocytes in a species-dependent manner. All lactobacilli showed a very significant bactericidal effect against BCG at low pH, but only isolates of L. plantarum and L. casei displayed such antimycobacterial activity at neutral pH. The genomes of these isolates revealed the presence of two-peptide bacteriocins whose precursor genes up-regulate in the presence of BCG cells. Furthermore, L. plantarum reduced significantly the BCG phagocytic intake, whereas L. casei had the opposite effect. L. salivarius had no significant influence on the phagocytic response to BCG.
Conclusions: Our in vitro results show that lactobacilli isolated from wild boar antagonize BCG as a consequence of their antimycobacterial activity and a competitive phagocytic response. These findings suggest that commensal bacteria could play a beneficial role in influencing the outcome of bTB dissemination. Further work with lactobacilli as a potential competitive pressure to control bTB will need to take into account the complex nature of the commensal microbiome, the specific immunity of the wild boar and the in vivo infection context with pathogenic strains of M. bovis.
Keywords: antagonism, Bacillus Calmette-Guerin, bovine tuberculosis, lactobacilli, Mycobacterium bovis, probiotics, wild boar
Introduction
Bovine tuberculosis (bTB) is a chronic infectious disease caused by Mycobacterium bovis that affects livestock production, leading to significant economic losses worldwide (Ayele et al., 2004; Naranjo et al., 2008). Despite decades-long eradication campaigns bTB is still very prevalent in some European countries including Spain, where wildlife reservoirs of Mycobacterium bovis have been confirmed (Corner, 2006; Naranjo et al., 2008). Wild ungulates such as the European wild boar (Sus scrofa) have been reported to show a high bTB prevalence across Spain (Parra et al., 2006). In this respect, the scientific evidence suggests that the cases of bTB infection occur more frequently in regions with higher densities of wild boar (Aranaz et al., 2004). Therefore, wild boar seems to be an additional obstacle for complete bTB eradication in Spain.
Together with epidemiological and ecological studies, a better knowledge on the pathology and transmission of M. bovis infection is essential to determine the significant role of wildlife as a bTB reservoir. Like any other species of the Mycobacterium tuberculosis complex, M. bovis cells are initially phagocytized by macrophages, where they are able to survive, replicate and disseminate into different anatomical parts of the body (Cosma et al., 2004). When the macrophages transport the bacteria into deeper tissues, additional macrophages gather around the individual infected foci to form granulomas, which are organized immune complexes of differentiated macrophages, lymphocytes, and neutrophils. Neutrophils are also present at early stages of the infection. Despite the formation of granulomas in response to M. bovis infection, bacterial persistence and reinfection occur, but with no clear symptoms, which is the central paradox of bTB immunity (Cosma et al., 2003). Some studies have revealed that a significant number of animals within a M. bovis-infected cattle group display no clinical signs, but present generalized bTB lesions at post-mortem examination (Menin et al., 2013). In this respect, wild boars are likely to be no different. Very recently, generalized bTB has been reported in wild boar, including cases with thoracic and abdominal lesions, usually in the bronchial and mesenteric lymph nodes (Martin-Hernando et al., 2007; Matos et al., 2016).
The absence of clinical symptoms in wild boar with generalized bTB, which usually shed a large amount of M. bovis cells to the environment (Santos et al., 2015), and the fact that bTB-like lesions are present in more than one anatomical region, complicate diagnosis, the understanding of the infection route, and, subsequently, disease control. It is assumed that M. bovis enters through the oral mucosa and tonsils via food, water or air borne, and disseminate from the mandibular lymph nodes (Martin-Hernando et al., 2007; Menin et al., 2013; Matos et al., 2016). However, the variable distribution of lesions suggests that; first, it is not possible to elucidate whether the respiratory or the digestive route is more relevant for bTB infection in wild boar; and, second, the immune response of each individual may have an influence on the outcome of the disease. Although the reasons for this variability in lesion distribution have not been accounted for conclusively, one explanation could be due to the contribution of the wild boar genetic variability (Acevedo-Whitehouse et al., 2005). Another possibility that has not been explored before is the influence of the host-beneficial commensal-primed immune barrier.
Pathogenic mycobacteria have evolved to evade host defenses in order to reach and exploit its replicative niche. However, this evasion may be unsuccessful if host commensal bacteria exert an extra immune-competitive pressure (Cambier et al., 2014). The host-commensal alliance forms a barrier that pathogens such as M. bovis must circumvent to survive and replicate. Commensal bacteria populate abundantly the mucosal surfaces of pigs and are composed of different species that predominantly belong to two dominant bacterial phyla: firmicutes and bacteroidetes (Xiao et al., 2016). In particular, one of the most abundant beneficial groups within firmicutes is the genus Lactobacillus (Mann et al., 2014). Lactobacilli are normal inhabitants of the intestinal tract of humans and mammals but also of the tonsillar crypts, nasopharynx, and upper respiratory tract, among others (Martens et al., 2018). Several species of Lactobacillus modulate immune responses as they interact regularly with epithelial cells and antigen-presenting cells such as macrophages (Mohamadzadeh et al., 2005; Rocha-Ramirez et al., 2017). The specific mechanisms of such immune modulations are unknown but it has been demonstrated that components of the cell wall and membrane of lactobacilli, such as pili (fimbriae), peptidoglycans, lipoteichoic acids, and exopolysaccharides play an important role in activating phagocytic innate immune cells (Hevia et al., 2015).
The functional involvement of lactobacilli in the modulation of immune responses is critical to maintain homeostasis, particularly in the gut (Diep et al., 2009; Gueimonde et al., 2013; Liu et al., 2014). Furthermore, Lactobacillus spp. are also essential to support a beneficial microbial balance in the host as they prevent against colonization of opportunistic pathogens (Martin et al., 2013). It is very well documented that lactobacilli produce antimicrobial metabolites such as organic acids, hydrogen peroxide, ethanol, and bacteriocins to compete for nutrients (Pessione, 2012); and some of these compounds can be active against M. bovis (Stedman et al., 2018). In particular, bacteriocins produced by Lactobacillus spp. have been reported to display antimicrobial activity against M. tuberculosis (Todorov et al., 2008, 2014). Bacteriocins are antimicrobial peptides that can be classified into three different classes depending on their chemical structure and size: (1) class I, post-translationally modified peptides (e.g. nisin); (2) class II, unmodified peptides smaller than 10 kDa, including the pediocin-like (class IIa); the two peptide- (class IIb); the leaderless (class IIc); and the single peptide-(class IId) bacteriocins; and (3) class III, unmodified peptides larger than 10 kDa with bacteriolytic (bacteriolysins) or non-lytic mechanism of action (Alvarez-Sieiro et al., 2016). Furthermore, it has been reported that bacteriocins from lactobacilli contribute to immunomodulatory effects on peripheral blood mononuclear cells and dendritic cells (Hegarty et al., 2016).
The fact that M. bovis may encounter lactobacilli in the gut or respiratory tract of the host while exploiting macrophages as its optimal niche for survival and replication and the evidence that lactobacilli are able to interact with macrophages and display antimycobacterial activity, suggest that lactobacilli could act as a key beneficial microbe against bTB dissemination as a consequence of an antagonistic competition for nutrients and macrophages. The aim of this study was to determine whether Lactobacillus spp. isolated from wild boar are capable of displaying this antagonistic role against the surrogate M. bovis strain Bacillus Calmette-Guerin (BCG). We isolated three Lactobacillus species from wild boar feces (L. plantarum, L. salivarius and L. casei) that were co-cultured with BCG to evaluate their potential antagonistic influence on the survival rate of M. bovis. To study their potential as macrophage competitors, porcine blood phagocytes were exposed to BCG in the presence of the three Lactobacillus species to quantify the BCG intake. Our in vitro results demonstrate that lactobacilli isolated from wild boar have the capacity to influence the phagocytic response to BCG and its survival in a species-dependent manner.
Materials and Methods
Isolation of Lactobacilli From Wild Boar Fecal Samples
Fecal samples were collected from a total of 30 wild boar from three fenced game estates located in mid-western Spain. These three states are surrounded by areas with a significant clinical history of bTB. In all cases, the fecal samples were taken using rectal swabs with AMIES transport medium (Deltalab) and stored at 4°C for a maximum of 1–2 days until further processing. The collected rectal swabs were immersed in sterile peptone water (Oxoid) to prepare serial dilutions that were spread onto De Man, Rogosa, and Sharpe (MRS) agar plates for the isolation of Lactobacillus spp. Plates were incubated under microaerophilic conditions at 37°C for 48–60 h and 50 colonies showing different morphologies (where possible) were then selected of each sample from plates containing 20–100 colonies. Individual colonies were inoculated to Thermo Scientific™ Nunc™ MicroWell™ 96-well plates with MRS broth at 37°C for 48 h under microaerophilic conditions.
Antimycobacterial Screening and Identification of Lactobacilli Isolates
Isolates propagated in MRS broth were tested against Mycobacterium smegmatis mc2155, a non-pathogenic fast-growing mycobacteria species that facilitates rapid antimycobacterial screening (Stedman et al., 2018). The MRS cultures present in the 96-well plates were replica plated with a Scienceware® 96-well replicator (Sigma-Aldrich) onto Tryptone Soya Agar (TSA, Oxoid) plates that had previously been swabbed with a culture of M. smegmatis mc2155 (Supplementary Figure S1). M. smegmatis was cultured in Tryptone Soya Broth (TSB, Oxoid) supplemented with 0.05% of Tween 80 (Sigma) in an orbital shaker at 37°C for 48 h. The TSA plates were then incubated at 37°C for 48 h and the cultures displaying antimycobacterial activity selected for colony isolation on MRS agar plates following an incubation of 48 h at 37°C. Colonies were identified using gram-staining (positive rods), oxidase/catalase tests (negative) and 16S rRNA sequencing (LGC Genomics) before a final selection of the most representative lactobacilli isolates comprising different strains and species from different animals belonging to different estates.
Growth Conditions for Lactobacilli Isolates and Mycobacterium Bovis Bacillus Calmette-Guerin Strains
Colonies from the selected lactobacilli isolates were grown in MRS broth/agar (Oxoid) at 37°C without any aeration for 24–48 h. Two Mycobacterium bovis Bacillus Calmette-Guérin (BCG) strains were used to determine the antagonistic effect of the lactobacilli isolates against M. bovis: BCG Pasteur and BCG ΔleuD pASOriMXF (Brosch et al., 2000; Stedman, 2017). Both BCG strains were cultured in Middlebrook 7H9 (Difco) broth supplemented with 10% Oleic acid-Albumin-Dextrose-Catalase enrichment (OADC, Sigma-Aldrich), 0.05% Tween 80 (Sigma-Aldrich), 0.2% glycerol and 40 μg/ml kanamycin at 37°C in an orbital shaker at 225 rpm for 5–7 days. BCG ΔleuD pASOriMXF was generated by transforming the BCG Pasteur ΔleuD strain with pASOriMXF as previously described (Stedman, 2017). pASOriMXF is a mycobacterial episomal vector that complements the leucine mutation to correct auxotrophy, and enable stable expression of GFP under the control of the constitutively expressed promoter pL5X (Borsuk et al., 2007). All bacterial strains were maintained as −80°C frozen stocks in their appropriate media with the addition of 15% glycerol.
Co-cultures of Lactobacilli and Bacillus Calmette-Guerin
Cell pellets of the selected lactobacilli and the BCG strains were obtained from cultures at their early stationary phase of growth following centrifugation at 8,000 rpm for 5 min. The BCG pellets were resuspended with each Lactobacillus pellet using Mueller-Hinton (MH) broth supplemented with 10% Oleic acid-Albumin-Dextrose-Catalase (OADC) enrichment, 0.1% Tween 80 (Tw80) and 0.2% glycerol (Gly) (MH-OADC-Tw80-Gly), to generate co-cultures at an initial concentration of 5 × 108 CFU/ml for BCG and 5 × 106 CFU/ml for the lactobacilli. In a previous study we reported that MH supplemented with OADC, tween, and glycerol is the optimal broth to support the growth of both lactobacilli and BCG when grown as mono-cultures for 48 h (Stedman et al., 2018). The co-cultures were then grown in an orbital shaker at 37°C for 48 h and samples were collected at 0, 24, and 48 h post-incubation to determine BCG survival rate.
Bacillus Calmette-Guerin Survival Rate in Co-cultures
The survival rate of BCG in co-cultures with the lactobacilli isolates was determined simultaneously by total bacterial counts and GFP expression. Total bacterial counts were calculated from co-cultures of the two selected BCG strains using a volume of 100 μl on mycobacteria-selective agar plates (Middlebrook 7H10 supplemented with 5% OADC and 0.2% glycerol). Bacterial counts were also carried out for lactobacilli on their corresponding selective MRS agar plates and presented as log10 CFU/ml. Colony forming units (CFU)/ml were calculated by dividing the number of colonies present on a plate by the dilution factor of the sample and the volume transferred to the selected plate. The GFP expression was monitored in co-cultures with the auxotrophy correction leucine auxotrophic strain BCG ΔleuD pASOriMXF, which maintains the GFP plasmid in broth without using any antibiotics (Stedman et al., 2018). Aliquots of 100 μl were transferred into Thermo Scientific Nunc MicroWell™ 96-well plates to measure fluorescence emission at 485/535 nm in a DTX 880 Multimode Detector microplate reader (Beckman Coulter). GFP expression was then indicated as fluorescence units (FU).
pH in Co-cultures
The pH of all co-cultures was measured using a Hanna pH meter at time points 0 and 48 h, starting from an initial pH of 7 for the media, to determine the effect of pH on the survival rate of BCG while co-culturing with lactobacilli samples. Furthermore, the survival rate of BCG was monitored as a monoculture at pH7 and pH4.5 in MH-OADC-Tw80-Gly to evaluate the influence of acidic pH on the growth of BCG. The monitoring was carried out based on bacterial counts (log10 CFU/ml) and GFP expression (FU) from the two BCG strains as explained above.
Accumulation of Anti-mycobacterial Metabolites From Lactobacilli in Mono-Cultures and Co-cultures With Bacillus Calmette-Guerin
In order to determine the effect of antimicrobial compounds produced by the lactobacilli isolates on BCG growth, cell pellets of BCG ΔleuD pASOriMXF were resuspended in cell-free supernatants collected from 24 h cultures of lactobacilli in MH-OADC-Tw80-Gly at a concentration of 5 × 108 CFU/ml. BCG pellets were also resuspended in cell-free supernatants obtained from 24 h co-cultures of lactobacilli and BCG at the same final concentration using the same supplemented MH broth to evaluate the influence of BCG on the production of antimycobacterial compounds by the lactobacilli isolates. For both sets of experiments cell pellets were obtained after centrifugation at 8,000 rpm for 5 min, and the pH of the supernatants was adjusted to 7 and 4.5 to consider any possible synergistic antimicrobial effects at low pH. The controls consisted of BCG cultures in cell-free supernatants obtained from 24 h-incubated MH-OADC-Tw80-Gly broth or 24 h-BCG monocultures in MH-OADC-Tw80-Gly. The survival rate of BCG was then determined as GFP expression (FU) as indicated above.
Nutrient Alteration Caused by Lactobacilli in Mono-Cultures and Co-cultures With Bacillus Calmette-Guerin
Additional GFP experiments with the collected supernatants indicated above were carried out at pH7 but mixed with MH-OADC-Tw80-Gly at a ratio of 1:1 to evaluate the influence of nutrient alteration on the survival rate of BCG. In this case, the supernatants for the controls were also adjusted to pH7 and supplemented with MH-OADC-Tw80-Gly at 1:1.
Influence of Bacillus Calmette-Guerin Metabolites on the Production of Anti-mycobacterial Metabolites by Lactobacilli
In order to test whether the antimycobacterial activity observed from co-cultures of BCG with our lactobacilli isolates could be due to an inducing effect from BCG metabolites we monitored the survival rate of BCG-GFP strain in supernatants derived from lactobacilli cultures in supernatants from BCG cultures. Briefly, BCG-GFP cells were resuspended in cell-free supernatants collected from cultures of lactobacilli after 24 h of incubation in supernatants from a 5 day-old BCG culture. Our controls consisted of BCG-GFP cells resuspended in cell-free supernatants obtained from 24 h cultures of lactobacilli in BCG supernatants supplemented with MH-OADC-Tw80-Gly at a ratio of 1:1, and also in MH-OADC-Tw80-Gly. For the three sets of experiments the pH of the supernatants was adjusted to 7. The survival rate of BCG was then determined as GFP expression (FU) as indicated above.
Genome Sequencing, Assembly, and Annotation of the Lactobacilli Isolates
The lactobacilli isolates were sent as pure isolated colonies on MRS agar plates to MicrobesNG, University of Birmingham, UK, where genome sequencing was carried out using the Illumina MiSeq platform. The DNA was extracted using the EZNA® Bacterial DNA kit (Omega Bio-Tek, USA) and the library preparation was carried out with the Nextera™ XT Library Prep Kit. The DNA from each isolate was sequenced using 2 × 250 bp paired-end reads and put through a standard analysis pipeline. The quality of the generated reads, which were first trimmed using Trimmomatic, was assessed using in-house scripts. Genomes were assembled from the paired-end reads using Shovill version 1.0.41 with SPAdes 3.13.0 as assembly module (Bankevich et al., 2012), using default settings. Assembly quality was assessed by N50 and L50, using Quast version 4.5 (Gurevich et al., 2013), and the genome assemblies were annotated using Prokka version 1.13 (Seemann, 2014). Sequencing reads, genome assemblies and metadata have been uploaded onto Genbank in BioProject PRJNA544176.
Identification of Genes and Clusters Associated With the Synthesis of Antimicrobial Metabolites and Macrophage Activation in Lactobacilli
The genome assemblies were uploaded on the online BAGEL3 software (van Heel et al., 2013) to identify gene clusters involved in the biosynthesis of bacteriocins. This software allows for a rapid and reliable identification of all classes of bacteriocin clusters, which are usually composed of genes encoding for the bacteriocin precursor (pre-bacteriocin), proteins involved in the transport and processing of the pre-bacteriocin as an active bacteriocin, posttranslational modification enzymes and immunity proteins. Furthermore, the genome annotations were used to confirm the presence or absence of fructose-6-phosphate aldolase and phosphoketolase, 2 enzymes that are involved in the 2 main glycolytic pathways in LAB: the Embden-Meyerhof pathway (EMP) and the phosphoketolase pathway (PKP), respectively (Papagianni, 2012a). In general, homofermentative LAB convert carbohydrates into lactate using EMP, whereas heterofermentative LAB produce lactate, acetate, ethanol, and carbon dioxide as antimicrobial metabolites via PKP. The genome annotations were also used to identify genes associated with hydrogen peroxide (H2O2) production, including genes that encode for pyruvate oxidase (Pox), lactate oxidase (Lox), and NADH oxidases. Genes encoding for NADH peroxidases were also included in the search for H2O2 production markers as H2O2 may accumulate is species that lack these hydrogen peroxide-scavenging enzyme (Hertzberger et al., 2014). We finally carried out a search on the genome annotations to detect cell wall and membrane compounds associated with macrophage activation via TLR and/or phagocytic receptors, including exopolysaccharides (EPS), fimbrial precursors, lipoteichoic acid (LTA), wall teichoic acid (WTA), and adhesins, as previously described for lactobacilli (Van Tassell and Miller, 2011; Sengupta et al., 2013; Hevia et al., 2015).
Level of Expression of Genes Encoding for Hypothetical Bacteriocin Precursors Identified in the Lactobacilli Genomes
The level of expression of transcripts from bacteriocin precursor genes was determined by Reverse Transcription-PCR (RT-PCR) analysis. Total RNA was extracted from cultures of lactobacilli in the absence or presence of BCG cells at two different concentrations (106 and 107 CFU/ml) using the High Pure RNA Isolation Kit (Roche Diagnostics Limited) as recommended by the manufacturer’s instructions. All culture samples were collected during exponential growth at an OD600nm of 0.6. RNA was treated with TURBO DNase (Ambion) to eliminate traces of contaminating genomic DNA and the DNA-free RNA was reverse transcribed using the SuperScript® III First-Strand Synthesis System from Invitrogen. Reverse transcription (RT)-PCR analyses were performed on the QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems) using the SYBR Master Mix (Applied Biosystems) and the specific primers for the selected transcripts (Table 1). Transcripts were amplified on a 96-well format plate by using three technical replicates of samples obtained from at least two biological replicates. We then used the ΔΔCT method to calculate the relative quantification of mRNA expression from cultures exposed to BCG cells by comparison with those with no BCG added. The levels of gene expression were normalized using the housekeeping transcripts gyrA and dnaG for each of the selected species as previously recommended (Rocha et al., 2015) and indicated as log2.
Table 1.
Primers used for the amplification of transcritps derived from the class II bacteriocin precursor genes of L. plantarum, L. casei, and L. salivarius.
| Gene | Bacteriocin | Primers | |
|---|---|---|---|
| plnE | Plantaricin E, L. plantarum | Fw: caatattccaggttgccgca | Rv: gaatgcctgcaactgaacca |
| plnF | Plantaricin F, L. plantarum | Fw: atttcaggtggcgttttcca | Rv: aatcctcggacagcgctaat |
| plnJ | Plantaricin J, L. plantarum | Fw: gccagcttcgccatcataaa | Rv: aggatttggatgtagtagatgca |
| plnK | Plantaricin K, L. plantarum | Fw: ttgaaccaccaagcacgg | Rv: ttgaagaattaactgctgacgc |
| A | Class IIb bacteriocin, L. casei | Fw: agttgtcaggggtttcaggt | Rv: ccgccgattatcccaaaagg |
| B | Class IIb bacteriocin, L. casei | Fw: gagccaagcgacgcaataaa | Rv: cgcctgcaacagttgtaaatg |
| Tα | Class IId bacteriocin, L. salivarius | Fw: gcaatcagaggaggaatggc | Rv: ccgatacaagccaatccacc |
| Tβ | Class IId bacteriocin, L. salivarius | Fw: gggaatggcattaattgggga | Rv: ggattaccgaaagctgcacc |
| gyrA | DNA topoisomerase, L. plantarum | Fw: tttaagtcgcaacaccgtgg | Rv: gattcctttggccgtacgac |
| dnaG | DNA primase, L. plantarum | Fw: agttggtagtcggtctggtg | Rv: cgcacctaaggatcagcaac |
| gyrA | DNA topoisomerase, L. paracasei | Fw: cttccacgcatgatgtcctg | Rv: cgccttcatgcacgttgata |
| dnaG | DNA primase, L. paracasei | Fw: cagttcggccaattgatcgt | Rv: cgactcgatccaggaatcca |
| gyrA | DNA topoisomerase, L. salivarius | Fw: gttttgccagcacgttttcc | Rv: tcccattacaattgcgccag |
| dnaG | DNA primase, L. salivarius | Fw: gcacaaagattcaacgtcgc | Rv: cgttctgctttctctgcctt |
gyrA and dnaG were selected as housekeeping transcripts as previously recommended (Rocha et al., 2015).
Phagocytosis Assay With Porcine Blood Cells
Whole fresh blood was collected in heparinized tubes from healthy pigs at the Pirbright Institute (UK), where all animal procedures are covered by a license issued by the UK Home Office under the Animal (Scientific Procedures) Act1986. About 100 μl of whole blood, which is equivalent to approximately 106 leukocytes, were mixed 1:1 with 10 mM EDTA (EDTA-PBS) and challenged with BCG in combination with lactobacilli at a multiplicity of infection (MOI) of 10 bacteria per 1 blood cell. The bacterial combinations were prepared in PBS and contained the GFP-expressing BCG strain ΔleuD pASOriMXF and each of the three Lactobacillus species. The same MOI was used for the preparation of control samples, which were obtained using monocultures of BCG ΔleuD pASOriMXF, BCG Pasteur and lactobacilli. The blood cells and bacteria were incubated in an orbital shaker at 37°C for 30 min and subsequently lysed with 1x RBC lysis solution (Biolegend) following incubation at room temperature for 15 min. The cells were then washed twice with EDTA-PBS, resuspended in PBS and acquired on the flow cytometer BD FACS Celesta. FACS sorting based on forward (FSC) and side (SSC) scatter was used to distinguish the main blood cell populations based on their size and granularity (lymphocytes vs. phagocytes), while the FITC channel let us measure the GFP levels in blood cells that bind (e.g. lymphocytes) and/or phagocytize (e.g. phagocytes including monocytes and PMNs such as neutrophils). The resulting SCC/GFP plot was then used to evaluate the influence of lactobacilli on the BCG intake by phagocytes.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). Data are mean ± SD, representing three biological replicates, unless indicated. Differences between time-points for the same samples were analyzed by the Student t-test.
Results
Lactic Acid Bacteria Isolated From Wild Boar Show Anti-mycobacterial Activity and Are Identified as Lactobacilli
A total of 16 strains isolated from wild boar feces on MRS selective agar displayed antimicrobial activity against M. smegmatis mc2155 as illustrated in Supplementary Figure S1. These 16 isolates were identified as L. plantarum, L. salivarius, L. paracasei, of which 6 were selected for further experiments as they represented 3 different species, with two strains of the same species (where possible) from at least two different animals and 3 different game estates. The six selected lactobacilli were L. plantarum C1, L. plantarum EML1, L. plantarum SA3, L. salivarius C2, L. salivarius C12, and L. paracasei SA5. Further species identification was carried out using StrainSeeker and ANItools (Han et al., 2016; Roosaare et al., 2017), two internet tools that allow for fast species identification by genome comparison with closely-related bacterial strains. With the exception of isolate SA5, the species of all isolates were confirmed. The genomes of our L. plantarum isolates form a phylogenetic clade with those of L. plantarum WCFS1 and B21 (isolates C1 and EML1) (Kleerebezem et al., 2003; Golneshin et al., 2015) and L. plantarum P-8 (isolate SA3) (Golneshin et al., 2015). We also observed two more clades between our isolates C2 and C12 and the L. salivarius strains JCM1046 and CECT5713 (Jimenez et al., 2010; Raftis et al., 2014), respectively. With regards to isolate SA5 this was finally determined to be L. casei instead of L. paracasei as its genome shares a phylogenetic clade with the genome of L. casei BL23 (Maze et al., 2010).
Lactobacilli Reduce Survival Rate of M. bovis Bacillus Calmette-Guerin and Lower pH in Co-cultures
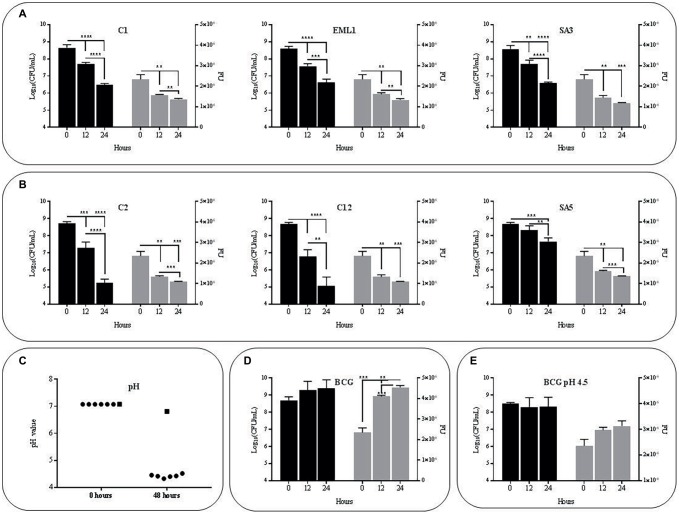
The selected isolates L. plantarum C1/EML1/SA3, L. salivarius C2/C12 and L. casei SA5 were cultured simultaneously with BCG Pasteur and the GFP-BCG strain ΔleuD pASOriMXF (Brosch et al., 2000; Stedman, 2017) to determine the influence of lactobacilli on the survival rate of M. bovis. We monitored the survival rate of BCG by measuring total BCG Pasteur counts and GFP emission from BCG ΔleuD pASOriMXF for 48 h, as illustrated in Figure 1. All the lactobacilli reduced BCG counts and GFP emission significantly after 24 and 48 h (Figures 1A,B). The pH of the co-cultures was also recorded in all the co-cultures after 48 h. We observed a pH decrease below 4.5, with no significant differences between co-cultures (Figure 1C). As pH of BCG monocultures slightly decreased from 7 to 6.8 we next tested whether acidic pH could account for the reduction observed in BCG viability. However, both total bacterial counts and GFP emission of BCG as a monoculture showed no detrimental changes at pH7 or pH4.5 after 24 and 48 h (Figures 1D,E). These results confirm that low pH may be insufficient on its own to cause a negative effect on BCG survival.
Figure 1.
Survival rate and acidity of BCG cultures after 48 h (h) of incubation in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%).The survival rate was monitored as bacterial counts (log10CFU/ml indicated with black bars) and GFP expression (FU indicated with gray bars), while acidity was measured as pH values. Data are mean ± SD with statistical analysis (Student’s t-test, **p < 0.01, ***p < 0.005, ****p < 0.001). (A) Survival rate of BCG in co-cultures with the L. plantarum strains C1, EML1 and SA3 after 0, 24 and 48 h of incubation. (B) Survival rate of BCG in co-cultures with the L. salivarius strains C2, C12 and L. casei SA5 after 0, 24 and 48 h of incubation. (C) Acidity of BCG co-cultures with lactobacilli strains C1, EML1, SA3, C2, C12 and SA5 (indicated as circles from left to right) and BCG monocultures (indicated as a square) after 0 and 48 h of incubation. (D) Survival rate of BCG mono-cultures after 0, 24 and 48 h of incubation from an initial pH of 7. (E) Survival rate of BCG mono-cultures after 0, 24 and 48 h of incubation from an initial pH of 4.5.
Metabolites From Lactobacilli Reduce Survival Rate of Bacillus Calmette-Guerin at Low pH
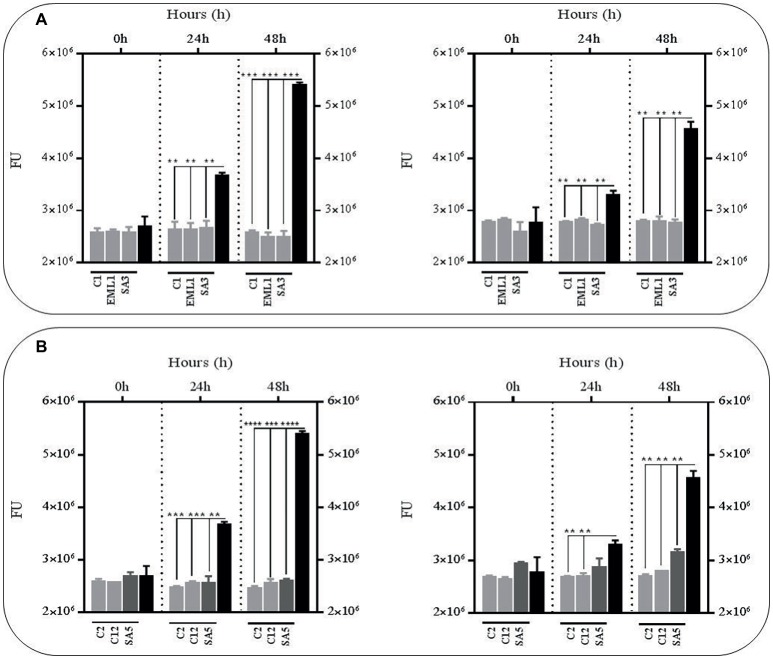
In order to test whether the antimicrobial effects observed against BCG in the presence of lactobacilli was due to an accumulation of toxic metabolites, we monitored the survival rate of the GFP-BCG strain in cell-free supernatants obtained from mono-cultures of lactobacilli and co-cultures of lactobacilli and BCG after 24 h of incubation (Figure 2). For these experiments the survival rate was recorded as GFP emission as the anti-mycobacterial effect observed in the co-cultures resulted in a very significant positive correlation between total bacterial counts and GFP emission (Supplementary Figure S2). Cell-free supernatants were collected from both mono-cultures of lactobacilli and co-cultures of lactobacilli and BCG to determine whether the presence of BCG may act as an inducer on the production of antimicrobial compounds by lactobacilli. We established 24 h as the collection time point for the supernatants since lactobacilli experience a significant log increase in bacterial counts over the first 24 h (Supplementary Figure S3). The bacterial counts recorded for all lactobacilli cultures, either as mono-cultures or co-cultures, were no significantly different. No significant differences were observed in the pH recorded for both lactobacilli culture conditions after 24 h either (data not shown). As illustrated in Figure 2, very significant reductions were observed with all the culture supernatants, either from mono-cultures or co-cultures with BCG, especially after 48 h. This demonstrated that the anti-mycobacterial activity of lactobacilli in co-cultures could be due to the combined effect of acidic pH and accumulation of antimicrobial metabolites derived from lactobacilli.
Figure 2.
Survival rate of BCG after 48 h (h) of incubation in acidic cell-free supernatants (pH 4.5) that were obtained from 24 h mono-cultures of lactobacilli (left hand side) or 24 h co-cultures of BCG with lactobacilli (right hand side) in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%). The BCG survival rate for both conditions was monitored as GFP expression (FU indicated with gray bars) and compared to their corresponding controls represented with black bars. The controls were BCG grown in cell-free supernatants obtained from a 24 h-incubated MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) (left hand side) and 24 h BCG monocultures in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) (right hand side), both at pH 4.5. Data are mean ± SD with statistical analysis (Student’s t-test, **p < 0.01, ***p < 0.005, ****p < 0.001). (A) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from mono-cultures of L. plantarum C1, EML1, SA3 (left hand side) or co-cultures of BCG with the L. plantarum strains (right hand side). (B) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from mono-cultures of L. salivarius C2 and C12 and L. casei SA5 (left hand side) or co-cultures of BCG with the strains of L. salivarius and L. casei (right hand side).
Metabolites From L. plantarum Co-cultures With Bacillus Calmette-Guerin Reduce the Survival Rate of Bacillus Calmette-Guerin Regardless of pH and Nutrient Supplementation
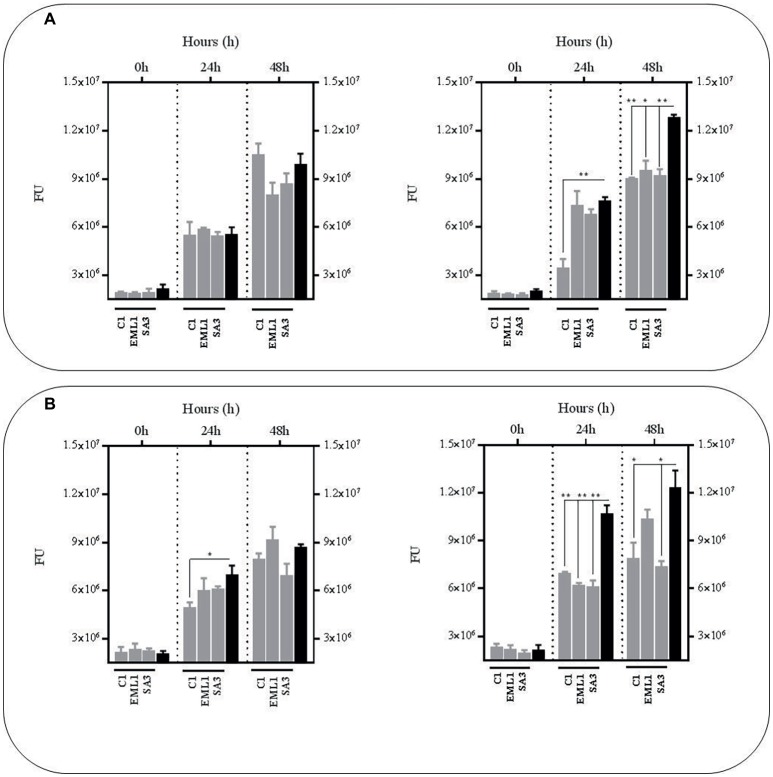
In order to confirm the anti-mycobacterial effect of metabolites produced by lactobacilli, we carried out the same experiments as described above with the supernatants, either from mono-cultures or co-cultures with BCG, but at pH 7 to exclude the positive antimicrobial effect of acidic pH. We also included a third experimental condition by supplementing the supernatants with fresh co-culture media to determine the influence of nutrient alteration caused by lactobacilli metabolism. Due to logistic reasons we started with the L. plantarum strains. As illustrated in Figure 3, no significant reductions were observed in BCG survival using the supernatants obtained from the L. plantarum mono-cultures, either with or without nutrient supplementation. However, the reduction was very significant when we used the supernatants from the co-cultures of the three L. plantarum strains with BCG, especially with supplementation after 24 h, but also after 48 h without any supplementation. These results suggest that our L. plantarum strains over-produce certain anti-mycobacterial metabolites in the presence of BCG. Furthermore, this induced antimicrobial activity seems to be stable at different ranges of pH and nutrient composition.
Figure 3.
Survival rate of BCG after 48 h (h) of incubation in neutralized cell-free supernatants (pH 7) that were obtained from 24 h mono-cultures of L. plantarum C1, EML1 and SA3 (left hand side) or 24 h co-cultures of BCG with the L. plantarum strains (right hand side) in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%). The BCG survival rate for both conditions was monitored as GFP expression (FU as gray bars) and compared to their corresponding controls represented with black bars, with or without nutrient supplementation. The controls were BCG grown in cell-free supernatants obtained from a 24 h-incubated MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) (left hand side) and 24 h BCG monocultures in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) (right hand side), both at pH 7. Data are mean ± SD with statistical analysis (Student’s t-test, *p < 0.05, **p < 0.01). (A) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from L. plantarum mono-cultures (left hand side) or co-cultures of BCG with the L. plantarum strains (right hand side). (B) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from L. plantarum mono-cultures (left hand side) or co-cultures of BCG with the L. plantarum strains (right hand side) that were supplemented with MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) at a ratio of 1:1. The supernatants for the controls were also supplemented with MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) at 1:1.
The Anti-mycobacterial Effect of Metabolites From L. salivarius and L. casei Depends on pH but Also on the Presence of Bacillus Calmette-Guerin if Derived From L. casei
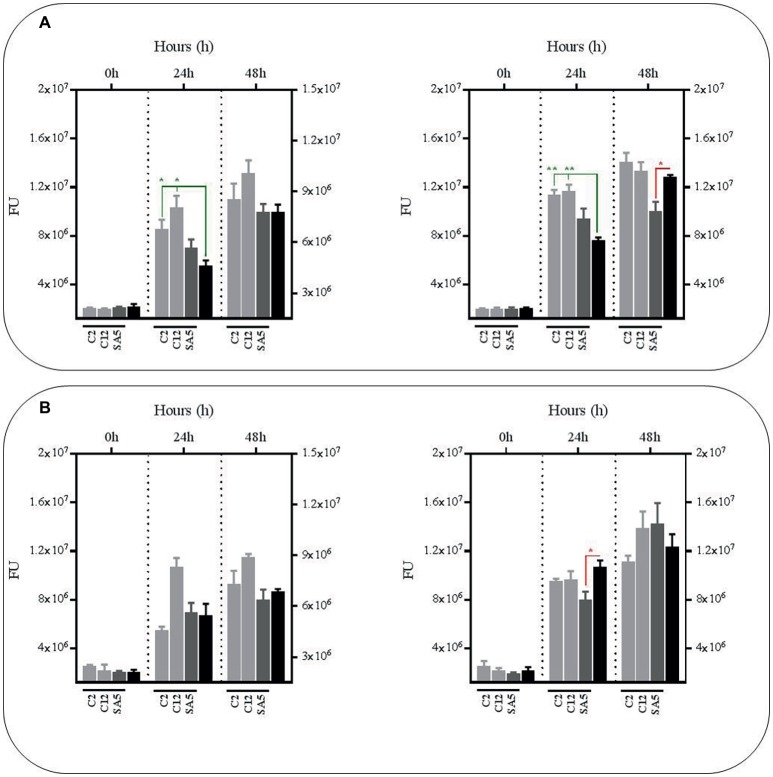
As observed with the L. plantarum supernatants no significant reductions were observed in BCG survival using the supernatants obtained from mono-cultures of L. salivarius and L. casei at pH7, either with or without nutrient supplementation (Figure 4). In fact, the BCG survival increased in the supernatants from the L. salivarius mono-cultures without supplementation. The same observation was recorded with the supernatants collected from the co-cultures of L. salivarius and BCG, confirming a pH-dependency on the anti-mycobacterial effect from the L. salivarius strains. On the contrary, the survival rate of BCG decreased slightly in the supernatants derived from co-cultures of L. casei and BCG, suggesting a role of BCG as an inducer on the production of antimicrobial compounds in L. casei. Nevertheless this induction seems to be significantly lower than that observed with L. plantarum.
Figure 4.
Survival rate of BCG after 48 h (h) of incubation in neutralized cell-free supernatants obtained from 24 h mono-cultures of L. salivarius C2 and C12 and L. casei SA5 (left hand side) or 24 h co-cultures of BCG with the strains of L. salivarius and L. casei (right hand side) in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%). The BCG survival rate for both conditions was monitored as GFP expression (FU as light gray bars for L. salivarius and dark gray bars for L. casei) and compared to their corresponding controls represented with black bars, with or without supplementation. The controls were BCG grown in cell-free supernatants obtained from a 24 h-incubated MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) (left hand side) and 24 h BCG monocultures in MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%), both at pH 7 (right hand side). Data are mean ± SD with statistical analysis (Student’s t-test, *p < 0.05, **p < 0.01) and significant GFP differences are indicated in green (increase) or red (decrease). (A) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from mono-cultures of L. salivarius and L. casei (left hand side) or co-cultures of BCG with the strains of L. salivarius and L. casei (right hand side). (B) Survival rate of BCG after 0, 24 and 48 h of incubation in supernatants from mono-cultures of L. salivarius and L. casei (left hand side) or co-cultures of BCG with the strains of L. salivarius and L. casei (right hand side) that were supplemented with MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) at a ratio of 1:1. The supernatants for the controls were also supplemented with MH broth with OADC (10%), Tween 80 (0.1%) and glycerol (0.2%) at 1:1.
Metabolites From Bacillus Calmette-Guerin Have No Influence on the Production of Anti-mycobacterial Metabolites by L. plantarum and L. casei
In order to test whether the induced antimycobacterial activity observed from co-cultures of BCG with the L. plantarum strains and L. casei SA5 was dependent on the presence of BCG metabolites rather than cells, we monitored the survival rate of BCG-GFP strain in cell-free supernatants derived from cultures of L. plantarum and L. casei that were propagated for 24 h in cell-free supernatants from 5 day-old BCG cultures. We also included two additional experimental conditions by supplementing the supernatants with fresh co-culture media but also by growing the lactobacilli in complete fresh co-culture broth to consider the possible negative influence of nutrient alteration in the growth of lactobacilli. The bacterial counts recorded for the 3 different types of lactobacilli cultures were not significantly different (Supplementary Figure S4). The pH of the supernatants from lactobacilli cultures propagated in BCG supernatants was slightly above 4.5 but no significant differences were observed when compared to the other lactobacilli cultures (data not shown). As illustrated in Supplementary Figure S5, no significant reductions were observed in BCG survival using the supernatants obtained from the L. plantarum cultures grown in supernatants from BCG cultures, either with or without nutrient supplementation. These data suggest that BCG metabolites have no inducing effect on the anti-mycobacterial activity from L. plantarum and L. casei. Subsequently, the higher antimicrobial effect observed in co-cultures of BCG with L. plantarum and L. casei when compared to mono-cultures could be due to the presence of BCG cells.
Lactobacilli Harbor Genes Associated With the Synthesis of Antimicrobial Metabolites
The genome annotations let us identify the genes encoding for fructose-6-phosphate aldolase and phosphoketolase in all the lactobacilli isolates, demonstrating their role as facultative heterofermenters. Therefore, our isolates are able to convert carbohydrates into lactate using the EMP pathway and/or produce lactate in combination with ethanol, acetate and carbon dioxide as antimicrobial metabolites via the PKP pathway. Furthermore, all the isolates harbor genes associated with H2O2 production, e.g. including Pox, Lox, and/or NADH oxidases as well as genes encoding for the H2O2-scavenging enzyme NADH peroxidase. Interestingly, the BAGEL3 genome analysis identified gene clusters involved in the hypothetical synthesis of class II and class III bacteriocins, including single-peptide bacteriocins in L. salivarius C2 (Table 2); two-peptide bacteriocins in all L. plantarum strains and L. casei SA5 (Table 3), and bacteriolysins in all lactobacilli isolates (Table 4). The two single-peptide bacteriocin precursors identified in the genome of L. salivarius C2 (Tα, Tβ) show a very high homology with two class II bacteriocins of L. salivarius BGH01 (Busarcevic and Dalgalarrondo, 2012), while the two-peptide bacteriocin precursors of the L. plantarum genomes (plnE, plnF) are identical to the plantaricin precursor genes plnE and plnF of L. plantarum C11 (Anderssen et al., 1998). The two-peptide bacteriocin precursors found in the genome of L. casei SA5 (A, B) were also identical to two hypothetical class II bacteriocins of Lactobacillus casei DPC6800 (Stefanovic et al., 2016) and Lactobacillus casei UCD174 (Broadbent et al., 2012). On the other hand, the bacteriolysin genes that we identified in all the genomes encode for enzymes that hydrolyse cell wall peptidoglycans between N-acetylmuramic acid and N-acetyl-D-glucosamide, or N-acetylmuramoyl and L-aa residues. These enzymes normally have target recognition site and a catalytic domain that shows homology to endopeptidases, muramidases, or amidases (Cotter et al., 2005). Furthermore, unlike the class II bacteriocin clusters, the bacteriolysins have no specific immunity genes that accompany the bacteriocin precursor genes as they rely on modifications of the producer cell wall to impart resistance. The identified bacteriolysins were very abundant in L. salivarius C2, followed by L. salivarius C12 and L. casei SA5. The L. plantarum strains only share two of them with the remainder of the isolates.
Table 2.
Proteins encoded by the hypothetical single peptide-bacteriocin cluster of Lactobacillus salivarius C2.
| ORF (gene) | Size (aa) | Function | Homologue | % | Reference |
|---|---|---|---|---|---|
| 1 (T2) | 87 | Putative protein | Lactobacillus salivarius CCUG47825 | 96 | (Harris et al., 2017) |
| 2 (T3) | 57 | Bacteriocin-like peptide | Lactobacillus salivarius UCC118 | 96 | (Flynn et al., 2002) |
| 3 (T4) | 85 | Bacteriocin-like peptide | Lactobacillus salivarius ATCC11741 | 96 | (Harris et al., 2017) |
| 4 (Ta) | 80 | Subunit A bacteriocin ThmA | Blp1a, Lactobacillus salivarius BGH01 | 96 | (Busarcevic and Dalgalarrondo, 2012) |
| 5 (T1 M1) | 59 | Immunity | Lactobacillus salivarius BGH01 | 82 | (Busarcevic and Dalgalarrondo, 2012) |
| 6 (Tb) | 69 | Subunit B bacteriocin TmhB | Blp1b, Lactobacillus salivarius BGH01 | 100 | (Busarcevic and Dalgalarrondo, 2012) |
| 7 (T1 M2) | 54 | Immunity | Lactobacillus salivarius BGH01 | 100 | (Busarcevic and Dalgalarrondo, 2012) |
| 8 (TIP) | 39 | Induction peptide | Lactobacillus salivarius ATCC11741 | 79 | (Harris et al., 2017) |
| 9 (TK) | 429 | Histidine kinase | Lactobacillus salivarius ATCC11741 | 87 | (Harris et al., 2017) |
| 10 (TR) | 264 | Response regulator | Lactobacillus salivarius UCC118 | 99 | (Flynn et al., 2002) |
| 11 (orf1) | 76 | Putative protein | Lactobacillus salivarius ATCC11741 | 99 | (Harris et al., 2017) |
| 12 (orf2) | 65 | Putative protein | Lactobacillus salivarius ATCC11741 | 100 | (Harris et al., 2017) |
| 13 (orf3) | 44 | Putative protein | Lactobacillus salivarius ATCC11741 | 100 | (Harris et al., 2017) |
| 14 (orf4) | 237 | Putative protein | Lactobacillus salivarius ATCC11741 | 98 | (Harris et al., 2017) |
| 15 (orf5) | 88 | Putative protein | Lactobacillus salivarius CCUG47825 | 99 | (Harris et al., 2017) |
| 16 (orf6) | 273 | Integrase | Lactobacillus salivarius UCC118 | 99 | (Flynn et al., 2002) |
Table 3.
Proteins encoded by the hypothetical two peptide-bacteriocin clusters of Lactobacillus plantarum C1, EML1 & SA3; and L. paracasei SA5.
| Strains | ORF (gene) | Size (aa) | Function | Homologue (100%) | Reference |
|---|---|---|---|---|---|
| C1/EML1/SA3 | 1 (W) | 228 | CAAX protease self-immunity | PlnW, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) |
| 2 (V) | 226 | CAAX protease self-immunity | PlnV, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 3(U) | 222 | CAAX family putative protein | PlnU, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 4 (T) | 149 | CAAX family putative protein | PlnT, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 5 (S) | 99 | Putative protein | PlnS, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 6 (H) | 458 | ABC-transporter accessory factor | PlnH, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 7 (G) | 716 | ABC transporter | PlnG, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 8 (E) | 56 | Plantaricin E | PlnE, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| 9 (F) | 52 | Plantaricin F | PlnF, Lactobacillus plantarum C11 | (Diep et al., 1996, 2009) | |
| SA5 | 1 (A) | 77 | Class IIb bacteriocin | Lactobacillus casei DPC6800 | (Stefanovic et al., 2016) |
| 2 (B) | 71 | Class IIb bacteriocin | Lactobacillus casei UCD174 | (Broadbent et al., 2012) | |
| 3(C) | 102 | Putative protein | Lactobacillus casei W56 | (Hochwind et al., 2012) | |
| 4 (AT) | 198 | Acetyltransferase | Lactobacillus casei W56 | (Hochwind et al., 2012) | |
| 5 (D) | 111 | Immunity | Lactobacillus casei DPC6800 | (Stefanovic et al., 2016) | |
| 6 (E) | 225 | Metalloprotease | Lactobacillus casei W56 | (Hochwind et al., 2012) | |
| 7 (F) | 110 | Putative protein | Lactobacillus casei W56 | (Hochwind et al., 2012) | |
| 8 (G) | 52 | Putative membrane protein | Lactobacillus casei BL23 | (Maze et al., 2010) |
Table 4.
Proteins encoded by the hypothetical bacteriolysin genes identified in the lactobacilli genomes.
| Protein | Gene | Size (aa) | Conserved catalytic domain | Lactobacillia | Homologue (100%) | Reference |
|---|---|---|---|---|---|---|
| Phage lysin | acm | 245–772 | M23 endopeptidase GH25-PlyB-like muramidase | C1, EML1, SA3 C2, C12, SA5 |
Lactobacillus sp. CBA3606 L. paracasei ATCC 25302 L. salivarius CECT 5713 |
NCBI Complete Genomes (Ward and Timmins, 1999) (Martin et al., 2006) |
| Lysozyme | lyc | 309–921 | GH25-LysA-like endolysin | C2, C12, SA5 |
L. reuteri TD1 L. casei 32G |
(Leonard et al., 2014) (Aktas et al., 2015) |
| Autolysin 1 | lytA_1 | 486 | C39 endopeptidase | C2 | L. salivarius cp400 | (Mackenzie et al., 2014) |
| Autolysin 2 | lytA_2 | 350–468 | PGRP-family amidase | C2, C12, SA5 |
L. salivarius ACS-116-V-Col5a L. casei 32G |
NCBI Complete Genomes (Aktas et al., 2015) |
| Amidase | lytC | 282–350 | MurNAc-LAA | C1, EML1, SA3 C2, C12, SA5 |
Lactobacillus sp. CBA3606 Lactobacillus sp. DS22_6 |
NCBI Complete Genomes |
| Toxin 1 | toxA_1 | 125 | Amidase | C2 | L. salivarius cp400 | (Mackenzie et al., 2014) |
| Toxin 2 | toxA_2 | 523 | M23 endopeptidase | C2, C12 | L. salivarius cp400 | (Mackenzie et al., 2014) |
Lactobacilli strains in which the genes have been identified.
The Genes Encoding for the Two-Peptide Bacteriocins of L. plantarum and L. casei Over-Express in the Presence of Bacillus Calmette-Guerin Cells
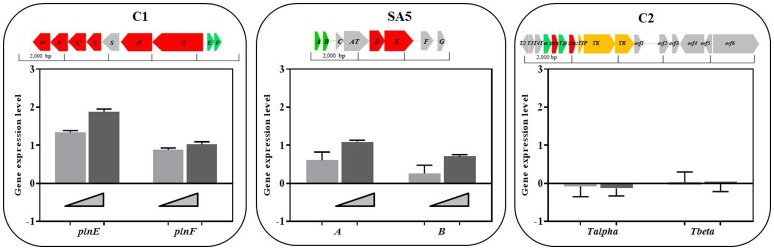
In order to determine whether the presence of BCG cells may regulate the level of expression of the identified class II bacteriocins we carried out an RT-PCR to quantify the amount of transcripts derived from the genes encoding for the hypothetical precursor bacteriocins in all L. plantarum strains, L. salivarius C2 and L. casei SA5 (indicated in green on top of Figure 5). The RNA was isolated from cultures of lactobacilli exposed to increasing concentrations of BCG cells during exponential growth. As illustrated in Figure 5, the level of expression of genes plnE and plnF was dependent on the amount of BCG cells used, with a very significant increase by comparison with L. plantarum cultures on their own. We observed similar results with A/B genes of L. casei SA5 although the increase was much lower. By contrast, the expression of genes Tα/Tβ in L. salivarius C2 was not affected by the presence of BCG cells (Figure 5). These data confirm not only the inducing effect of BCG cells on the anti-microbial activity displayed by L. plantarum and L. casei in co-cultures with BCG but also the possible role of two-peptide bacteriocins is such antimicrobial activity against BCG.
Figure 5.
Class II bacteriocin gene clusters identified in the genome of the lactobacilli strains and level of expression of genes encoding for the hypothetical precursor bacteriocins in lactobacilli cultures exposed to increasing concentrations of BCG cells. The nomenclature for the bacteriocin clusters follows specific recommendations (Diep et al., 2009; O’Shea et al., 2011) and represents precursor bacteriocins (green), posttranslational modification enzymes (blue), transport/immunity proteins (red) and other hypothetical proteins (gray). C1. Two peptide-bacteriocin cluster of L. plantarum strains C1 (as a representative of all L. plantarum strains) and level of expression of its corresponding precursor bacteriocin genes plnE and plnF in cultures exposed to BCG cells at 106 CFU/ml (light gray bars) and 107 CFU/ml (dark gray bars). SA5. Two peptide-bacteriocin cluster of L. casei SA5 and level of expression of its corresponding precursor bacteriocin genes A and B in cultures exposed to increasing concentrations of BCG cells at 106 CFU/ml (light gray bars) and 107 CFU/ml (dark gray bars). C2. Single-peptide bacteriocin cluster of L. salivarius C2 and level of expression of its corresponding precursor bacteriocin genes Tα and Tβ in cultures exposed to BCG cells at 106 CFU/ml (light gray bars) and 107 CFU/ml (dark gray bars).
Lactobacilli Influence Bacillus Calmette-Guerin Phagocytosis in a Species-Dependent Manner
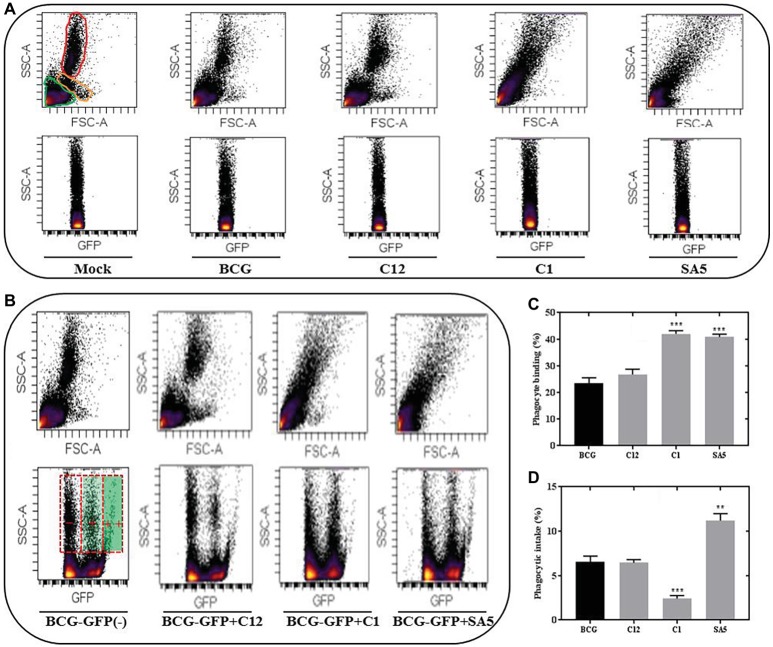
Representative FSC-SSC plots indicating the presence of lymphocytes, monocytes and PMNs (neutrophils) in porcine blood are illustrated in Figure 6A. Unlike BCG and L. salivarius, the presence of L. plantarum and L. casei alters significantly the blood scatter profile, which complicated the identification of monocytes and neutrophils. Nevertheless, the different distribution of lymphocytes and phagocytes that we observed on the SCC-GFP plots under all bacterial conditions, let us distinguish between blood cells that are positive for BCG binding or BCG intake based on the GFP intensity. Lymphocytes and some phagocytes were found to be GFP positive but the highest GFP intensity was only observed in the phagocytic cells (Figure 6B). L. salivarius (C12) had no significant effect on the BCG binding (Figure 6C); neither on the phagocytic response to BCG (Figure 6D). However, the percentage of phagocytes positive for BCG binding and BCG intake changed significantly in the presence of L. plantarum (C1) and L. casei (SA5) Although both lactobacilli increased the binding between BCG and phagocytes (Figure 6C) their effect on BCG phagocytosis was completely different (Figure 6D). L. plantarum reduced the BCG intake, whereas L. casei caused the opposite effect.
Figure 6.
Lactobacilli interact with porcine blood cells, interfering with BCG phagocytosis in a species-dependent manner. (A) SSC/FSC plot areas representing porcine blood cells and their corresponding SSC/GFP areas when exposed to non-stimulated conditions (mock), BCG, L. salivarius C12, L. plantarum C1 and L. casei SA5. For the mock experimental conditions the blood cells are indicated in green (lymphocytes), orange (monocytes) and red (PMNs). (B) SSC/GFP plot areas illustrating the response of porcine blood cells to BCG-GFP alone (−) or in combination (+) with L. salivarius C12, L. plantarum C1 and L. casei SA5. The SSC/FSC areas for each of the conditions are included above and the GFP intensity recorded from phagocytes is indicated on the BCG-GFP experimental condition as negative (−), positive (+) and very positive (++). (C) Percentage of phagocytes that bind BCG when tested alone (black bar) or in combination (gray bars) with L. salivarius C12, L. plantarum C1 and L. casei SA5. Data are mean ± SD with Student’s t-test statistical analysis (***p < 0.001). (D) Percentage of phagocytes that phagocytize BCG when tested alone (black bar) or in combination (gray bars) with L. salivarius C12, L. plantarum C1 and L. casei SA5. Data are mean ± SD with Student’s t-test statistical analysis (**p < 0.01, ***p < 0.001).
Discussion
To the best of our knowledge, this is the second study reporting the isolation of Lactobacillus spp. from wild boar. The only previous publication is focused on the antibiotic susceptibility of lactobacilli isolates and the authors found Lactobacillus species are different from this study, including L. mucosae, L. reuteri, L murinus, and L. fermentus (Klose et al., 2014). In our study we describe, for the first time, that wild boar carry Lactobacillus spp. such as L. plantarum, L. salivarius and L. casei that exert antimycobacterial activity when they compete against M. bovis BCG in co-cultures. Our results are consistent with previous studies that found that BCG and M. bovis are inhibited by lactobacilli isolated from badger feces or present in fermented milk products (Mariam, 2009; Macuamule et al., 2016; Stedman et al., 2018). We initially suspected that the antimicrobial activity that our lactobacilli isolates display against BCG was due to acidic pH but low pH was insufficient to induce a decrease in the survival of BCG when grown as a mono-culture as previously reported (de la Rua-Domenech, 2006; Stedman et al., 2018). In fact, M. bovis naturally resist acidic pH due to their the ability to adopt intracellular homeostasis (Rao et al., 2001).
Overall the lactobacilli isolates that we have isolated in this study seem to produce metabolites that display a very significant antimicrobial activity against BCG, but only at low pH, suggesting a synergistic effect at acidity conditions. All our isolates contain genes associated with the synthesis of lactate and acetate, which inhibit active transport in other bacteria by causing interference on the membrane potential; as well as ethanol, CO2 and hydrogen peroxide, additional antimicrobial compounds that may prevent bacterial growth by creating a hostile environment (Pessione, 2012). It has been well-documented that the antibacterial activity of lactobacilli is multifactorial, including lowering of the pH with simultaneous production of lactic acid and of non-lactic acid metabolites (Fayol-Messaoudi et al., 2005). These antimicrobial metabolites could even act in synergy since lactic acid permeabilizes the outer membrane of bacteria, allowing the non-lactic acid metabolites to enter the bacterial cells (Alakomi et al., 2000). This potential synergistic effect between acidic pH and antimicrobial metabolites was further supported by the lack of antimycobacterial activity that we observed at neutral pH. All the lactobacilli mono-cultures provided with metabolites that were incapable of inhibiting the growth of BCG at pH 7.
The reasons why metabolites from the lactobacilli mono-cultures display no activity against BCG at pH7 could also lie on the fact that some bacteriocins are only fully functional at acidic conditions. The genome of all the lactobacilli isolates showed genes related to the biosynthesis of bacteriolytic class III bacteriocins known as bacteriolysins, especially in L. salivarius. Bacteriolysins are enzymes that have been adapted to the host habitat of acidification, with an optimal bactericidal activity that ranges between pH values of 4 and 6 (Sable and Lortal, 1995; Ribelles et al., 2012). This could also explain the absence of antimicrobial activity that we recorded against BCG using metabolites from all the L. salivarius co-cultures at pH7. However, neutral pH had no influence on the antimycobacterial activity observed with the metabolites derived from co-cultures of BCG with L. plantarum and L. casei. This remarkable observation suggests that, first, such antimycobacterial activity is dependent on the lactobacilli species and is not associated with low pH; and, second, BCG triggers its own bactericidal effect. Interestingly, we found that the strains of L. plantarum and L. casei carry gene clusters associated with the production of two-peptide bacteriocins, also known as class IIb bacteriocins, which consist of two different peptides that confer optimal antimicrobial activity if both peptides are present in equal amounts (Nissen-Meyer et al., 2010). Two-peptide bacteriocins often display an antibacterial activity that is very stable at a very broad range of pH (1–11); and the genes that encode for the two peptides are next to each other in the same operon where other genes associated with processing, transport and immunity are located.
The genes that encode for the two-peptide bacteriocin that we identified in the genome of the three L. plantarum strains are identical to the two genes of operon plnEF. Genes plnE and plnF allow the expression of two peptides, plantaricins E and F, that interact together to form a helix-helix structure that binds to a specific membrane protein of the target bacteria, presumably a transporter of the APC superfamily, leading to membrane leakage and cell death (Ekblad et al., 2016; Oppegard et al., 2016). The remainder of the genes that we detected in the L. plantarum genomes are identical to operon plnWVUTSHG, which are thought to be involved in transport, processing and self-protection (Diep et al., 2009). The operons plnEF and plnWVUTSHG are frequently found in many L. plantarum strains, along with other two-peptide bacteriocin clusters such as plantaricin JK, although it has been reported to be on their own in certain isolates (Saenz et al., 2009; Tai et al., 2015). Furthermore, we detected a gene cluster involved in the hypothetical synthesis of a novel two-peptide bacteriocin in L. casei. The gene cluster is composed of two genes that encode for 2 bacteriocins that show certain similarity with thermophilin A (Ward and Somkuti, 1995) and carnocin CP51 (Herbin et al., 1997), followed by a galactoside O-acetyltransferase gene prior to two additional genes involved in immunity and transport. Carnocin CP51 is expressed with the complementary peptide carnocin CP52 in Carnobacterium and thermophilin A glycosylates as it occurs with some plantaricins (Diep et al., 2009).
As a very interesting fact we observed that the two gene operons that encode for the two-peptide bacteriocins in L. plantarum and L. casei over-express in the presence of BCG cells in a dose-dependent manner. These results could explain why metabolites that derive from co-cultures of BCG with L. plantarum and L. casei, show anti-mycobacterial activity. Neither monocultures of these two Lactobacillus spp. or their metabolites after propagation in supernatants containing BCG metabolites displayed any antimycobacterial activity. In fact, the regulation of plantaricin production had been previously described in co-cultures of L. plantarum (Maldonado et al., 2004b; Maldonado-Barragan et al., 2013). In agreement with our transcriptional data, a previous study has reported that operon of the two-peptide bacteriocin plantaricin NC8 of L. plantarum NC8 is up-regulated in broth only after cultivation with other gram-positive bacteria or the addition of heat-killed cells from some of the inducing bacteria (Maldonado et al., 2004a). Whether two-peptide bacteriocins such as plantaricins are involved in the antimycobacterial activity that we have observed in this study is a question that remains unanswered. Based on our results it is too early to speculate about the possible role of plantaricins as antimicrobials against mycobacteria; mainly because of their relatively narrow inhibitory spectra, which includes other species of lactobacilli as well as gram-positive closely related bacteria such as Pediococcus (Diep et al., 2009). However, this potential anti-mycobacterial role is worthy of further investigation due to the promising contribution of plantaricins to probiotic functionality when colonizing the gut (Maldonado-Barragan et al., 2013) and the fact that class II bacteriocins have antimicrobial activity against M. tuberculosis (Sosunov et al., 2007).
Unlike the two-peptide bacteriocins, the two identified single-peptide bacteriocins of L. salivarius showed no transcriptional up-regulation in the presence of BCG cells, which is in accordance with the evidence that metabolites from L. salivarius have no influence on the survival of BCG at pH7. The two bacteriocins that we found in the genome of L. salivarius show similarities with multiple class IIb bacteriocins previously described in other L. salivarius strains (O’Shea et al., 2011) but also with bacteriocins blp1α/blp1β of L. salivarius BGHO1 (Busarcevic and Dalgalarrondo, 2012) and ThmA/ThmB (Thermophilin 13) of Streptococcus thermophilus (Marciset et al., 1997). Our two L. salivarius bacteriocins contain the double-glycine leader sequence of class II bacteriocins but, in contrast to two-peptide bacteriocins (class IIb), their operons carry immunity genes for each of the bacteriocin precursors. As these bacteriocins also lack the YGNGV-C consensus sequence of class IIa bacteriocins the bacteriocin cluster was classified as a single-peptide bacteriocin (class IId). The evidence that bacteriocins blp1α/blp1β are produced only after culturing in a very chemically defined medium and that bacteriocins ThmA/ThmB function without a bacterial membrane receptor may also explain the absence of antimycobacterial activity observed with the L. salivarius metabolites at neutral pH.
Another important aspect that is worth mentioning is the fact that nutrient deprivation caused by lactobacilli metabolism seems to have no influence on the survival of BCG. The viability of BCG that we have recorded following incubation in pH7-adjusted supernatants from lactobacilli cultures, remains unaffected. In addition, lactobacilli grow well in the presence of BCG metabolites, suggesting that the metabolic pathways of Lactobacillus and M. bovis have no cross-interference. Lactobacilli, as many other lactic acid bacteria, have a relatively simple metabolism that converts sugars to pyruvate via the glycolytic pathway, generating energy (Papagianni, 2012b), whereas mycobacteria shows a very complex, but flexible, central carbon metabolism that generates energy from glycolysis, gluconeogenesis, the pentose phosphate pathway, and the TCA pathway depending on the resources available (Cumming and Steyn, 2015). These metabolic facts, together with the evidence that acidic pH has no in vitro anti-mycobacterial effect, reinforce our hypothesis that lactobacilli exert antimicrobial activity against BCG through synergistic mechanisms that include combination of acidity with different metabolites such as organic acids, hydrogen peroxide, ethanol and bacteriocins.
We finally determined the phagocytic response to BCG in the presence of lactobacilli. Isolates of L. plantarum and L. casei had a significantly opposite effect on BCG phagocytosis. L. plantarum decreased the BCG intake, whereas L. casei increased the phagocytic response, suggesting a completely different interaction between these two species and the phagocytes. Therefore, we believe that L. plantarum and L. casei may be recognized by different phagocytic receptors that, to a greater or lesser extent, could also be involved in the recognition of BCG. Phagocyte internalization is clearly beneficial to the survival of M. tuberculosis complex species, which may enter macrophages via different receptor molecules, including complement receptors involved in the classical and alternative pathways such as C1 and C-type lectin receptors (Pieters, 2008; Marakalala et al., 2018). In this respect, very recent studies have described that lactobacilli are able to express adhesins that bind to C-type lectin receptors (Bene et al., 2017). Interestingly, the genomes of our L. plantarum isolates have revealed the presence of cna, a gene that encodes a collagen adhesin (cna) and that is absent in the L. casei genome. As this collagen adhesive protein has also been reported to block the C1-dependent complement activation (Kang et al., 2013), the antagonism that L. plantarum shows against BCG intake could be caused by competition not only for lectin receptors but also for the classical complement activation pathway.
The reason why L. casei increases BCG phagocytosis is intriguing and could be due to many different factors including the expression of spaD, a fimbrial protein gene that we have identified in its genome. This protein shows a very high similarity with the backbone-pilin subunit spaD, and fimbriae (or pili) of lactobacilli have been reported to facilitate phagocytosis in macrophages via integrin CD11b/CD18 (Vargas Garcia et al., 2015), also known as complement receptor 3 (CR3). On the other hand, CR3 may act as a negative regulator of C-type lectin receptors (Zhang et al., 2018) and seems not to alter the progression of M. tuberculosis infection (Hu et al., 2000). Therefore, fimbriae could promote BCG macrophage uptake through indirect positive collaboration with main bTB phagocytic receptors such as lectins. Phagocytic receptors that could have no significant influence on BCG intake might include sensors for sialic acid recognition as both L. plantarum and casei contain the serin-rich adhesin gene sraP in their corresponding genomes (Kline et al., 2009).
Isolates of L. salivarius seem to have no interaction with phagocytes as they caused no significant effects on the BCG intake, neither on the BCG binding. By contrast, L. plantarum and L. casei clearly stimulated phagocytosis, as explained above, as well as BCG binding. These two lactobacilli species also altered significantly the porcine blood profile. Lactobacilli are known to activate macrophages via Toll-like Receptors (TLR) and our 3 Lactobacillus species contain the genes ltaS1 and ltaS2 involved in the synthesis of lipoteichoic acid (LTA), a major constituent of the cell wall that acts as a TLR2 stimulator (Sengupta et al., 2013). However, L. plantarum and L. casei also harbor genes encoding other components of the cell wall and the cytoplasmic membrane that are absent in L. salivarius, such as the L. casei spaD-like fimbrial protein and the genes tagBFGH associated with the biosynthesis of cell wall teichoic acids (WTA) in L. plantarum. Fimbriae and WTA are capable to activate macrophages via TLR5 and 2, respectively (Ganguli et al., 2015; Hevia et al., 2015; Yu et al., 2015) and, under our experimental conditions, their contribution towards phagocyte stimulation could be much more significant than LTA. TLR activation may collaborate with phagocytic receptors with subsequent implications on the uptake process (Gordon, 2016).
Conclusions
We started from the premise that commensal bacteria such as lactobacilli could play an important, and hitherto underappreciated role in the outcome of bTB in wild boar. In our study we have found that lactobacilli isolated from wild boar influence the viability of M. bovis BCG and modify its phagocytic intake in a species-dependent manner. In particular, isolates of L. plantarum showed antimicrobial and immunomodulatory properties that could antagonize M. bovis survival. The genome of these isolates revealed the presence of two-peptide bacteriocins and a collagen adhesive protein that could act as antimycobacterials and innate immunomodulators; a very important question that remains to be elucidated. Our preliminary results have been generated from reductionist in vitro assays, but bring positive prospects with regards to the potential use of lactobacilli as an additional competitive pressure to control bTB in wild boar. In this respect, in vitro and in vivo work with pathogenic M. bovis will be needed to further explore this possibility. Oral administration of lactobacilli with antimycobacterial activity could reduce the gut burden of M. bovis, thus reducing the risk of transmission of bTB between domestic and wild animals, provided that fecal shedding is one of the main excretion routes of mycobacteria (Santos et al., 2015).
Ethics Statement
All experiments included in this study involve no animals and the sampling procedures were performed under safe and protective legal regulations. Fecal samples were collected from 30 wild boar of which 20 were live animals and the remainder 10 dead after being hunted in legal recreational events on which we did not take any participation at all. The samples from the 20 live animals were taken from young wild boars aged between 4 and 8 months using feeding-traps under safe and protective legal regulations in accordance with the required animal husbandry procedures that meet the guidelines set by the appropriate Regional College of Veterinary Surgeons. The samples from the 10 dead animals were randomly collected among others aging between 1 and 3 years old by registered veterinary surgeons at official certified abattoirs. The porcine blood was collected in heparinized tubes from healthy pigs at the Pirbright Institute (UK), where all animal procedures are covered by a license issued by the UK Home Office under the Animal (Scientific Procedures) Act1986.
Author Contributions
MB and TC planned, performed experiments, and edited the manuscript. FM designed experiments, aided in data analysis, and aided in preparing and editing the manuscript. WG-J and PF-L conducted the sampling and edited the manuscript. JR and DR designed experiments and aided in preparing and editing the manuscript. JG-M designed and performed experiments, aided in data analysis, and prepared and edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to express our gratitude to Dr. Arnoud Van Vliet (School of Veterinary Medicine, University of Surrey) for helping us with the assembly and annotation of the genomes.
Funding. This study was funded by the following Spanish institutions: Fundación Tatiana Pérez de Guzmán el Bueno, Ministerio de Economía, Industria y Competitividad and CDTi (Centro para el Desarrollo Tecnológico Industrial; IDI-20170896).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01663/full#supplementary-material
References
- Acevedo-Whitehouse K., Vicente J., Gortazar C., Hofle U., Fernandez-De-Mera I. G., Amos W. (2005). Genetic resistance to bovine tuberculosis in the Iberian wild boar. Mol. Ecol. 14, 3209–3217. 10.1111/j.1365-294X.2005.02656.x, PMID: [DOI] [PubMed] [Google Scholar]
- Aktas B., De Wolfe T. J., Tandee K., Safdar N., Darien B. J., Steele J. L. (2015). The effect of Lactobacillus casei 32G on the mouse cecum microbiota and innate immune response is dose and time dependent. PLoS One 10:e0145784. 10.1371/journal.pone.0145784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakomi H. L., Skytta E., Saarela M., Mattila-Sandholm T., Latva-Kala K., Helander I. M. (2000). Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66, 2001–2005. 10.1128/aem.66.5.2001-2005.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Sieiro P., Montalban-Lopez M., Mu D., Kuipers O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. 10.1007/s00253-016-7343-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderssen E. L., Diep D. B., Nes I. F., Eijsink V. G., Nissen-Meyer J. (1998). Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 64, 2269–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranaz A., De Juan L., Montero N., Sanchez C., Galka M., Delso C., et al. (2004). Bovine tuberculosis (Mycobacterium bovis) in wildlife in Spain. J. Clin. Microbiol. 42, 2602–2608. 10.1128/JCM.42.6.2602-2608.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayele W. Y., Neill S. D., Zinsstag J., Weiss M. G., Pavlik I. (2004). Bovine tuberculosis: an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 8, 924–937. PMID: [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bene K. P., Kavanaugh D. W., Leclaire C., Gunning A. P., Mackenzie D. A., Wittmann A., et al. (2017). Lactobacillus reuteri surface mucus adhesins upregulate inflammatory responses through interactions with innate C-type lectin receptors. Front. Microbiol. 8:321. 10.3389/fmicb.2017.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsuk S., Mendum T. A., Fagundes M. Q., Michelon M., Cunha C. W., Mcfadden J., et al. (2007). Auxotrophic complementation as a selectable marker for stable expression of foreign antigens in Mycobacterium bovis BCG. Tuberculosis 87, 474–480. 10.1016/j.tube.2007.07.006, PMID: [DOI] [PubMed] [Google Scholar]
- Broadbent J. R., Neeno-Eckwall E. C., Stahl B., Tandee K., Cai H., Morovic W., et al. (2012). Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533. 10.1186/1471-2164-13-533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R., Gordon S. V., Buchrieser C., Pym A. S., Garnier T., Cole S. T. (2000). Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busarcevic M., Dalgalarrondo M. (2012). Purification and genetic characterisation of the novel bacteriocin LS2 produced by the human oral strain Lactobacillus salivarius BGHO1. Int. J. Antimicrob. Agents 40, 127–134. 10.1016/j.ijantimicag.2012.04.011 [DOI] [PubMed] [Google Scholar]
- Cambier C. J., Falkow S., Ramakrishnan L. (2014). Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509. 10.1016/j.cell.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Corner L. A. (2006). The role of wild animal populations in the epidemiology of tuberculosis in domestic animals: how to assess the risk. Vet. Microbiol. 112, 303–312. 10.1016/j.vetmic.2005.11.015, PMID: [DOI] [PubMed] [Google Scholar]
- Cosma C. L., Humbert O., Ramakrishnan L. (2004). Superinfecting mycobacteria home to established tuberculous granulomas. Nat. Immunol. 5, 828–835. 10.1038/ni1091, PMID: [DOI] [PubMed] [Google Scholar]
- Cosma C. L., Sherman D. R., Ramakrishnan L. (2003). The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 57, 641–676. 10.1146/annurev.micro.57.030502.091033 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Hill C., Ross R. P. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- Cumming B. M., Steyn A. J. (2015). Metabolic plasticity of central carbon metabolism protects mycobacteria. Proc. Natl. Acad. Sci. USA 112, 13135–13136. 10.1073/pnas.1518171112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rua-Domenech R. (2006). Human Mycobacterium bovis infection in the United Kingdom: incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis 86, 77–109. 10.1016/j.tube.2005.05.002, PMID: [DOI] [PubMed] [Google Scholar]
- Diep D. B., Havarstein L. S., Nes I. F. (1996). Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178, 4472–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep D. B., Straume D., Kjos M., Torres C., Nes I. F. (2009). An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 30, 1562–1574. 10.1016/j.peptides.2009.05.014, PMID: [DOI] [PubMed] [Google Scholar]
- Ekblad B., Kyriakou P. K., Oppegard C., Nissen-Meyer J., Kaznessis Y. N., Kristiansen P. E. (2016). Structure-function analysis of the two-peptide bacteriocin plantaricin EF. Biochemistry 55, 5106–5116. 10.1021/acs.biochem.6b00588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayol-Messaoudi D., Berger C. N., Coconnier-Polter M. H., Lievin-Le Moal V., Servin A. L. (2005). pH-, Lactic acid-, and non-lactic acid-dependent activities of probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 71, 6008–6013. 10.1128/AEM.71.10.6008-6013.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn S., Van Sinderen D., Thornton G. M., Holo H., Nes I. F., Collins J. K. (2002). Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148, 973–984. 10.1099/00221287-148-4-973 [DOI] [PubMed] [Google Scholar]
- Ganguli K., Collado M. C., Rautava J., Lu L., Satokari R., Von Ossowski I., et al. (2015). Lactobacillus rhamnosus GG and its SpaC pilus adhesin modulate inflammatory responsiveness and TLR-related gene expression in the fetal human gut. Pediatr. Res. 77, 528–535. 10.1038/pr.2015.5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golneshin A., Adetutu E., Ball A. S., May B. K., Van T. T., Smith A. T. (2015). Complete genome sequence of Lactobacillus plantarum strain B21, a bacteriocin-producing strain isolated from Vietnamese fermented sausage Nem Chua. Genome Announc 3. 10.1128/genomeA.00055-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. (2016). Phagocytosis: an immunobiologic process. Immunity 44, 463–475. 10.1016/j.immuni.2016.02.026 [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Sanchez B., G de Los Reyes-Gavilán C., Margolles A. (2013). Antibiotic resistance in probiotic bacteria. Front. Microbiol. 4:202. 10.3389/fmicb.2013.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. 10.1093/bioinformatics/btt086, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han N., Qiang Y., Zhang W. (2016). ANItools web: a web tool for fast genome comparison within multiple bacterial strains. Database 2016. 10.1093/database/baw084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. M. B., Bourin M. J. B., Claesson M. J., O’toole P. W. (2017). Phylogenomics and comparative genomics of Lactobacillus salivarius, a mammalian gut commensal. Microb. Genom. 3:e000115. 10.1099/mgen.0.000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty J. W., Guinane C. M., Ross R. P., Hill C., Cotter P. D. (2016). Bacteriocin production: a relatively unharnessed probiotic trait? F1000Res. 5:2587. 10.12688/f1000research.9615.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbin S., Mathieu F., Brule F., Branlant C., Lefebvre G., Lebrihi A. (1997). Characteristics and genetic determinants of bacteriocin activities produced by Carnobacterium piscicola CP5 isolated from cheese. Curr. Microbiol. 35, 319–326. 10.1007/s002849900262, PMID: [DOI] [PubMed] [Google Scholar]
- Hertzberger R., Arents J., Dekker H. L., Pridmore R. D., Gysler C., Kleerebezem M., et al. (2014). H(2)O(2) production in species of the Lactobacillus acidophilus group: a central role for a novel NADH-dependent flavin reductase. Appl. Environ. Microbiol. 80, 2229–2239. 10.1128/AEM.04272-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia A., Delgado S., Sanchez B., Margolles A. (2015). Molecular players involved in the interaction between beneficial bacteria and the immune system. Front. Microbiol. 6. 10.3389/fmicb.2015.01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwind K., Weinmaier T., Schmid M., Van Hemert S., Hartmann A., Rattei T., et al. (2012). Draft genome sequence of Lactobacillus casei W56. J. Bacteriol. 194:6638. 10.1128/JB.01386-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Mayadas-Norton T., Tanaka K., Chan J., Salgame P. (2000). Mycobacterium tuberculosis infection in complement receptor 3-deficient mice. J. Immunol. 165, 2596–2602. 10.4049/jimmunol.165.5.2596 [DOI] [PubMed] [Google Scholar]
- Jimenez E., Martin R., Maldonado A., Martin V., Gomez De Segura A., Fernandez L., et al. (2010). Complete genome sequence of Lactobacillus salivarius CECT 5713, a probiotic strain isolated from human milk and infant feces. J. Bacteriol. 192, 5266–5267. 10.1128/JB.00703-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Ko Y. P., Liang X., Ross C. L., Liu Q., Murray B. E., et al. (2013). Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 288, 20520–20531. 10.1074/jbc.M113.454462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Boekhorst J., Van Kranenburg R., Molenaar D., Kuipers O. P., Leer R., et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100, 1990–1995. 10.1073/pnas.0337704100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K. A., Falker S., Dahlberg S., Normark S., Henriques-Normark B. (2009). Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592. 10.1016/j.chom.2009.05.011 [DOI] [PubMed] [Google Scholar]
- Klose V., Bayer K., Kern C., Goelß F., Fibi S., Wegl G. (2014). Antibiotic resistances of intestinal lactobacilli isolated from wild boars. Vet. Microbiol. 168, 240–244. 10.1016/j.vetmic.2013.11.014 [DOI] [PubMed] [Google Scholar]
- Leonard M. T., Valladares R. B., Ardissone A., Gonzalez C. F., Lorca G. L., Triplett E. W. (2014). Complete genome sequences of Lactobacillus johnsonii strain N6.2 and Lactobacillus reuteri strain TD1. Genome Announc 2. 10.1128/genomeA.00397-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Pang H., Zhang H., Cai Y. (2014). “Biodiversity of lactic acid bacteria” in Lactic Acid Bacteria (Dordrecht: Springer; ), 103–203. [Google Scholar]
- Mackenzie D. A., Mclay K., Roos S., Walter J., Swarbreck D., Drou N., et al. (2014). Draft genome sequence of a novel Lactobacillus salivarius strain isolated from piglet. Genome Announc. 2. 10.1128/genomeA.01231-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macuamule C., Wiid I., Van Helden P., Tanner M., Witthuhn R. (2016). Effect of milk fermentation by kefir grains and selected single strains of lactic acid bacteria on the survival of Mycobacterium bovis BCG. Int. J. Food Microbiol. 217, 170–176. 10.1016/j.ijfoodmicro.2015.10.024 [DOI] [PubMed] [Google Scholar]
- Maldonado A., Jimenez-Diaz R., Ruiz-Barba J. L. (2004a). Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific Gram-positive bacteria is mediated by an autoinduction mechanism. J. Bacteriol. 186, 1556–1564. 10.1128/jb.186.5.1556-1564.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado A., Ruiz-Barba J. L., Jimenez-Diaz R. (2004b). Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Arch. Microbiol. 181, 8–16. 10.1007/s00203-003-0606-8 [DOI] [PubMed] [Google Scholar]
- Maldonado-Barragan A., Caballero-Guerrero B., Lucena-Padros H., Ruiz-Barba J. L. (2013). Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 33, 40–47. 10.1016/j.fm.2012.08.009 [DOI] [PubMed] [Google Scholar]
- Mann E., Schmitz-Esser S., Zebeli Q., Wagner M., Ritzmann M., Metzler-Zebeli B. U. (2014). Mucosa-associated bacterial microbiome of the gastrointestinal tract of weaned pigs and dynamics linked to dietary calcium-phosphorus. PLoS One 9:e86950. 10.1371/journal.pone.0086950, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala M. J., Martinez F. O., Pluddemann A., Gordon S. (2018). Macrophage heterogeneity in the immunopathogenesis of tuberculosis. Front. Microbiol. 9:1028. 10.3389/fmicb.2018.01028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciset O., Jeronimus-Stratingh M. C., Mollet B., Poolman B. (1997). Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272, 14277–14284. [DOI] [PubMed] [Google Scholar]
- Mariam S. H. (2009). Interaction between lactic acid bacteria and Mycobacterium bovis in Ethiopian fermented milk: insight into the fate of M. bovis. Appl. Environ. Microbiol. 75, 1790–1792. 10.1128/AEM.01943-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens K., Pugin B., De Boeck I., Spacova I., Steelant B., Seys S. F., et al. (2018). Probiotics for the airways: potential to improve epithelial and immune homeostasis. Allergy 73, 1954–1963. 10.1111/all.13495 [DOI] [PubMed] [Google Scholar]
- Martin R., Jimenez E., Olivares M., Marin M. L., Fernandez L., Xaus J., et al. (2006). Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 112, 35–43. 10.1016/j.ijfoodmicro.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Martin R., Miquel S., Ulmer J., Kechaou N., Langella P., Bermudez-Humaran L. G. (2013). Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb. Cell Factories 12:71. 10.1186/1475-2859-12-71, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Hernando M. P., Hofle U., Vicente J., Ruiz-Fons F., Vidal D., Barral M., et al. (2007). Lesions associated with Mycobacterium tuberculosis complex infection in the European wild boar. Tuberculosis 87, 360–367. 10.1016/j.tube.2007.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Matos A. C., Andrade S., Figueira L., Matos M., Pires M. A., Coelho A. C., et al. (2016). Mesenteric lymph node granulomatous lesions in naturally infected wild boar (Sus scrofa) in Portugal–histological, immunohistochemical and molecular aspects. Vet. Immunol. Immunopathol. 173, 21–26. 10.1016/j.vetimm.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Maze A., Boel G., Zuniga M., Bourand A., Loux V., Yebra M. J., et al. (2010). Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J. Bacteriol. 192, 2647–2648. 10.1128/JB.00076-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menin A., Fleith R., Reck C., Marlow M., Fernandes P., Pilati C., et al. (2013). Asymptomatic cattle naturally infected with Mycobacterium bovis present exacerbated tissue pathology and bacterial dissemination. PLoS One 8:e53884. 10.1371/journal.pone.0053884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M., Olson S., Kalina W. V., Ruthel G., Demmin G. L., Warfield K. L., et al. (2005). Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA 102, 2880–2885. 10.1073/pnas.0500098102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo V., Gortazar C., Vicente J., De La Fuente J. (2008). Evidence of the role of European wild boar as a reservoir of Mycobacterium tuberculosis complex. Vet. Microbiol. 127, 1–9. 10.1016/j.vetmic.2007.10.002, PMID: [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Oppegard C., Rogne P., Haugen H. S., Kristiansen P. E. (2010). Structure and mode-of-action of the two-peptide (Class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2, 52–60. 10.1007/s12602-009-9021-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegard C., Kjos M., Veening J. W., Nissen-Meyer J., Kristensen T. (2016). A putative amino acid transporter determines sensitivity to the two-peptide bacteriocin plantaricin JK. Microbiology 5, 700–708. 10.1002/mbo3.363, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea E. F., O’connor P. M., Raftis E. J., O’toole P. W., Stanton C., Cotter P. D., et al. (2011). Production of multiple bacteriocins from a single locus by gastrointestinal strains of Lactobacillus salivarius. J. Bacteriol. 193, 6973–6982. 10.1128/JB.06221-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni M. (2012a). Metabolic engineering of lactic acid bacteria for the production of industrially important compounds. Comput. Struct. Biotechnol. J. 3:e201210003. 10.5936/csbj.201210003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni M. (2012b). Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Factories 11:50. 10.1186/1475-2859-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra A., Garcia A., Inglis N. F., Tato A., Alonso J. M., Hermoso De Mendoza M., et al. (2006). An epidemiological evaluation of Mycobacterium bovis infections in wild game animals of the Spanish Mediterranean ecosystem. Res. Vet. Sci. 80, 140–146. 10.1016/j.rvsc.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Pessione E. (2012). Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infect. Microbiol. 2:86. 10.3389/fcimb.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. (2008). Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe 3, 399–407. 10.1016/j.chom.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Raftis E. J., Forde B. M., Claesson M. J., O’toole P. W. (2014). Unusual genome complexity in Lactobacillus salivarius JCM1046. BMC Genomics 15:771. 10.1186/1471-2164-15-771, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M., Streur T. L., Aldwell F. E., Cook G. M. (2001). Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147, 1017–1024. 10.1099/00221287-147-4-1017, PMID: [DOI] [PubMed] [Google Scholar]
- Ribelles P., Rodriguez I., Suarez J. E. (2012). LysA2, the Lactobacillus casei bacteriophage A2 lysin is an endopeptidase active on a wide spectrum of lactic acid bacteria. Appl. Microbiol. Biotechnol. 94, 101–110. 10.1007/s00253-011-3588-5, PMID: [DOI] [PubMed] [Google Scholar]
- Rocha D. J., Santos C. S., Pacheco L. G. (2015). Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek 108, 685–693. 10.1007/s10482-015-0524-1, PMID: [DOI] [PubMed] [Google Scholar]
- Rocha-Ramirez L. M., Perez-Solano R. A., Castanon-Alonso S. L., Moreno Guerrero S. S., Ramirez Pacheco A., Garcia Garibay M., et al. (2017). Probiotic Lactobacillus strains stimulate the inflammatory response and activate human macrophages. J. Immunol. Res. 2017:4607491. 10.1155/2017/4607491, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosaare M., Vaher M., Kaplinski L., Mols M., Andreson R., Lepamets M., et al. (2017). StrainSeeker: fast identification of bacterial strains from raw sequencing reads using user-provided guide trees. PeerJ 5:e3353. 10.7717/peerj.3353, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable S., Lortal S. (1995). The lysins of bacteriophages infecting lactic acid bacteria. Appl. Microbiol. Biotechnol. 43, 1–6. [DOI] [PubMed] [Google Scholar]
- Saenz Y., Rojo-Bezares B., Navarro L., Diez L., Somalo S., Zarazaga M., et al. (2009). Genetic diversity of the pln locus among oenological Lactobacillus plantarum strains. Int. J. Food Microbiol. 134, 176–183. 10.1016/j.ijfoodmicro.2009.06.004, PMID: [DOI] [PubMed] [Google Scholar]
- Santos N., Almeida V., Gortazar C., Correia-Neves M. (2015). Patterns of Mycobacterium tuberculosis-complex excretion and characterization of super-shedders in naturally-infected wild boar and red deer. Vet. Res. 46:129. 10.1186/s13567-015-0270-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sengupta R., Altermann E., Anderson R. C., Mcnabb W. C., Moughan P. J., Roy N. C. (2013). The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediat. Inflamm. 2013:237921. 10.1155/2013/237921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunov V., Mischenko V., Eruslanov B., Svetoch E., Shakina Y., Stern N., et al. (2007). Antimycobacterial activity of bacteriocins and their complexes with liposomes. J. Antimicrob. Chemother. 59, 919–925. 10.1093/jac/dkm053 [DOI] [PubMed] [Google Scholar]
- Stedman A. (2017). BCG as a vaccine vehicle to deliver porcine immunity to african swine fever virus.
- Stedman A., Maluquer De Motes C., Lesellier S., Dalley D., Chambers M., Gutierrez-Merino J. (2018). Lactic Acid Bacteria isolated from European badgers (Meles meles) reduce the viability and survival of Bacillus Calmette-Guerin (BCG) vaccine and influence the immune response to BCG in a human macrophage model. BMC Microbiol. 18:74. 10.1186/s12866-018-1210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic E., Casey A., Cotter P., Cavanagh D., Fitzgerald G., Mcauliffe O. (2016). Draft genome sequence of Lactobacillus casei DPC6800, an isolate with the potential to diversify flavor in cheese. Genome Announc. 4. 10.1128/genomeA.00063-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H. F., Foo H. L., Abdul Rahim R., Loh T. C., Abdullah M. P., Yoshinobu K. (2015). Molecular characterisation of new organisation of plnEF and plw loci of bacteriocin genes harbour concomitantly in Lactobacillus plantarum I-UL4. Microb. Cell Factories 14:89. 10.1186/s12934-015-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov S. D., Botes M., Guigas C., Schillinger U., Wiid I., Wachsman M. B., et al. (2008). Boza, a natural source of probiotic lactic acid bacteria. J. Appl. Microbiol. 104, 465–477. 10.1111/j.1365-2672.2007.03558.x, PMID: [DOI] [PubMed] [Google Scholar]
- Todorov S. D., Franco B. D., Wiid I. J. (2014). In vitro study of beneficial properties and safety of lactic acid bacteria isolated from Portuguese fermented meat products. Benef. Microbes 5, 351–366. 10.3920/BM2013.0030 [DOI] [PubMed] [Google Scholar]
- van Heel A. J., De Jong A., Montalban-Lopez M., Kok J., Kuipers O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453. 10.1093/nar/gkt391, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tassell M. L., Miller M. J. (2011). Lactobacillus adhesion to mucus. Nutrients 3, 613–636. 10.3390/nu3050613, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas Garcia C. E., Petrova M., Claes I. J., De Boeck I., Verhoeven T. L., Dilissen E., et al. (2015). Piliation of Lactobacillus rhamnosus GG promotes adhesion, phagocytosis, and cytokine modulation in macrophages. Appl. Environ. Microbiol. 81, 2050–2062. 10.1128/AEM.03949-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. J., Somkuti G. A. (1995). Characterization of a bacteriocin produced by Streptococcus thermophilus ST134. Appl. Microbiol. Biotechnol. 43, 330–335. [DOI] [PubMed] [Google Scholar]
- Ward L. J., Timmins M. J. (1999). Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 29, 90–92. 10.1046/j.1365-2672.1999.00586.x, PMID: [DOI] [PubMed] [Google Scholar]
- Xiao L., Estelle J., Kiilerich P., Ramayo-Caldas Y., Xia Z., Feng Q., et al. (2016). A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 1:16161. 10.1038/nmicrobiol.2016.161 [DOI] [PubMed] [Google Scholar]
- Yu X., Jaatinen A., Rintahaka J., Hynonen U., Lyytinen O., Kant R., et al. (2015). Human gut-commensalic Lactobacillus ruminis ATCC 25644 displays sortase-assembled surface piliation: phenotypic characterization of its fimbrial operon through in silico predictive analysis and recombinant expression in Lactococcus lactis. PLoS One 10:e0145718. 10.1371/journal.pone.0145718, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Lee W. B., Kang J. S., Kim L. K., Kim Y. J. (2018). Integrin CD11b negatively regulates Mincle-induced signaling via the Lyn-SIRPalpha-SHP1 complex. Exp. Mol. Med. 50:e439. 10.1038/emm.2017.256, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.