Abstract
Background
Although EGFR mutant tumors exhibit low response rates to immune checkpoint blockade overall, some EGFR mutant tumors do respond to these therapies; however, there is a lack of understanding of the characteristics of EGFR mutant lung tumors responsive to immune checkpoint blockade.
Patients and methods
We retrospectively analyzed de-identified clinical and molecular data on 171 cases of EGFR mutant lung tumors treated with immune checkpoint inhibitors from the Yale Cancer Center, Memorial Sloan Kettering Cancer Center, University of California Los Angeles, and Dana Farber Cancer Institute. A separate cohort of 383 EGFR mutant lung cancer cases with sequencing data available from the Yale Cancer Center, Memorial Sloan Kettering Cancer Center, and The Cancer Genome Atlas was compiled to assess the relationship between tumor mutation burden and specific EGFR alterations.
Results
Compared with 212 EGFR wild-type lung cancers, outcomes with programmed cell death 1 or programmed death-ligand 1 (PD-(L)1) blockade were worse in patients with lung tumors harboring alterations in exon 19 of EGFR (EGFRΔ19) but similar for EGFRL858R lung tumors. EGFRT790M status and PD-L1 expression did not impact response or survival outcomes to immune checkpoint blockade. PD-L1 expression was similar across EGFR alleles. Lung tumors with EGFRΔ19 alterations harbored a lower tumor mutation burden compared with EGFRL858R lung tumors despite similar smoking history.
Conclusions
EGFR mutant tumors have generally low response to immune checkpoint inhibitors, but outcomes vary by allele. Understanding the heterogeneity of EGFR mutant tumors may be informative for establishing the benefits and uses of PD-(L)1 therapies for patients with this disease.
Keywords: epidermal growth factor receptor, immune checkpoint blockade, non-small-cell lung cancer
Key Message
The results of this study suggest that EGFR mutant subtypes of non-small-cell lung cancer have distinct response to immune checkpoint inhibitors and distinct tumor mutation burdens. This knowledge may be useful in selecting patients with a higher likelihood of response to immunotherapy.
Introduction
Epidermal growth factor receptor (EGFR) mutant lung cancers represent a distinct subset of non-small-cell lung cancer (NSCLC) with broad molecular and clinical heterogeneity. Recurrent alterations in exons 18–21 are commonly observed [1–3] and most, but not all, confer sensitivity to EGFR tyrosine kinase inhibitors (TKIs) [4–6]. Even the most common EGFR TKI-sensitizing alleles, EGFR L858R (EGFRL858R) and EGFR exon 19 deletions (EGFRΔ19), have differences in outcomes with TKIs [7, 8]. Despite initial responsiveness to EGFR TKIs, acquired resistance is routine [4, 9–11]. The inevitability of resistance has raised hopes of a role for immune checkpoint inhibitors (ICIs), with the potential for more durable responses; however, in contrast to preclinical studies [12], clinical evidence suggests that EGFR mutant lung cancers rarely derive benefit from treatment with ICIs [13–16]. Rates of positivity for potential predictors of response to ICIs, such as tumor mutation burden (TMB) and concurrent programed death-ligand 1 (PD-L1) plus CD8+ tumor infiltrating lymphocyte expression, are low [17]. Yet recent studies have emerged, such as ATLANTIC and IMpower150, that have shown more encouraging results for PD-(L)1 blockade in EGFR mutant lung cancers [18, 19].
We hypothesized that the molecularly heterogeneous features of EGFR mutant lung cancers may provide insight into the outcomes with ICIs and improve understanding of the determinants of response in these tumors [20]. To test this, we established a multi-institutional consortium and examined the molecular and clinical features of 171 EGFR mutant lung cancer cases treated with ICIs. A cohort of 212 patients with EGFR wild-type NSCLC (previously published) treated with ICIs was used for comparison. Due to limited sequencing data available for ICI-treated EGFR mutant cases in this study, we examined a separate cohort of 383 patients with EGFR mutant lung cancer (irrespective of treatment history) to examine the relationship between TMB and EGFR mutation subtype.
Methods
Cohorts of EGFR mutant lung cancers
Following IRB approval at each respective institution, patients with EGFR mutant lung cancer treated with PD-(L)1 blockade therapy were identified (Yale Cancer Center n = 37, Memorial Sloan Kettering Cancer Center n = 67, University of California Los Angeles n = 35, Dana Farber Cancer Institute n = 32). Patients were treated as part of a clinical trial (n = 97; 56.7%) or standard-of-care (n = 74; 43.3%). Due to the retrospective nature of this study, scan intervals were not uniform between all patients. Patients were included who received anti-PD-(L)1 alone or in combination with anti-cytotoxic T-cell lymphocyte-4 (anti-CTLA-4), and this treatment was their first exposure to ICIs. In a subset of patients (n=15), ICIs were added to continuation of EGFR TKIs at TKI resistance. In EGFRL858R and EGFRΔ19 cases treated with ICIs before EGFR TKIs, this was due to the absence of information regarding their EGFR alteration at the time of treatment (n = 7), because the patient was enrolled on a specific clinical trial (n = 1) or because the tumor had a baseline EGFRT790M mutation and was treated with anti-PD-1 plus anti-CTLA-4 therapy (n = 1). TMB was studied in data from a cohort of 383 patients with EGFR mutant lung cancer, irrespective of treatment exposure, collected from three sources: (i) The Cancer Genome Atlas (n = 53), (ii) Yale University (n = 17), and (iii) Memorial Sloan Kettering Cancer Center (n = 313). TMB was calculated as the total number of non-synonymous mutations divided by the coding region captured for each individual platform (see supplementary Methods, available at Annals of Oncology online).
Results
Distinct EGFR subtypes have different outcomes with immune checkpoint blockade
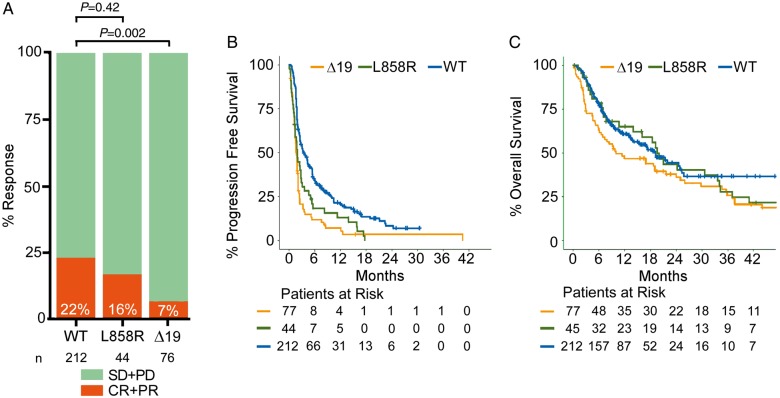
We investigated the impact of varying EGFR alleles on outcomes with ICIs (anti-PD-1 or anti-PD-L1, with or without CTLA-4 blockade) in our cohort of 171 EGFR mutant cases from four institutions (Table 1), focusing particularly on those 126 patients with tumors with the two most common EGFR mutation subtypes [EGFRL858R(n = 46) or EGFRΔ19 (n = 80)] (supplementary Figure S1, available at Annals of Oncology online). These cases were evaluated and compared with 212 patients with EGFR wild-type (WT) NSCLC treated with ICIs [21]. EGFRΔ19tumors had a significantly lower overall response rate (ORR) compared with EGFR WT tumors (5 of 76, 7% versus 47 of 212, 22%, respectively, P = 0.002), whereas EGFRL858Rtumors had similar response rates compared with EGFR WT tumors (7 of 44, 16%, versus 47 of 212, 22%, respectively, P = 0.42) (Figure 1A). Progression-free survival (PFS) was significantly reduced in both EGFRΔ19[(WT versus EGFRΔ19) HR (hazard ratio) 0.449, 95% CI (confidence interval) 0.338–0.595, log-rank P < 0.001] and EGFRL858R[(WT versus EGFRL858R) HR 0.578, 95% CI 0.412–0.811, log-rank P = 0.001] subtypes compared with EGFR WT (Figure 1B). Overall survival (OS) in the EGFRΔ19 group was reduced whereas EGFRL858R tumors had similar OS compared with the EGFR WT subgroup (HR 0.69, 95% CI 0.493–0.965, log-rank P = 0.03; HR 0.917, 95% CI 0.597–1.409, log-rank P = 0.69, respectively) (Figure 1C). Overall, these data suggest that patients with EGFRΔ19 mutant tumors, in particular, have a significantly reduced benefit of treatment with ICIs.
Table 1.
Characteristics of patients with EGFR mutant tumors treated with immune checkpoint inhibitors
| Characteristics | EGFRΔ19 (n = 80) | EGFRL858R (n = 46) | EGFR20Ins (n = 28) | EGFRG719 (n = 7) | EGFRL861Q (n = 5) | EGFROther (n = 5) | All EGFR cases (n = 171) |
|---|---|---|---|---|---|---|---|
| Smoking | |||||||
| Ever—no. (%) | 27 (33.8) | 20 (43.5) | 10 (35.7) | 6 (85.7) | 2 (40) | 3 (60) | 68 (39.8) |
| Never—no. (%) | 53 (66.3) | 26 (56.5) | 18 (64.3) | 1 (14.3) | 3 (60) | 2 (40) | 103 (60.2) |
| Pack-year (median) | 0 | 0 | 0 | 27 | 0 | 20 | 0 |
| Pack-year (range) | 0–40 | 0–115 | 0–27 | 0–40 | 0–10 | 0–76 | 0–115 |
| Pack-year data—Not available—no. (%) | 0 (0) | 1 (2.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.6) |
| Prior lines of therapy—no. (%) | |||||||
| 0–2 lines | 29 (36.3) | 21 (45.7) | 14 (50) | 4 (57.1) | 4 (80) | 3 (60) | 75 (43.9) |
| 3+ lines | 51 (63.8) | 25 (54.3) | 14 (50) | 3 (42.9) | 1 (20) | 2 (40) | 96 (56.1) |
| Drug target—no. (%) | |||||||
| PD-1 | 66 (82.5) | 36 (78.3) | 24 (85.7) | 7 (100) | 3 (60) | 4 (80) | 140 (81.9) |
| PD-L1 | 5 (6.3) | 7 (15.2) | 1 (3.6) | 0 (0) | 1 (20) | 1 (20) | 15 (8.8) |
| PD-(L)1 + CTLA-4 | 9 (11.3) | 3 (6.5) | 3 (10.7) | 0 (0) | 1 (20) | 0 (0) | 16 (9.4) |
| Progression-free survival (PFS) | |||||||
| Median | 1.6 | 1.9 | 1.9 | 4.8 | 1.3 | 2.6 | 1.8 |
| Range | 0–40.5 | 0.1–17.7 | 0.2–6.4 | 1.7–37.6 | 0.9–5.1 | 1.2–8.7 | 0–40.5 |
| Not available—no. (%) | 3 (3.8) | 2 (4.3) | 2 (7.1) | 1 (14.3) | 0 (0) | 0 (0) | 8 (4.7) |
| Overall survival (OS) | |||||||
| Median | 9.4 | 12.1 | 5.5 | 29.0 | 5.2 | 11.4 | 9.4 |
| Range | 0.1–71 | 0.3–63 | 0.6–73.3 | 2.2–64.8 | 0.9–13.5 | 5.2–19.0 | 0.1–73.3 |
| Not available—no. (%) | 3 (3.8) | 1 (2.2) | 2 (7.1) | 3 (42.9) | 0 (0) | 0 (0) | 9 (5.3) |
| Best response—no. (%) | |||||||
| Complete/partial response | 5 (6.3) | 7 (15.2) | 3 (10.7) | 2 (28.6) | 0 (0) | 0 (0) | 17 (9.9) |
| Stable disease | 13 (16.3) | 10 (21.7) | 6 (21.4) | 3 (42.9) | 1 (20) | 1 (20) | 34 (19.9) |
| Progressive disease | 58 (72.5) | 27 (58.7) | 18 (64.3) | 2 (28.6) | 4 (80) | 4 (80) | 113 (66.1) |
| Not available | 4 (5) | 2 (4.3) | 1 (3.6) | 0 (0) | 0 (0) | 0 (0) | 7 (4.1) |
| EGFRT790M before ICI—no. (%) | |||||||
| Yes | 37 (46.3) | 17 (37.0) | 0 (0) | 1 (14.3) | 0 (0) | 0 (0) | 55 (32.2) |
| No | 38 (47.5) | 29 (63.0) | 27 (96.4) | 6 (85.7) | 5 (100) | 5 (100) | 110 (64.3) |
| Not available | 5 (6.3) | 0 (0) | 1 (3.6) | 0 (0) | 0 (0) | 0 (0) | 6 (3.5) |
| EGFR TKI before ICI—no. (%) | |||||||
| Yes | 74 (92.5) | 43 (93.5) | 7 (25) | 4 (57.1) | 3 (60) | 2 (40) | 133 (77.8) |
| No | 6 (7.5) | 3 (6.5) | 21 (75) | 3 (42.9) | 2 (40) | 3 (60) | 38 (22.2) |
| Not available | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| PD-L1 expression—no. (%) | |||||||
| <1% | 19 (23.8) | 11 (23.9) | 6 (21.4) | 1 (14.3) | 0 (0) | 1 (20) | 38 (22.2) |
| >1% | 10 (12.5) | 14 (30.4) | 7 (25.0) | 4 (57.1) | 0 (0) | 0 (0) | 35 (20.5) |
| Not available | 51 (63.8) | 21 (45.7) | 15 (53.6) | 2 (28.6) | 5 (100) | 4 (80) | 98 (57.3) |
EGFR TKI, EGFR tyrosine kinase inhibitor; ICI, immune checkpoint inhibitor; PD-L1, programed death-ligand 1.
Figure 1.
Response, progression-free survival, and overall survival of EGFRL858Rand EGFRΔ19 mutant tumors to immune checkpoint blockade. (A) Response rate in tumors with EGFRΔ19 (n = 76) or EGFRL858R(n = 44) mutations, and wild-type for EGFR (WT) (n = 212). Overall response rate is indicated on each bar in white. Statistics were calculated using Fisher’s exact test. (B) Progression-free survival in tumors with EGFRΔ19 (n = 77) (HR 0.449, 95% CI 0.338–0.595, log-rank P < 0.001) or EGFRL858R(n = 44) (HR 0.578, 95% CI 0.412–0.811, log-rank P = 0.001) alterations compared with lung tumors that are EGFR wild-type (n = 212). (C) Overall survival in tumors with EGFRΔ19 (n = 77) (HR 0.69, 95% CI 0.493–0.965, log-rank P = 0.03) or EGFRL858R(n = 45) (HR 0.917, 95% CI 0.597–1.409, log-rank P = 0.69) alterations compared with lung tumors that are EGFR wild-type (n = 212). HR, hazard ratio; CI, confidence interval.
Clinicopathologic features associated with outcomes in EGFR mutant lung cancers
We examined the effect of clinical and pathologic features on response to ICIs in patients with EGFRL858Rand EGFRΔ19 mutant lung cancers. ORR, PFS, and OS were all significantly improved in patients who had received 0–2 prior lines of therapy compared with those with 3+ lines of therapy (ORR: 9 of 47, 19%, versus 3 versus 73, 4%, P = 0.01) (PFS: HR 2.267, 95% CI 1.499–3.427, log-rank P < 0.001) (OS: HR 1.845, 95% CI 1.204–2.826, log-rank P = 0.004) (Figure 2A–C). When examined independently, this difference in survival was statistically significant in the EGFRΔ19cohort but not within the EGFRL858Rgroup (supplementary Figure S2A–F, available at Annals of Oncology online). Smoking history was assessed in patients with EGFRL858Rand EGFRΔ19 mutant lung cancers and positively associated with response rate (P = 0.01), but not significantly for PFS or OS outcomes (log-rank P = 0.06, P = 0.23, respectively). Among patients with tumors resistant to EGFR TKIs, the presence or absence of EGFRT790M had no impact on the benefit from treatment with ICIs (Figure 2D–F), irrespective of EGFR allele (supplementary Figure S3, available at Annals of Oncology online).
Figure 2.
Clinicopathologic features associated with response, progression-free survival, and overall survival of EGFRL858Rand EGFRΔ19 mutant tumors. (A) Response rate of tumors with 0–2 (n = 47) or ≥3 (n = 73) prior lines of therapy, P = 0.01. (B) Progression-free survival with 0–2 (n = 46) or ≥3 (n = 75) prior lines of therapy (HR 2.267, 95% CI 1.499–3.427, log-rank P < 0.001). (C) Overall survival with 0–2 (n = 48) or ≥3 (n = 74) prior lines of therapy (HR 1.845, 95% CI 1.204–2.826, log-rank P = 0.004). (D) Response rate in tumors harboring EGFRT790M (T790M+, n = 52) or negative for EGFRT790M (T790M−, n = 56) that had prior EGFR tyrosine kinase inhibitor (EGFR TKI) treatment, P = 0.21. (E) Progression-free survival in tumors harboring EGFRT790M (n = 52) or negative for EGFRT790M (n = 57) that had prior EGFR TKI treatment (HR 1.348, 95% CI 0.905–2.007, log-rank P = 0.15). (F) Overall survival in tumors harboring EGFRT790M (n = 50) or negative for EGFRT790M (n = 60) that had prior EGFR TKI treatment (HR 0.878, 95% CI 0.574–1.343, log-rank P = 0.55). (G) Response rate in tumors with <1% PD-L1 expression (n = 28) or ≥1% PD-L1 expression (n = 23), P > 0.99. (H) Progression-free survival in tumors with <1% PD-L1 expression (n = 29) or ≥1% PD-L1 expression (n = 22) (HR 1.370, 95% CI 0.761–2.466, log-rank P = 0.29). (I) Overall survival in tumors with <1% PD-L1 expression (n = 30) or ≥1% PD-L1 expression (n = 21) (HR 1.747, 95% CI 0.913–3.342, log-rank P = 0.084). Statistical analysis for response rate used Fisher’s exact test and statistical analysis for Kaplan–Meier plots used the log-rank test. CI, confidence interval; CR, complete response; HR, hazard ratio; PD-L1, programed death-ligand 1; PR, partial response; SD, stable disease; PD, progressive disease.
We also evaluated whether tumor PD-L1 expression was associated with response to ICIs in 73 cases for which staining was available. First, we observed in agreement with published literature [22], that there was no difference in PD-L1 expression by EGFR allele (supplementary Figure S4A, available at Annals of Oncology online). We also noted that PD-L1 expression did not correlate to smoking status in EGFR mutant cases. There was no association between the efficacy of ICIs in tumors with ≥1% or <1% PD-L1 positive staining (ORR: 3 of 23, 13%, versus 4 of 28, 14%, P > 0.99) (PFS: HR 1.370, 95% CI 0.761–2.466, log-rank P = 0.29) (OS: HR 1.747, 95% CI 0.913–3.342, log-rank P = 0.084) in EGFRΔ19and EGFRL858R cases (Figure 2G–I, supplementary Figure S4B, available at Annals of Oncology online), irrespective of EGFR subtype (supplementary Figure S4C–I, available at Annals of Oncology online). In EGFRΔ19and EGFRL858R tumors, we also noted no association between the efficacy of ICIs and PFS or OS in patients with ≥50% (n = 4) or <50% (n = 47) tumor PD-L1 expression, although this comparison was underpowered to make a conclusive association. Due to lack of TMB data in PD-L1 stained cases, we were unable to assess the correlation between TMB and PD-L1 expression, but we acknowledge previous studies in lung cancer showing that PD-L1 expression and TMB are largely uncorrelated [23–25].
EGFRΔ19 mutant lung cancers have a lower tumor mutation burden compared with EGFRL858R mutant lung cancers
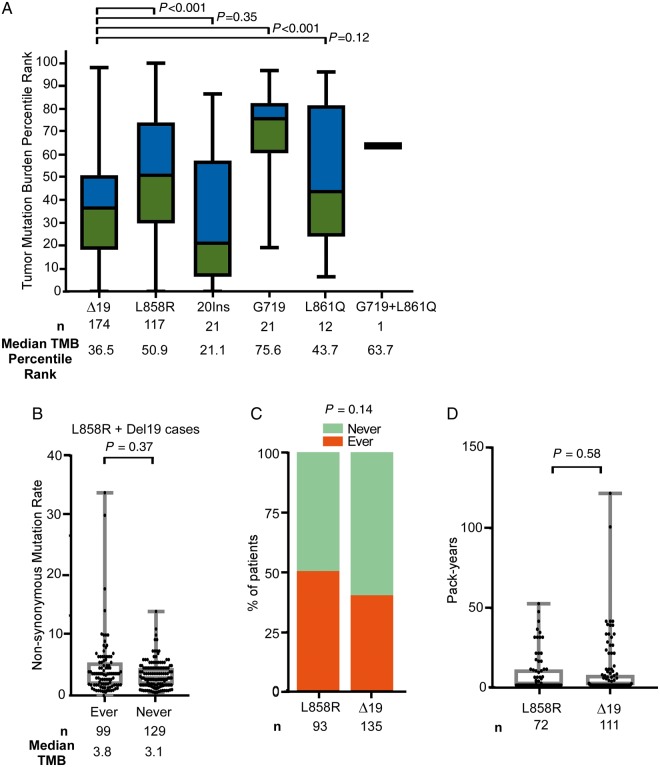
Due to the reported association between TMB and response to ICIs, we investigated the TMB across EGFR mutation subtypes in lung cancer [26]. A lack of sequencing data available from our cohort of 171 EGFR mutant tumors treated with immunotherapy led us to compile data from a cohort of 383 sequenced cases of EGFR mutant lung cancer from YCC, MSKCC, and TCGA, irrespective of treatment history (Table 2). Across all EGFR mutation subtypes, the median TMB was 3.8 non-synonymous mutations/megabase (Mb) with a mean TMB of 5.6 non-synonymous mutations/Mb. This is notably less than the median TMB observed in unselected NSCLC cases (7.4 non-synonymous mutations/Mb by MSK-IMPACT) and the TMB cut-off associated with improved outcomes with immunotherapy in NSCLC (10 non-synonymous mutations/Mb) [21, 25, 27]. TMB was significantly lower in EGFRΔ19 tumors compared with EGFRL858R tumors (Figure 3A, supplementary Figure S5, available at Annals of Oncology online). EGFRΔ19mutant tumors had similar TMB compared with EGFR20Ins (P = 0.35) and EGFRL861Q tumors, while the TMB in the EGFRG719 group was higher than in EGFRΔ19tumors (P < 0.001) (Figure 3A).
Table 2.
Characteristics of cases included in the tumor mutation burden analysis
| Characteristics | EGFRΔ19 | EGFRL858R | EGFR20Ins | EGFRG719 | EGFRL861Q | EGFRG719 + EGFRL861Q | EGFROther | All EGFR cases |
|---|---|---|---|---|---|---|---|---|
| Yale Cancer Center | ||||||||
| Number of cases with TMB | 12 | 5 | 0 | 0 | 0 | 0 | 0 | 17 |
| TMB median | 1.8 | 2.5 | n/a | n/a | n/a | n/a | n/a | 2.0 |
| TMB range | 0.1–4.1 | 2.0–4.1 | n/a | n/a | n/a | n/a | n/a | 0.1–4.1 |
| Smoking (ever/never)—no. (%) | 7/5 (58.3/41.7) | 4/1 (80/20) | n/a | n/a | n/a | n/a | n/a | 11/6 (64.7/35.3) |
| Smoking (ever/never)—data not available—no. (%) | 0 (0) | 0 (0) | n/a | n/a | n/a | n/a | n/a | 0 (0) |
| Smoking (pack-year)—range | 0–120 | 0–30 | n/a | n/a | n/a | n/a | n/a | 0–120 |
| Smoking (pack-year)—median | 1.5 | 10 | n/a | n/a | n/a | n/a | n/a | 4.5 |
| Smoking (pack-year)—data not available—no. (%) | 0 (0) | 0 (0) | n/a | n/a | n/a | n/a | n/a | 0 (0) |
| Memorial Sloan Kettering Cancer Center | ||||||||
| Number of cases with TMB | 139 | 90 | 19 | 18 | 9 | 1 | 37 | 313 |
| TMB median | 3.8 | 4.7 | 2.8 | 7.3 | 3.8 | 5.7 | 11.3 | 4.1 |
| TMB range | 0.9–30.2 | 0.9–17.9 | 0.9–9.2 | 2.8–22.6 | 1.9–10.2 | n/a | 0.9–91.8 | 0.9–91.8 |
| Smoking (ever/never)—no. (%) | 39/62 (28.1/44.6) | 28/40 (31.1/44.4) | 3/10 (15.8/52.6) | 12/2 (66.7/11.1) | 6/2 (66.7/22.2) | 1/0 (100/0) | 13/7 (35.1/18.9) | 102/123 (32.6/39.3) |
| Smoking (ever/never)—data not available—no. (%) | 38 (27.3) | 22 (24.4) | 6 (31.6) | 4 (22.2) | 1 (11.1) | 0 (0) | 17 (45.9) | 88 (28.1) |
| Smoking (pack-year)—range | 0–99 | 0–51 | 0–67.5 | 0–47.3 | 0–15 | n/a | 0–108 | 0–108 |
| Smoking (pack-year)—median | 0 | 0 | 0 | 6.3 | 6.5 | 30 | 18.5 | 0 |
| Smoking (pack-year)—data not available—no. (%) | 40 (28.8) | 23 (25.6) | 6 (31.6) | 4 (22.2) | 1 (11.1) | 0 (0) | 17 (45.9) | 91 (29.1) |
| The Cancer Genome Atlas | ||||||||
| Number of cases with TMB | 23 | 22 | 2 | 3 | 3 | n/a | n/a | 53 |
| TMB median | 1.3 | 1.6 | 1.5 | 2.2 | 3.0 | n/a | n/a | 1.4 |
| TMB range | 0.7–11.9 | 0.7–33.9 | 1.3–1.7 | 1.0–3.0 | 1.3–6.3 | n/a | n/a | 0.7–33.9 |
| Smoking (ever/never)—no. (%) | 7/15 (30.4/65.2) | 14/6 (63.6/27.3) | 0/1 (0/50) | 3/0 (100/0) | 2/1 (66.7/33.3) | n/a | n/a | 26/23 (49.1/43.4) |
| Smoking (ever/never)—data not available—no. (%) | 1 (4.3) | 2 (9.1) | 1 (50) | 0 (0) | 0 (0) | n/a | n/a | 4 (7.5) |
| Smoking (pack-year)—range | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Smoking (pack-year)—median | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Smoking (pack-year)—data not available—no. (%) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TMB, tumor mutation burden; n/a, not applicable.
Figure 3.
Characterization of EGFR allele-specific tumor mutation burden (TMB) and smoking history. (A) TMB was calculated for EGFR mutant tumors harboring deletions in exon 19 [(Δ19) (n = 174)], mutations in exon 21 [L858R (n = 117) and L861Q (n = 12)], insertions in exon 20 [(20ins) (n = 21)], mutations in exon 18 [(G719) (n = 21)], or co-mutations at positions G719 and L861Q (n = 1). Data were combined from Memorial Sloan Kettering Cancer Center, the Yale Cancer Center, and The Cancer Genome Atlas cohorts. Data were transformed within each cohort to within-cohort percentile rank to permit unified analysis, and median TMB percentile rank is indicated. (B) TMB in EGFRL858Rand EGFRΔ19 mutant tumors from patients with ever (n = 99) or never (n = 129) smoking status (median 3.8 versus 3.1, P = 0.37). (C) Percentage of ever and never smokers within the EGFRL858Rand EGFRΔ19 mutant tumors groups (P = 0.14). (D) Pack-years in EGFRL858Rand EGFRΔ19 mutant tumors groups (P = 0.58). Statistics were calculated using the Fisher’s exact test.
We examined whether smoking history accounted for the differences in TMB in each allele. As expected, there was an association between ever smoking status and higher TMB in all EGFR mutant tumors (data not shown), but this was less evident when interrogating only EGFRL858Rand EGFRΔ19cases (Figure 3B). Smoking status and pack-years were not different based on the specific EGFR allele (Figure 3C and D) suggesting that there is a difference in TMB between the two most common genetic subtypes of EGFR mutant lung cancer that is not simply reflective of differential smoking exposure.
Discussion
Despite the success of EGFR TKIs in EGFR mutant lung cancer, all patients eventually develop acquired resistance to these therapies. ICIs have recently emerged as a therapeutic approach in lung cancer with the potential for durable responses but current data suggest that there is limited efficacy in EGFR-driven cancers [13–16]. For example, the ImmunoTarget group assessed response to ICIs across various molecular subgroups of lung cancer and found that tumors with KRAS, BRAF, or MET exon 14 alterations were more likely to derive benefit than cases with EGFR, ALK, and RET alterations [28, 29, 30]. Yet, some EGFR mutant tumors do respond to ICIs [18, 19]. In this study, we assembled the largest cohort of EGFR mutant cases treated with ICIs to retrospectively interrogate how genetic, molecular, and clinical factors impact response and survival in this subset of lung cancer. Using this multi-institutional collection of patients, we identified allele-specific differences in response to immune checkpoint inhibition. EGFRL858R tumors had a similar response rate and OS outcomes to an EGFR wild-type lung cancer population, while EGFRΔ19cases did substantially worse. Of note, we did observe substantially worse PFS between both EGFRL858Rand EGFRΔ19lung cancer cases compared with EGFR wild-type lung cancer cases. The underlying cause for this discrepancy is unknown, but it may be reflective of the variable scanning intervals represented by this multi-institutional cohort composed of both on-trial and off-trial cases. A recent report evaluating outcomes of 27 patients with EGFR mutant tumors on ICIs found the best ORR in cases with less common EGFR alterations, such as G719X and exon 20 insertions, highlighting potential differences between EGFR alleles [31].
The outcomes on ICIs contrast with those on EGFR TKIs, where EGFRL858Rtumors have a worse durability of response to EGFR TKIs compared with EGFRΔ19 tumors, highlighting the context specificity of genotypic responses to different therapeutic agents [32–34]. One limitation of our study was the lack of sufficient sequencing data to directly compare TMB to response in our cohort of 171 EGFR mutant patients treated with ICI. To address this, we employed a separate cohort of 383 EGFR mutant cases with sequencing data available in which we found that EGFRΔ19 tumors had substantially fewer non-synonymous mutations compared with EGFRL858Rtumors [35] aligning with the immunotherapy response data. At present, it is unclear what is driving the difference in the TMB between these alleles. It is possible that the increased mutation burden in EGFRL858Rtumors reflects the generally more advanced age of patients with EGFRL858R at diagnosis compared with patients with EGFRΔ19alterations [36, 37]. This association would suggest that a clock-like mutational process is at play in EGFR mutant tumors, but additional studies are needed to validate this hypothesis [38]. In addition, recent work has found that p53 alterations are associated with EGFR mutant lung cancer with higher TMB possibly suggesting a more genetically unstable and aggressive tumor state [35].
We also found that outcomes of patients treated with ICIs were not affected by EGFRT790M status or PD-L1 expression levels before immunotherapy. Although we found that fewer prior lines of therapy were associated with increased response to ICI, we unequivocally support the guidance that EGFR TKIs should be the preferred first line treatment option for patients with EGFR mutant lung cancer (irrespective of TMB or PD-L1). This guidance is based on substantially higher response rates to EGFR TKIs, the overall low rates of response to PD-(L)1 blockade in this subset of lung cancer, lack of efficacy of PD-L1 blockade in PD-L1+, TKI naïve, EGFR mutant lung cancer [16], and risk of synergistic toxicity with initial PD-1 blockade followed by osimertinib [39, 40].
This study combined data from multiple institutions, which has advantages and disadvantages. A major advantage is that by pooling data we were able to examine a larger cohort than we would have done individually; however, there is also heterogeneity in the analytical tools used at different institutions, although we aimed to normalize data to the size of the exome sequenced. Another possible limitation of this study was the inclusion of cases treated with different single agent ICIs [e.g. PD-1 (n = 140) and PD-L1 (n = 15)] or combinations of ICIs [e.g. PD-1 + CTLA-4 (n = 15) or PD-L1 + CTLA-4 (n = 1)]. It is possible that these treatment subsets might display unique survival outcomes that are masked by combining the cases.
In summary, our analysis revealed that EGFR mutant tumors have differing responses to ICIs and underlying molecular profiles. These data serve as a foundation for further investigating which patients with EGFR mutant disease have a higher likelihood of benefitting from immunotherapies, in particular when combined with chemotherapy or antiangiogenesis agents. Studies in animal models of EGFR mutant lung cancer with varying baseline mutations and TMB will also be valuable tools for evaluating such approaches. More broadly, our data provide rationale for evaluating genomic and molecular subsets within tumor types with lower TMB to better understand which features are associated with successful outcomes with ICIs.
Funding
This work was supported by National Institutes of Health/National Cancer Institute (NIH/NCI) grants P50CA196530 (RSH, KP, SG, SBG, DZ), P01CA129243 (ML), R01CA195720 (KP), R01CA208403 (EBG), and F32CA210516 (KH), the Leslie H. Warner Fellowship to KH, the Italian Association for Cancer Research (AT), a Department of Defense Lung Cancer Research Program Idea Award W81XWH-17-1-0351 (KP), the Diane and David Heller Foundation, the Ginny and Kenneth Grunley Fund for Lung Cancer Research, the Brown Performance Group Fund for Innovation in Cancer Informatics (FSV), and the Druckenmiller Center for Lung Cancer Research at MSKCC. YCC and MSKCC shared resources used in this manuscript were in part supported by NIH/NCI Cancer Center Support Grants P30CA016359 and P30CA008748. MDH is a Damon Runyon Clinical Investigator supported in part by the Damon Runyon Cancer Research Foundation (CI-98-18) and is a member of the Parker Institute for Cancer Immunotherapy. Gilead Sciences, Inc. supported the sequencing of a subset of the Yale Cancer Center specimens.
Disclosure
HAY is a consultant for AstraZeneca and has received travel support from Eli Lilly. Her institution, Memorial Sloan Kettering has received research funding from Astellas Pharma, AstraZeneca, Daiichi, Eli Lilly, Novartis, and Pfizer for clinical trials she is involved in. She is listed as an inventor on a patent application submitted for pulsatile use of erlotinib to treat or prevent brain metastases. AL receives research support (to UCLA) from Daiichi, Calithera, Dracen, and AstraZeneca; is on the advisory board for AstraZeneca and Bristol–Myers Squibb; is also a consultant for Leica Biosystems and his wife is an employee of and owns stock from Boston Scientific. AT is currently an employee of MSD, however, these studies were conducted when she was at Yale. BSH is a consultant for Boehringer Ingelheim and previously owned stock in Abbvie. GJR receives collaborative research funding (to MSKCC) from Novartis, Roche, Pfizer, Mirati, Merck, and Takeda; has consulted for Merck and Roche; and has received travel support from Merck. MEA has received speaker and consulting fees from Invivoscribe and Biocartis. ML receives research support from LOXO Oncology and Helsinn Therapeutics, and he is an ad hoc advisory board member at AstraZeneca, Bristol-Myers Squibb, Merck, Takeda, and Bayer. RSH receives research support from AstraZeneca, Eli Lilly and Company, and Merck and Company; is a paid consultant to Abbvie Pharmaceuticals, ARMO Biosciences, AstraZeneca, Biodesix, Bristol-Myers Squibb, Eli Lilly and Company, EMD Serrano, Genentech/Roche, Genmab, Halozyme, Heat Biologics, Infinity Pharmaceuticals, Loxo Oncology, Merck and Company, Nektar, Neon Therapeutics, NextCure, Novartis, Pfizer, Sanofi, Seattle Genetics, Shire PLC, Spectrum Pharmaceuticals, Symphogen, Tocagen and Tesaro; is a scientific advisory board member at Neon Therapeutics, Infinity Pharmaceuticals, and NextCure; is a board member (non-executive/independent) at Junshi Pharmaceuticals. SBG receives research funding from AstraZeneca and research support (to Yale) from MedImmune, AstraZeneca, Spectrum, Genentech, Pfizer, Bristol–Myers Squibb, and Immunogen, and is a paid consultant for AstraZeneca, Boehringer Ingelheim, Bristol–Myers Squibb, Eli Lilly, Genentech, Amgen, and Spectrum. MMA has received research funding from Bristol-Myers Squibb, Eli Lilly, Genentech, and AstraZeneca, and he is a compensated consultant or has received honoraria from Abbvie, Ariad, AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Merck, Genentech, Syndax, Nektar, Blueprint, and Maverick. JK is a consultant for Arterys. EBG has received research support (to UCLA) from AstraZeneca, Bristol-Myers Squibb, Genentech, Eli Lilly, Merck, Neon Pfizer, Dynavax, Iovance, Mirati, Novartis, and has received honoraria from Dracen. SG has received research support (to Yale) from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Iovance, and is a consultant for Bristol-Myers Squibb and Nektar. MDH receives research funding from Bristol-Myers Squibb; is paid consultant to Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Janssen, Nektar, Syndax, Mirati, and Shattuck Labs; receives travel support/honoraria from AstraZeneca and Bristol-Myers Squibb; and a patent has been filed by MSK related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from PGDx. KP receives or has received research funding through her institution from AstraZeneca, Kolltan, Roche, Gilead, and Symphogen; consults or receives honoraria from Takeda, NCCN, Novartis, Merck, AstraZeneca, Tocagen, Maverick Therapeutics, and Dynamo Therapeutics; and is a co-inventor on a patent licensed to Molecular MD for EGFR T790M mutation testing (through MSKCC). All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Lynch TJ, Bell DW, Sordella R. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350(21): 2129–2139. [DOI] [PubMed] [Google Scholar]
- 2. Paez JG, Janne PA, Lee JC. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304(5676): 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3. Pao W, Miller V, Zakowski M. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004; 101(36): 13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maemondo M, Inoue A, Kobayashi K. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362(25): 2380–2388. [DOI] [PubMed] [Google Scholar]
- 5. Mitsudomi T, Morita S, Yatabe Y. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010; 11(2): 121–128. [DOI] [PubMed] [Google Scholar]
- 6. Rosell R, Moran T, Queralt C. et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361(10): 958–967. [DOI] [PubMed] [Google Scholar]
- 7. Lee CK, Wu YL, Ding PN. et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus chemotherapy in EGFR-mutant lung cancer: a meta-analysis. J Clin Oncol 2015; 33(17): 1958–1965. [DOI] [PubMed] [Google Scholar]
- 8. Yang JC, Wu YL, Schuler M. et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015; 16(2): 141–151. [DOI] [PubMed] [Google Scholar]
- 9. Pao W, Miller VA, Politi KA. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2(3): e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi S, Boggon TJ, Dayaram T. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352(8): 786–792. [DOI] [PubMed] [Google Scholar]
- 11. Thress KS, Paweletz CP, Felip E. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015; 21(6): 560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akbay EA, Koyama S, Carretero J. et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013; 3(12): 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borghaei H, Paz-Ares L, Horn L. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015; 373(17): 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garon EB, Rizvi NA, Hui R. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015; 372(21): 2018–2028. [DOI] [PubMed] [Google Scholar]
- 15. Lee CK, Man J, Lord S. et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol 2018; 4(2): 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lisberg A, Cummings A, Goldman JW. et al. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor (TKI) naive patients with advanced NSCLC. J Thorac Oncol 2018; 13(8): 1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gainor JF, Shaw AT, Sequist LV. et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016; 22(18): 4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garassino MC, Cho BC, Kim JH. et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol 2018; 19(4): P521–P536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reck MSM, Cappuzzo F, Orlandi F. et al. Primary PFS and safety analyses of a randomized phase III study of carboplatin + paclitaxel +/− bevacizumab, with or without atezolizumab in 1L non-squamous metastatic NSCLC (IMPOWER150). Ann Oncol 2017; 28(Suppl 11): mdx760.002. [Google Scholar]
- 20. Yu HA, Suzawa K, Jordan E. et al. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res 2018; 24(13): 3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rizvi H, Sanchez-Vega F, La K. et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018; 36(7): 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toki MI, Mani N, Smithy JW. et al. Immune marker profiling and programmed death ligand 1 expression across NSCLC mutations. J Thorac Oncol 2018; 13(12): 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carbone DP, Reck M, Paz-Ares L. et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017; 376(25): 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hellmann MD, Callahan MK, Awad MM. et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with Ipilimumab in small-cell lung cancer. Cancer Cell 2018; 33: 853–861.e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hellmann MD, Ciuleanu TE, Pluzanski A. et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018; 378(22): 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizvi NA, Hellmann MD, Snyder A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348(6230): 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ready N, Hellmann MD, Awad MM. et al. First-line nivolumab plus Ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019; 37(12): 992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazieres J, Drilon AE, Mhanna L. et al. Efficacy of immune-checkpoint inhibitors (ICI) in non-small cell lung cancer (NSCLC) patients harboring activating molecular alterations (ImmunoTarget). J Clin Oncol 2018; 36(Suppl 15): 9010. [Google Scholar]
- 29. Mazieres J, Drilon A, Lusque A. et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019; 30(8): 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gautschi O, Drilon A, Milia J. et al. MA04.03 immunotherapy for non-small cell lung cancers (NSCLC) with oncogenic driver mutations: new results from the global IMMUNOTARGET registry. J Thorac Oncol 2018; 13(10): S367. [Google Scholar]
- 31. Yamada T, Hirai S, Katayama Y. et al. Retrospective efficacy analysis of immune checkpoint inhibitors in patients with EGFR-mutated non-small cell lung cancer. Cancer Med 2019; 8(4): 1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riely GJ, Pao W, Pham D. et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12(3 Pt 1): 839–844. [DOI] [PubMed] [Google Scholar]
- 33. Paz-Ares L, Tan EH, O'Byrne K. et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: overall survival data from the phase IIb LUX-Lung 7 trial. Ann Oncol 2017; 28(2): 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, Sheng J, Kang S. et al. Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: a meta-analysis. PLoS One 2014; 9(9): e107161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Offin M, Rizvi H, Tenet M. et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res 2018; 25(3): 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dogan S, Shen R, Ang DC. et al. Molecular epidemiology of EGFR and KRAS mutations in 3, 026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012; 18(22): 6169–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee B, Lee T, Lee SH. et al. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6, 595 lung cancers. Oncotarget 2016; 7(17): 23874–23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alexandrov LB, Jones PH, Wedge DC. et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015; 47(12): 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oshima Y, Tanimoto T, Yuji K, Tojo A. EGFR-TKI-associated interstitial pneumonitis in nivolumab-treated patients with non-small cell lung cancer. JAMA Oncol 2018; 4(8): 1112–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schoenfeld AJ, Arbour KC, Rizvi H. et al. Severe immune-related adverse events are common with sequential PD-(L)1 blockade and osimertinib. Ann Oncol 2019; 30(5): 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.