Abstract
Since the update of the 4th edition of the WHO Classification of Central Nervous System (CNS) Tumors published in 2016, particular molecular characteristics are part of the definition of a subset of these neoplasms. This combined ‘histo-molecular’ approach allows for a much more precise diagnosis of especially diffuse gliomas and embryonal CNS tumors. This review provides an update of the most important diagnostic and prognostic markers for state-of-the-art diagnosis of primary CNS tumors. Defining molecular markers for diffuse gliomas are IDH1/IDH2 mutations, 1p/19q codeletion and mutations in histone H3 genes. Medulloblastomas, the most frequent embryonal CNS tumors, are divided into four molecularly defined groups according to the WHO 2016 Classification: wingless/integrated (WNT) signaling pathway activated, sonic hedgehog (SHH) signaling pathway activated and tumor protein p53 gene (TP53)-mutant, SHH-activated and TP53-wildtype, and non-WNT/non-SHH-activated. Molecular characteristics are also important for the diagnosis of several other CNS tumors, such as RELA fusion-positive subtype of ependymoma, atypical teratoid rhabdoid tumor (AT/RT), embryonal tumor with multilayered rosettes, and solitary fibrous tumor/hemangiopericytoma. Immunohistochemistry is a helpful alternative for further molecular characterization of several of these tumors. Additionally, genome-wide methylation profiling is a very promising new tool in CNS tumor diagnostics. Much progress has thus been made by translating the most relevant molecular knowledge into a more precise clinical diagnosis of CNS tumors. Hopefully, this will enable more specific and more effective therapeutic approaches for the patients suffering from these tumors.
Keywords: CNS tumor, molecular pathology, glioma, medulloblastoma, embryonal tumor, integrated diagnosis
Key Message
State-of-the-art pathological diagnosis of primary CNS tumors is increasingly based on a combined ‘histo-molecular’ approach. This review provides an update on the diagnostic and prognostic value of key molecular markers for these tumors. Translation into more specific and effective therapeutic approaches has started and will hopefully lead to improved patient survival in the near future.
Introduction
Up until the 4th edition of World Health Organization (WHO) Classification of Central Nervous System (CNS) Tumors that was published in 2007 [1], definitions of CNS tumor entities were mainly based on histological characteristics and resemblance with a supposed cell type of origin. This approach was increasingly supplied by panels of immunohistochemical markers giving information on differentiation and proliferation. Although many microscopy-based diagnoses were and still are rather robust, review panels have revealed considerable diagnostic inter-observer variation with a danger of detrimental consequences for patients [2, 3]. This situation prompted the identification and implementation of more robust diagnostic markers.
The tremendous increase in knowledge of the molecular characteristics of CNS tumors during the last decade has allowed for a paradigm shift. In the update of the 4th edition of the WHO classification CNS tumors published in 2016 [4], molecular aberrations are part of the definition of particular brain tumor entities for the first time. Especially, the classification of the most frequent primary neoplasms of the CNS parenchyma itself, the diffuse gliomas, has undergone major restructuring based on the status of a few key molecular aberrations. Similarly, major changes have been introduced in the classification of medulloblastomas and some other embryonal tumors. This situation brings new challenges for the work-up of these tumors. Meanwhile, technology continues to develop along with reduced costs of molecular diagnostic platforms. This, combined with the possibility to make a ‘molecular diagnosis’ based on immunohistochemical analysis, brings a state-of-the-art, integrated morphological and molecular diagnosis of CNS tumors within reach of an increasing number of centers.
In this review, the most significant developments with respect to molecular diagnosis of primary tumors of the CNS are highlighted, with a strong focus on markers conveying diagnostic and/or prognostic information. An overview of these markers is given in Tables 1–3. Some of these diagnostic and/or prognostic markers may provide leads for specific therapeutic management, an aspect that is briefly covered in this review as well. For more detailed information on purely predictive markers for the efficacy of particular therapeutic approaches such as targeted treatment, the reader is referred to other reviews [5–8]. For a recent overview of the molecular diagnostic tools that may be used, see our recent review on this topic [9].
Table 1.
Genetic aberrations presented in alphabetical order for gliomas
| Genetic aberration | Diagnostic (D), prognostic (P) and therapeutic/predictive (T) value |
|---|---|
| ATRX mutation | D Frequently present in IDH-mutant astrocytic tumors |
| (Alpha-thalassemia/mental retardation syndrome X) | |
| BRAF V600E mutation | D Present in 65%–75% of pleomorphic xanthoastrocytomas, 25%–60% of gangliogliomas, and ∼50% of epithelioid glioblastomas |
| (B-raf) | D Also found in dysembryoplastic neuroepithelial tumors, SEGAs, pilocytic astrocytomas |
| T Possible therapeutic target | |
| CDKN2A/B homozygous deletion | D Frequent feature in pleomorphic xanthoastrocytomas |
| (Cyclin-dependent kinase inhibitor 2A/B) | D Occurs in IDH-wildtype astrocytic tumors with piloid features |
| P Associated with aggressive course in IDH-mutant diffuse astrocytic tumors | |
| CIC mutation | D Present in majority of (but not specific for) oligodendroglial tumors |
| (Homolog of capicua drosophila) | |
| EGFR amplification/EGFRvIII | D High copy number amplification common in IDH-wildtype glioblastomas (∼40%) |
| (Epidermal growth factor receptor) | D EGFRvIII present in about half of EGFR-amplified glioblastomas |
| T Possible therapeutic target | |
| FUBP1 mutation | D Present in a subset of oligodendrogliomas |
| (Far upstream element binding protein) | |
| H3 G34 mutation | D Occurs most often in high-grade, IDH-wildtype tumors in the cerebral hemisphere in young patients with glial or embryonal histology |
| [H3 Histone Family Member 3A (H3F3A)] | |
| H3 K27M mutation | D Required for the diagnosis ‘diffuse midline glioma (DMG), H3 K27M-mutant’ |
| [H3 Histone Family Member 3A (H3F3A) or Histone Cluster 1 H3 Family Member B/C (HIST1H3B/C)] | D Occasionally also found in other tumors such as posterior fossa ependymomas, gangliogliomas, pilocytic astrocytomas. |
| P Signifies poor prognosis in DMG, H3 K27M-mutant (mean survival of +/− 9 months for both pediatric and adult patients); prognostic meaning in other tumors less clear | |
| T Potentially predictive of effect of EZH2 inhibitors | |
| IDH1/IDH2 mutation | D Frequent in WHO grade II and III astrocytomas (>80%), oligodendrogliomas and ‘secondary’ glioblastomas |
| (Isocitrate dehydrogenase1/2) | P IDH-mutant status of astrocytic tumor signifies better prognosis compared with that of IDH-wildtype astrocytic tumor with the histologically same WHO grade |
| T IDH1 R132H mutation may represent a promising target for mutation specific vaccination | |
| KIAA1549-BRAF gene fusion | D Present in ∼70% of pilocytic astrocytomas |
| (KIAA1549, uncharacterized; abbreviation for BRAF listed above) | D Also found in diffuse DLGNT, pilomyxoid astrocytoma and ganglioglioma |
| D Rare in other gliomas | |
| MGMT promoter hypermethylation (O-6-methylguanine–DNA methyltransferase) | P Reported as independent favorable prognostic factor in glioblastomas (irrespective of treatment) |
| T Predictive for response to temozolomide | |
| RELA fusion to C11orf95 | D Defining feature for the diagnosis ‘ependymoma, RELA fusion-positive’ |
| (V-rel avian reticuloendotheliosis viral oncogene homolog A) | T C11orf95-RELA fusion protein potential therapeutic target |
| (C11orf95, uncharacterized) | |
| TERT promoter mutation | D Present in almost all IDH-mutant, 1p/19q-codeleted oligodendrogliomas |
| (Telomerase reverse transcriptase) | D Frequent in IDH-wildtype GBM |
| D/PTERT promoter mutation in histologically lower-grade, IDH-wildtype astrocytoma indicates aggressive behavior (‘molecular glioblastoma’) | |
| TP53 mutation | D Frequent in IDH-mutant astrocytic tumors (>80%), but also quite frequent in IDH-wildtype diffuse gliomas; very infrequent in oligodendrogliomas |
| (Tumor protein p53) | |
| YAP1 fusion | D Present in some supratentorial ependymomas, primarily in children |
| (Yes-associated protein 1) | P Generally favorable prognosis |
| T Potential therapeutic target | |
| 1p/19q codeletion | D Required for diagnosis of ‘canonical’ oligodendroglioma (as it is the complete codeletion of these arms that counts, ideally the molecular test allows for discriminating complete from partial loss of 1p and 19q) |
| [Short arm of chromosome 1(1p)] | |
| [Long arm of chromosome 19 (19q)] |
Table 2.
Genetic aberrations presented in alphabetical order for embryonal CNS tumors
| Genetic aberration | Diagnostic (D) and prognostic (P) value |
|---|---|
| APC mutation (may be germline) | D May occur in WNT-activated medulloblastomas |
| (Adenomatous polyposis coli) | |
| BCOR exon 15 internal tandem duplication | D Described in subgroup of CNS embryonal tumors: ‘BCOR-altered neuroepithelial tumor (BCOR-NET)’; N.B. Non-embryonal pediatric CNS tumors, esp. pediatric high-grade gliomas may show other BCOR (or BCORL1) alterations such as fusion, truncating mutation |
| (BCL6, corepressor/BCL6, corepressor like 1) | |
| BRCA2 mutation (may be germline) | D May occur in SHH-activated medulloblastoma and non-WNT/non-SHH medulloblastoma. |
| (Breast cancer 2 gene) | |
| Chromosome 6 monosomy | D Present in ∼85% of WNT-activated medulloblastomas |
| CIC-NUTM1 gene fusion or CIC frameshift deletion | D Characteristic of subgroup of CNS embryonal tumors described as Ewing’s sarcoma family tumor with CIC alteration (EFT-CIC) |
| (For CIC mutation, see Table 1, Genetic aberrations in gliomas) | |
| (NUT midline carcinoma family member 1) | |
| CTNNB1 mutation | D Present in 90% of WNT-activated medulloblastomas |
| (Catenin beta-1) | P Children with WNT-activated medulloblastomas generally have a good prognosis |
| C19MC (19q13.42) alteration (amplification or fusion with TTYH1) | D High level amplicon is detected in majority of embryonal tumors with multilayered rosettes/ETMRs (specific and sensitive diagnostic marker for these tumors). |
| (Tweety family member 1) | |
| DICER1 mutation (may be germline) | D Predisposing event to the development of a pituitary blastoma. |
| (Dicer 1, ribonuclease III) | |
| FOXR2 fusion with different gene fusion partners | D Defining feature of subgroup of CNS embryonal tumors: ‘CNS neuroblastoma with FOXR2 activation’ |
| (Forkhead box R2) | |
| MN1 with different gene fusion partners [Meningioma (disrupted in balanced translocation)1] | D Defining feature of subgroup of CNS embryonal tumors described as ‘high-grade neuroepithelial tumor with MN1 alteration’ (HGNET-MN1) |
| P Better prognosis than other CNS embryonal tumors | |
| PALB2 (may be germline) | D May occur in SHH-activated medulloblastoma and non-WNT/non-SHH medulloblastoma. |
| (Partner and localizer of BRCA2) | |
| PTCH1 (may be germline) | D May occur in SSH-activated medulloblastoma |
| (Patched 1) | |
| SMARCB1/SMARCA4 loss (may be germline) | D Required for diagnosis of atypical teratoid/rhabdoid tumor (AT/RT) |
| (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily B, member 1) | |
| (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4) | |
| SUFU mutation (may be germline) | D May occur in SSH-activated medulloblastoma |
| (Suppressor of fused homolog) | |
| GAB1 (GRB2 associated binding protein 1) | D Surrogate marker for activated hedgehog signaling seen in SSH-activated medulloblastoma |
| TP53 mutation (may be germline) | D Discriminates medulloblastoma, SSH-activated & TP53- mutant vs. SHH-activated & TP53-wildtype |
| (Tumor protein p53) | |
| P Presence of a TP53 mutation in SSH-activated medulloblastoma indicates poor prognosis |
Table 3.
Genetic aberrations presented in alphabetical order for ‘other’ (i.e. non-glial, non-embryonal) CNS tumors
| Genetic aberration | Diagnostic (D), and prognostic (P) value |
|---|---|
| AKT1 mutation | D Associated with meningothelial and transitional variants of meningioma |
| (AKT serine/threonine kinase 1) | |
| BRAF V600E mutation | D Present in > 90% of papillary craniopharyngiomas |
| (B-raf) | |
| CDKN2 inactivation | D Combined CDKN2 and NF1 inactivation is frequent in malignant peripheral nerve sheath tumors (MPNSTs) |
| (Cyclin-dependent kinase inhibitor 2A) | |
| CTNNB1 mutation | D Present in >90% of adamantinomatous craniopharyngiomas |
| (Catenin beta 1) | |
| DICER1 mutation (may be germline) | D Frequent in intracranial sarcomas with rhabdomyosarcoma-like features in children |
| (Dicer 1, ribonuclease III) | |
| GNAQ/GNA11 hotspot mutation | D Frequent in primary melanocytic tumors of the CNS in adults (and uveal melanomas, but very infrequent in skin melanomas; therefor very helpful in differential diagnosis with metastatic cutaneous melanoma) |
| (Guanine nucleotide-binding protein) | |
| KLF4 mutation | D Characteristic of secretory meningiomas |
| (Kuppel like factor 4) | |
| NAB2-STAT6 gene fusion | D Typically found in CNS solitary fibrous tumors/hemangiopericytomas (CNS SFTs/HPCs); STAT6 staining of tumor cell nuclei is a very reliable immunohistochemical surrogate marker for presence of NAB2-STAT6 fusion |
| (NGFI-A Binding Protein 2) | |
| (Signal Transducer and Activator of Transcription 6) | |
| NF1 inactivation | D Combined NF1 and CDKN2 inactivation is frequent in malignant peripheral nerve sheath tumors (MPNSTs) |
| (Neurofibromin 1) | |
| NRAS mutation | D Occurs in primary melanocytic tumors of the CNS, especially in children |
| (Neuroblastoma RAS viral oncogene homolog) | |
| SMARCE1 mutation (may be germline) | D Associated with clear cell meningiomas |
| (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily E, member 1) | |
| TERT promoter mutation | P Signifies more aggressive clinical behavior in meningiomas |
| (Telomerase reverse transcriptase) | |
| TRAF7 mutation | D Characteristic of secretory meningiomas |
| (TNF receptor associated factor 7) |
Gliomas
Gliomas comprise a very heterogeneous group of primary CNS tumors, originally classified according to their microscopic similarity with or presumed origin of non-neoplastic glial (precursor) cells (e.g. astrocytes—astrocytoma; oligodendroglial cells—oligodendroglioma; ‘glioblast’—glioblastoma). Gliomas are traditionally divided into two major categories: ‘diffuse’ gliomas and ‘non-diffuse’ gliomas. Diffuse gliomas are characterized by tumor cell migration over large distances into the CNS parenchyma, thereby precluding curative surgical resection. Diffuse gliomas have for decades been diagnosed as diffuse astrocytomas and oligodendrogliomas, or as tumors with a mixed astrocytic and oligodendroglial phenotype (oligoastrocytomas). In addition, a malignancy grade was assigned based on the presence or the absence of marked mitotic activity, necrosis and/or florid microvascular proliferation. In contrast to diffuse gliomas, non-diffuse gliomas are generally much more circumscribed. Examples from this category are pilocytic astrocytoma and different variants of ependymoma. Now, molecular information helps to categorize glial tumors into different diffuse and non-diffuse glioma entities as explained below.
Discovery of 1p/19q codeletion as a marker for oligodendroglial tumors
In 1994, it was reported that many oligodendroglial tumors show loss of heterozygosity (LOH) for the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q) [10]. Soon after, it became clear that 1p/19q codeletion is associated with sensitivity to procarbazine–lomustine–vincristine (PCV) chemotherapy and improved outcome [11]. Since then, testing for the presence/absence of this codeletion has increasingly been used for recognition of this subset of diffuse gliomas.
Discovery of isocitrate dehydrogenase mutations
The discovery of point mutations in the isocitrate dehydrogenase 1 and 2 (IDH1/IDH2) genes by large scale next-generation sequencing (NGS) in glioblastomas [12], and soon after also in lower grade diffuse gliomas [13–16], has been a major driver of classifying diffuse gliomas on a molecular basis. IDH1/IDH2 mutations were found at low frequency in glioblastomas but at much higher frequencies in WHO grade II and III diffuse astrocytomas, oligodendrogliomas, and oligoastrocytomas. The glioblastomas with IDH1/IDH2 mutations were later on considered to be ‘secondary’ glioblastomas originating from such lower grade diffuse gliomas, and the IDH-wildtype glioblastomas as ‘de novo’ or ‘primary’ glioblastomas [12, 14, 17]. Patients with an IDH-mutant glioblastoma generally showed substantially longer overall survival than those with IDH-wildtype glioblastoma [12]. This prognostic impact of IDH mutation was later confirmed for WHO grade II and III diffuse gliomas [13–16]. In fact, the impact of IDH mutation on survival was so pronounced that the overall survival for patients with IDH-wildtype anaplastic astrocytoma (WHO grade III) was found to be worse than for patients with IDH-mutant glioblastoma (WHO grade IV) [18]. IDH mutations are considered to be the initiating event in the oncogenesis of IDH-mutant gliomas [19]. The mutant IDH protein is a tumor-specific neoantigen/immunogenic epitope and may represent a promising therapeutic target, especially the IDH1 R132H mutation, which accounts for ∼90% of the IDH mutations in gliomas [16, 20, 21]. Mutation-specific antibodies allow for a very reliable detection of IDH1 R132H protein [17, 18].
Impact of 1p/19q codeletion and IDH mutations on WHO classification
Based on the above described findings, the following three major categories of diffuse gliomas have been defined in the WHO 2016 Classification of CNS tumors:
diffuse astrocytic tumors (astrocytoma/anaplastic astrocytoma/glioblastoma), IDH-wildtype;
diffuse astrocytic tumors (astrocytoma/anaplastic astrocytoma/glioblastoma), IDH-mutant;
oligodendroglial tumors (oligodendroglioma/anaplastic oligodendroglioma), IDH-mutant and 1p/19q-codeleted.
The armamentarium required to adequately diagnose diffuse gliomas has thus become more complex. Recognizing that molecular testing cannot always be carried out due to lack of resources or suboptimal quality/quantity of the tissue samples, a ‘not otherwise specified’ (NOS) category has been introduced in the WHO 2016 Classification for cases in which relevant molecular information is not available because molecular testing could not (successfully) be carried out [22].
Other molecular markers in diffuse gliomas - TERT promoter, ATRX and TP53 mutations
Almost all IDH-mutant, 1p/19q-codeleted oligodendroglial tumors have activating mutations in the telomerase reverse transcriptase gene (TERT) promoter region [23–25], making this genetic aberration a valuable diagnostic marker in the right context. However, these mutations are also frequent in IDH-wildtype glioblastomas [24]. In fact, in a histologically lower-grade, diffuse, IDH-wildtype astrocytoma the presence of a TERT promoter mutation and/or of epidermal growth factor receptor (EGFR) gene amplification and/or of combined gain of whole chromosome 7 plus loss of whole chromosome 10 signifies behavior of the tumor as of glioblastoma (WHO grade IV) [23, 24]. Unlike IDH-mutant and 1p/19q-codeleted oligodendrogliomas, IDH-mutant astrocytic tumors frequently carry an alpha-thalassemia/mental retardation syndrome X-linked gene (ATRX) and a tumor protein p53 gene (TP53) mutation [26–28]. Loss of nuclear ATRX immunohistochemical staining (IHC) is a strong predictor of presence of ATRX mutation [29], while strong and extensive nuclear staining of tumor cell nuclei for tumor protein p53 (p53) signifies presence of a TP53 mutation.
Oligoastrocytomas
For decades, unequivocal histopathological delineation of oligoastrocytoma from astrocytoma or oligodendroglioma remained very difficult [2, 3]. Accumulation of molecular knowledge has now revealed that, at the molecular level, ‘real oligoastrocytomas’ are very rare. The WHO 2016 Classification still encompasses a diagnosis of (anaplastic) oligoastrocytoma, NOS. In the very rare cases in which both an IDH-mutant (and ATRX/TP53-mutant) astrocytic and an IDH-mutant, 1p/19q-codeleted component can be demonstrated, one may want to add that this denotes a molecularly-proven ‘dual genotype’ oligoastrocytoma [30–32].
Diffuse midline glioma, H3 K27M-mutant
Another new entity in the WHO 2016 Classification is ‘diffuse midline glioma, H3 K27M-mutant’. This entity must harbor a K27M mutation in either the H3 Histone Family Member 3A (H3F3A) or Histone Cluster 1 H3 Family Member B/C (HIST1H3B/C) gene, have a glial phenotype, be located in the midline, and show a diffuse growth pattern. Both the morphological and molecular parts of the definition are important, since H3 K27M mutations are not exclusive to midline gliomas. Recent studies have identified H3 K27M mutations in, e.g. a subset of posterior fossa ependymomas [33] and rarely in gangliogliomas [34] and (anaplastic) pilocytic astrocytomas [35, 36]. H3 K27M mutation in these tumors seems to implicate more aggressive behavior.
Diffuse midline gliomas occur primarily in children, but may occur in adults as well [37]. Most of the tumors previously diagnosed as diffuse intrinsic pontine glioma are H3 K27M-mutant and thus belong to the ‘diffuse midline glioma, H3 K27M-mutant’ entity. This tumor carries a very poor prognosis, with a 2-year survival rate below 10% [38, 39] and a mean survival of ∼9 months [40, 41]. Presence of H3 K27M mutation can now also reliably be demonstrated using immunohistochemistry [37, 38].
RELA fusion-positive ependymoma
Until the WHO 2016 Classification, ependymal tumors were classified based on morphology, but the correlation between malignancy grade as assessed by histopathological examination and clinical behavior remained unclear [42, 43]. Based on DNA methylation profiling analysis, nine distinct molecular subgroups of ependymal tumors were reported (three in each of the following compartments: supratentorial, posterior fossa, and spinal canal) [44]. In the supratentorial compartment, ‘ependymoma, v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA) fusion-positive’ was considered to be so distinct that it was designated a separate entity in the WHO 2016 Classification. These tumors are characterized by oncogenic fusions between RELA, the principal effector of canonical nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, and C11orf95 (an uncharacterized gene). RELA fusion-positive ependymomas represent the majority of pediatric supratentorial ependymal tumors but can also occur in adults. L1 cell adhesion molecule (L1CAM) and cyclin D1 expression, as detected by immunohistochemistry, are useful but non-specific surrogate markers for RELA fusion-positive ependymomas [45].
Other molecular markers in gliomas
Detection of the B-raf proto-oncogene (BRAF) V600E mutation can be of value in the diagnosis of CNS tumors, its contribution depending on the exact differential diagnostic context. BRAF V600E mutation occurs in about half of all epithelioid glioblastomas [46, 47], pleomorphic xanthoastrocytomas and gangliogliomas, and in a smaller subset of subependymal giant cell astrocytomas (SEGAs), pilocytic astrocytomas and dysembryoplastic neuroepithelial tumors. Demonstration of BRAF V600E mutation in a tumor may provide a useful therapeutic target [48, 49].
The oncogenic KIAA1549 and B-raf proto-oncogene (KIAA1549-BRAF) fusion is present in ∼70% of pilocytic astrocytomas and has high differential diagnostic value as it is only found in the rare diffuse leptomeningeal glioneuronal tumor (DLGNT) and very rarely in other gliomas [50].
Most oligodendroglial tumors harbor drosophila homolog of capicua gene (CIC) mutations, and a smaller subset of far upstream element binding protein gene (FUBP1) mutations [28, 51, 52]. These mutations may be of differential diagnostic value but their prognostic meaning is so far unclear.
Glioblastomas were the first tumors for which an epigenetic biomarker came into clinical use. The DNA repair enzyme O-6-methylguanine-DNA methyltransferase (MGMT) removes the alkyl groups and thereby repairs the mutagenic DNA lesions, whereby DNA damage and apoptosis are prevented. Accordingly, promoter methylation of the MGMT gene has been found to be a useful predictive marker for the responsiveness to temozolomide [53]. Most clinical information on the impact of hypermethylation of the MGMT promoter focusses on glioblastomas. Its implication for other (diffuse) gliomas is much less clear, also because in previous studies of patients with oligodendrogliomas PCV rather than temozolomide was used as chemotherapy, and treatment of histologically lower grade astrocytomas often did not include chemotherapy.
Emerging glioma entities
It is expected that more subgroups of gliomas will emerge as distinct entities in the near future. High-grade IDH-wildtype gliomas with an H3 G34 mutation (or, dependent on the nomenclature used, H3 G35 mutation) occur most often in the cerebral hemispheres in adolescents and young adult patients and may histologically show glioblastoma as well as embryonal tumor histology. While the microscopic phenotype is not associated with a clear difference in prognosis, presence of MGMT promoter methylation and lack of oncogene amplification has been reported to be associated with longer survival [54].
Another group of gliomas that may deserve its own ‘entity’ in CNS tumor classification are high-grade, IDH-wildtype astrocytic tumors with piloid features that relatively frequently occur in the posterior fossa of adult patients. Molecularly, these often show cyclin-dependent kinase inhibitor 2A/B gene (CDKN2A/B) deletion, mitogen-activated protein kinase pathway gene alteration and, somewhat less frequently, ATRX mutation [55].
In the overarching category of ependymomas, some new entities are emerging as well. Yes-associated protein 1 gene (YAP1) fusion-positive supratentorial ependymomas occur primarily in children and generally have a favorable prognosis [44]. Regarding ependymal tumors in the posterior fossa, based on methylation profiling analysis, group-A and group-B ependymomas can be identified, with, respectively, relatively poor and good prognosis. Recently, loss of global H3 K27 trimethylation (H3 K27me3), which can be detected by immunohistochemistry, has been reported to be very powerful tool for discriminating group A posterior fossa ependymomas from the group B tumors, the latter showing retained nuclear H3 K27me3 expression [56].
CNS embryonal tumors
In the WHO 2016 Classification, the term ‘primitive neuroectodermal tumor’ (PNET) has been replaced by ‘CNS embryonal tumor’, partly to avoid further confusion with non-CNS PNETs, and partly because the term PNET was increasingly used as a poorly defined waste basket. CNS embryonal tumors predominantly occur in children and are histologically characterized by very high cellularity with densely packed and poorly differentiated small cells that generally show a limited amount of cytoplasm, variable nuclear pleomorphism and marked mitotic activity. This category encompasses medulloblastomas, embryonal tumors with multilayered rosettes (ETMRs), atypical teratoid/rhabdoid tumors (AT/RTs) and a heterogeneous group of other embryonal CNS tumors.
Medulloblastomas
The vast majority of embryonal tumors in the posterior fossa are medulloblastomas. The histologic medulloblastoma subtypes (classic, desmoplastic/nodular, extensive nodularity, large cell/anaplastic) described in the WHO 2016 Classification have not substantially changed compared with the WHO 2007 Classification [4, 57]. More recent studies, however, revealed particular molecular medulloblastoma subgroups and have showed that molecular and histological data provide complementary diagnostic information [4, 58, 59]. The WHO 2016 Classification proposes an integrated ‘histo-molecular’ diagnosis of medulloblastomas and lists four molecular groups:
WNT-activated
These tumors encompass ∼10% of all medulloblastomas, and generally show the classic, but occasionally the large cell/anaplastic, phenotype. Over 90% of wingless/integrated (WNT)-activated medulloblastomas carry a beta-catenin gene (CTNNB1) mutation. Less frequently, mutations in other components of the WNT-signaling pathway, such as the axis inhibition protein 1 gene (AXIN1) and adenomatous polyposis coli gene (APC), are found. The defect in the WNT-signaling pathway results in nuclear accumulation of beta-catenin as can be demonstrated by immunohistochemistry. About 85% of the tumors in this group show monosomy for chromosome 6. Children with WNT-activated medulloblastomas generally have a good prognosis, but in adults the prognosis may be less favorable [60, 61].
Sonic hedgehog-activated and TP53-wildtype
About 30% of all medulloblastomas belong to the Sonic hedgehog (SHH)-activated group and the vast majority of these are TP53-wildtype. The nodular/desmoplastic and the much less frequently occurring extensive nodularity histologic subtype are almost exclusively found in this molecular group. These tumors occur predominantly in infants and adulthood and are considered low risk. Especially in younger patients, patched 1 gene (PTCH1) or suppressor of fused [(SUFU), negative regulator of hedgehog signaling] gene germline mutations may be found.
SHH-activated and TP53-mutant
A small percentage of SHH-activated medulloblastomas is TP53-mutant. These tumors occur predominantly in childhood, often show the large cell/anaplastic phenotype and have a poor prognosis. Up to half of the patients in this group have a TP53 germline mutation [62].
Non-WNT/non-SHH
This last category encompasses the ‘group 3 and group 4’ molecular categories as recognized in multiple studies. Group 3 and group 4 medulloblastomas, representing, respectively, ∼20% and 40% of all medulloblastomas, occur especially in infancy/childhood. Because in many centers, demonstration of these subcategories is still difficult due to a lack of easily accessible diagnostic tools, groups 3 and 4 are still included under the umbrella of the non-WNT/non-SHH molecular category in the WHO 2016 Classification. Histologically, these medulloblastomas are almost always of the classic or large cell/anaplastic phenotype.
Assessment of the molecular subtype of medulloblastoma can often be achieved by performing immunohistochemistry for (surrogate) markers like beta-catenin, GRB2 associated binding protein 1 (GAB1), YAP1, p53, homeobox protein OTX2 (OTX2) and/or low-affinity nerve growth factor receptor (p75NGFR) [63]. Across different histologic and molecular medulloblastoma groups, v-Myc avian myelocytomatosis viral oncogene homolog (MYC) and/or v-Myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog (MYCN) are often amplified and may provide prognostic information, but the exact prognostic impact appears to be subgroup dependent [64, 65]. In ∼5% of the children diagnosed with medulloblastoma, a germline mutation accounts for the development of the tumor. This is most frequently seen in the SHH-activated subgroup (TP53, SUFU, PTCH1), but can also be found in the WNT-activated subgroup (APC) and rarely based on partner and localizer of BRCA2 (PALB2) or BRCA2, DNA repair associated gene (BRCA2) germline mutation (in SHH-activated and non-WNT/non-SHH subgroups) [66]. In order to improve outcome and reduce side-effects, a molecularly driven, risk-adapted treatment approach is crucial and may necessitate further subgrouping of medulloblastomas [65, 67–69].
Atypical teratoid/rhabdoid tumors
AT/RT was already introduced as an entity in previous WHO classifications, but in the WHO 2016 Classification demonstration of an underlying defect in SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin, subfamily B, member 1 gene (SMARCB1) or, rarely, SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily A, member 4 gene (SMARCA4) is now required for the diagnosis of canonical AT/RT. The products of these genes are essential components of the SWI/SNF chromatin remodeling complex. Defect function of SMARCB1 or SMARCA4 results in lack of nuclear staining for the intact integrase interactor 1 (INI1) or Brahma-related gene 1 (BRG1) protein, respectively [70]. In the WHO 2016 Classification, tumors with AT/RT phenotype but with INI1 and BRG1 nuclear staining are now designated CNS embryonal tumor with rhabdoid features. Further molecular subgrouping of AT/RTs may become clinically relevant, e.g. because of differences in therapeutic targets in these subgroups [71, 72].
Embryonal tumors with multilayered rosettes
‘ETMR, C19MC-altered’ has been introduced in the WHO 2016 Classification as a separate entity. In the past, these tumors were generally diagnosed as ependymoblastoma, medulloepithelioma, or embryonal tumor with abundant neuropil and true rosettes [73]. The C19MC alteration generally consists of a focal high-level amplicon of chromosome 19q13.42, covering a large microRNA cluster that can be detected by fluorescence in situ hybridization (FISH) or high-resolution cytogenetic techniques [74–76]. Strong and diffuse lin-28 homolog A (LIN28A) cytoplasmic immunostaining of tumor cells is a highly sensitive surrogate marker for ‘ETMR, C19MC-altered’, but medulloepitheliomas lacking the C19MC alteration and some other CNS tumors (e.g. gliomas, AT/RTs and germ cell tumors) can be LIN28A positive as well [77, 78]. ETMRs in which the C19MC status is not tested or demonstrated are designated in the WHO 2016 Classification as ‘ETMR, NOS’, or, in case of the medulloepithelioma phenotype, as medulloepithelioma.
Embryonal tumors of the pineal and pituitary region
Compared with other embryonal CNS tumors, pineoblastomas are reported to have fewer cytogenetic alterations. RB transcriptional corepressor 1 (RB1) mutations (+/− germline defect) and Dicer 1, ribonuclease III (DICER1) mutations are linked to pineoblastoma [79, 80]. Pituitary blastoma is an extremely rare embryonal tumor of the pituitary gland, with DICER1 mutation as a key predisposing event [81]. Recently, it was reported that intracranial sarcomas with rhabdomyosarcoma-like features in children often carry a DICER1 mutation as well (in some patients, a germline mutation without evidence of a cancer-related syndrome at the time of diagnosis) [82]. Pineal anlage tumors are very rare pineal tumors with an embryonal component combined with heterologous differentiation (e.g. skeletal muscle, chondroid differentiation) and often contain melanin. So far, no distinctive diagnostic molecular features of this tumor have been identified.
Other embryonal CNS tumors
Apart from the abovementioned embryonal tumors, the WHO 2016 Classification lists CNS neuroblastoma, CNS ganglioneuroblastoma and CNS embryonal tumor NOS. Meanwhile, detailed molecular (including methylation) analysis of tumors previously diagnosed as CNS PNET has revealed that some of these tumors could be reclassified as glioblastoma, ependymoma or Ewing sarcoma, and four new subgroups with recurrent gene fusions [83, 84]:
CNS neuroblastoma with forkhead box R2 (FOXR2) activation (NB-FOXR2), typically showing FOXR2 fusions.
High-grade neuroepithelial tumor with meningioma 1 gene (MN1) alteration (HGNET-MN1), often carrying an MN1 fusion that can be identified by FISH using an MN1 break apart probe. These tumors may have an astroblastoma-phenotype and are reported to be associated with a somewhat less grim prognosis compared with other embryonal CNS tumors.
Ewing sarcoma family tumor with CIC alteration (EFT-CIC), typically characterized by structural variants involving CIC that can be detected by break-apart FISH (in case of CIC-NUTM1 fusion) or RNA sequencing (in case of CIC frameshift deletion) and positive NUT Midline Carcinoma Family Member 1 (NUTM1) nuclear immunohistochemistry as a surrogate marker.
BCL6 corepressor (BCOR)-altered neuroepithelial tumor (BCOR-NET), characterized by typically an internal tandem repeat in the BCOR gene.
Further study is necessary to assess the exact clinical significance of such a refined classification of ‘other embryonal CNS tumors’. Also, since e.g. EFT-CIC and BCOR-NET are not limited to the CNS, these tumors may in fact represent malignant mesenchymal tumors/sarcomas [64].
‘Other’ (non-glial, non-embryonal) primary CNS tumors
The group of ‘other’ primary CNS tumors encompasses a very heterogeneous collection of neoplasms, including meningiomas (malignant) peripheral nerve sheath tumors, primary melanocytic tumors of the CNS, and craniopharyngiomas. Hematologic tumors and neoplasms of the soft tissues and bone occur elsewhere in the body as well and are beyond the scope of this review. Further information on these tumors the can be found in the respective WHO classifications [85, 86]. Pituitary adenomas, by far the most frequent pituitary tumors, are dealt with in the WHO Classification of endocrine neoplasms and are also not further discussed. Immunohistochemical transcription termination factor 1 (TTF1) nuclear staining is very helpful for the diagnosis of primary neurohypophyseal tumors including granular cell tumor, pituicytoma and spindle cell oncocytoma. Meanwhile, molecular diagnostics so far does not yet play an important role in the clinical diagnosis of most pituitary neoplasms [87]. Also, the diagnosis of the heterogeneous group of primary CNS germ cell tumors generally does not yet require molecular diagnostics.
Regarding ‘meningiomas’, there is now increasing evidence that presence of a TERT promoter mutation signifies more aggressive clinical behavior [88, 89]. Furthermore, some mutations are clearly associated with particular histological phenotypes (secretory meningioma-combined kruppel like factor 4 gene (KLF4) and TNF receptor associated factor 7 (TRAF7) mutations [90, 91]; clear cell meningioma—SMARCE1 (germline) mutation [92]; meningothelial and transitional meningioma—AKT serine/threonine kinase 1 gene (AKT1) mutations [93, 94]). Recently, DNA methylation profiling was reported to allow for better prediction of tumor recurrence/prognosis compared with WHO grading [95]. This potentially influences the clinical follow-up plan and whether patients should be offered radiotherapy. However, according to the WHO 2016 Classification, molecular analysis is not yet required for the diagnosis of meningiomas. Re-evaluation of previous clinical trials combined with information obtained by future clinical studies is necessary to address more precisely how DNA methylation profiling and other molecular alterations can help to improve the therapeutic management of these patients [96].
High-grade ‘malignant peripheral nerve sheath tumors’ (MPNSTs) frequently show combined inactivation of neurofibromin gene 1 (NF1), CDKN2A, and of the polycomb repressive complex 2 (PRC2) complex, irrespective if it concerns sporadic, radiation-induced or NF1-associated tumors. Loss of H3 K27me3 nuclear staining is now used as an important aid in the diagnosis of MPNSTs [97–99]. For cases with a challenging differential diagnosis between a benign and malignant nerve sheath tumor, this marker helps to increase the number of patients being treated based on the correct diagnosis [100]. Other immunohistochemical markers that may be helpful in this realm are neurofibromin (the product of NF1) [97, 101], EGFR, CDKN2A (p16), SRY-Box 10 (SOX10) [102] and in some cases INI-1 [103, 104].
As ‘CNS solitary fibrous tumors/hemangiopericytomas’ (SFTs/HPCs), like SFTs elsewhere in the body, typically show gene fusion between NGFI-A Binding Protein 2 gene (NAB2) and signal transducer and activator of transcription 6 gene (STAT6) (NAB2-STAT6), they are now considered as tumors that may show differences in histology but belong to the same entity. In order to ‘smoothen’ the transition towards a new classification, these tumors are still listed in the WHO 2016 Classification as SFT/HPC, rather than just SFT as is done in the WHO classification of soft tissue tumors. NAB2-STAT6 fusion results in aberrant accumulation of STAT6 protein in tumor cell nuclei, which can reliably be demonstrated by simple STAT6 immunohistochemistry.
In adult patients, activating GNAQ or GNA11 hotspot mutations are frequent in ‘primary melanocytic tumors of the CNS’ (melanocytomas, melanomas). Thereby, these CNS tumors closely resemble uveal melanomas at the molecular level. Demonstration of guanine nucleotide-binding protein gene (GNAQ or GNA11) mutations in a melanocytic CNS tumor very strongly favors a primary CNS tumor over metastasis of cutaneous melanoma. Especially in children, primary melanocytic CNS tumors relatively frequently harbor a neuroblastoma RAS viral oncogene homolog (NRAS) mutation, especially so in the context of neurocutaneous melanosis [105–108].
Most ‘craniopharyngiomas’ are of the adamantinomatous subtype, and more than 90% of these tumors carry a CTNNB1 mutation, resulting in aberrant nuclear beta-catenin expression that can be demonstrated by immunohistochemistry. In contrast, the vast majority of craniopharyngiomas of the much less frequent papillary subtype carry the BRAF V600E mutation, which can be demonstrated by IHC for the mutant protein as well and may be used as a therapeutic target [109–112].
Discussion
Conclusions and future perspectives
Current neuro-oncological practice is increasingly dependent on molecular diagnostics of tumor tissue. To provide the best patient care possible, it is important to carefully select the assays used as well as to monitor their validity and accuracy. Simpler techniques such as Sanger sequencing, FISH and LOH analysis can provide very valuable molecular information but have their shortcomings. For example, for the detection of 1p/19q codeletion in diffuse gliomas preferably a platform is used that allows for discriminating partial 1p and/or 19q losses from the clinically relevant, complete 1p/19q codeletion [9].
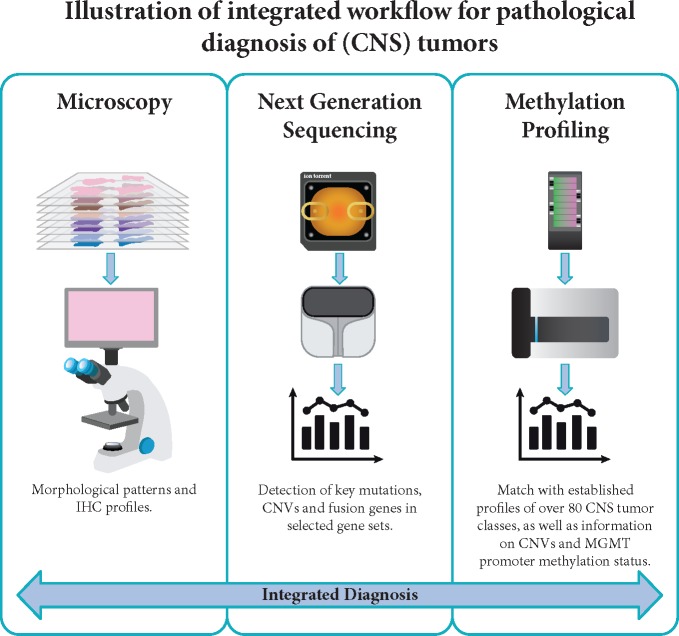
The rapidly growing number of mutations to detect and the increased possibilities for targeted therapies has propelled the development of NGS panels, where multiple mutations can be detected in a single analysis. Some of these panels allow for simultaneous detection of fusions and chromosomal copy number aberration, as well [9]. More recently, genome-wide methylation profiling has been reported as a very valuable tool for CNS tumor diagnostics [113, 114]. Indeed, in an increasing number of laboratories, advanced setups have been established with integrated diagnostic workflows covering microscopy-based methods, NGS and genome-wide methylation profiling (Figure 1). In addition, recent advances in neuro-imaging with techniques that assess, e.g. IDH status and 1p/19q-codeletion, are emerging and are playing an increasingly important role in diagnosis of CNS tumors [115, 116].
Figure 1.
The integrated diagnostic workflow used in CNS tumor diagnostics depicted here is based on novel molecular platforms for next-generation sequencing (NGS) and genome-wide DNA methylation profiling besides conventional microscopy. Microscopy for standard histological evaluation includes panels of immunohistochemical staining (IHC) and in some laboratories also FISH analyses. NGS panels with selected genes allow for the detection of mutations, copy number variations (CNVs) and gene fusions. Genome-wide DNA methylation profiling is a novel approach with high potential as a support tool for a more refined and robust classification of CNS tumors.
Acknowledging that the WHO Classification is meant to be used world-wide, it is important to keep a balance between incorporation of the latest molecular findings into a classification and the fact that in many places around the world testing for such aberrations is not possible. Indeed, the ‘NOS’ categories in the updated WHO Classification allow a WHO diagnosis based on histopathological analysis alone for tumors that ideally are further characterized at the molecular level.
Meanwhile, further elucidation of the molecular underpinnings of CNS tumors is occurring at a rapid pace and can be expected to allow for an even more precise and objective diagnosis of a substantial subset of these tumors in the near future. In 2016, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxomy (cIMPACT-NOW consortium) consisting of expert-neuropathologists and a clinical advisory panel, was established with the goal of facilitating implementation of such novel, relevant molecular information into the clinical diagnosis of CNS tumors and into future classifications of these neoplasms [117]. This consortium has already published recommendations on how to use the term NOS versus ‘not elsewhere classified’ in the context of CNS tumor diagnostics according to the WHO 2016 Classification [22], a clarification of the diagnosis of H3 K27M-mutant gliomas and diffuse low grade and anaplastic, IDH-mutant astrocytomas [118].
Acknowledging that, with the introduction of molecularly defined subgroups of diffuse gliomas the traditionally used microscopic criteria for grading of these neoplasms might not suffice anymore [119, 120], the recently published cIMPACT-NOW update 3 explains that EGFR amplification, TERT promoter mutation, and/or combined gain of complete chromosome 7 and loss of complete chromosome 10 can be used to make a diagnosis of ‘molecular glioblastoma’. These tumors are designated as WHO grade IV based on the molecular parameters [121]. This supports treatment of IDH-wildtype anaplastic astrocytoma and potentially also IDH-wildtype diffuse astrocytoma having these molecular alterations as glioblastomas, although these tumors histologically appear as WHO grade III and II tumor, respectively. Also, new insights are emerging with regard to how to improve grading within the category of IDH-mutant diffuse astrocytic tumors, with homozygous CDKN2A/B loss as a molecular marker strongly associated with aggressive clinical behavior in this category [122]. Very recently, a cIMPACT-NOW update 4 has been published dealing with the indolent clinical behavior and rare anaplastic progression of diffuse IDH-wt/H3-wt gliomas with either a BRAF V600E mutation, an FGFR alteration, or an MYB or MYBL1 rearrangement. These diffuse gliomas mainly present in children but sometimes in adults. Identification of these molecular alterations warrant different approaches to the post-operative management of a WHO grade II diffuse glioma and, for some patients, even targeted therapies [123]. Detection of homozygous deletion at the CDKN2A/B locus is a molecular marker that should direct the neuropathologist away from a diagnosis of ‘pediatric-type’ diffuse glioma [123].
Of note, current neuro-oncological treatment guidelines are still generally based on studies and experiences dating from the time before the availability of detailed molecular information. It is essential that treatment guidelines and neuro-oncology practices are soon re-evaluated in light of this more precise diagnostic information. Importantly, novel trials like the N2M2 (NOA20) phase I/II trial offering molecularly matched targeted therapies to patients with IDH-wildtype non-MGMT promoter hypermethylated glioblastomas take the molecular status of the tumor into account and investigate the value of novel targeted drugs and radiotherapy in this context [124].
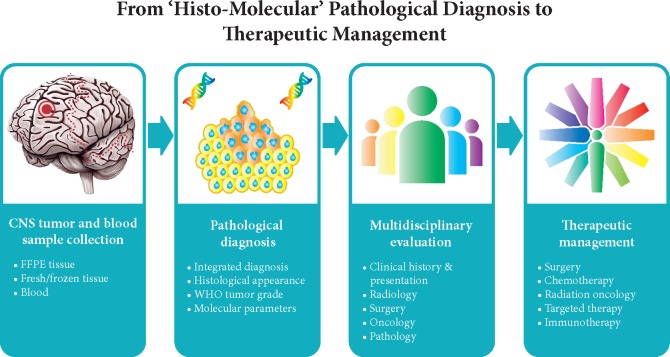
In conclusion, enormous progress has been made by the elucidation of the molecular underpinnings of CNS tumors and by translating this information into a more precise clinical diagnosis. For conveying the essence of the molecular findings to clinicians using a layered reporting format of the conclusion has been proposed [59] as given in Table 4. More recently, the International Collaboration on Cancer Reporting (ICCR) has established guidelines about how to structure a pathology report that encompasses both histopathological and molecular information using the layered diagnostic approach (http://www.iccr-cancer.org/datasets/published-datasets/central-nervous-system). Evaluation of the molecular information in multidisciplinary teams will further facilitate optimal use of molecular diagnostics of CNS tumors in clinical practice (Figure 2). Hopefully, in this way, an integrated ‘histo-molecular’ diagnosis of CNS tumors will boost more specific and effective therapeutic approaches for patients that suffer from these tumors.
Table 4.
Structure of four-layered conclusion in the pathology report on CNS tumors with three examples
| Four layers | Contents of the four layers | Example 1 | Example 2 | Example 3 |
|---|---|---|---|---|
| 1. Integrated diagnosis | Diagnosis based on integration of all tissue-based (especially histological and molecular) information |
|
Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma (WHO grade IV) | Ependymoma, RELA fusion-positive |
| 2. Histological diagnosis | Classification of tumor based on (immuno)histochemical evaluation | Diffuse astrocytoma | Anaplastic astrocytoma | Ependymoma |
| 3. WHO grade | ‘Standard’ histological WHO tumor grade | WHO grade II | WHO grade III | WHO grade II |
| 4. Molecular information | Most important data from molecular analyses (e.g. sequencing, FISH, methylation profiling) |
|
|
C11orf95-RELA fusion |
Now that the definition of some CNS tumors is based on a combination of histological and molecular features, a layered reporting format of the conclusion in the pathology report helps to convey not only the message of the ‘integrated diagnosis’, but also provides in a nutshell the most relevant information on the ‘building blocks’ used to reach this diagnosis. Of note, the WHO grade in layer 3 is based on standard histological evaluation. In some situations this grade may be overruled by information obtained by molecular analysis (WHO grade IV instead of WHO grade III in the integrated diagnosis in example 2), in other cases, the WHO grade may be left out in the integrated diagnosis as assigning an unequivocal WHO grade is (still) difficult (example 3).
Figure 2.
The pathological diagnosis of CNS tumors is a multi-step process starting with tumor tissue and in some cases also blood samples being analyzed with multiple tests to provide an integrated diagnosis. Evaluation and discussion of the pathological diagnosis by a multidisciplinary board of specialists from radiology, surgery, oncology, and (neuro)pathology is crucial for translating the findings into optimal therapeutic management for individual patients.
Funding
None declared.
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Louis DN, Ohgaki H, Wiestler OD. et al. WHO Classification of Tumours of the Central Nervous System, 4th edition Lyon: International Agency for Research on Cancer (IARC; ) 2007. [Google Scholar]
- 2. Kros JM, Gorlia T, Kouwenhoven MC. et al. Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol 2007; 66(6): 545–551. [DOI] [PubMed] [Google Scholar]
- 3. van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathol 2010; 120(3): 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Louis DN, Ohgaki H, Wiestler OD. et al. World Health Organization Histological Classification of Tumours of the Central Nervous System. Lyon: International Agency for Research on Cancer (IARC; ) 2016. [Google Scholar]
- 5. Berghoff AS, Stefanits H, Woehrer A. et al. Clinical neuropathology practice guide 3-2013: levels of evidence and clinical utility of prognostic and predictive candidate brain tumor biomarkers. Clin Neuropathol 2013; 32(05): 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berghoff AS, Bartsch R, Wohrer A. et al. Predictive molecular markers in metastases to the central nervous system: recent advances and future avenues. Acta Neuropathol 2014; 128(6): 879–891. [DOI] [PubMed] [Google Scholar]
- 7. Simonelli M, Persico P, Perrino M. et al. Checkpoint inhibitors as treatment for malignant gliomas: “A long way to the top”. Cancer Treat Rev 2018; 69: 121–131. [DOI] [PubMed] [Google Scholar]
- 8. Passiglia F, Caglevic C, Giovannetti E. et al. Primary and metastatic brain cancer genomics and emerging biomarkers for immunomodulatory cancer treatment. Semin Cancer Biol 2018; 52: 259–268. [DOI] [PubMed] [Google Scholar]
- 9. Priesterbach-Ackley LP, Wesseling P, Snijders TJ. et al. Molecular Tools for the Pathologic Diagnosis of CNS Tumors 2018. Neuro-oncology Practice. [DOI] [PMC free article] [PubMed]
- 10. Reifenberger J, Reifenberger G, Liu L. et al. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol 1994; 145: 1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 11. Cairncross JG, Ueki K, Zlatescu MC. et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998; 90(19): 1473–1479. [DOI] [PubMed] [Google Scholar]
- 12. Parsons DW, Jones S, Zhang X. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321(5897): 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balss J, Meyer J, Mueller W. et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 2008; 116(6): 597–602. [DOI] [PubMed] [Google Scholar]
- 14. Ichimura K, Pearson DM, Kocialkowski S. et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 2009; 11(4): 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009; 174(4): 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan H, Parsons DW, Jin G. et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360(8): 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohgaki H, Burger P, Kleihues P. Definition of primary and secondary glioblastoma–response. Clin Cancer Res 2014; 20(7): 2013.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartmann C, Hentschel B, Wick W. et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 2010; 120(6): 707–718. [DOI] [PubMed] [Google Scholar]
- 19. Barthel FP, Wesseling P, Verhaak R. Reconstructing the molecular life history of gliomas. Acta Neuropathol 2018; 135(5): 649–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schumacher T, Bunse L, Pusch S. et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 2014; 512(7514): 324–327. [DOI] [PubMed] [Google Scholar]
- 21. Waitkus MS, Diplas BH, Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell 2018; 34(2): 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Louis DN, Wesseling P, Paulus W. et al. cIMPACT-NOW update 1: not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol 2018; 135(3): 481–484. [DOI] [PubMed] [Google Scholar]
- 23. Arita H, Narita Y, Fukushima S. et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol 2013; 126(2): 267–276. [DOI] [PubMed] [Google Scholar]
- 24. Killela PJ, Reitman ZJ, Jiao Y. et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA 2013; 110(15): 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koelsche C, Sahm F, Capper D. et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol 2013; 126(6): 907–915. [DOI] [PubMed] [Google Scholar]
- 26. Sahm F, Reuss D, Koelsche C. et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 2014; 128(4): 551–559. [DOI] [PubMed] [Google Scholar]
- 27. Eckel-Passow JE, Lachance DH, Molinaro AM. et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 2015; 372(26): 2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki H, Aoki K, Chiba K. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 2015; 47(5): 458–468. [DOI] [PubMed] [Google Scholar]
- 29. Reuss DE, Sahm F, Schrimpf D. et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol 2015; 129(1): 133–146. [DOI] [PubMed] [Google Scholar]
- 30. Huse JT, Diamond EL, Wang L, Rosenblum MK. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true “oligoastrocytoma”? Acta Neuropathol 2015; 129(1): 151–153. [DOI] [PubMed] [Google Scholar]
- 31. Wilcox P, Li CC, Lee M. et al. Oligoastrocytomas: throwing the baby out with the bathwater? Acta Neuropathol 2015; 129(1): 147–149. [DOI] [PubMed] [Google Scholar]
- 32. Barresi V, Lionti S, Valori L. et al. Dual-genotype diffuse low-grade glioma: is it really time to abandon oligoastrocytoma as a distinct entity? J Neuropathol Exp Neurol 2017; 76(5): 342–346. [DOI] [PubMed] [Google Scholar]
- 33. Gessi M, Capper D, Sahm F. et al. Evidence of H3 K27M mutations in posterior fossa ependymomas. Acta Neuropathol 2016; 132(4): 635–637. [DOI] [PubMed] [Google Scholar]
- 34. Joyon N, Tauziede-Espariat A, Alentorn A. et al. K27M mutation in H3F3A in ganglioglioma grade I with spontaneous malignant transformation extends the histopathological spectrum of the histone H3 oncogenic pathway. Neuropathol Appl Neurobiol 2017; 43(3): 271–276. [DOI] [PubMed] [Google Scholar]
- 35. Hochart A, Escande F, Rocourt N. et al. Long survival in a child with a mutated K27M-H3.3 pilocytic astrocytoma. Ann Clin Transl Neurol 2015; 2(4): 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez FJ, Brosnan-Cashman JA, Allen SJ. et al. Alternative lengthening of telomeres, ATRX loss and h3-k27m mutations in histologically defined pilocytic astrocytoma with anaplasia. Brain Pathol 2019; 29: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Solomon DA, Wood MD, Tihan T. et al. Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol 2016; 26(5): 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buczkowicz P, Bartels U, Bouffet E. et al. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol 2014; 128(4): 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khuong-Quang DA, Buczkowicz P, Rakopoulos P. et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 2012; 124(3): 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim HW, Park T, Quiring S, Barrett D. The anti-human trafficking collaboration model and serving victims: providers' perspectives on the impact and experience. J Evid Inf Soc Work 2018; 15: 185–202. [DOI] [PubMed] [Google Scholar]
- 41. Karremann M, Gielen GH, Hoffmann M. et al. Diffuse high-grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro Oncol 2018; 20(1): 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venkatramani R, Dhall G, Patel M. et al. Supratentorial ependymoma in children: to observe or to treat following gross total resection? Pediatr Blood Cancer 2012; 58(3): 380–383. [DOI] [PubMed] [Google Scholar]
- 43. Ellison DW, Kocak M, Figarella-Branger D. et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed 2011; 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pajtler KW, Witt H, Sill M. et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 2015; 27(5): 728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parker M, Mohankumar KM, Punchihewa C. et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 2014; 506(7489): 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Broniscer A, Tatevossian RG, Sabin ND. et al. Clinical, radiological, histological and molecular characteristics of paediatric epithelioid glioblastoma. Neuropathol Appl Neurobiol 2014; 40(3): 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kleinschmidt-DeMasters BK, Aisner DL, Birks DK, Foreman NK. Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol 2013; 37(5): 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Myung JK, Cho H, Park CK. et al. Analysis of the BRAF(V600E) mutation in central nervous system tumors. Transl Oncol 2012; 5: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kieran MW. Targeting BRAF in pediatric brain tumors. Am Soc Clin Oncol Educ Book 2014; 34: e436–e440. [DOI] [PubMed] [Google Scholar]
- 50. Rodriguez FJ, Schniederjan MJ, Nicolaides T. et al. High rate of concurrent BRAF-KIAA1549 gene fusion and 1p deletion in disseminated oligodendroglioma-like leptomeningeal neoplasms (DOLN). Acta Neuropathol 2015; 129(4): 609–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sahm F, Koelsche C, Meyer J. et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol 2012; 123(6): 853–860. [DOI] [PubMed] [Google Scholar]
- 52. Yi KS, Sohn CH, Yun TJ. et al. MR imaging findings of extraventricular neurocytoma: a series of ten patients confirmed by immunohistochemistry of IDH1 gene mutation. Acta Neurochir 2012; 154(11): 1973–1979; discussion 1980. [DOI] [PubMed] [Google Scholar]
- 53. Esteller M, Garcia-Foncillas J, Andion E. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000; 343(19): 1350–1354. [DOI] [PubMed] [Google Scholar]
- 54. Korshunov A, Capper D, Reuss D. et al. Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol 2016; 131(1): 137–146. [DOI] [PubMed] [Google Scholar]
- 55. Reinhardt A, Stichel D, Schrimpf D. et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol 2018; 136(2): 273–291. [DOI] [PubMed] [Google Scholar]
- 56. Panwalkar P, Clark J, Ramaswamy V. et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol 2017; 134(5): 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Louis DN, Ohgaki H, Wiestler OD. et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114(2): 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Taylor MD, Northcott PA, Korshunov A. et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012; 123(4): 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Louis DN, Perry A, Burger P. et al. International Society Of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014; 24(5): 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gajjar A, Pfister SM, Taylor MD, Gilbertson RJ. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin Cancer Res 2014; 20(22): 5630–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao F, Ohgaki H, Xu L. et al. Molecular subgroups of adult medulloblastoma: a long-term single-institution study. Neuro Oncol 2016; 18(7): 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhukova N, Ramaswamy V, Remke M. et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol 2013; 31(23): 2927–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pfister S, Remke M, Benner A. et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 2009; 27(10): 1627–1636. [DOI] [PubMed] [Google Scholar]
- 64. Pickles JC, Hawkins C, Pietsch T, Jacques TS. CNS embryonal tumours: WHO 2016 and beyond. Neuropathol Appl Neurobiol 2018; 44: 151–162. [DOI] [PubMed] [Google Scholar]
- 65. Schwalbe EC, Lindsey JC, Nakjang S. et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 2017; 18(7): 958–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Waszak SM, Northcott PA, Buchhalter I. et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol 2018; 19(6): 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cavalli FMG, Remke M, Rampasek L. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 2017; 31(6): 737–754. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Northcott PA, Buchhalter I, Morrissy AS. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017; 547(7663): 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robinson GW, Rudneva VA, Buchhalter I. et al. Risk-adapted therapy for young children with medulloblastoma (SJYC07): therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol 2018; 19(6): 768–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miller S, Ward JH, Rogers HA. et al. Loss of INI1 protein expression defines a subgroup of aggressive central nervous system primitive neuroectodermal tumors. Brain Pathol 2013; 23(1): 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Torchia J, Golbourn B, Feng S. et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell 2016; 30(6): 891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johann PD, Erkek S, Zapatka M. et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 2016; 29(3): 379–393. [DOI] [PubMed] [Google Scholar]
- 73. Korshunov A, Sturm D, Ryzhova M. et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol 2014; 128(2): 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li M, Lee KF, Lu Y. et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 2009; 16(6): 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pfister S, Remke M, Castoldi M. et al. Novel genomic amplification targeting the microRNA cluster at 19q13.42 in a pediatric embryonal tumor with abundant neuropil and true rosettes. Acta Neuropathol 2009; 117(4): 457–464. [DOI] [PubMed] [Google Scholar]
- 76. Kleinman CL, Gerges N, Papillon-Cavanagh S. et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet 2014; 46(1): 39–44. [DOI] [PubMed] [Google Scholar]
- 77. Korshunov A, Ryzhova M, Jones DT. et al. LIN28A immunoreactivity is a potent diagnostic marker of embryonal tumor with multilayered rosettes (ETMR). Acta Neuropathol 2012; 124(6): 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Spence T, Sin-Chan P, Picard D. et al. CNS-PNETs with C19MC amplification and/or LIN28 expression comprise a distinct histogenetic diagnostic and therapeutic entity. Acta Neuropathol 2014; 128(2): 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Miller S, Rogers HA, Lyon P. et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol 2011; 13(8): 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. de Kock L, Sabbaghian N, Druker H. et al. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol 2014; 128(4): 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. de Kock L, Sabbaghian N, Plourde F. et al. Pituitary blastoma: a pathognomonic feature of germ-line DICER1 mutations. Acta Neuropathol 2014; 128(1): 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Koelsche C, Mynarek M, Schrimpf D. et al. Primary intracranial spindle cell sarcoma with rhabdomyosarcoma-like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 2018; 136(2): 327–337. [DOI] [PubMed] [Google Scholar]
- 83. Sturm D, Orr BA, Toprak UH. et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 2016; 164(5): 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gessi M, Gielen GH, Hammes J. et al. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J Neurooncol 2013; 112(1): 67–72. [DOI] [PubMed] [Google Scholar]
- 85. Fletcher CDM, Bridge JA, Hogendoorn PCW. et al. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: International Agency for Research on Cancer (IARC; ) 2013. [Google Scholar]
- 86. Swerdlow SHC, Campo E, Harris NL. et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (IARC; ) 2017. [Google Scholar]
- 87. Lopes M. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathol 2017; 134(4): 521–535. [DOI] [PubMed] [Google Scholar]
- 88. Goutagny S, Nault JC, Mallet M. et al. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol 2014; 24(2): 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sahm F, Schrimpf D, Olar A. et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst 2015; 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Clark VE, Erson-Omay EZ, Serin A. et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 2013; 339(6123): 1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Reuss DE, Piro RM, Jones DT. et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol 2013; 125(3): 351–358. [DOI] [PubMed] [Google Scholar]
- 92. Smith MJ, O'Sullivan J, Bhaskar SS. et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet 2013; 45(3): 295–298. [DOI] [PubMed] [Google Scholar]
- 93. Brastianos PK, Horowitz PM, Santagata S. et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 2013; 45(3): 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sahm F, Bissel J, Koelsche C. et al. AKT1E17K mutations cluster with meningothelial and transitional meningiomas and can be detected by SFRP1 immunohistochemistry. Acta Neuropathol 2013; 126(5): 757–762. [DOI] [PubMed] [Google Scholar]
- 95. Sahm F, Schrimpf D, Stichel D. et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 2017; 18(5): 682–694. [DOI] [PubMed] [Google Scholar]
- 96. Suppiah S, Nassiri F, Bi WL. et al. Molecular and translational advances in meningiomas. Neuro Oncol 2019; 21(Suppl 1): i4–i17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rohrich M, Koelsche C, Schrimpf D. et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol 2016; 131: 877–887. [DOI] [PubMed] [Google Scholar]
- 98. Prieto-Granada CN, Wiesner T, Messina JL. et al. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol 2016; 40(4): 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schaefer IM, Fletcher CD, Hornick JL. Loss of H3K27 trimethylation distinguishes malignant peripheral nerve sheath tumors from histologic mimics. Mod Pathol 2016; 29(1): 4–13. [DOI] [PubMed] [Google Scholar]
- 100. Sahm F, Reuss DE, Giannini C. WHO 2016 classification: changes and advancements in the diagnosis of miscellaneous primary CNS tumours. Neuropathol Appl Neurobiol 2018; 44(2): 163–171. [DOI] [PubMed] [Google Scholar]
- 101. Reuss DE, Habel A, Hagenlocher C. et al. Neurofibromin specific antibody differentiates malignant peripheral nerve sheath tumors (MPNST) from other spindle cell neoplasms. Acta Neuropathol 2014; 127(4): 565–572. [DOI] [PubMed] [Google Scholar]
- 102. Pekmezci M, Reuss DE, Hirbe AC. et al. Morphologic and immunohistochemical features of malignant peripheral nerve sheath tumors and cellular schwannomas. Mod Pathol 2015; 28(2): 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jo VY, Fletcher CD. Epithelioid malignant peripheral nerve sheath tumor: clinicopathologic analysis of 63 cases. Am J Surg Pathol 2015; 39(5): 673–682. [DOI] [PubMed] [Google Scholar]
- 104. Jo VY, Fletcher C. SMARCB1/INI1 loss in epithelioid schwannoma: a clinicopathologic and immunohistochemical study of 65 cases. Am J Surg Pathol 2017; 41(8): 1013–1022. [DOI] [PubMed] [Google Scholar]
- 105. Kusters-Vandevelde HV, Klaasen A, Kusters B. et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system. Acta Neuropathol 2010; 119: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Koelsche C, Hovestadt V, Jones DT. et al. Melanotic tumors of the nervous system are characterized by distinct mutational, chromosomal and epigenomic profiles. Brain Pathol 2015; 25(2): 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kinsler VA, Thomas AC, Ishida M. et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol 2013; 133(9): 2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kusters-Vandevelde HV, Kusters B, van Engen-van Grunsven AC. et al. Primary melanocytic tumors of the central nervous system: a review with focus on molecular aspects. Brain Pathol 2015; 25: 209–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim JH, Paulus W, Heim S. BRAF V600E mutation is a useful marker for differentiating Rathke's cleft cyst with squamous metaplasia from papillary craniopharyngioma. J Neurooncol 2015; 123(1): 189–191. [DOI] [PubMed] [Google Scholar]
- 110. Larkin SJ, Preda V, Karavitaki N. et al. BRAF V600E mutations are characteristic for papillary craniopharyngioma and may coexist with CTNNB1-mutated adamantinomatous craniopharyngioma. Acta Neuropathol 2014; 127(6): 927–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Schweizer L, Capper D, Holsken A. et al. BRAF V600E analysis for the differentiation of papillary craniopharyngiomas and Rathke's cleft cysts. Neuropathol Appl Neurobiol 2015; 41(6): 733–742. [DOI] [PubMed] [Google Scholar]
- 112. Brastianos PK, Taylor-Weiner A, Manley PE. et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet 2014; 46(2): 161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Capper D, Jones DTW, Sill M. et al. DNA methylation-based classification of central nervous system tumours. Nature 2018; 555(7697): 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Capper D, Stichel D, Sahm F. et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol 2018; 136(2): 181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhou H, Chang K, Bai HX. et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol 2019; 142(2): 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang B, Chang K, Ramkissoon S. et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. NEUONC 2017; 19(1): 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Louis DN, Aldape K, Brat DJ. et al. Announcing cIMPACT-NOW: the Consortium to inform molecular and practical approaches to CNS tumor taxonomy. Acta Neuropathol 2017; 133(1): 1–3. [DOI] [PubMed] [Google Scholar]
- 118. Louis DN, Giannini C, Capper D. et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol 2018; 135(4): 639–642. [DOI] [PubMed] [Google Scholar]
- 119. Olar A, Wani KM, Alfaro-Munoz KD. et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol 2015; 129(4): 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Reuss DE, Mamatjan Y, Schrimpf D. et al. IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol 2015; 129(6): 867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Brat DJ, Aldape K, Colman H. et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 2018; 136(5): 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Shirahata M, Ono T, Stichel D. et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol 2018; 136(1): 153–166. [DOI] [PubMed] [Google Scholar]
- 123. Ellison DW, Hawkins C, Jones DTW. et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF(V600E) mutation. Acta Neuropathol 2019; 137(4): 683–687. [DOI] [PubMed] [Google Scholar]
- 124. Wick W, Dettmer S, Berberich A. et al. N2M2 (NOA-20) phase I/II trial of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed non-MGMT hypermethylated glioblastoma. Neuro Oncol 2019; 21(1): 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]