Abstract
Diatoms are unicellular algae and evolved by secondary endosymbiosis, a process in which a red alga-like eukaryote was engulfed by a heterotrophic eukaryotic cell. This gave rise to plastids of remarkable complex architecture and ultrastructure that require elaborate protein importing, trafficking, signaling and intracellular cross-talk pathways. Studying both plastids and mitochondria and their distinctive physiological pathways in organello may greatly contribute to our understanding of photosynthesis, mitochondrial respiration and diatom evolution. The isolation of such complex organelles, however, is still demanding, and existing protocols are either limited to a few species (for plastids) or have not been reported for diatoms so far (for mitochondria). In this work, we present the first isolation protocol for mitochondria from the model diatom Thalassiosira pseudonana. Apart from that, we extended the protocol so that it is also applicable for the purification of a high-quality plastids fraction, and provide detailed structural and physiological characterizations of the resulting organelles. Isolated mitochondria were structurally intact, showed clear evidence of mitochondrial respiration, but the fractions still contained residual cell fragments. In contrast, plastid isolates were virtually free of cellular contaminants, featured structurally preserved thylakoids performing electron transport, but lost most of their stromal components as concluded from Western blots and mass spectrometry. Liquid chromatography electrospray-ionization mass spectrometry studies on mitochondria and thylakoids, moreover, allowed detailed proteome analyses which resulted in extensive proteome maps for both plastids and mitochondria thus helping us to broaden our understanding of organelle metabolism and functionality in diatoms.
Keywords: Chloroplast, Organelle isolation, Photosynthesis, Proteomics, Respiration, Thylakoids
Introduction
Diatoms (Bacillariophyceae) are unicellular, photosynthetic algae that represent one of the most predominant and diverse phytoplankton groups in marine and freshwater ecosystems, making them equally relevant for aquatic biomass production and marine carbon fixation (Round et�al. 2007). The complex architecture of diatom plastids reflects their convoluted evolution by secondary endosymbiosis, in which a eukaryotic red alga progenitor, that already possessed plastids, was taken up by a eukaryotic host cell. Such secondary plastids of diatoms strongly differ from primary plastids of green algae, land plants, red algae and glaucophytes, because they possess four instead of two plastid envelope membranes, with the outermost being connected to the endoplasmic reticulum (Cavalier-Smith 2000). Depending on the species, plastids of diatoms can be highly diverse in number and size. Centric diatoms possess plastids ranging from 2 to 3 per cell (like in Thalassiosira pseudonana) up to several hundreds (as in Odontella sinensis) (Round et�al. 2007). The thylakoids of diatoms are typically organized in membrane stacks of three that are arranged in parallel in a highly ordered manner, usually surrounding a pyrenoid (Cavalier-Smith 2000). Diatom thylakoids also do not show the spatial separation of grana and stroma lamellae that is characteristic for primary plastids of the green lineage (Andersson and Anderson 1980). Furthermore, each plastid possesses a so-called ‘girdle lamella’, a triple thylakoid structure that encircles the center thylakoids being located close to the four plastid envelope membranes (Cavalier-Smith 2000).
Diatom mitochondria, in contrast, derived directly from the secondary host cell (Martin et�al. 2015, Roger et�al. 2017). Prevailing in all eukaryotes, mitochondria are ascribed to a single endosymbiotic event, in which an α-proteobacterial endosymbiont may have been integrated into an archaeal host that is also the progenitor of eukaryotes (Martin et�al. 2015, Roger et�al. 2017). Typically, diatom mitochondria sizes range from approximately 0.5 to 1 �m in diameter and they feature tubular-type cristae (Werner 1977). The absolute number per cell, however, differs between diatom groups and species. Thus, T. pseudonana, like other centric diatoms, possesses few oval mitochondria per cell, while pennate diatoms like Phaeodactylum tricornutum have tubular-like mitochondria of variable shape (e.g. visible in R�o B�rtulos et�al. 2018). Interestingly, the mitochondria of diatoms possess some unique metabolic features, like a partial mitochondrial glycolysis, that is only found in stramenopiles (R�o B�rtulos et�al. 2018), and the bacterial Entner–Doudoroff pathway (Fabris et�al. 2012). Along with their conventional role in energy maintenance, diatom mitochondria are believed to play an important role in the regulation of the carbon flux. Apparently, mitochondria and plastids are tightly coupled metabolically by constantly shuttling energy and reducing equivalents between both organelles (Bailleul et�al. 2015), which may be facilitated by some spatial interconnections between diatom mitochondria and plastids (Flori et�al. 2017).
Albeit the first diatom genome was sequenced in 2004 (Armbrust et�al. 2004), and numerous elaborate biochemical and molecular studies on diatoms have been performed (Kroth et�al. 2008, Prihoda et�al. 2012), diatom plastids and mitochondria are still poorly understood, not least because of a lack of methods to study isolated organelles. Therefore, isolation or enrichment of organelles is one missing tool to broaden our understanding of diatom photosynthesis, mitochondrial respiration, organellar metabolic pathways, metabolite shuttling, organellar DNA and RNA as well as protein import. Additionally, organelle fractionation techniques may render possible potential ‘-omics’ approaches such as metabolomics, lipidomics, glycomics or proteomics that may lead to a more conclusive picture when performed on organelles instead of on complex cellular systems.
In this article, we report the first organelle isolation protocol, which is applicable for the isolation of high-quality plastids and mitochondria fractions from the model diatom T. pseudonana. Obtained fractions were analyzed thoroughly by selected physiological and molecular methods in order to assess the quality of the isolates. We further provide refined sets of proteome data that might serve as references for further studies on diatom physiology and evolution and add experimental evidence for the existing organelle targeting prediction pipelines.
Results
Mitochondria and plastids isolation from T. pseudonana
The protocol presented here is modified from a plastid isolation technique published previously (Schober et�al. 2018). By utilizing the mutant line TIG19 (preTPI-GAPDH::GFP; see Supplementary Fig. S1 and Materials and Methods section) that expresses and targets GFP to the mitochondria, we were able to discriminate mitochondria, plastids and other organelles based both on mitochondria-targeted GFP and plastid derived chlorophyll fluorescence. This allowed us to establish a routine workflow that assures high and reproducible fraction purities (Fig. 1).
Fig. 1.
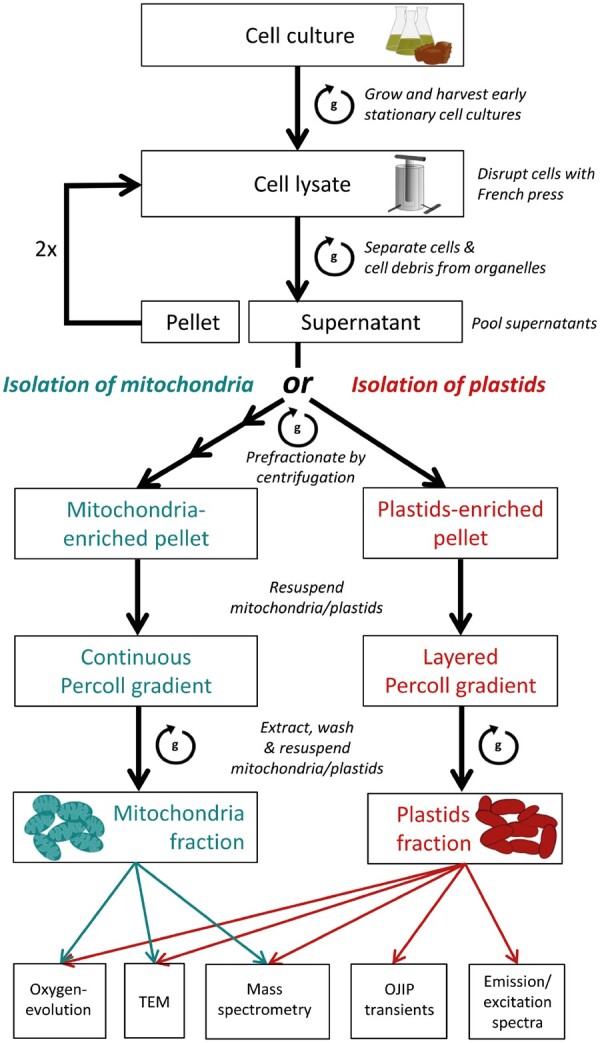
Workflow of mitochondria or plastids isolation from T. pseudonana. Thalassiosira pseudonana Tig19 cells were harvested by centrifugation and disrupted at a constant working pressure of 90 MPa using a French Press. Unopened cells and large cell fragments were sedimented by a gentle centrifugation step at 300�g and again passed twice through the French Press. All resulting supernatants were pooled (= crude organelle suspensions) and purified from residual cells and large-sized contaminants by differential centrifugation. The crude organelle extract was then either loaded to a continuous (mitochondria isolation) or a layered Percoll density gradient (plastids isolation) in order to purify the organelles from persistent contaminants. Mitochondria or plastids of the highest quality, as assessed by epifluorescence microscopy, where extracted from the gradient and organelles were washed by centrifugation in a large volume of reaction buffer to remove residual Percoll. The quality of final mitochondria or PF were then assessed by the methods listed below.
Based on our experience, best organelle separation and highest metabolic rates were achieved when early-stationary phase cultures of T. pseudonana with 5–7 � 106 cells/ml and an electrical volume diameter of 4.7–5.3 �m (Coulter Counter) were used (best working culture parameters are given in the Methods section and Supplementary Fig. S1). These culture parameters in combination with a constant French Press working pressure of 90 MPa during cell rupture were found to be crucial for preserving the structural integrity of the organelles (Fig. 2A, B).
Fig. 2.
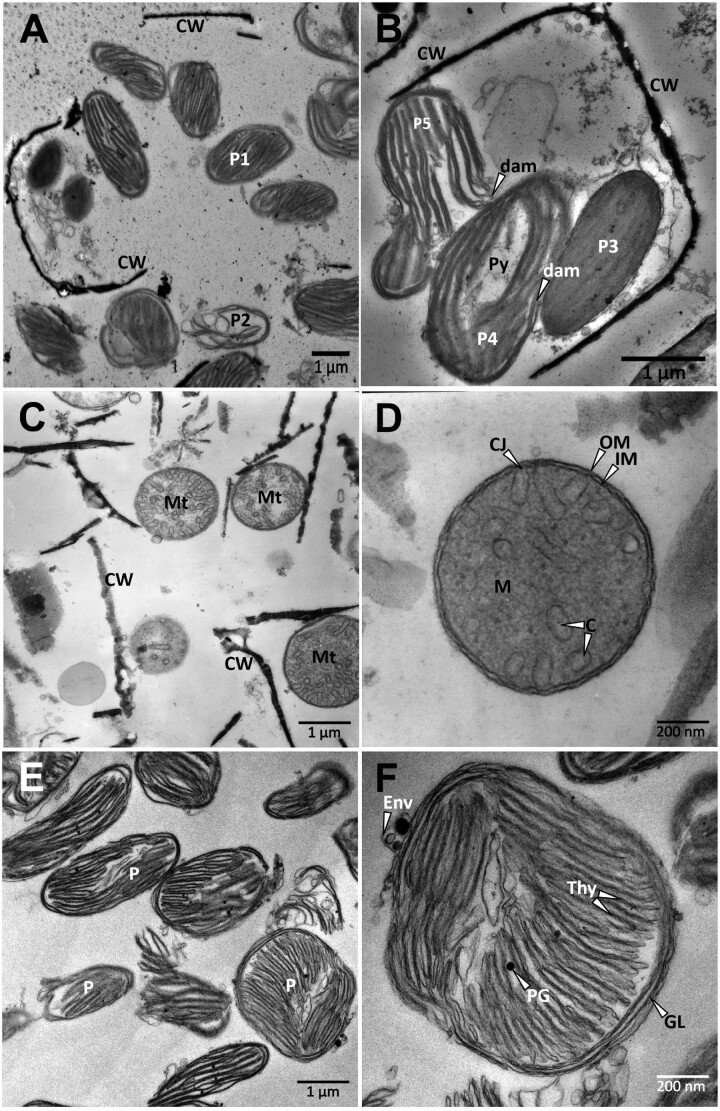
Transmission electron micrographs of French Press eluate and the final mitochondria and PF. (A, B) Representative image of French Press eluate. (A) Eluate comprises residual cell debris and plastids of various qualities. (B) Representation of different plastid qualities as resulted upon cell rupture with a French Press. P1–5, stages of plastid damages; Py, pyrenoid; CW, cell wall; dam, organelle damages. (C) Representative overview of the final mitochondria fraction. Mt, mitochondrion; CW, cell wall. (D) View of a single mitochondrion. C, cristae; CJ, cristae junction; IM, inner membrane; OM, outer membrane; M, matrix. (E) Representative overview of the final PF. P, plastid (F) View of a single isolated plastid. Env, envelope membrane; Thy, thylakoids; PG, plastoglobule; GL, girdle lamella.
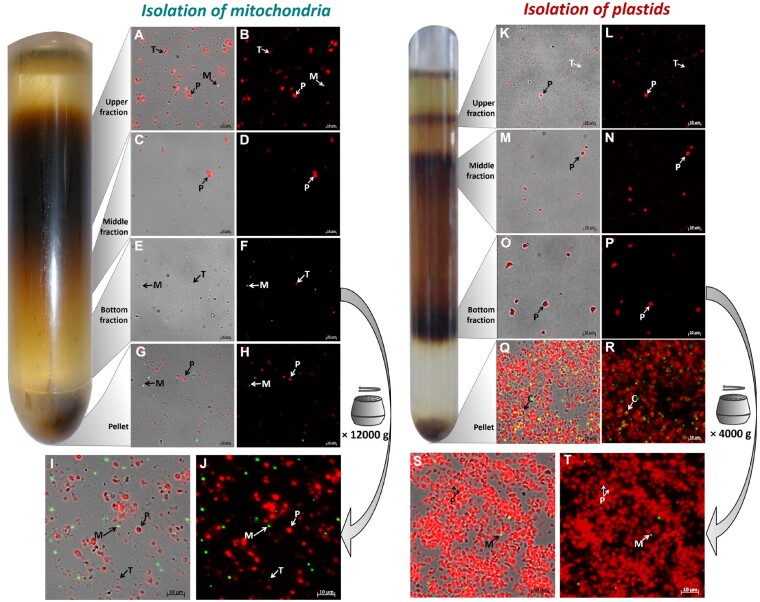
The majority of plastids in Fig. 2A (e.g. plastid P1) still looked vital, while plastids like P2 seem to be ruptured. All stages of plastid damages can be seen in Fig.�2B. The consistent dark color of P3 (Fig. 2B) suggests that the plastid contained its stroma. Some plastids (Fig. 2B, P4), in contrast, lost stroma and pyrenoids, whereas the plastid P5 (Fig. 2B) completely broke apart (arrow). To remove residual cellular debris and to further separate intact from fractured organelles, mitochondria were loaded onto a continuous Percoll gradient, while plastids were separated on a three-phased layered Percoll gradient, respectively. After centrifugation, a typical band pattern harboring different organelle populations, as shown in Fig. 3 (left) for mitochondria and Fig. 3 (right) for plastids, had formed. The upper fraction of both gradients mainly harbored fragmented thylakoid membranes and tiny, mostly ruptured plastids (Fig. 3A, B and K, L), whereas the pellet predominantly consisted of residual cells and cell debris (Fig. 3G, H and Q, R). Therefore, these fractions were discarded. The middle fractions contained organelles of medium size and variable shape (Fig. 3C, D and M, N), while plastids or mitochondria of the highest homogeneity and largest size accumulated in the bottom fractions (Fig. 3E, F and O, P). The bottom fractions therefore were extracted from the gradient and were washed in order to remove residual Percoll. Fluorescence microscopy images of the resulting final fractions are shown in Fig.�3I, J (mitochondria) and Fig.�3S, T (plastids), respectively. Obviously, the final mitochondrial fraction (MF) still contained a large proportion of thylakoid fragments, as visible by the emitted chlorophyll autofluorescence (Fig. 3I, J). Separation efficiency, however, was limited by the nature of the organelle sample and could not be improved further by tested protocol modifications or purification strategies (such as different density gradient media, osmotic conditions or separation strategies).
Fig. 3.
Fluorescence microscopic analysis of plastids or mitochondria after Percoll-density centrifugation. (Left) Mitochondria separated by a continuous Percoll density gradient. Pre-fractionated cell lysate was separated by density centrifugation on a continuous Percoll gradient. Resulting bands were analyzed by fluorescence microscopy (A–H). Mitochondria that accumulated in the bottom fraction (= highest quality and purity; E, F) were extracted and subsequently washed by centrifugation at 12,000�g (I, J). (Right) Plastids separated on a layered Percoll density-gradient. Pre-fractionated cell lysate was separated by centrifugation on a layered Percoll density-gradient [from top to bottom: 10%, 20% and 30%]. Resulting bands were analyzed by fluorescence microscopy (K–R). Plastids that accumulated in the bottom fraction (= highest quality and purity; O, P) were extracted and subsequently washed by centrifugation at 4,000�g (S, T). Chlorophyll autofluorescence is shown in red. GFP fluorescence is depicted in green. M, mitochondrion; P, plastid; C, intact cell; T, thylakoid.
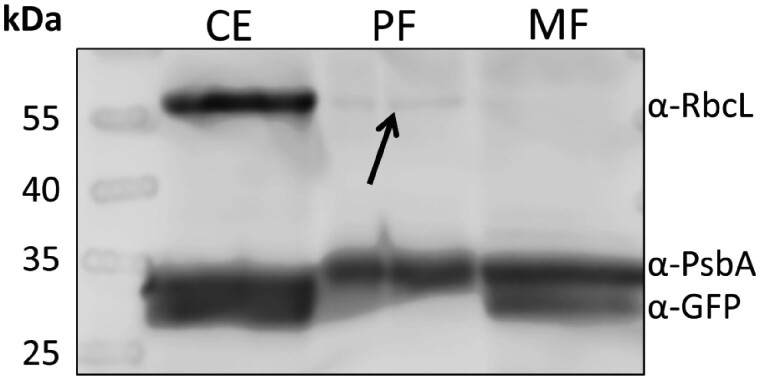
Transmission electron microscopy (TEM) of the final MF (Fig.�2C, D) and of Tig19 cells (Supplementary Fig. S2A, B) indicates that the outer and the inner mitochondrial membrane as well as the mitochondrial matrix remained structurally intact. The Western blot of the MF showed a strong signal of the D1 protein (α-PsbA, Fig. 4), which is in line with the chlorophyll autofluorescence signal (Fig. 3I, J). However, no such large and stacked thylakoid structures as in the PF fraction (Fig. 2E) could be observed in the MF fraction by TEM (Fig. 2C) indicating, that thylakoid fragments co-isolated with the mitochondria are only small and strongly disrupted parts of the thylakoid system.
Fig. 4.
Western blot analysis of the final mitochondria (MF) and plastids fraction (PF). 0.5 �g of total protein per lane were tested with three different compartment marker antibodies: α-RbcL, antibody against Rubisco large subunit [marker for plastid stroma]; α-PsbA, antibody against photosystem II protein D1 [marker for thylakoids]; α-GFP, antibody against green fluorescent protein [marker for mitochondrial matrix proteins]. CE, Tig19 cell extract; PF, plastids fraction; MF, mitochondria fraction.
The final plastids fraction (PF) contains only low amounts of mitochondrial or cellular contaminations (cell wall fragments, lipid bodies or peroxisomes) as illustrated by TEM (Fig. 2E) as well as a low emission of mitochondrial GFP fluorescence (Fig. 3S, T). Accordingly, in Western blot analyses we could not detect GFP (α-GFP, refer Fig. 4) in the PF. TEM further shows that the thylakoid membrane organization remained intact. Most plastid envelopes, though, were lost during the isolation process, while some plastids show residual parts of ruptured envelopes (Env, Fig.�2F) that remained attached to the plastid. Also, the pyrenoid was not present (Fig.�2E, F). This is in line with Western blots showing a weaker signal of the stromal marker α-RbcL (Rubisco large subunit, Fig. 4, indicated by an arrow) in the PF than in the Tig19 cell extract (CE) fraction, indicating that only a small proportion of stromal and/or pyrenoidal proteins were preserved during the isolation process. The strong signal of the D1 subunit of PSII in the PF immunoblot (α-PsbA) is in line with TEM, suggesting that the thylakoid membranes are still intact.
Oxygen consumption of the MF
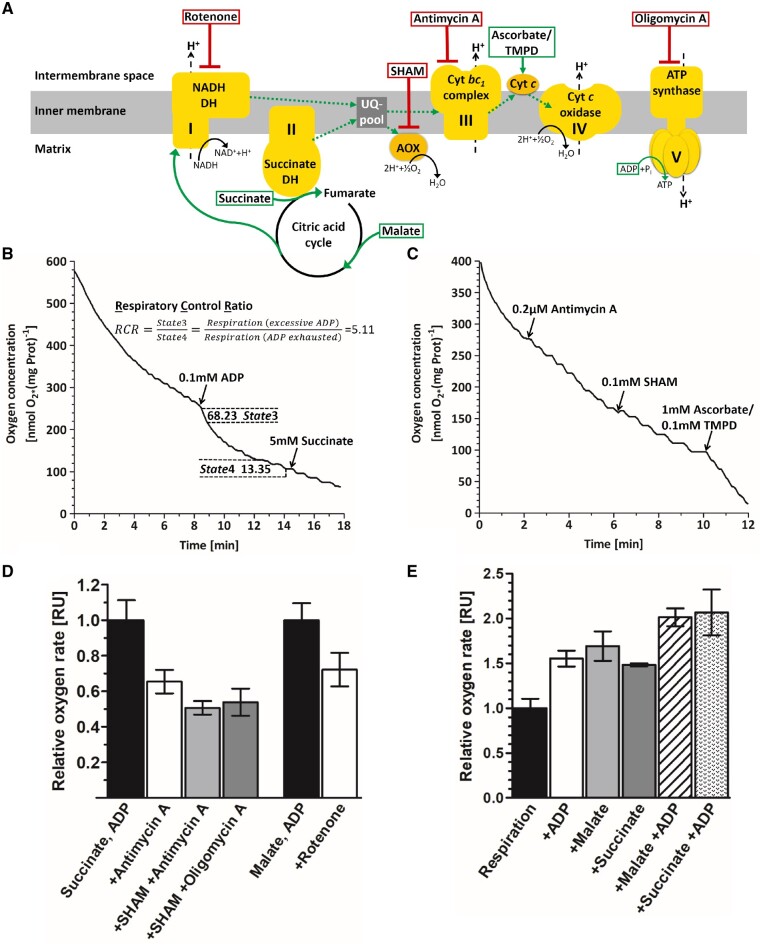
In order to assess the capacity of the MF to perform mitochondrial respiration, we measured oxygen evolution with a Clark-type oxygen electrode. To account for possible interfering oxygen production by thylakoid fragments (Fig.�3I, J), measurements were performed in complete darkness. We furthermore addressed the integrity and the coupling state of the mitochondria by estimating the respiratory control ratio (RCR) according to Chance and Williams (1955). The RCR is calculated as the ratio of active respiration (State3, not substrate-limited while ADP is available in excess) to respiration in which the ADP is exhausted (ADP-limited, State4) and rates the degree of coupling from 1 (uncoupled) to 10 (fully coupled; usually only observed in vivo) between respiration and phosphorylation (Chance and Williams 1955). In the representative RCR measurement (Fig. 5B), respiration rates were tripled from initially 22.12 nmol O2�(mg Prot�min)−1 to 68.23 nmol O2�(mg Prot�min)−1 (State3) when 0.1 mM ADP (first arrow) were added. At minute 13, most ADP was phosphorylated, leading to a drop in the oxygen rate to 13.35 nmol O2�(mg Prot�min)−1 (State4). Generally, in six independent measurements, we obtained RCR values ranging from 1.4 to 5.1, agreeing with literature values of 2–4 obtained for Aspergillus niger (Watson and Smith 1967) or 1.1–1.4 for skunk cabbage (Storey and Bahr 1969) and suggesting that most preparations showed fairly intact mitochondrial respiration being coupled to phosphorylation.
Fig. 5.
Analysis of mitochondrial respiration by Clark electrode measurements. (A) Schematic representation of the mitochondrial respiratory chain and phosphorylation complexes (yellow). The direction of the electron flow (dotted green arrows) is stimulated by different substrates (green boxes). Application of inhibitors at the corresponding site of action are indicated by red boxes. Complex I, NADH dehydrogenase; complex II, succinate dehydrogenase; complex III, cytochrome bc1 reductase; complex IV, cytochrome c reductase; complex V, ATP synthase; UQ-pool, ubiquinone pool; AOX, alternative oxidases; cyt c, cytochrome c. (B, C) Exemplary oxygen measurements. Mitochondrial respiration (solid line) is plotted as time-resolved oxygen consumption and was measured in complete darkness. (B) Addition of ADP (first arrow) uncouples phosphorylation from the mitochondrial respiratory chain, resulting in increased oxygen consumption. Reintroduction of the substrate succinate (second arrow) did not increase respiration rates, illustrating that substrates were not limited during the measurement. The respiratory control ratio (RCR = 5.11) shows tight coupling of respiration to phosphorylation. State3 (active respiration), maximum speed of respiration when both substrate and ADP are available in excess. State4 (ADP-limiting state), speed of the respiratory chain when substrate is available, but all ADP has been phosphorylated. (C) Inhibition of complex III by Antimycin A only becomes evident when Alternative oxidases (AOX) are inhibited similarly. Ascorbate and TMPD bypasses the inhibition leading to increased oxygen depletion. (D) Examination of mitochondrial respiration upon application of different inhibitors. Mitochondrial respiration was saturated with substrate (succinate or malate) and ADP before different inhibitors were applied. Data represent the means of at least two replicates � SEM. (E) Relative effects of substrates on the respiration rates normalized to endogenous respiration, data represent the means of at least two replicates � SEM.
For further functional assessment of the MF, classical electron transport chain inhibition experiments were performed (Fig. 5C, D). Inhibition of complex III with Antimycin A in the presence of succinate as a substrate (Fig. 5C; first arrow and Fig. 5D) lowered respiration rates slightly; this effect, however, could be intensified by addition of salicylhydroxamic acid (SHAM) to inhibit oxygen production by the alternative oxidases (AOX) (Fig. 5C; second arrow and Fig. 5D). Addition of ascorbate in combination with tetramethyl-p-phenylenediamine (TMPD) bypassed the inhibition, leading to a drastic increase of oxygen rates (Supplementary Fig. S4A). Respiration was also inhibited by Oligomycin A, an inhibitor of the ATP synthase (Fig.�5D), and Rotenone, inhibitor of complex I (Fig. 5D), when malate was used as a substrate. Accessory introduction of succinate, a substrate of complex II, abolished this effect (Supplementary Fig. S4B). The capability to perform mitochondrial respiration with malate as sole substrate moreover suggests the presence of a functional citric acid cycle and/or a malate/oxaloacetate shuttle, while stimulatory effects of ADP strongly suggest coupling between the respiratory chain and phosphorylation (Fig. 5E).
Oxygen evolution of the PF
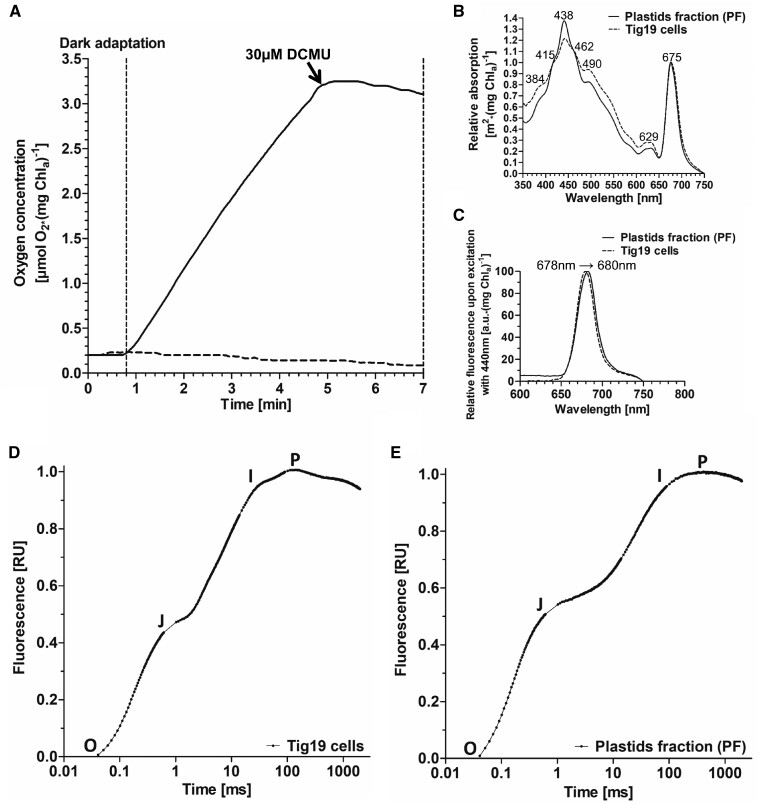
In order to test the capacity of the isolated thylakoids to perform electron transport, we investigated the light-dependent electron transport capacity (ETC) in the presence of the artificial Hill electron acceptor potassium ferricyanide (K3[Fe(CN)6]) (Fig. 6A). While the oxygen level remained stable in the dark, a rapid oxygen increase was observed when thylakoids were exposed to 200 �E m−2 s−1 (Fig. 6A, solid line) with oxygen evolution rates of up to 32 �mol O2�(mg chlorophyll�h)−1. To verify that the oxygen production measured here can exclusively be ascribed to the photosynthetic ETC, 30 �M 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added subsequently to the sample (Fig. 6A, black arrow). DCMU blocks the plastoquinone binding site of PSII, and we observed an immediate inhibition of oxygen production (Fig. 6A, solid line).
Fig. 6.
Oxygen evolution, absorption, fluorescence emission spectra and O-J-I-P fluorescence transients of isolated plastids and cells. (A) Oxygen evolution of plastids preparations. Prior to every measurement, isolated plastids were adapted to the dark for 2 min. Thirty-five micrograms of isolated plastids (corresponding to chlorophyll a) were illuminated with 200 �E m−2 s−1 in the presence of 40 mM potassium ferricyanide (solid line). DCMU treatment blocked the plastoquinone binding pocket QB which immediately abolished oxygen production. Dark control (dashed line). (B) Absorption spectrum of Tig19 cells (dashed line) compared to plastids fraction (PF; solid line). (C) Fluorescence emission spectrum of Tig19 cells (dashed line) compared to plastids fraction (PF; solid line). (D, E) O-J-I-P fluorescence transients of T. pseudonana cells and plastids fraction (PF). Untreated Tig19 cells or plastids corresponding to 1 �g/ml chlorophyll a were adapted to the dark for 10 min prior to measurements. Curves were double normalized between O and P [VOP = (FT−Fo)/Fm−Fo)] and plotted on a logarithmic scale.
Spectral and photophysiological characterization of the PF
The absorption spectrum of the PF in the visible light range (Fig. 6B, solid line) mimics well the curve characteristics observed for Tig19 cells (dotted line). Slight deviations are likely due to altered scattering and self-shading properties in the PF. The absorption shoulder at 629 nm, indicating chlorophyll c bound to the peripheral light harvesting complexes (Lepetit et�al. 2007), is not visible in the fluorescence emission spectrum (Fig.�6C), demonstrating an efficient energy transfer from chlorophyll c to chlorophyll a within the antennae. Similarly, chlorophyll a fluorescence peaking at only slightly shifted wavelengths compared to whole cells indicates energetic coupling between the light harvesting complexes and the photosystems.
Photochemical activity of the PF
In order to study the energy transfer within the PSII towards PSI, we performed O-J-I-P-fluorescence induction curves of the plastids and dark-adapted cells (Fig. 6D, E). The graphs originate in point O, in which QA is fully oxidized and hence fluorescence is very low, but rises towards J as a consequence of the saturating pulse until QA is reduced. In a functional electron transport chain, maximum fluorescence will not be reached at the J-phase, as downstream electron acceptors like the plastoquinone pool, the cytochrome b6f-complex, and the whole photosystem I site lead to a partial re-oxidation of QA during the saturating pulse. By a progressive reduction of acceptor site molecules in the following JIP phase during the ongoing saturating flash, QA will become fully reduced and maximum fluorescence level will be reached. These graph characteristics observed for the plastid fraction are very similar to those of Tig19 cells. However, the differentiation between the I and P phase, which is visible in cells (Fig. 6D) and which indicates electron transfer towards photosystem I and the final acceptors, is less pronounced in the PF (Fig. 6E). The vital parameter Qy (≙Fv/Fm), the maximum photosynthetic efficiency of photosystem II, reached approximately 70% (0.51; three independent preparations) of the intact cell value (0.73; n = 3) in the PF.
Organelle proteomics
Organelle fractionation may facilitate comparative subcellular proteomics. Therefore, MF, PF as well as Tig19 cell lysates were separated by SDS-PAGE and whole lanes were analyzed by liquid chromatography electrospray-ionization mass spectrometry (LC-ESI-MS/MS). The resulting peptide profiles were compared to an optimized protein catalog in order to provide sets of evidence-driven organellar proteomes.
Mitochondria fraction. Out of 325 proteins that have been identified within MF, 196 proteins (listed in Fig. 7A) could be assigned to functional groups that are either mitochondrial or associated to mitochondrial pathways as judged by T. pseudonana KEGG pathway maps (organism code: tps), while 130 of the detected proteins were predominantly plastidic contaminations (for details refer Supplementary Table S1).
Fig. 7.
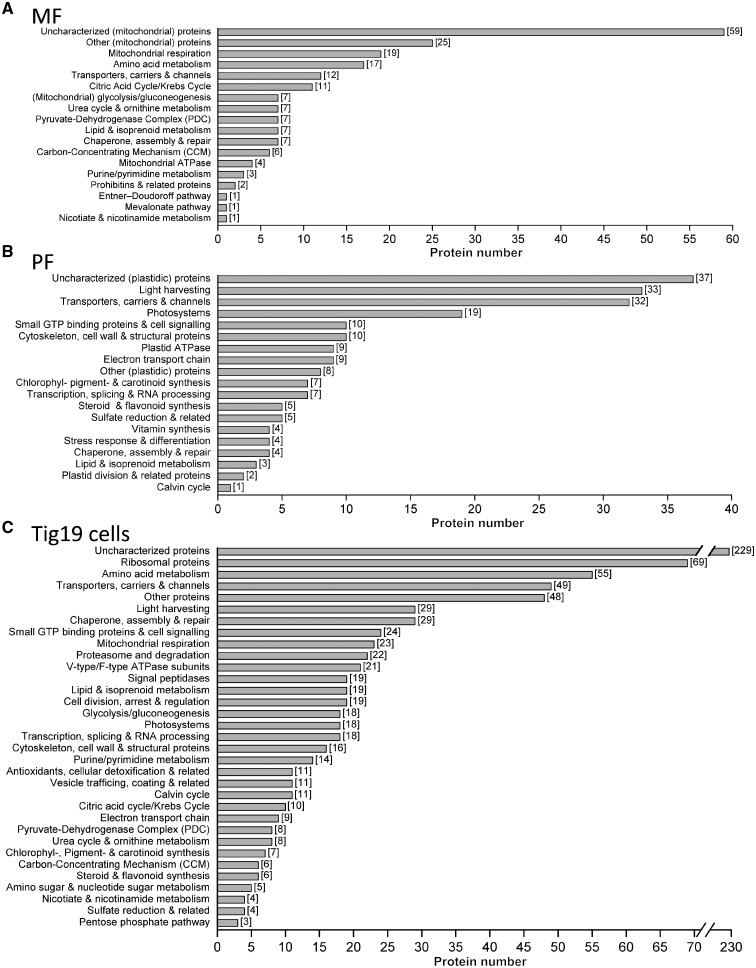
Mass Spectrometry profile of proteins within mitochondria (MF), plastids (PF) and Tig19 cells categorized in functional groups. All proteins detected within the MF (325), PF (217) and Tig19 cells (842) were annotated according to the workflow described in the Materials and Methods section and in Supplementary Fig. S5. All proteins annotated this way were manually compiled in functional groups by following the T. pseudonana KEGG pathways reference maps (three-letter code: tps) and database annotations. Bar charts of the MF (A) and PF (B) do not include proteins that were assigned as ‘cellular contaminants’ (MF: 129 proteins; PF: 8 proteins; refer Supplementary Tables S1 and S2). Bar charts of Tig19 cells (C) represent all evident proteins detected.
The largest protein category comprises 19 proteins that are directly involved in respiratory chain function, covering all major complexes required for functionality (Fig. 7A; KEGG pathways in Supplementary File S1). The TCA cycle is represented by 11 out of 15 known T. pseudonana proteins, which is consistent with the competence to metabolize malate during oxygen measurements observed here. Other soluble proteins of the mitochondrial glycolysis/gluconeogenesis (7), the Carbon-Concentrating Mechanism (CCM) (6) or chaperones/proteins involved in assembly and repair (7) were identified as well, arguing for preserved matrix structures. The MF proteome also contains several membrane-associated proteins, e.g. transporters, carriers and channels (12), proteins involved in the lipid (7) or amino acid (17) metabolism (KEGG pathways in Supplementary File S1). Additionally, four enzymes of the diatom-specific urea cycle (Allen et�al. 2011), namely mitochondrial carbamoyl phosphate synthase (CPSase (Thaps3a_40323), urease (Thaps3a_25118), urease accessory protein (Thaps3a_7461) and glutamate dehydrogenase (Thaps3a_26192) were detected.
Plastids fraction. We detected 217 proteins in the PF fraction, of which 209 proteins (listed in Fig. 7B) could be assigned to functional groups (as judged by KEGG pathway reference maps), that are related to plastid functionality (for details refer Supplementary Table S2). There is also indication that the PF lacks most stromal proteins, but features many membrane integrated and -associated proteins of the photosynthetic electron transport chain (9), light harvesting complexes (32), photosystem components (19) or subunits of the plastid ATPase (9) (Fig. 7B; KEGG pathways in Supplementary File S2). The detection of 10 proteins belonging to the Rab-type GTPase family might be related to vesicle trafficking between (chloroplast) endoplasmic reticulum ((c)ER) and Golgi apparatus and is in line with TEM micrographs (Fig. 2E, F) that show that some plastids retained parts of the chloroplast ER.
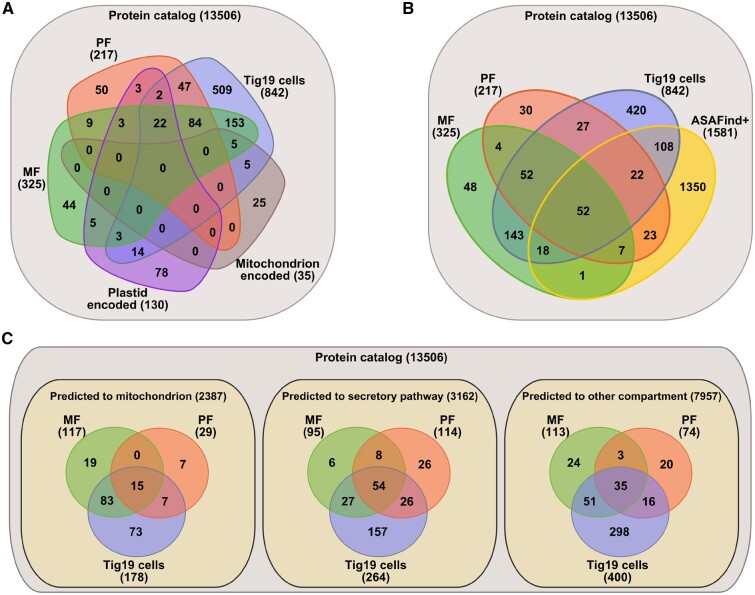
Organelle prediction analyses. Organellar proteins obtained from PF, MF and Tig19 cells were analyzed comparatively, and in the following used to benchmark the plastid-targeting prediction pipeline ‘ASAFind’ and the universal cell-targeting prediction tool ‘TargetP’ (here for mitochondria) (Fig. 8). Comparative proteomics (Fig. 8A) reveals 53 (red (50) plus overlap of red and purple (3)) PF and 49 (green (44) plus overlap of green and purple (5)) MF proteins that have not been detected in any other fraction compared here. The targeting prediction analysis of the PF (Fig. 8B) moreover suggests that 104 (intersections of PF with ASAFind+: 52 + 22 + 7 + 23) out of all 217 proteins were predicted to possess a plastid-targeting signal (not accounting for false positives). This leaves 113 (intersections 4 + 30 + 52 + 27) proteins in the PF which have not been predicted to be plastid imported. Out of these 113 proteins, 26 (Supplementary Table S2) were found to be plastid-encoded, justifying the predicted absence of a plastid-targeting signal. We cannot claim that all of the residual 87 are true plastid proteins, but our protein annotation suggests that most proteins are relevant for plastid functionality; e.g. several Lhc and three Lhc-like proteins, photosystem components PsaE, PsaF, PsbA, a putative PSII repair protein (Thaps3a_3853) and several transporters, like the Tic-22 like protein or the triose phosphate translocators Tpt1 and Tpt8.
Fig. 8.
Comparison and targeting prediction of proteins detected within the mitochondria, plastids and Tig19 cell fractions. (A) Venn diagram representation of absolute protein numbers detected in the MF (325), PF (217) and Tig19 (842) compared to proteins encoded on organelle genomes. Intersections describe identical protein hits within overlapping sets. (B) Comparison of protein identified in the individual fractions to ASAfind positive proteins. ASAFind+ thereby represents all proteins within the protein catalog that have been predicted (with high and low confidence) to be plastid-targeted. (C) TargetP 1.1 prediction analysis of proteins identified in the respective fraction.
Discussion
Isolation protocol optimization
Numerous protocols exist for the isolation of primary plastids and mitochondria of land plants, such as pea or spinach (Lyttleton 1962, Palmer 1967, Stankovic and Walker 1977, Schwitzguebel and Siegenthaler 1984, Beston et�al. 1990, Vishwakarma and Gupta 2017) or green algae (Mendiola-Morgenthaler et�al. 1985, Eriksson et�al. 1995, Mason et�al. 2006). The protocols are usually simple, yielding organelles of consistent quality. Other isolation approaches bypass conventional tissue or cell extraction by isolating organelles from cell-wall deficient mutant lines (Pollock et�al. 2003) or protoplasts (Edwards et�al. 1978, Schnell and Lefebvre 1993). Those techniques, however, demand the availability of respective mutant lines or specific cell wall degrading enzymes. The first protocols on the isolation of photosynthetically active thylakoids were reported for the pennate diatoms P. tricornutum (Murata et�al. 1979) and Cylindrotheca fusiformis (Martinson et�al. 1998). Both pennate species are well suited for organelle isolation, as they either do not possess a silica cell wall or allow protoplast formation, respectively. Also for the centric diatoms Chaetoceros gracilis (Nagao et�al. 2007) and Cyclotella meneghiniana (Kansy et�al. 2017), the isolation of photoactive thylakoids is feasible. So far, the preparation of physiologically and structurally intact plastids has only been reported for the diatoms O. sinensis and Coscinodiscus granii (Wittpoth et�al. 1998). These species are especially suited as they easily break due to their large cell size while accommodating several hundreds of plastids. However, for both species no sequenced genomes are available as well as no established transformation protocols or genome-editing tools.
Here, we present the first procedure for isolation of mitochondria and modified it towards the applicability for preparation of PFs. Optimization of the isolation procedure in more than 60 individual preparations revealed that cell culture densities and average cell sizes in combination with a constant working pressure of 90 MPa are essential for a thorough cell rupture by the French Press. Other cell rupture approaches like osmotic shock, enzymatic digestion or homogenization with glass beads were also tested, but proved unsuccessful. Additionally, we found that accurately adjusted osmolalities of the preparation buffers are critical for a successful organelle isolation. Already slight deviations of 50-100 mOsm/kg caused drastic deformations of the plastids (Supplementary Fig. S3). In line, the osmolality had been previously shown to alter the multilamellar organization of diatom thylakoids in intact diatom cells (Szabo et�al. 2008). Also the concentration of divalent cations in preparation buffers, especially Mg2+, was demonstrated to influence pigment organization as well as thylakoid membrane appression (Szabo et�al. 2008, J�ger and B�chel 2019).
Purity and quality of mitochondria
Our isolated mitochondria are structurally intact (TEM); however, the preparations still contain some residual contaminations with cell wall fragments and plastid membranes (Figs.�2–5). Regarding mitochondria function, our experiments (Fig. 5) indicated a full connectivity of all major complexes: (i) complex I contributed to respiration, until it was inhibited by Rotenone. (ii) the reducing equivalents required for complex I could be provided by malate, suggesting a functional citric acid cycle and/or a functional malate/oxaloacetate shuttle, both of which were detected in the proteomic data. (iii) Inhibiting complex I by Rotenone and subsequent addition of succinate increased respiration, demonstrating that complex II is also functional. (iv) Succinate dependent respiration was reduced by Antimycin A, an inhibitor of complex III when it was used in combination with SHAM to inhibit the AOX. (v) Ascorbate and TMPD circumvented this inhibition by feeding electrons to the cytochrome c site (Fig. 6C, D). As both, complex III and cytochrome c can only transfer electrons to oxygen via complex IV, these findings demonstrate a linear electron transport chain from complex I/II to complex IV and eventually oxygen. Finally our oxygen measurements also confirmed a functional coupling of mitochondria’s electron transport chain with phosphorylation (Fig. 5B, E), suggesting a functional ATP synthase (complex V).
Purity and quality of plastids
A meaningful measure for the physiological integrity of plastids is their capability to perform CO2-dependent photosynthesis autonomously. However, only plastids which still possess intact envelope membranes, and thereby harbor stromal components essential for electron transport (such as ferredoxin), are able to accomplish photosynthesis with CO2 as the only substrate (Wittpoth 1996). This is not the case in the PF isolated here, as most of the plastid envelopes were damaged throughout the isolation process and consequently lacked stroma and the pyrenoid as judged by TEM (Fig. 2E, F), Western blots (Fig. 4) and proteome analysis (Fig. 7B).
The thylakoids, however, are structurally intact and are almost free from contaminants as shown by immunoblotting (Fig. 4), fluorescence (Fig. 3, right) and TEM (Fig. 2E, F) as well as by proteomic analyses (Fig. 7B). Oxygen measurements (Fig. 6A) further demonstrated reproducibly high photosynthetic rates of up to 32 �mol O2�(mg chlorophyll�h)−1. These rates are comparable to C. gracilis thylakoids (30–40 �mol O2�(mg chlorophyll�h)−1) (Nagao et�al. 2007) measured with potassium ferricyanide at comparable light intensities. The high photosynthetic rates measured are moreover corroborated by the O-J-I-P analyses (Fig. 6E), which resemble those of cells (Fig. 6D). The less pronounced inflections between I and P of the PF compared to the well-defined IP-phase of cells might be caused by inefficient electron transfer to the terminal acceptors ferredoxin and NADP+ (Stirbet and Govindjee 2011). Maximum quantum yields of 0.51 demonstrate a good physiological condition (≙70% of intact cells) which is clearly higher than the 0.39 measured in previous thylakoid preparations of P. tricornutum (Szabo et�al. 2008). These parameters suggest physiological intactness of the thylakoids corroborated by the spectroscopic features obtained via absorption and fluorescence emission spectra a (Fig. 6B, C).
Proteome targeting prediction
In total, we identified 325 MF and 217 PF proteins (Fig.�8A), of which 49 (MF) and 53 (PF) were exclusively found in their respective fraction and not in the whole cell lysate. This suggests that isolation and concentration of organelles itself greatly improved the detection of putatively lowly expressed organelle proteins, leading to a better coverage of the organellar proteomes. Interestingly, only 19 out of the 49 MF exclusive proteins were predicted to feature a mitochondrial targeting signal, while only 4 of the non-predicted proteins were organelle encoded. For the PF, just 23 out of 53 fraction exclusive proteins were predicted to be targeted to the plastids, with 3 proteins found to be plastid encoded. These examples highlight that prediction analyses alone may not be sufficient and need to be verified by other methods, such as localization of fluorescently tagged fusion proteins.
Photosystems, light harvesting and photoprotection
The proteins identified in the PF cover most of the PSII reaction center proteins, e.g. the core proteins D1 (PsbA) and D2 (PsbD), the core antenna CP43 (PsbC) and CP47 (PsbB) and cytochrome b559 (PsbE) (Supplementary Table S2 and Supplementary File S2). Also, peripheral PSII proteins like the PSII stability and assembly proteins PsbH and Psb27, the OEC components PsbO, Psb, PsbV, as well as an uncharacterized protein (Thaps3a_3853) with similarity to a putative PSII repair protein were identified. The only PSII membrane proteins that have not been detected are PsbM and Psb28, both proposed to be involved in PSII dimerization (Jiroutov� et�al. 2010, Kawakami et�al. 2011) and PsbZ which in higher plants mediates coupling of core antenna to PSII (Swiatek et�al. 2001). Both mechanisms are likely to play a role in photoacclimation (Nymark et�al. 2009) or responses to abiotic stressors such as iron-limitation (Allen et�al. 2008), which might explain the absent expression in our plastids derived from cells grown under low light and full nutrient supply. Interestingly, also in other proteomic studies on T. pseudonana and P. tricornutum (Grouneva et�al. 2011, Grouneva et�al. 2016) grown under moderate conditions, these proteins were not detected, arguing for the hypothesis of stress-driven expression. The presence of both PSI core (PsaA, PsaB) and all known peripheral reaction center subunits (PsaC, PsaD, PsaE, PsaF and PsaL), required to provide electron flow to ferredoxin (Supplementary Table S2 and Supplementary File S2) could be confirmed. Moreover, all mapped T. pseudonana subunits (PetA, PetB, PetC and PetD) of the cytochrome b6f complex, and six out of eight plastid ATP synthase subunits were identified in the PF. With respect to the photosynthetic electron transport, two isoforms (Thaps3a_4586 and Thaps3a_4914) of the ferredoxin-NADP+ reductase (FNR/PetH), but neither ferredoxin (Fd/PetF) nor cytochrome c6 (cyt c6/PetJ) were found. Instead, an uncharacterized ferredoxin protein containing a 2Fe-2S Rieske domain (Thaps3a_29842) was detected.
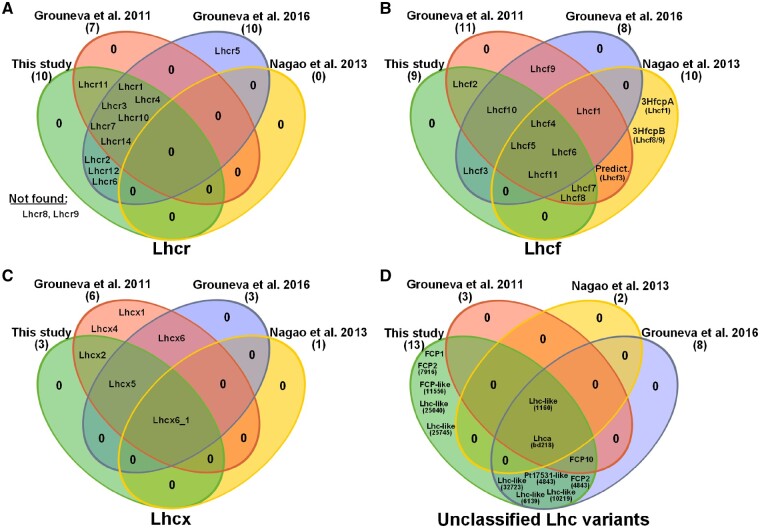
One of the protein groups that showed a remarkably high protein coverage was the light harvesting complex protein (LHC) family. This group is divided into three major sub groups: the red alga-like Lhcr, predominantly comprising PSI-specific antennae; Lhcf, that serve primarily as PSII antennae, and Lhcx proteins, mainly playing a role in energy dissipation via NPQ (Lepetit et�al. 2012, B�chel 2015). We identified 35 Lhc and Lhc-like proteins (33 within the PF and 2 exclusively in MF/CL), and compared them to existing proteome studies performed on diatoms (Fig.�9). Lhcr proteins detected here (Fig. 9A) are almost identical to the ones reported in proteomes published by Grouneva and colleagues (Grouneva et�al. 2011, Grouneva et�al. 2016). The only PSI-specific antennae protein that was solely detected in the Grouneva et�al. (2016) study is the Lhcr5. A study published by Dong et�al. (2016) suggested Lhcr5 to be involved in dissipation of heat through binding of diatoxanthin (Ddt) after it showed increased expression upon 10 h of high light (HL). This would agree with the absence of Lhcr5 in the Grouneva et�al. (2011) data and the study presented here, as both were performed on low light cultured cells, eventually leading to expression levels below the detection limit of LC-ESI-MS/MS. A similar explanation seems to be valid for Lhcx-class proteins 4 and 6 that were also hypothesized to act as photoprotectors that are upregulated during HL-conditions (Zhu and Green 2010, Dong et�al. 2016). With respect to Lhc proteins that could not be attributed to one of the three major classes, therefore termed as ‘unclassified Lhc variants’, it strikes that most proteins were shared exclusively between the Grouneva et�al. (2016) and our study. Besides the high comparability of the two studies, we could accessorily report the detection of three FCP (FCP1, FCP2 and FCP-like 11556) and two Lhc-like (25040 and 25745) proteins that have not been found in any of the studies compared here.
Fig. 9.
Comparison of Lhc proteins identified in recent proteome studies. Lhcr (A), Lhcf (B), Lhcx (C) and unclassified Lhc variants (D) identified in this study compared to recent proteome studies published by Grouneva et al. (2016), Grouneva et al. (2011) and Nagao et al. (2013). Intersections illustrate Lhc proteins that were identified in all studies of the respective intersection-forming ovals.
Particular features of diatom metabolism
Mitochondrial glycolysis. Diatoms feature special metabolic traits that are not present in other photosynthetic eukaryotes. Glycolysis, for instance, the major energy source of eukaryotic catabolism, is partially performed in the mitochondria of stramenopiles (R�o B�rtulos et�al. 2018), instead of being restricted to the cytosol as in most eukaryotes. A search in our mitochondrial proteome returned all enzymes required to perform the second half of glycolysis: two isoforms of the triosephosphate isomerase/glyceraldehyde-3-phosphate dehydrogenase (TPI: GAPDH), the phosphoglycerate kinase (PGK), the phosphoglycerate mutase (PGM), an enolase (ENO) and the pyruvate kinase (PYK). This strengthens the previously proposed localization of parts of the glycolysis to the mitochondria (Kroth et�al. 2008), which recently was verified experimentally in P. tricornutum by R�o B�rtulos et�al. (2018).
Entner–Doudoroff pathway. The Entner–Doudoroff pathway (EDP) is an ancient alternative route to the classical glycolytic reactions that is usually restricted to prokaryotes, but also found in mitochondria of diatoms (Fabris et�al. 2012). It catalyzes the metabolic breakdown of glucose to pyruvate via the two enzymes 6-phosphogluconate dehydratase (EDD) and 2-keto-3-deoxyphosphogluconate aldolase (EDA) after an intermediary conversion step via 6-phosphogluconate (6PG). Both enzymes possess coding genes in the pennate and centric diatoms P. tricornutum and T. pseudonana (Kroth et�al. 2008, Fabris et�al. 2012). We detected protein signatures of both enzymes required to operate a functional EDP in our proteomic data, with EDA (Thaps3a_25880) being expressed in both the MF and the Tig19 cell lysate, while EDD (Thaps3a_27997) was solely evident in the Tig19 lysate, implying that the EDP might indeed be active in the diatom metabolism.
Carbon concentrating mechanism. The operation of a carbon concentrating mechanism (CCM) actively increases the accessibility of CO2 or bicarbonate required for photosynthetic carbon fixation (Matsuda et�al. 2011). In the model diatoms T. pseudonana and P. tricornutum, the presence of biochemical C4 mechanism is still under discussion (Clement et�al. 2016; Ewe et�al. 2018), but a biophysical carbon concentrating mechanism (CCM) is present and apparently also takes place in the mitochondria (Clement et�al. 2016). We found isoforms of the major key players for a putative C4 metabolism, the mitochondrial phosphoenolpyruvate carboxylase (PEPC2) and mitochondrial phosphoenolcarboxykinase 1 (PEPCK1) as well as three carbonic anhydrases γCA4, ζCA1 and δCA1. Moreover, we detected the malate dehydrogenase 1 (MDH1) and an oxoglutarate/malate translocator.
Import of nucleotides, amino acids and metabolites. In contrast to primary plastids of higher plants, complex plastids of diatoms lack a plastidic nucleotide biosynthesis pathway (Armbrust et�al. 2004, Ast et�al. 2009) and the oxidative pentose-phosphate pathway (Michels et�al. 2005) to provide nucleotide precursors, thus relying on the import of nucleotides from the cytosol. Specifically adapted nucleotide import systems are therefore indispensable for maintaining the chloroplasts’ nucleotide and energy metabolism. The PF proteome confirmed the presence of the ADP/ATP carrier proteins NTT1 and NTT3 (Ast et�al. 2009) and the adenine nucleotide translocator ATP/ADP translocase ANT1. Both NTTs are most likely localized in the inner membranes and feature a broad substrate specificity towards other nucleotides beside ADP/ATP (Ast et�al. 2009), suggesting a minor relevance for other metabolic tasks apart from the energy maintenance. The ANT1 protein, moreover, might be an AXER (ATP/ADP exchanger in the ER membrane) orthologue, recently described by Klein et�al. (2018), that was shown to be involved in maintaining energy shuttling across the chloroplast membranes.
The triose phosphate translocators Tpt1—recently reported to be integrated in the (c)ER membrane (Moog et�al. 2015)–and Tpt8—whose localization in the (c)ER membrane or the periplastidal membrane is still discussed controversially (Moog et�al. 2015)—were also identified. Besides, we obtained evidence for the presence of unspecific amino acid (Thaps3a_23142 and Thaps3a_25887) and protein (SecA, SecY, Tic110 and Tic22-like) transport in the PF, suggesting that the inner and outer plastidic envelopes show at least partial preservation in our isolates.
Thioredoxins and redox signaling. Redox regulation via thioredoxins is a ubiquitous feature present in all sorts of cellular processes, but is of major importance for the regulation of photosynthesis in higher plants (Gelhaye et�al. 2004, Meyer et�al. 2005). To which extent these small redox proteins contribute to the regulation of diatom photosynthesis is still contentious (Weber et�al. 2009). We detected six novel uncharacterized proteins with thioredoxin domains (Supplementary Table S3; see Thioredoxin and Related Proteins), sharing approximately 40–90% of sequence similarity to known thioredoxins and one additional thioredoxin-reductase. All of the mentioned proteins are not predicted to be plastid targeted and were interestingly only disclosed in the Tig19 cell lysate fraction and neither in the PF nor in the MF, arguing for a redox regulatory role rather in the cytosol or the nucleus instead of serving in serving in regulation of photosynthesis.
Concluding remarks and the role of GreenCut proteins
Our work provides a novel tool to investigate mitochondrial respiration and photosynthesis of diatoms in organello, which will help us to broaden our understanding of diatom metabolism and its compartmentalization. Studies that may profit from the described methodology might address questions concerning organelle-specific metabolite uptake and release, pigment composition, post-transcriptional modifications or investigation of organellar DNA and gene regulation. Also, comparative proteomics, metabolomics, transcriptomics or lipidomics on organelles derived from cultures grown with varying nutrition or illumination are imaginable and thus might help us to shed further light on organellar fate and functionality in diatoms.
Moreover, we would like to raise the idea that organelle proteomes provided here might be used to improve meta-analyses that benefit from the report of experimental data sets. One example for a large meta-study is the so-called ‘GreenCut proteins’ analysis (Merchant et�al. 2007, Karpowicz et�al. 2011). The ‘GreenCut’ includes all Chlamydomonas reinhardtii plastid proteins that are nucleus-encoded and only found in photosynthetic organism. With respect to the PF isolated here, we report 30 proteins (see Supplementary Table S2; indicated by colors) that have been reported as ‘GreenCut proteins’ (Merchant et�al. 2007, Karpowicz et�al. 2011). Within these 30 proteins, 15 were previously attributed to the subgroup termed ‘ViridiCut’ (proteins that are exclusively found in the green linage of plantae, but are not found in red algae or diatom). Therefore, these proteins should be re-assigned to the ‘DiatomCut’ (Merchant et�al. 2007, Karpowicz et�al. 2011), proteins that are only found in the green lineage and at least one diatom. As some of these proteins might be essential for photosynthetic organisms and the function of some is still unknown, future work should concentrate on revealing their role.
Materials and Methods
Cloning and generation of the Tig19 mutant line
The T. pseudonana mutant line Tig19 (Schober et�al. 2018) expresses GFP fused to the pre-sequence of mitochondrial triosephosphate isomerase/glyceraldehyde-3-phosphate dehydrogenase (preTPI-GAPDH::GFP) and thus promotes fluorescence microscopic tracking of mitochondria during organelle isolation. Initial parts of the TPI-GAPDH pre-sequence (short: TIG; JGI Protein ID: 28239) were amplified from T. pseudonana cDNA using the TripleMaster PCR System (Eppendorf; Hamburg, Germany) with the oligonucleotides TIG28239_fw: TAGGTACCAAGATGTTATCAAACACTG and TIG28239_rev: AAGGTACCGTTGCATTTCCAGTTTC. The resulting 142 bp PCR fragment was subcloned into the pGEM-T cloning Vector (#A1360, Promega; Mannheim, Germany). Digestion of both flanking KpnI restriction sites allowed cloning into the T. pseudonana GFP expression vector pTpFcpGFP Vector (Poulsen et�al. 2006). Correct insertion was verified by Sanger sequencing (GATC; Konstanz, Germany) and resulting vector was co-transformed together with the selection plasmid pTpfcpNAT into T. pseudonana wild type strain CCMP 1335 [National Center for Marine Algae and Microbiota (NCMA); Maine, USA] as described by Poulsen et�al. (2006). Cell line number 19 (TIG19) was used for further experiments.
Cell culture
Cells were grown axenically in modified F/2 growth medium according to Guillard and Lorenzen (but with only halve of the original sea salt content, enriched with 250 �M NaSiO3x5H2O and 5 �M H2SeO3) (Guillard and Lorenzen 1972). Four liters of batch-cultures were inoculated sterile from pre-culture and kept under standard atmospheric pressure at 20�C. During the whole cultivation period, cells were aerated with filtered ambient air and illuminated with constant light intensities of 40 �E m−2 s−1 (16 h light-8 h dark cycle).
Preparation of crude organelle suspensions
Utilized preparation media are indicated in Table�1. Their composition was modified based on the recipes employed by Wittpoth et�al. (1998). All following steps were performed at 4�C while organelles were kept on ice and in the dark wherever applicable. Four liters of early-stationary T. pseudonana Tig19 culture was harvested at cell densities between 5–7 � 106 cells/ml and an spherical volume diameter of 4.7–5.3 �m (see Supplementary Fig. S1) determined using a Coulter Counter (Beckman Coulter; Krefeld, Germany, as described by Rottberger et�al. 2013). Cells were resuspended in 9 ml isolation buffer (IB; Table 1) and resulting cell suspension was loaded to a pre-chilled French Press mini cell. Cell rupture at 90 MPa was performed with an SLM Aminco cell press (Laurier Research Instruments; ON, Canada) by releasing the fragmented cells gently to avoid shear forces. The homogenate was diluted and cautiously inverted for 30 s in a total volume of approximately 20 ml of washing buffer (WB; Table 1). Cells and cell debris were sedimented by centrifugation using a swing-out bucket rotor TS-5.1-500 (Beckman Coulter; Krefeld, Germany) at 300�g for 10 min. To increase organelle yield, sedimented cells and cell debris were again resuspended in 9 ml of WB and processed with the French Press followed by centrifugation at 300�g for 10 min. This step was typically repeated 3 times during mitochondria isolation and once when plastids were isolated. Resulting supernatants (=crude organelle suspensions) containing mostly plastids and mitochondria of varying qualities were collected, pooled and kept in the dark on ice.
Table 1.
Buffers
| Isolation buffer (IB) | Washing buffer (WB) | Reaction buffer (RB) | Mitochondria percoll solution (MPS) | Plastid percoll solution (PPS) | |||
|---|---|---|---|---|---|---|---|
| (10%) | (20%) | (30%) | |||||
| Sorbitol (M) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| HEPES-KOH; pH8 (mM) | 50 | 30 | 30 | 30 | 30 | 30 | 30 |
| EDTA (mM) | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| MgCl2 (mM) | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| KCl (mM) | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| MnCl2 (mM) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| PVP 40 (K30) (% w/v) | 1 | 1 | 0.1 | 0.1 | 0.1 | 0.1 | |
| Adjust pH | 7.2–7.5 | 7.2–7.5 | 7.2–7.5 | 7.2–7.5 | 7.2–7.5 | 7.2–7.5 | 7.2–7.5 |
| Adjust osmolality (mOsm/kg) | 730–750 | 730–750 | 730–750 | 930–990 | 830 | 935 | 1055 |
| Add fresh | |||||||
| BSA; defatted (% w/v) | 0.5 | 0.5 | |||||
| L-Cysteine (% w/v) | 0.1 | 0.1 | |||||
| K2HPO4 (mM) | 5 | ||||||
| Glutathione (mM) | 1 | ||||||
| Percoll (% v/v) | 28 | 10 | 20 | 30 | |||
Isolation of mitochondria
Starting from a pooled organelle extract (see Fig. 1), suspensions were centrifuged with the swing-out bucket rotor TS-5.1-500 (Beckman Coulter) at 500�g, 4�C for 10 min to remove residual cells and cell debris with the pellet. The supernatant was carefully transferred to two SS34 centrifuge tubes and mitochondria were pelleted at 12,000�g, 4�C for 10 min. Each pellet was gently resuspended in 2–4 ml WB (see Table 1) before being pooled to a single SS34 centrifuge tube. The organelle extract was topped up to approximately 30 ml with WB and centrifugation at 500�g with a Sorvall RC6Plus centrifuge (Thermo Fisher Scientific; Karlsruhe, Germany) using the corresponding SS34 fixed-angle rotor. The supernatant was carefully collected, again centrifuged at 12,000�g, and the pellet was resuspended in 2–4 ml of WB using a fine paint-brush. A maximum of 1–2 ml of organelle suspension was loaded on top of two continuous Percoll gradients each. Both gradients have been adjusted preliminarily by centrifugation of Mitochondria Percoll Solution (MPS; Table 1) for 40 min at 40,000�g, 4�C in a Sorvall RC6Plus (Thermo Fisher Scientific) using rotor SS34 without using the rotor break. The loaded gradients were centrifuged for 30 min at 40,000�g, 4�C (without using the rotor break) (see Fig. 3). The separated bands were evaluated by fluorescence microscopy and mitochondria of highest purity were extracted. Percoll was removed by diluting with 30 ml reaction buffer (RB; Table 1) followed by centrifugation at 12,000�g, 4�C for 10 min. The supernatant was discarded and the pellet was again dissolved carefully with a fine paint-brush in 2–4 ml of RB.
Isolation of plastids
Pooled organelle extract (see Fig. 1) was diluted with WB (see Table 1) in a total volume of approximately 30 ml and plastids were collected by centrifugation at 6,000�g and 4�C for 10 min with a Sorvall RC6Plus centrifuge (Thermo Fisher Scientific). The supernatant was discarded and the pellet was dissolved gently in 2–4 ml WB using a fine paint-brush. A maximum of 1–2 ml organelle suspension was loaded to layered Percoll density gradients, consisting of 1 ml 10%, 5 ml 20% and 3 ml 30% (from top to bottom) Plastid Percoll Solution (PPS; Table 1). Separation of organelles by density centrifugation was performed in an Optima LE-80K Ultra Centrifuge (Beckman Coulter) with corresponding SW40Ti rotor at 14,400�g and 4�C for 30 min. The band formed at the boundary of the 20% and 30% phase (see Fig. 3), comprising the plastids with highest quality, was extracted without dispersing the 30% phase. The PF was washed through dilution in 30 ml WB and subsequently centrifuged at 4,000�g, 4�C for 10 min. The supernatant was discarded and the pellet containing the final PF was dissolved carefully with a fine paint-brush in 2–4 ml of RB (see Table 1).
Preparation of cell- and organelle lysates for immunoblotting and mass spectrometry
Cell- or plastid material was pelleted at 10,000�g, 4�C for 10 min and resuspended in ice-cold 75 mM Tris–HCl; pH 7.6 supplemented with double concentrated Protease-Inhibitor (cOmplete EDTA-free, Roche Applied Science; Penzberg, Germany). Glass beads with three different diameters (0.1–0.11 mm, 0.25–0.3 mm and 1 mm) were added to the suspension to perform mechanical disruption by applying 6 cycles � 30 s at power setting 6.5 with a reciprocating Savant FastPrep FP120 cell disruptor (MP Biomedicals; CA, USA). Homogenate was kept on ice for 5 min in between every cycle. After homogenization, 0.5% v/v SDS was added to each samples followed by incubation on ice for 5 min. If desired, membrane fraction was separated by two subsequent 30 min centrifugation steps at 18,000�g at 4�C.
Determination of chlorophyll a content
Chlorophyll a (Chl a) content of either cells or isolated plastids was carried out spectrophotometrically with an Ultrospec 2100 Pro (GE healthcare, Munich, Germany) according to the method described by Jeffrey and Humphrey (1975).
Determination of protein content
Protein yields were determined colorimetrically according to the Bradford protein assay kit (catalog #B6916, Sigma Aldrich; Munich, Germany, Bradford 1976, Compton and Jones 1985).
Optical spectrometry
Ambient temperature absorption spectra of dark acclimated plastids/cells were determined in 1 ml WB (see Table 1) using an Ultrospec 8000 Spectrophotometer (GE healthcare). Relative absorbance was recorded at wavelength ranging from 350 to 750 nm with a bandwidth set to 1 nm. Fluorescence emission spectra were measured with a LS50B luminescence/fluorescence spectrophotometer using an excitation wavelength of 440 nm (Perkin Elmer; Hamburg, Germany). Plastids/cells corresponding to 20 �g Chl a were diluted in a total volume of 1 ml WB (see Table�1). Scan speed was kept at constant 150 nm/min.
Fluorescence and light microscopy
Plastids and mitochondria isolation was monitored by fluorescence and light microscopy during the whole isolation process. Particular attention was paid to the French Press eluates and the Percoll gradient fractions. Microscopy was performed with a BX51 epifluorescence microscope (Olympus; Tokyo, Japan) at excitation and emission wavelengths (for GFP) of 395 nm and 509 nm, respectively. Chlorophyll autofluorescence was observed at wavelengths of 465 nm (excitation) and 673 nm (emission). Photos were taken with an AxioCam MRm digital camera system and Axio Vision 40′ V.4.6.3.0 Documentation software (Carl Zeiss; Jena, Germany). Three-dimensional models were reconstructed as described in R�o B�rtulos et�al. (2018) from pictures taken with a confocal laser scanning microscope (cLSM-510META, Carl Zeiss) using a Plan-Neofluar 40x/1.3 Oil DIC objective.
Transmission electron microscopy
Three-staged fixation of samples was carried out by first mixing the samples dissolved in isolation buffer with double-concentrated fixation buffer (4% glutardialdehyde, 200 mM sodium cacodylate, 20 mM CaCl2 and 20 mM MgCl2, buffered in 50 mM HEPES; pH 7) at the ratio of 1:1. After incubation of samples for 15 min on ice, fixation was completed by replacing the buffer twice with 1�fixation buffer. Each step was followed by a subsequent incubation step of 15 min on ice. Samples were then washed 3 times for 10 min in cacodylate buffer (100 mM sodium cacodylate, 10 mM CaCl2 and 10 mM MgCl2, buffered in 50 mM HEPES; pH 7). Staining of the biological material was performed in 1% osmium tetroxide (OsO4) and cacodylate buffer for 1 h, while permanently keeping the samples in the dark, on ice. To remove excessive osmium tetroxide, samples were again washed 3 � 15 min in cacodylate buffer. Dehydration of samples was achieved by passage through a consecutive ethanol series of ice-cold 15% (30 min), 30% (30 min), 50% (overnight), 70% (1 h), 90% (1 h), 96% (2 � 30 min) and absolute ethanol (2 � 30 min). Preliminary embedding took place overnight at 4�C in a solution composed of Spurr’s resin (Spurr 1969) and absolute ethanol, mixed at the ratio of 1:1. Final embedding was performed in pure Spurr’s resin by incubating samples 3 times for 2 h at 4�C. Polymerization was achieved by incubation of samples at 65�C for 3 d. Polymerized blocks were cut in thin sections of about 70–75 �m with a Leica Ultracut R ultramicrotome (Leica; Solms, Germany). In the following, thin sections were applied to copper grids and contrasted with 2% uranyl acetate and 0.4% lead citrate. Microscopic analysis was carried out with a TEM 912 Omega (Carl Zeiss). The pictures were taken with a CCD-sensor camera (TRS, 2,048 � 2,048 for Zeiss EM 906–922, Carl Zeiss).
Oxygen measurements
Oxygen evolution was measured with the Clark-type (Clark et�al. 1953) oxygen-electrode system Chlorolab 2 (Hansatech Instruments Ltd; Norfolk, UK). Prior to every experiment, the luminous source was adjusted, and the oxygen electrode was calibrated under consideration of daily atmospheric pressure. Mitochondrial respiration (mitochondria fraction) was measured in complete darkness in order to avoid light-driven oxygen production performed by residual thylakoidal contaminants. Therefore, 0.3–0.5 ml of mitochondria suspension (typically corresponding to 0.1–0.15 mg/ml protein in the inhibition experiments; and 0.374 mg/ml protein for the RCR measurements) were measured in 1 ml RB (see Table 1) supplemented with 5 mM dipotassium phosphate (K2HPO4) and 1 mM glutathione. Light-dependent oxygen evolution (PF) was determined in the same buffer, but complemented with 40 mM of the artificial electron-acceptor potassium ferricyanide (K3[Fe(CN)6]). Therefore, plastid material corresponding to 35 �g Chl a (diluted in RB) was treated by 2 min of dark adaptation, before being illuminated with 200 �E m−2 s−1. The temperature was kept at constant 20�C during each measurement. If not stated otherwise, final concentrations of substrates and inhibitors applied were: 5 mM malate, 5 mM succinate, 0.3 mM ADP, 1 mM ascorbate, 0.1 mM TMPD, 0.2 �M Antimycin A, 0.1 �M Oligomycin A, 1 �M Rotenone and 0.1 mM SHAM.
O-J-I-P fluorescence transients
The O-J-I-P fluorescence transient measuring routine describes the stepwise increase in fluorescence during electron transport rate saturation upon application of a saturation multi turnover pulse, from the minimum fluorescence Fo (O) via the inflection points J and I towards the maximum fluorescence Fm (P), and is characteristic for the reduction of the PSII electron acceptor site components (Neubauer and Schreiber 1987, Schreiber and Neubauer 1987, Strasser and Srivastava 1995). O-J-I-P transients were measured using an AquaPen AP-100 (Photon Systems Instruments; Drasov, Czech Republic). Therefore, cells or isolated plastids, diluted in 1 ml RB to contain a final Chl a concentration of 1 �g/ml (see Table 1), were dark adapted for 10 min prior every experiment. When plastids were measured, 5 mM of dipotassium phosphate was added to the RB. The saturating pulse at 450 � 10 nm used to induce the fluorescence transients had an intensity of 1,500 �E m−2 s−1.
SDS-PAGE and immunoblotting
Proteins were separated on 12% SDS-polyacrylamide gels (Fling and Gregerson 1986) by following the method described by Laemmli (1970). Gels were stained with the Coomassie-based dye Instant Blue (Expedeon; Heidelberg, Germany). Proteins were transferred to a 0.1 �m nitrocellulose membrane (Amersham Protran, GE Healthcare) using a ‘semi-dry’ Trans-Blot Turbo system (Bio-Rad Laboratories; CA, USA). After a successful blotting control with 0.5% Ponceau S and 1% acetic acid for 1 min, membranes were blocked for 1 h in Roti Block (Carl Roth; Karlsruhe, Germany) at room temperature (RT). Blocked membranes were washed twice for 5 min (RT) in TN buffer (10 mM Tris–HCl pH 8, 150 mM NaCl). Thereafter, simultaneous 2� h incubation steps with three commercial antibodies were performed at RT. Therefore, anti-GFP (#A6455 Thermo Fisher Scientific), anti-PsbA (#AS05 084 Agrisera; V�nnas, Sweden) and anti-RbcL (#AS03 037 Agrisera) antibodies were diluted 1:3,000, 1:10,000 and 1: 2,500, respectively, in 10 ml Roti Block. Membranes were washed twice for 5 min in TN buffer before an analogous incubation step for 1� h with the anti-rabbit-HRP secondary antibody (#AS09 602; Agrisera) diluted 1:10,000 in Roti Block was performed. The membrane was again washed twice for 5 min in TN buffer before it was covered with Roti-Lumin plus (Carl Roth). Luminescence was detected using an Odyssey Imaging system (Li-Cor, Biotechnology; Bad Homburg, Germany).
Mass spectrometry and in silico data evaluation
Proteins were separated and stained as described above. Mitochondria samples were analyzed as whole lanes, while plastid and cell samples were cut into four separate gel slices (0–25 kDa, 25–35 kDa, 35–55 kDa, 55-rest) that were measured individually. Proteins samples were measured with an LTQ Orbitrap Discovery mass spectrometer (Thermo Fisher Scientific) by LC-ESI-MS/MS at the proteomics service facility (University of Konstanz).
Data analyses
A workflow of the annotation procedure is illustrated in Supplementary Fig. S5. Mass spectrometric raw data were matched against the optimized, de-replicated T. pseudonana (Thaps) protein catalog (Supplementary Table S4) based on the United States Department of Energy Joint Genome Institute (JGI; https://jgi.doe.gov/) sequencing project for T. pseudonana v3.0 (Armbrust et�al. 2004) compiled by Gruber et�al. (2015). For this study, the supplementary file tpj12734-sup-0013-datasets1_tmp.fas of Gruber et�al. (2015) consisting of ‘Thaps3a_ID’ (assembly) and ‘Thaps3u_ID’ (unmapped sequences) was modified in the following way: (i) transgenes only present in the mutant line Tig19 were included into the protein catalog [marked ‘Thapst_ID’, these were nourseothricin acetyltransferase (GenBank: AAC60439.1) and enhanced green fluorescent protein (GenBank: AAB02576.1)]; (ii) mitochondria-encoded proteins were appended to the file (marked ‘Thapsm_ID’, NCBI Reference Sequence: NC_007405.1, Oudot-Le Secq and Green 2011); (iii) plastid-encoded proteins were appended to the file (marked ‘Thapsp_ID’, GenBank: EF067921.1, Oudot-Le Secq et�al. 2007); (iv) protein IDs that referred to gene models with identical coding sequence and gene coordinates were manually collapsed to one ID (the first of the ones listed) per sequence. Protein models that encoded identical proteins on different genomes (e.g. nuclear and plastid) were kept as separate protein catalog entries, but were run with double-accessions during mass spectrometry data evaluation. The classification of proteins into metabolic pathway-groups was carried out based on annotations provided by JGI (Grigoriev et�al. 2012, Nordberg et�al. 2014) and National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). KEGG ontology (KO) tags designation was performed with ‘GhostKOALA’ (Kanehisa et�al. 2016b), while mapping of KEGG pathways against the T. pseudonana reference maps (Three-letter KEGG organism code: tps) was carried out with ‘KEGG Mapper – Search Pathway’ (Kanehisa and Goto 2000; Kanehisa et�al. 2016a, Kanehisa et�al. 2017). Orthologues of ‘GreenCut proteins’ were identified with ‘pblast’ (Altschul et�al. 1990) by blasting proteins detected within the PF against the NCBI C. reinhardtii (taxid: 3055) protein catalog. Hits that generated blast scores of at least 80 were reviewed manually and marked as ‘GreenCut protein’ if listed in the C. reinhardtii ‘GreenCut2 resource’ protein list published by Karpowicz et�al. (2011). Plastid-targeting predictions were performed using the ASAFind pipeline as described in Gruber et�al. (2015). Proteins that are potentially targeted to the mitochondria, the secretory or other pathways were predicted using the TargetP 1.1 with default parameters (Emanuelsson et�al. 2007) following the strategies described in Gruber and Kroth (2014) and Gruber and Kroth (2017).
Supplementary Material
Acknowledgments
We thank Doris Ballert for performing biolistic bombardment and for routine cell cultivation, Dr. Joachim Hentschel, Dr. Michael Laumann, Dr. Paavo Bergman and Lauretta Nejedli of the Electron Microscopy Centre (EMC) as well as the Bioimaging Centre (BIC) of the University of Konstanz for sample preparation and kind assistance during transmission electron and confocal fluorescence microscopy. Further we grateful acknowledge Dr. Andreas Marquardt from the University of Konstanz Proteomics Centre (PC) for the mass spectrometric analysis of organelle and cell fractions.
Funding
The Graduate School Chemical Biology (KoRS-CB) to A.F.S.; the University of Konstanz; the German Research Foundation DFG [Grants no. LE 3358/3–1 to B.L., KR 1661/6–1 to P.G.K.]; the Baden-W�rttemberg Elite program and the Zukunftskolleg (to B.L.); as well as Gordon and Betty Moore Foundation [Grant GBMF 4966 ‘DiaEdit’ to C.R.B. and P.G.K.].
Disclosures
The authors have no conflicts of interests to declare.
References
- Allen A.E., Dupont C.L., Oborn�k M., Hor�k A., Nunes-Nesi A., McCrow J.P., et al. (2011) Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207. [DOI] [PubMed] [Google Scholar]
- Allen A.E., Laroche J., Maheswari U., Lommer M., Schauer N., Lopez P.J., et al. (2008) Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 105: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andersson B., Anderson J.M. (1980) Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta (BBA) Bioenerg. 593: 427–440. [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., Berges J.A., Bowler C., Green B.R., Martinez D., Putnam N.H., et al. (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- Ast M., Gruber A., Schmitz-Esser S., Neuhaus H.E., Kroth P.G., Horn M., et al. (2009) Diatom plastids depend on nucleotide import from the cytosol. Proc. Natl. Acad. Sci. USA 106: 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B., Berne N., Murik O., Petroutsos D., Prihoda J., Tanaka A., et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369. [DOI] [PubMed] [Google Scholar]
- Beston H., Ulf B., AnnaCarin E., Elzbieta G. (1990) Large‐scale purification procedure of spinach leaf mitochondria—isolation and immunological studies of the F1-ATPase. Physiol. Plant 78: 367–373. [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- B�chel C. (2015) Evolution and function of light harvesting proteins. J. Plant Physiol. 172: 62–75. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2000) Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5: 174–182. [DOI] [PubMed] [Google Scholar]
- Chance B., Williams G.R. (1955) Respiratory enzymes in oxidative phosphorylation: IV. The respiratory chain. J. Biol. Chem. 217: 429–438. [PubMed] [Google Scholar]
- Clark L.C. Jr., Wolf R., Granger D., Taylor Z. (1953) Continuous recording of blood oxygen tensions by polarography. J. Appl. Physiol. 6: 189–193. [DOI] [PubMed] [Google Scholar]
- Clement R., Dimnet L., Maberly S.C., Gontero B. (2016) The nature of the CO2-concentrating mechanisms in a marine diatom, Thalassiosira pseudonana. New Phytol. 209: 1417–1427. [DOI] [PubMed] [Google Scholar]
- Compton S.J., Jones C.G. (1985) Mechanism of dye response and interference in the Bradford protein assay. Anal. Biochem. 151: 369–374. [DOI] [PubMed] [Google Scholar]
- Dong H.-P., Dong Y.-L., Cui L., Balamurugan S., Gao J., Lu S.-H., et al. (2016) High light stress triggers distinct proteomic responses in the marine diatom Thalassiosira pseudonana. BMC Genomics 17: 994–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G.E., Robinson S.P., Tyler N.J., Walker D.A. (1978) Photosynthesis by isolated protoplasts, protoplast extracts, and chloroplasts of wheat: influence of orthophosphate, pyrophosphate, and adenylates. Plant Physiol. 62: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Gardestrom P., Samuelsson G. (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol. 107: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewe D., Tachibana M., Kikutani S., Gruber A., Rio Bartulos C., Konert G., et al. (2018) The intracellular distribution of inorganic carbon fixing enzymes does not support the presence of a C4 pathway in the diatom Phaeodactylum tricornutum. Photosynth. Res. 137: 263–280. [DOI] [PubMed] [Google Scholar]
- Fabris M., Matthijs M., Rombauts S., Vyverman W., Goossens A., Baart G.J.E. (2012) The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70: 1004–1014. [DOI] [PubMed] [Google Scholar]
- Fling S.P., Gregerson D.S. (1986) Peptide and protein molecular weight determination by electrophoresis using a high-molarity tris buffer system without urea. Anal. Biochem. 155: 83–88. [DOI] [PubMed] [Google Scholar]
- Flori S., Jouneau P.-H., Bailleul B., Gallet B., Estrozi L.F., Moriscot C.et al. (2017) Plastid thylakoid architecture optimizes photosynthesis in diatoms. Nat. Commun. 8: 15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E., Rouhier N., Jacquot J.-P. (2004) The thioredoxin h system of higher plants. Plant Physiol. Biochem. 42: 265–271. [DOI] [PubMed] [Google Scholar]
- Grigoriev I.V., Nordberg H., Shabalov I., Aerts A., Cantor M., Goodstein D., et al. (2012) The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40: D26–D32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouneva I., Muth-Pawlak D., Battchikova N., Aro E.-M. (2016) Changes in relative thylakoid protein abundance induced by fluctuating light in the diatom Thalassiosira pseudonana. J. Proteome Res. 15: 1649–1658. [DOI] [PubMed] [Google Scholar]
- Grouneva I., Rokka A., Aro E.M. (2011) The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J. Proteome Res. 10: 5338–5353. [DOI] [PubMed] [Google Scholar]
- Gruber A., Kroth P.G. (2014) Deducing intracellular distributions of metabolic pathways from genomic data. InPlant Metabolism. Edited by Sriram G. pp. 187–211. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Gruber A., Kroth P.G. (2017) Intracellular metabolic pathway distribution in diatoms and tools for genome-enabled experimental diatom research. Philos. Trans. R Soc. B 372: 20160402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A., Rocap G., Kroth P.G., Armbrust E.V., Mock T. (2015) Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 81: 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard R.R.L., Lorenzen C.J. (1972) Yellow-green algae with chlorophyllide C1, 2. J. Phycol. 8: 10–14. [Google Scholar]
- J�ger S., B�chel C. (2019) Cation-dependent changes in the thylakoid membrane appression of the diatom Thalassiosira pseudonana. Biochim. Biophys. Acta (BBA) Bioenerg. 1860: 41–51. [DOI] [PubMed] [Google Scholar]
- Jeffrey S.W., Humphrey G.F. (1975) New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz 167: 191–204. [Google Scholar]
- Jiroutov� K., Kořen� L., Bowler C., Oborn�k M. (2010) A gene in the process of endosymbiotic transfer. PLoS One 5: e13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45: D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K. (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428: 726–731. [DOI] [PubMed] [Google Scholar]
- Kansy M., Gurowietz A., Wilhelm C., Goss R. (2017) An optimized protocol for the preparation of oxygen-evolving thylakoid membranes from Cyclotella meneghiniana provides a tool for the investigation of diatom plastidic electron transport. BMC Plant Biol. 17: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz S.J., Prochnik S.E., Grossman A.R., Merchant S.S. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286: 21427–21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Umena Y., Iwai M., Kawabata Y., Ikeuchi M., Kamiya N., et al. (2011) Roles of PsbI and PsbM in photosystem II dimer formation and stability studied by deletion mutagenesis and X-ray crystallography. Biochim. Biophys. Acta (BBA) Bioenerg. 1807: 319–325. [DOI] [PubMed] [Google Scholar]
- Klein M.-C., Zimmermann K., Schorr S., Landini M., Klemens P.A.W., Altensell J., et al. (2018) AXER is an ATP/ADP exchanger in the membrane of the endoplasmic reticulum. Nat. Commun. 9: 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroth P.G., Chiovitti A., Gruber A., Martin-Jezequel V., Mock T., Parker M.S., et al. (2008) A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lepetit B., Goss R., Jakob T., Wilhelm C. (2012) Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth. Res. 111: 245–257. [DOI] [PubMed] [Google Scholar]
- Lepetit B., Volke D., Szabo M., Hoffmann R., Garab G., Wilhelm C., et al. (2007) Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry 46: 9813–9822. [DOI] [PubMed] [Google Scholar]
- Lyttleton J.W. (1962) Isolation of ribosomes from spinach chloroplasts. Exp. Cell Res. 26: 312–317. [DOI] [PubMed] [Google Scholar]
- Martin W.F., Garg S., Zimorski V. (2015) Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. B 370: 20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson T.A., Ikeuchi M., Plumley F.G. (1998) Oxygen-evolving diatom thylakoid membranes. Biochim. Biophys. Acta (BBA) Bioenerg. 1409: 72–86. [DOI] [PubMed] [Google Scholar]
- Mason C.B., Bricker T.M., Moroney J.V. (2006) A rapid method for chloroplast isolation from the green alga Chlamydomonas reinhardtii. Nat. Protoc. 1: 2227–2230. [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Nakajima K., Tachibana M. (2011) Recent progresses on the genetic basis of the regulation of CO2 acquisition systems in response to CO2 concentration. Photosynth. Res. 109: 191–203. [DOI] [PubMed] [Google Scholar]
- Mendiola-Morgenthaler L., Leu S., Boschetti A. (1985) Isolation of biochemically active chloroplasts from chlamydomonas. Plant Sci. 38: 33–39. [Google Scholar]
- Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y., Reichheld J.P., Vignols F. (2005) Thioredoxins in Arabidopsis and other plants. Photosyn. Res. 86: 419–433. [DOI] [PubMed] [Google Scholar]
- Michels A.K., Wedel N., Kroth P.G. (2005) Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol. 137: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog D., Rensing S.A., Archibald J.M., Maier U.G., Ullrich K.K. (2015) Localization and evolution of putative triose phosphate translocators in the diatom Phaeodactylum tricornutum. Genome Biol. Evol. 7: 2955–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N., Kume N-A., Okada Y., Hori T. (1979) Preparation of girdle lamella-containing chloroplasts from the diatom Phaeodactylum tricornutum. Plant Cell Physiol. 20: 1047-1053 [Google Scholar]
- Nagao R., Ishii A., Tada O., Suzuki T., Dohmae N., Okumura A., et al. (2007) Isolation and characterization of oxygen-evolving thylakoid membranes and photosystem II particles from a marine diatom Chaetoceros gracilis. Biochim. Biophys. Acta (BBA) Bioenerg. 1767: 1353–1362. [DOI] [PubMed] [Google Scholar]
- Nagao R., Takahashi S., Suzuki T., Dohmae N., Nakazato K., Tomo T. (2013) Comparison of oligomeric states and polypeptide compositions of fucoxanthin chlorophyll a/c-binding protein complexes among various diatom species. Photosynth. Res. 117: 281–288. [DOI] [PubMed] [Google Scholar]
- Neubauer C., Schreiber U. (1987) The polyphasic rise of chlorophyll fluorescence upon onset of strong continuous illumination: I. Saturation characteristics and partial control by the photosystem II acceptor side. Z. Naturforsch. C 42: 1246–1247. [Google Scholar]
- Nordberg H., Cantor M., Dusheyko S., Hua S., Poliakov A., Shabalov I., et al. (2014) The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucl. Acids Res. 42: D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M., Valle K.C., Brembu T., Hancke K., Winge P., Andresen K., et al. (2009) An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One 4: e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudot-Le Secq M.P., Green B.R. (2011) Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476: 20–26. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq M.P., Grimwood J., Shapiro H., Armbrust E.V., Bowler C., Green B.R. (2007) Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277: 427–439. [DOI] [PubMed] [Google Scholar]
- Palmer J.M. (1967) Rapid isolation of active mitochondria from plant tissue. Nature 216: 1208.4294737 [Google Scholar]
- Pollock S.V., Colombo S.L., Prout D.L. Jr, Godfrey A.C., Moroney J.V. (2003) Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO(2) atmosphere. Plant Physiol. 133: 1854–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen N., Chesley P.M., Kr�ger N. (2006) Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 42: 1059–1065. [Google Scholar]
- Prihoda J., Tanaka A., de Paula W.B.M., Allen J.F., Tirichine L., Bowler C. (2012) Chloroplast-mitochondria cross-talk in diatoms. J. Exp. Bot. 63: 1543–1557. [DOI] [PubMed] [Google Scholar]
- R�o B�rtulos C., Rogers M.B., Williams T.A., Gentekaki E., Brinkmann H., Cerff R., et al. (2018) Mitochondrial glycolysis in a major lineage of eukaryotes. Genome Biol. Evol. 10: 2310–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger A.J., Mu�oz-G�mez S.A., Kamikawa R. (2017) The origin and diversification of mitochondria. Curr. Biol. 27: R1177–R1192. [DOI] [PubMed] [Google Scholar]
- Rottberger J., Gruber A., Kroth P. (2013) Analysing size variation during light-starvation response of nutritionally diverse chrysophytes with a Coulter counter. Algol. Stud. 141: 37–51. [Google Scholar]
- Round F.E., Crawford R.M., Mann D.G. (2007) 59. Biology of diatoms. InDiatoms: Biology and Morphology of the Genera. pp. 1–125. Cambridge University Press, New York, NY. [Google Scholar]
- Schnell R.A., Lefebvre P.A. (1993) Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober A.F., Flori S., Finazzi G., Kroth P.G., B�rtulos C.R. (2018) Isolation of plastid fractions from the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum. In Plastids: Methods and Protocols. Edited by Mar�chal E. pp. 189–203. Springer US, New York, NY. [DOI] [PubMed] [Google Scholar]
- Schreiber U., Neubauer C. (1987) The polyphasic rise of chlorophyll fluorescence upon onset of strong continuous illumination: II. Partial control by the photosystem II donor side and possible ways of interpretation. Z. Naturforsch. C 42: 1255. [Google Scholar]
- Schwitzguebel J.-P., Siegenthaler P.-A. (1984) Purification of peroxisomes and mitochondria from spinach leaf by percoll gradient centrifugation. Plant Physiol. 75: 670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr A.R. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26: 31–43. [DOI] [PubMed] [Google Scholar]
- Stankovic Z.S., Walker D.A. (1977) Photosynthesis by isolated pea chloroplasts: some effects of adenylates and inorganic pyrophosphate. Plant Physiol. 59: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirbet A. and Govindjee (2011) On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and photosystem II: basics and applications of the OJIP fluorescence transient. J. Photochem. Photobiol. B 104: 236–257. [DOI] [PubMed] [Google Scholar]
- Storey B.T., Bahr J.T. (1969) The respiratory chain of plant mitochondria. II. Oxidative phosphorylation in skunk cabbage mitochondria. Plant Physiol. 44: 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R.J., Srivastava A. (1995) Polyphasic chlorophyll A fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 61: 32–42. [Google Scholar]
- Swiatek M., Kuras R., Sokolenko A., Higgs D., Olive J., Cinque G., et al. (2001) The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13: 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo M., Lepetit B., Goss R., Wilhelm C., Mustardy L., Garab G. (2008) Structurally flexible macro-organization of the pigment-protein complexes of the diatom Phaeodactylum tricornutum. Photosynth. Res. 95: 237–245. [DOI] [PubMed] [Google Scholar]
- Vishwakarma A., Gupta K.J. (2017) Isolation and structural studies of mitochondria from pea roots. InPlant Respiration and Internal Oxygen: Methods and Protocols. Edited by Jagadis Gupta K. pp. 87–95. Springer; New York, NY. [DOI] [PubMed] [Google Scholar]
- Watson K., Smith J.E. (1967) Oxidative phosphorylation and respiratory control in mitochondria from Aspergillus niger. Biochem. J. 104: 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Gruber A., Kroth P.G. (2009) The presence and localization of thioredoxins in diatoms, unicellular algae of secondary endosymbiotic origin. Mol. Plant 2: 468–477. [DOI] [PubMed] [Google Scholar]
- Werner D. (1977) The biology of diatoms. InBotanical Monographs,Vol. 13. Edited by Werner D. pp. 85–87. University of California Press, Berkeley and New York, NY. [Google Scholar]
- Wittpoth C. (1996) Isolierung und Characterisierung von Plastiden aus Kieselalgen. Dissertation. Heinrich-Heine Universit�t D�sseldorf, D�sseldorf, NRW.
- Wittpoth C., Kroth P.G., Weyrauch K., Kowallik K.V., Strotmann H. (1998) Functional characterization of isolated plastids from two marine diatoms. Planta 206: 79–85. [Google Scholar]
- Zhu S.-H., Green B.R. (2010) Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim. Biophys. Acta (BBA) Bioenerg. 1797: 1449–1457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.