Abstract
It is increasingly common in oncology practice to perform tumour sequencing using large cancer panels. For pathogenic sequence variants in cancer susceptibility genes identified on tumour-only sequencing, it is often unclear whether they are of somatic or constitutional (germline) origin. There is wide-spread disparity regarding both the extent to which systematic ‘germline-focussed analysis’ is carried out upon tumour sequencing data and for which variants follow-up analysis of a germline sample is carried out. Here we present analyses of paired sequencing data from 17 152 cancer samples, in which 1494 pathogenic sequence variants were identified across 65 cancer susceptibility genes. From these analyses, the European Society of Medical Oncology Precision Medicine Working Group Germline Subgroup has generated (i) recommendations regarding germline-focussed analyses of tumour-only sequencing data, (ii) indications for germline follow-up testing and (iii) guidance on patient information-giving and consent.
Keywords: sequencing, panel, susceptibility, predisposition, gene, germline
Key Message
A sizeable proportion of the pathogenic variants in cancer susceptibility genes that are observed on tumour-only analysis are of germline origin, but this fraction is highly variable between genes. Pragmatic germline-focussed tumour analysis, in which variants of low variant allele frequency are excluded and analysis is restricted to certain genes, will ensure that the majority of highly actionable germline variants are identified but that the number of variants for which germline follow-up is required remains modest.
Introduction
Tumour testing, until recently, typically comprised genotyping of specific hotspot mutations in oncogenes. These analyses seldom gave insights into patient germline status [1, 2]. Over the last 5 years, evolution of next-generation sequencing (NGS) technology, growth in molecular oncology, emergence of molecularly stratified basket/umbrella trials and vigorous marketing from commercial providers, have fuelled expansion of ‘large’ tumour sequencing panels targeting the full coding region of hundreds of genes [3–5]. Whilst paired tumour-germline analysis may be carried out in major academic cancer centres, tumour-only analysis is the more typical current clinical standard [6].
As well as influencing cancer management and therapy, a substantial proportion of genes included on these large tumour panels confer heritable predisposition to cancer, the so-called cancer susceptibility genes (CSGs). A subset of variants identified in CSGs are ‘pathogenic’ that is when present in the constitutional DNA, they will confer an elevated risk of developing one or more cancer types. When such a variant is detected in a tumour, it is unclear whether the variant is constitutional in origin (i.e. is present in all or most tissues, including the germline) or it has been somatically acquired (i.e. not in the germline but present in the tumour ± surrounding tissue). We refer to these as ‘tumour-detected pathogenic variants of potential germline origin’, or ‘tumour-detected pathogenic variants’.
Having identified that clinical practice in this area is widely disparate, the European Society of Medical Oncology (ESMO) Precision Medicine Working Group (PMWG) convened a subgroup tasked with addressing germline management of tumour-detected pathogenic variants, specifically:
to identify the key issues relevant to laboratory and clinical management,
to undertake analyses of relevant tumour and germline data,
-
to generate consensus recommendations applicable in routine clinical-laboratory services regarding:
extent of germline-focussed analyses to be carried out as routine,
when follow-up testing in a germline sample should be undertaken,
patient information and consent.
Key issues relevant to germline-focussed tumour analysis:
The ESMO PMWG germline subgroup identified and considered the following issues within their analyses and recommendations:
‘On-tumour’ and ‘Off-tumour’ associations of gene with tumour type
CSGs have been established, through prior research, as conferring predisposition to specific tumour types; these associations we hereafter refer to as ‘on-tumour’. Hence, for BRCA1, a pathogenic variant identified in a breast tumour would be described as on tumour, as presence of a germline BRCA1 mutation confers elevated risk of breast cancer. A BRCA1 pathogenic variant identified in a testicular seminoma would be defined as ‘off-tumour’ as presence of a germline BRCA1 mutation does not confer elevated risk of testicular seminoma. Such a finding off-tumour would typically be termed a ‘secondary’ finding (if sought deliberately), or an ‘incidental’ finding (if happened upon by chance during data review) [7].
Clinical Actionability
Quantification of ‘clinical actionability’ for pathogenic variants in CSGs has not been formally established but would widely be agreed to encompass the penetrance (risk) of the associated cancers, the ‘severity’ of the cancer and the availability of clinical management options that mitigate the increased cancer risk (screening, surgical prophylaxis, chemoprophylaxis and lifestyle modification), along with the quality of evidence underpinning each of these estimates [8].
For more frequently detected CSGs such as BRCA1/BRCA2/MLH1/MSH2, evidence regarding cancer risk and the impact of screening/interventions are relatively well established. For less frequently detected CSGs, e.g. FLCN or FH, whilst robust evaluations of clinical and economic impact are lacking, clinical management is relatively consistent. For other CSGs such as DICER1 and BAP1, evidence regarding penetrance and efficacy of interventions is sparse and management varies widely between centres.
The American College of Medical Genetics (ACMG) has assembled a set of 25 CSGs of higher actionability for which they have advised analysis for and return of pathogenic variants, regardless of context of ascertainment [7, 9, 10].
An additional aspect of actionability relates to the impact on management of the current cancer of a germline pathogenic variant. For example, identification of a germline BRCA1 pathogenic variant in the on-tumour setting (breast or ovarian cancer) may influence the choice of chemotherapy (platinum), eligibility for targeted agents (PARP inhibitors) and/or primary surgical management (bilateral mastectomy in localised unilateral disease). In genomically selected basket trials the germline status may influence eligibility for targeted drugs even in the off-tumour setting. However, for many CSGs, identification of a germline pathogenic variant has no impact on management of a cancer once diagnosed in the on- or off-tumour setting.
Penetrance (risk) of cancer
Early estimates of cancer penetrance (risk) for CSGs were typically derived from linkage analysis using large multicase families. Subsequent studies based on ascertainment from more modest family clusters or unselected incident cancer cases have revealed progressively lower estimates of penetrance for many CSGs. Analyses of population data suggest penetrance may be lower still if the germline pathogenic variant is ascertained completely agnostic to phenotype [11]. Impact of preventive clinical interventions will be predicated on the estimates of penetrance used for the clinical or cost-effectiveness analyses.
Tumour heterogeneity and contamination with normal tissue
A heterozygous germline pathogenic variant in a CSG would be anticipated to be present in 50% of alleles across body tissues, including a tumour. On occasion, the variant allele frequency (VAF) of a somatically-acquired mutation may exceed 50% due to Loss of Heterozygosity of the opposite allele or tumour aneuploidy. Most typically, somatic variants are detected at VAF <50% due to a combination of clonal heterogeneity, contamination with lymphocytes/non-tumour cells and/or tumour aneuploidy. It is important to note that compared with hybrid-capture-based NGS strategies such as the one used for this analysis, PCR-based NGS strategies may not yield as consistent a VAF, especially in FFPE specimens with poor DNA quality and yield.
Threshold for triggering germline follow-up testing
To control the volume of germline tests triggered, we required a working threshold of ‘likelihood of germline origin’. Historically, a threshold of 10% (based on personal and/or family history of cancer) has widely been adopted in the UK National Health Service for BRCA1/2 detection on clinical testing [12].
Workflows, logistics and cost
Tumour molecular genetic analysis is frequently carried out in molecular pathology laboratories geographically or administratively distinct from those offering germline testing for CSGs. Resources in these molecular pathology laboratories are focussed on interpretation of somatic genetics and expertise in germline interpretation may be lacking [13].
Consent and patient education
Explicit consent is not routinely sought ahead of molecular genetic analysis of the tumour, being a test undertaken alongside histopathological examination to characterise the malignant tissue in order to inform immediate management. Furthermore, while tumour testing for treatment selection is most often requested by oncologists, some molecular genetic tumour tests are only initiated downstream of patient contact once the pathologist has carried out histological examination.
Conversely, a germline pathogenic CSG variant is often not relevant for current cancer management, but may have implications for future health, family members, reproductive decision-making and insurance. Accordingly it remains conventional practice that explicit information and consent-taking precede such analyses [14, 15].
Methods
ESMO PMWG germline subgroup
The Germline Subgroup convened by the ESMO PMWG comprised representation from medical oncology, surgical oncology, clinical cancer genetics, molecular pathology and medical law. The Group met five times to develop these recommendations (supplementary note, available at Annals of Oncology online).
Case series
We utilised the largest available dataset of paired tumour-normal sequencing, comprising 17 152 unselected cancer patients who presented to Memorial Sloan Kettering Cancer Center between 2014 and 2017 in whom clinical sequencing of both germline (blood) and tumour samples had been successfully carried out using the MSK-IMPACT assay (the MSK dataset) [4, 16–18]. All patients in this cohort had consented to somatic and/or germline testing in the context of tumour-normal sequencing using an institutional review board-approved protocol. For the purpose of this analysis, all genetic data were anonymised.
Sequencing
Samples were sequenced to a median depth of 741× (tumour) and 470× (normal). Tumour samples exhibiting somatic hypermutation (defined as >95th percentile of mutational burden) were analysed separately. All patients received clinical MSK-IMPACT sequencing using either the first or generation of the panel design interrogating 341 and 410 genes, respectively. Data were extracted for 65 genes associated with germline susceptibility to invasive cancers. For 64 genes, the mode of inheritance was autosomal dominant; MUTYH (autosomal recessive) was also included on account of being on the ‘ACMG secondary findings gene list’ (supplementary Table S1, available at Annals of Oncology online) [7].
Calling and variant classification
Joint variant calling was carried out in the tumour and the germline samples to generate optimal somatic calls. These were summed with the germline-only calls to generate tumour variant frequencies [16, 17]. Variants retained for analysis were those predicted to cause protein truncation in genes acting via loss-of-function and/or classified as germline 4-Likely Pathogenic or 5-Pathogenic (any star rating) based on the ClinVar variant classification resource [19]. We excluded low penetrance alleles assigned within ClinVar as ‘risk-factors’, such as APC c.3920T>A (p.Ile1307Lys) and VHL c.598C>T (p.Arg200Trp). Inclusion of founder mutations did not substantially alter germline conversion rates for the respective genes (BRCA1, BRCA2, ATM) and these founder mutations were therefore retained for subsequent analyses (supplementary Table S2, available at Annals of Oncology online).
Gene annotation
A panel of five clinical (medical) geneticists specialising in cancer susceptibility was convened (supplementary note, available at Annals of Oncology online). Annotation was carried out independently for the 65 genes for (i) association with each individual cancer type [20], (ii) actionability (high/non-high), (iii) penetrance [high (RR > 4)/intermediate (RR = 2–4)] [21] and (iv) robustness of implication pathogenic variants in cancer susceptibility (clinical—grade or not). A status was assigned to each gene based on the majority decision (supplementary Table S1, available at Annals of Oncology online).
High-actionability CSGs (HA-CSGs) were defined as those of a level of actionability by which return of pathogenic germline variants would be appropriate in the off-tumour as well as on-tumour context. We established as a set of HA-CSGs, the 25 genes recommended for secondary findings by the ACMG and identified five additional genes as being of equivalent actionability to these 25 (PALB2, RAD51C, RAD51D, BRIP1, SDHA). These genes are under current review for inclusion in the ACMG secondary findings gene list [22–26].
From the remaining 35 genes, we established a set of 27 ‘standard-actionability’ genes (SA-CSGs), namely those of high penetrance, for which the actionability of monoallelic pathogenic variants was agreed to be ‘clinical grade’, for which return of pathogenic germline variants in the on-tumour context was agreed to be appropriate.
Calculation of germline conversion rate and clinical recommendations
We defined a metric of ‘germline conversion rate’ (number of pathogenic variants of true germline origin× 100/total number of tumour-detected pathogenic variants), which we calculated for each gene (i) for all genes and (ii) stratified by gene actionability, context (on-tumour/off-tumour) and/or patient age. We recommended for germline follow-up only genes (i) of high/standard actionability (ii) for which a total of ≥2 mutations of true germline origin were detected across relevant tumour types (iii) for which the germline conversion rate exceeded 10% for that group defined by context and patient characteristics.
Results
Gross analyses of paired tumour-germline data
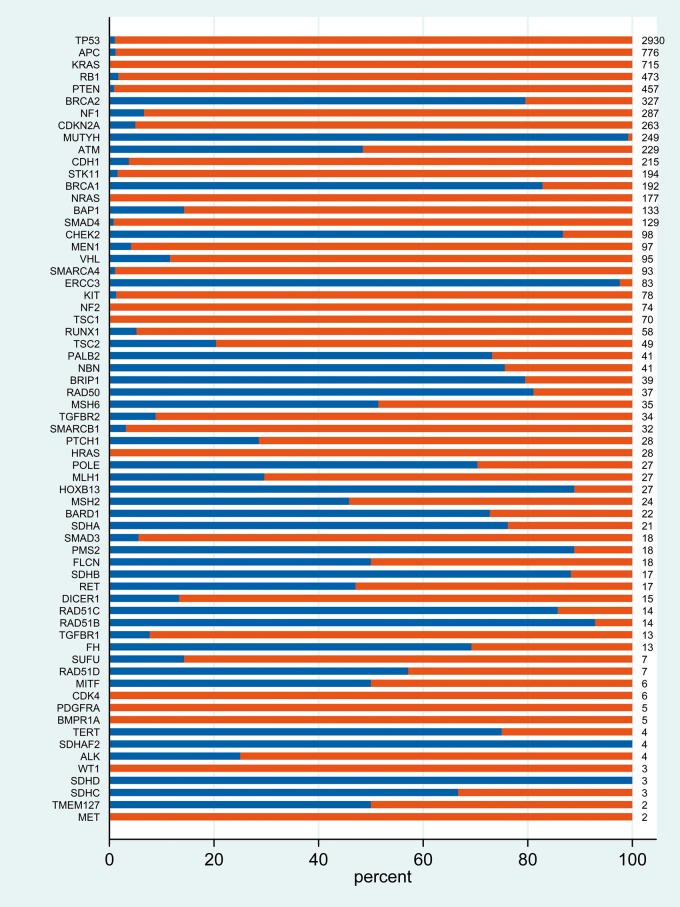
In total, 17 152 tumour-germline pairs were successfully analysed of which 830 were excluded due to somatic hypermutation. The remaining 16 322 paired samples were distributed by tumour type as per supplementary Table S3, available at Annals of Oncology online. Across the 65 genes analysed in 16 322 tumours, in total 1 997 499 tumour variants were identified (of which 1 959 587 were of true germline origin) (Table 1). Removal of common polymorphisms [≥1% minor allele frequency (MAF)] reduced the number of tumour variants to 79 342 (of which 53 388 were of true germline origin). We then applied an automated variant ‘pathogenicity filter’, retaining only variants predicted to cause loss of function of the protein and/or classified in ClinVar as Likely Pathogenic (class 4) or Pathogenic (class 5). A total of 17 075 tumour-detected pathogenic variants were retained [of which 1494 (8.7%) were of true germline origin]. Having examined the distribution of tumour-detected VAF for variants of true germline origin, we then removed variants of tumour VAF < 0.3 (SNVs) or VAF <0.2 (small insertions/deletions) (Figure 1). Application of this ‘VAF filter’ reduced the number of tumour-detected pathogenic variants from 17 075 to 9222, and true germline pathogenic variants from 1494 to 1442 (Figure 2).
Table 1.
Summary of number of (i) tumour detected and (ii) true germline variants detected with application of (A) serial filters (B) gene/context/age criteria, from data on 65 genes for 16 322 tumours (MSK dataset)
| All tumours |
Associated tumours |
Non-associated tumours |
||||||
|---|---|---|---|---|---|---|---|---|
| Tumour detected | True germline | Tumour detected | True germline | Tumour detected | True germline | |||
| (A) Application of serial filters to MSK data on 65 genes for 16 322 tumours: number of variants | 1 997 499 | 1 959 587 | ||||||
| Retained: MAF ≤0.01 | 79 342 | 53 388 | ||||||
| Retained: LP/P/truncating | 17 075 | 1494 | ||||||
| Retained: VAF ≥0.3 (SNV) or ≥0.2 (insdel) | All | 9222 | 1442 | 2904 | 454 | 6305 | 983 | |
| HA-CSGs (AD) | 6141 | 677 | 2259 | 326 | 3882 | 351 | ||
| SA-CSGs (AD) | 2372 | 213 | 539 | 37 | 1820 | 176 | ||
| Other | 709 | 547 | 106 | 91 | 603 | 456 | ||
| (B) Application of ESMO-PWG recommendations for gene/context/age criteria based on 10% germline conversion: number of variants | HA-CSGs (AD) | all ages (18 genes) | 851 | 615 | 410 | 300 | 441 | 315 |
| age <30 (APC, RB1) | 63 | 10 | 37 | 4 | 26 | 6 | ||
| age <30, on-tumour only (TP53) | 59 | 7 | 59 | 7 | n/a | n/a | ||
| Total | 973 | 632 | 506 | 311 | 467 | 321 | ||
| SA-CSGs (AD) | all ages, on tumour only (BAP1, FH, FLCN, POLE), | 60 | 17 | 60 | 17 | n/a | n/a | |
| age <30, on tumour only (NF1) | 9 | 4 | 9 | 4 | n/a | n/a | ||
| Total | 69 | 21 | 69 | 21 | n/a | n/a | ||
| Grand total | 1042 | 653 | 575 | 332 | 467 | 321 | ||
HA-CSGs, high actionability genes; SA-CSGs, standard actionability CSGs; AD, autosomal dominantly inherited; other, CSGs of recessive inheritance, intermediate penetrance and/or non-clinical grade actionability.
Figure 1.
Distribution of variant allele frequency observed in the tumour for variants of true germline origin which were (i) small insertion/deletions (ii) SNVs.
Figure 2.
Distribution of germline and somatic pathogenic variants detected upon tumour analysis. Only variants classified pathogenic/likely pathogenic AND above VAF threshold are included (blue, germline origin; red, somatic origin; numbers, total number of pathogenic variants observed in tumour).
Recommendations, see also Box 1
|
Analyses by gene: actionability and on/off tumour association
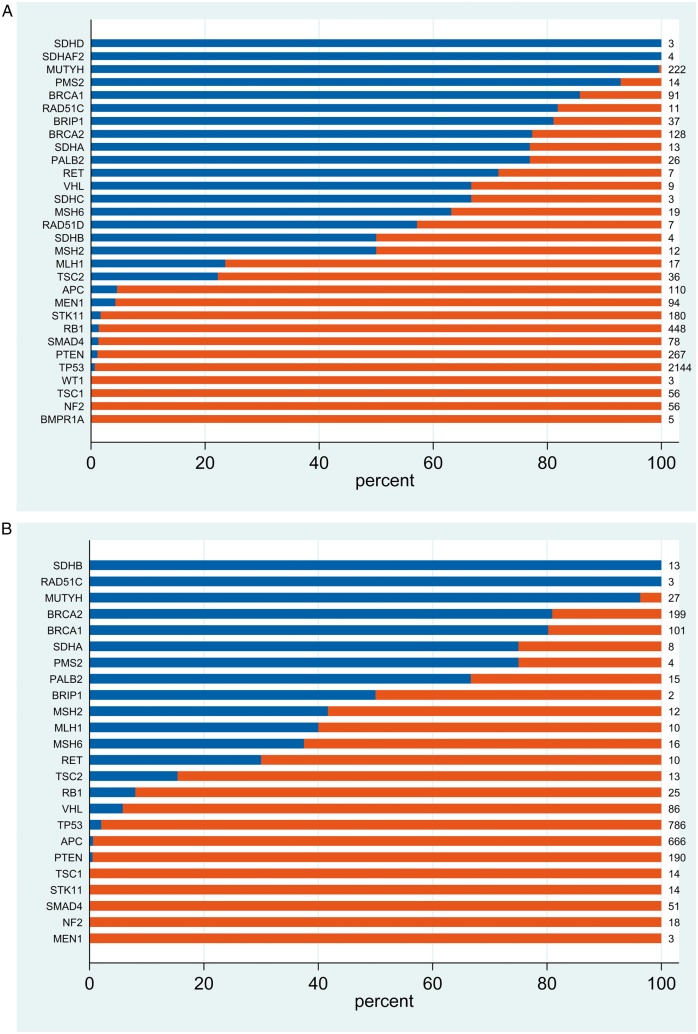
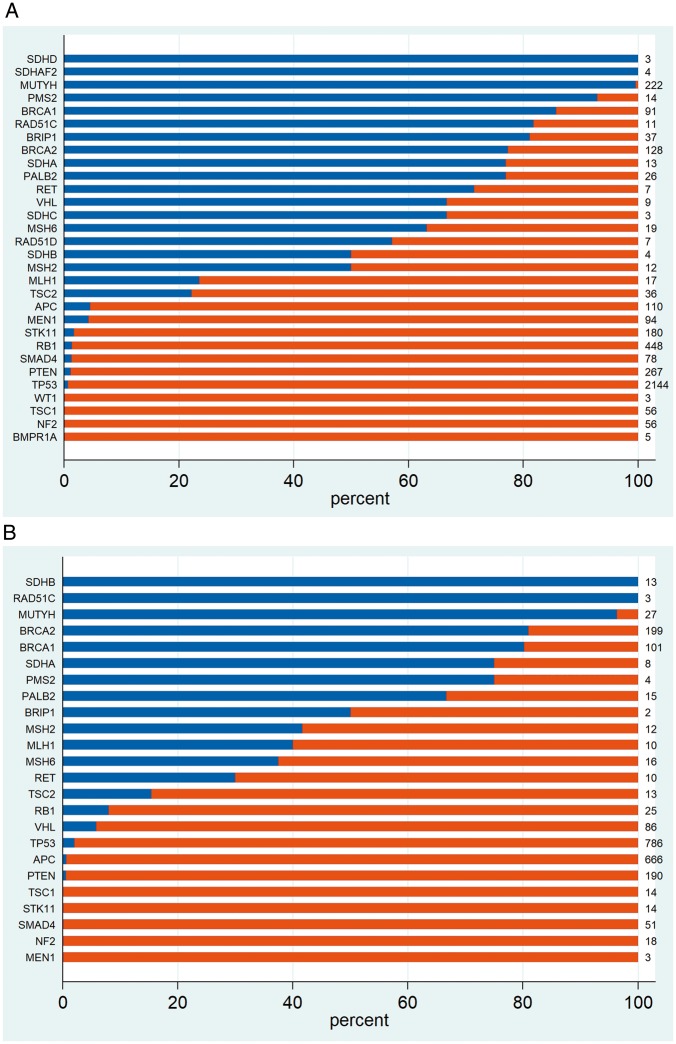
We went on to examine germline conversion rate stratified by gene actionability, context (on-tumour/off-tumour) and/or patient age. We first examined the germline conversion rate in the 30 HA-CSGs. In the off-tumour setting, 19 of 30 genes surpassed the 10% threshold (Figure 3A). In the on-tumour setting, for 14 of those 19 genes the ‘germline conversion rate’ was likewise >10% and for 4 genes (SDHAF2, SDHD, SDHC and RAD51D) no pathogenic sequence variants were observed in associated tumours (Figure 3B). For VHL, 67% (6 of 9) pathogenic variants observed off-tumour were of germline origin whilst in the on-tumour setting this figure was only 6% (5 of 86). This paradoxical observation is explained by the high rate of somatic VHL mutation in renal cancers (79 of 82 tumour-detected pathogenic variants being somatic; supplementary Figure S1, available at Annals of Oncology online). Similarly, reflecting the high somatic APC mutation rate in colorectal cancer (CRC), only 2 of 637 (0.3%) tumour-detected pathogenic APC variants observed in CRC were of germline origin (supplementary Figure S2, available at Annals of Oncology online).
Figure 3.
Distribution of germline and somatic pathogenic variants detected upon tumour analysis for 30 high-actionability CSGs. (A) Off-tumour and (B) on-tumour.
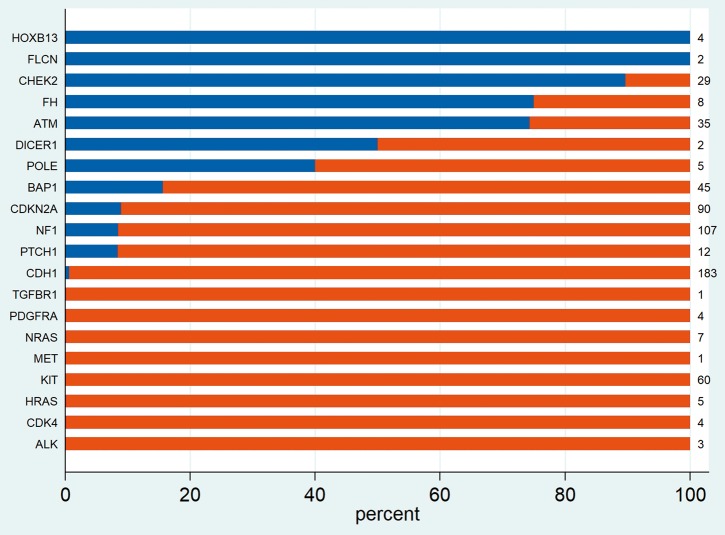
For the 27 standard actionability CSGs analysed, a >10% on-tumour germline conversion rate was attained for genes BAP1, FLCN, POLE and FH and a near 10% rate for CDKN2A (8.9%, 8 of 90) and NF1 (9 of 107, 8.4%). Notably intermediate penetrance genes CHEK2, ATM and HOXB13 also had a >10% germline conversion rate (Figure 4).
Figure 4.
Distribution of germline and somatic pathogenic variants detected upon tumour analysis for non-high actionability CSGs in associated tumours (on-tumour).
Analyses of TP53: the most frequently somatically mutated CSG
We went on to perform more in depth analysis of tumour-detected TP53 pathogenic variants, on account of their high frequency and the significant clinical implications of diagnosis of Li-Fraumeni syndrome. Overall, 2930 tumour-detected TP53 pathogenic variants were detected across 16 322 tumours, with an overall germline conversion rate of 1% (30/2930). This rate was modestly higher in the on-tumour setting (2%, 16 of 786) than off-tumour (0.7%, 14 of 2144) (supplementary Figure S3a, available at Annals of Oncology online). To explore whether enrichment for germline origin was age-associated, we retained the 1273 tumours arising at age <30 years, in which there were 174 tumour-detected TP53 pathogenic variants. The distribution of the 11 true germline variants amongst the 174 tumour-detected TP53 pathogenic variants included 4 of 31 (12.9%) for breast cancers, 2 of 20 (10%) for soft tissue sarcomas, 1 of 7 (14.3%) for bone tumours and 1 of 57 (1.8%) for gliomas and 3 of 60 (5%) in non-associated tumours (supplementary Figure 3b, available at Annals of Oncology online). The overall TP53 germline conversation rate in the on-tumour setting age <30 years was 6.8% (8 of 117), but with the exclusion of glioma, this rate was improved to 7 of 59 (11.7%). We also explored whether there was enrichment of the TP53 germline conversation rate if only selected TP53 mutations were included but did not find evidence for this (supplementary Table S4, available at Annals of Oncology online).
Analyses by patient age at tumour occurrence
Through restriction to tumours arising age <30 years, the overall germline conversion rate was improved to >10% for high-actionability genes RB1 (7 of 35, 20%) and APC (3 of 28, 10.7%), and in the on-tumour context for the standard actionability gene NF1 (4 of 9, 44.4%) (supplementary Figures S4 and S5, available at Annals of Oncology online).
Examining further each individual gene for specific scenarios of elevated germline conversion rate by each tumour type, and by all ages, age <30 and age <5, we did not identify any additional scenarios in which a germline conversion rate of >10% was achieved.
Analyses in hypermutated samples
In separate analysis of the 830 samples in which the mutational burden was above the 95th centile, the germline conversion rate was >10% for the majority of HA-CSGs and notably all four mismatch repair genes (MLH1, MSH2, MSH6, PMS2) (supplementary Figure S6, available at Annals of Oncology online). Of the 14 tumour-detected POLE pathogenic variants in these samples, only one was of germline origin.
Summary of rate of detection of variants of true germline origin
Overall, in our tumour-focussed germline analysis of the MSK dataset, restricting firstly by VAF and secondly to the 27 genes yielding germline conversion rate >10% as per specified gene/context/age, the number of tumour-detected pathogenic variants requiring ‘germline follow-up’ could be reduced by 94% from 17 075 to 1042 such that germline follow-up would only be required in 6.4% of tumours (1042/16 322) (Table 1).
Of these 1042 variants, 653 (62.7%) were of true germline origin. These 653 pathogenic germline variants comprised 615 pathogenic variants detected age-unselected in HA-CSGs (300 on-tumour, 315 off-tumour), 17 on-tumour pathogenic variants in BAP1/FH/FLCN/POLE, 7 on-tumour pathogenic TP53 variants in tumours arising age <30 years and 14 pathogenic variants in APC, RB1 and NF1 in tumours arising age <30 years.
Of the 689 true germline variants in dominant HA-CSGs amongst the original 17 075 tumour-observed pathogenic variants, 677 remain following VAF filtering and 45 are ‘thrown out’ via application of the 10% germline conversion gene/context/age criteria [MEN1(4), PTEN(4), SMAD4(1), STK11(3) VHL(3), TP53(23), APC(6), RB1(1)]. Of the 37 true germline variants arising on-tumour across SA-CSG genes, 21 are detected and 16 are ‘missed’. Overall, of the 789 (1442–653) true germline variants that are ‘missed’, 547 are in genes acting recessively (namely MUTYH), of intermediate penetrance and/or of non-clinical-grade cancer association. A total of 176 variants are ‘missed’ because they occur off-tumour in SA-CSGs and we chose not to identify them.
Discussion
Through our large-scale analysis of paired somatic/germline data, the ESMO PMWG germline subgroup sought to develop recommendations regarding germline-focussed analysis of tumour-only sequencing data in order to optimise detection of true germline variants in genes of clinical utility, whilst avoiding excessive diversion of effort and resources towards ‘germline follow-up testing’ of vast numbers of variants.
The first issue we explored was restriction based on VAF. For this MSK dataset, we found that crude ‘pan-tumour’ VAF thresholds (20% for small insertions/deletions, 30% for SNVs) enabled reduction by almost half the number of tumour-detected variants requiring follow-up (17 075 to 9222) whilst losing only a tiny proportion of true germline variants (52 of 1494, 3.5%) and even smaller proportion of variants in dominant HA-CSGs (12 of 689, 1.7%). This filter near doubles the germline conversion rate from 8.7% (1494 of 17 075) to 15.6% (1442 of 9222). However, the VAFs used in this analysis may need to be re-evaluated by laboratories using PCR-based NGS methodologies rather than hybridisation-based methods.
Next, to reduce further the number of variants requiring follow-up, we recommend exclusion from germline-focussed tumour analysis of gene/context/age scenarios in which the germline conversion rate is <10%. Thus we highlight the 27 genes of >10% germline conversion rate (Box 1). Following filters for pathogenicity and VAF, restriction of germline-focussed tumour analysis to just these 27 genes (as per gene/context/age recommendations) enables reduction by 88.7% of the number of tumour observed variants requiring follow-up testing (from 9222 to 1042).
Box 1.
Recommendations for genes to be included for germline-focussed analysis and triggering of germline sample laboratory confirmation
| Any tumour type | Associated tumour type only | ||
|---|---|---|---|
| Tumour arising any age |
|
|
|
| Tumour arising age <30 only |
|
|
|
Renal tumours to be excluded.
MUTYH should be included for germline-focussed tumour analysis but reporting and germline follow-up testing should only be performed on detection of two pathogenic variants.
Brain tumours to be excluded.
Although their germline conversion rate is high, we do not recommend germline-focussed tumour analysis for intermediate penetrance genes, such as CHEK2 and ATM, as strategies are not well agreed regarding management of risk within families. We recognised nevertheless that in healthcare settings in which germline analysis for these genes is routinely offered, there would be a consistency in inclusion of these genes in germline-focussed tumour analysis. Indeed, detection of CHEK2 c.1100delC may offer greater clinical utility to the family when ascertained in an isolated 60-year-old breast cancer case than in a multiplex breast cancer family.
We recommend germline-focussed tumour analysis in the off-tumour context is restricted to HA-CSGs, as per ACMG guidance regarding return of secondary findings. Nevertheless, for some genes on the ACMG secondary findings gene list, estimates are highly uncertain regarding penetrance outside of phenotype-driven ascertainment. Some centres may argue the merit of a narrower set of HA-CSGs [27, 28]. For the remainder of genes (of standard actionability) we recommended that germline-focussed tumour analysis be restricted to the on-tumour setting, such that germline pathogenic variants identified would be aetiologically pertinent with regard to the tumour type in which they were ascertained. For paediatric patients, special consideration will be required regarding return of either (i) a germline pathogenic variant deemed causative of the early onset cancer, (ii) an off-tumour pathogenic variant conferring risk only for adult onset cancers.
The germline conversion rates presented have been derived from the MSK dataset. When applying these filters and criteria in other settings, the frequency of germline variants detected will be predicated on (i) the distribution of different tumour types, (ii) the genes included on the panel, (iii) the purity of the tumours and (iv) the accuracy of tumour VAF estimation. Furthermore, a number of CSGs were not included on the MSK-IMPACT panel; overall, the subgroup agreed that for these more ‘obscure’ CSGs, whilst evaluation would be useful, these genes would at best be of standard actionability and generally (i) the frequency/contribution to overall cancer susceptibility of pathogenic germline variants is low (ii) the penetrance for cancer is poorly characterised (iii) evidence is limited regarding the efficacy of clinical interventions in carriers of pathogenic variants [20].
Local clinical workflows will need to evolve to encompass patient education, patient consent, acquisition of the germline sample and return of germline results. Some centres may elect as routine to acquire a germline sample and provide up-front consent to all individuals in whom tumour-only testing is carried out (potentially as an ‘opt-out’). Alternatively, a two-stage approach may be preferred, whereby germline consultation and acquisition of the germline sample is only triggered on detection of a tumour-detected pathogenic variant. Telephone consultation, postal blood-packs and/or saliva sampling may mitigate otherwise problematic increases in clinical workload associated with this new burden of germline follow-up. If adjacent normal (or tumour-poor) tissue is available, testing in this for the tumour-detected pathogenic variant could provide a pragmatic means of triaging out variants of low germline likelihood.
In conclusion, identification of a pathogenic variant in a CSG can offer significant opportunity for the prevention and early detection of future cancers in the patient as well as their family, and may also influence management of the current cancer. Pragmatic, strategic germline-focussed tumour analysis can offer a high yield of true germline findings [63% true germline yield from follow-up of 6.4% of tumours (MSK dataset)].
The remit of the current recommendations was to guide germline-focussed tumour analysis of the tumour panels already in current use. An urgent priority for debate by clinicians and policy makers is consideration as to whether tumour panel content should be designed a priori to include genes selected for their germline utility, not just for CSGs but perhaps also those relating to pharmacogenomics.
Supplementary Material
Acknowledgements
The Germline Subgroup meetings were supported by the ESMO Precision Medicine Working Group.
Funding
CT, HH, CL and KS are supported by the Cancer Research UK Catalyst Award CanGene-CanVar (grant number C61296/A26688). ML and DM's work at MSKCC was supported in part by Cancer Center Support Grant NIH P30 CA008748.
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Dias-Santagata D, Akhavanfard S, David SS. et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med 2010; 2(5): 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meric-Bernstam F, Brusco L, Shaw K. et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. JCO 2015; 33(25): 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia EP, Minkovsky A, Jia Y. et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 2017; 141(6): 751–758. [DOI] [PubMed] [Google Scholar]
- 4. Cheng DT, Mitchell TN, Zehir A. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015; 17(3): 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frampton GM, Fichtenholtz A, Otto GA. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013; 31(11): 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell 2017; 168(4): 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Green RC, Berg JS, Grody WW. et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15(7): 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turnbull C, Sud A, Houlston RS. Cancer genetics, precision prevention and a call to action. Nat Genet 2018; 50(9): 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015; 17: 68–69. [DOI] [PubMed] [Google Scholar]
- 10. Kalia SS, Adelman K, Bale SJ. et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017; 19(2): 249–255. [DOI] [PubMed] [Google Scholar]
- 11. Loveday C, Josephs K, Chubb D. et al. p.Val804Met, the most frequent pathogenic mutation in RET, confers a very low lifetime risk of medullary thyroid cancer. J Clin Endocrinol Metab 2018; 103(11): 4275–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer (CG164), National Institute for Health and Care Excellence, 2013. [PubMed]
- 13. Millner LM, Strotman LN. The future of precision medicine in oncology. Clin Lab Med 2016; 36(3): 557–573. [DOI] [PubMed] [Google Scholar]
- 14. Foulkes WD, Knoppers BM, Turnbull C. Population genetic testing for cancer susceptibility: founder mutations to genomes. Nat Rev Clin Oncol 2016; 13(1): 41–54. [DOI] [PubMed] [Google Scholar]
- 15. Roche MI, Berg JS. Incidental findings with genomic testing: implications for genetic counseling practice. Curr Genet Med Rep 2015; 3(4): 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandelker D, Zhang L, Kemel Y. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 2017; 318(9): 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nat Med 2017; 23(6): 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng DT, Prasad M, Chekaluk Y. et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics 2017; 10(1): 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landrum MJ, Lee JM, Riley GR. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucl Acids Res 2014; 42(D1): D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rahman N. Realizing the promise of cancer predisposition genes. Nature 2014; 505(7483): 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Easton DF, Pharoah PD, Antoniou AC. et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015; 372(23): 2243–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Antoniou AC, Casadei S, Heikkinen T. et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371(6): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loveday C, Turnbull C, Ruark E. et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet 2012; 44(5): 475–476. author reply 476 [DOI] [PubMed] [Google Scholar]
- 24. Loveday C, Turnbull C, Ramsay E. et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 2011; 43(9): 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pelttari LM, Heikkinen T, Thompson D. et al. RAD51C is a susceptibility gene for ovarian cancer. Hum Mol Genet 2011; 20(16): 3278–3288. [DOI] [PubMed] [Google Scholar]
- 26. Ramus SJ, Song H, Dicks E. et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst 2015; 107. doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunter JE, Irving SA, Biesecker LG. et al. A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med 2016; 18(12): 1258–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berg JS, Foreman AK, O'Daniel JM. et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med 2016; 18(5): 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.