Short abstract
Objective
A relationship between polymorphisms rs1128503 and rs1045642 in the multidrug resistance 1 gene (MDR1) and susceptibility to hepatocellular carcinoma (HCC) has been reported but is inconclusive. This study was performed to explore the significance of MDR1 polymorphisms rs1128503 and rs1045642 in screening and diagnosis of HCC.
Methods
Studies of association analyses between MDR1 gene polymorphisms rs1128503 and rs1045642 and HCC were selected from three foreign language databases (PubMed, Cochrane, and Embase) and three Chinese databases (Wanfang, China National Knowledge Infrastructure, and China Knowledge Network) and subjected to meta-analysis.
Results
We found no significant relationship between the rs1128503 polymorphism and susceptibility to HCC in 4 cohorts and no significant relationship between the rs1045642 polymorphism and susceptibility to HCC in 3 cohorts.
Conclusions
There was no relationship between polymorphisms rs1128503 or rs1045642 of the MDR1 gene and susceptibility to HCC.
Keywords: Hepatocellular carcinoma (HCC), meta-analysis, multidrug resistance 1 (MDR1), polymorphism, susceptibility, P-glycoprotein
Introduction
Hepatocellular carcinoma (HCC) has a high degree of malignancy and a poor prognosis. The highest worldwide incidences of HCC are in Asia and Africa; about 75% of HCCs occur in Asia.1 Chronic hepatitis B and hepatitis C virus infections are major risk factors for HCC, but only 10% of individuals infected with these viruses eventually develop HCC.2,3 Therefore, genetic and environmental factors may be involved in the occurrence and development of HCC.
The human multidrug resistance 1 gene (MDR1) is located on the long arm of chromosome 7 and contains 28 exons. The intron and exon junctions conform to the classical A/G rule and have a full length of 4.5 kb. An open reading frame encoding a 1280-amino acid polypeptide is glycosylated to form a 170-kDa membrane glycoprotein (P-glycoprotein), which plays a physiological role in protecting cells from toxin and metabolite damage.4 Recent studies have found that MDR1 polymorphism is not only an important genetic factor affecting the response of cancer patients to chemotherapy drugs but is also related to patients’ susceptibility to disease and clinical manifestations.5–7
A relationship between polymorphism rs1128503 or rs1045642 of the MDR1 gene and susceptibility to HCC has been reported but the conclusions are inconsistent. Therefore, we performed this study to objectively evaluate the relationship between HCC and MDR1 polymorphisms rs1128503 and rs1045642 by meta-analysis. We aimed to explore the significance of MDR1 polymorphisms rs1128503 and rs1045642 in HCC screening and diagnosis.
Material and methods
Inclusion and exclusion criteria
The inclusion criteria for this study were as follows: (1) a case control study; (2) clinical study to evaluate the relationship between rs1128503 and rs1045642 polymorphisms of MDR1 and the risk of HCC; and (3) sufficient data, including the number of subjects and gene frequency.
The exclusion criteria for this study were as follows: (1) meeting summary, case report, or review article; (2) relationship between MDR1 rs1128503 and rs1045642 polymorphisms and HCC risk was not detected; or (3) a study with repeatedly reported data or unclear data.
Literature retrieval
Three foreign language databases, PubMed, Cochrane, and Embase, and three Chinese databases, Wanfang, China National Knowledge Infrastructure (CNKI), and China Knowledge Network, were comprehensively searched by the method of retrospection. The retrieval date ended on August 23, 2018. We used the following combined keywords and MeSH terms: “ABCB1, C3435T, C1236T, rs1128503, rs1045642, MDR1, MDR-1, p-glycoprotein, P-gp” and “polymorphism, SNP, variation, variants, locus, mutation” and “liver cancer, liver tumor, liver tumour, liver malignance, liver carcinoma, liver neoplasm, hepatocellular carcinoma, HCC, intrahepatic cholangiocarcinoma, ICC, hepato-cholangio-carcinoma, HCC-CC, hepatoma”.
Literature extraction and filtering and evaluation of data quality
Evaluation of the extracted publications was carried out by two independent researchers; if there was disagreement, a third researcher was included in the evaluation until consensus was reached. The retrieved publications were screened according to the preset inclusion and exclusion criteria, reviewing title, abstract, and full text systematically. Data extracted included first author, publication year, country, number of subjects and gene distribution, type of adverse reactions, source of controls, ethnicity, and Hardy-Weinberg equilibrium (HWE) test. The quality of the included studies was assessed using the Newcastle-Ottawa scale (NOS).
Statistical analysis
Statistical analysis was performed using Stata 13.0 (StataCorp LLC, College Station, TX, USA) for data processing, and heterogeneity among the studies was analyzed using the Q test and P-value, and heterogeneity was evaluated by I2. When P ≥ 0.1 or I2 ≤ 50%, there was no statistical heterogeneity among the studies and the fixed-effect model was used for combined analysis. When P < 0.1 or I2 > 50%, there was statistical heterogeneity among studies, and the combined analysis was performed using the random-effect model. The odds ratio (OR) value and 95% confidence interval (CI) were analyzed as the combined effect value with a test level α = 0.05. Potential publication bias was analyzed by using Egger’s test, and sensitivity analysis was performed if necessary.
Results
Literature search and screening results
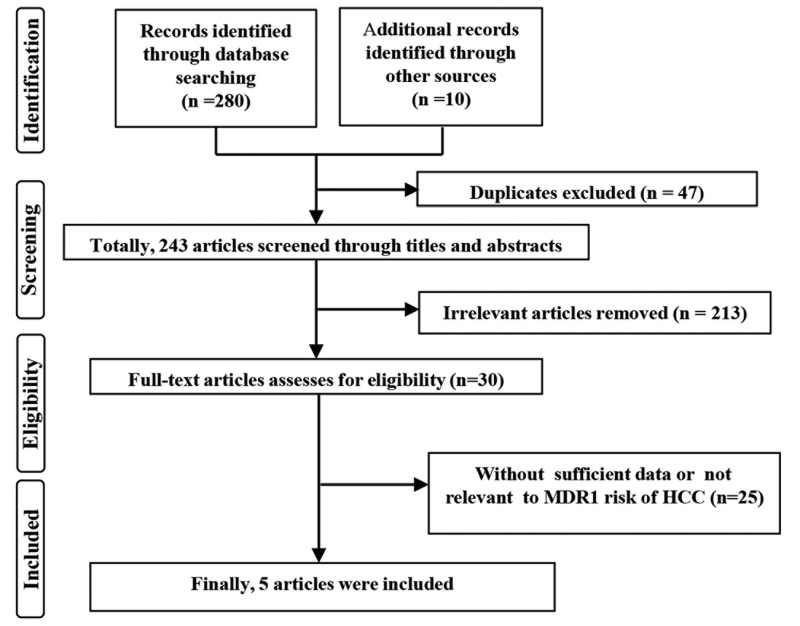
According to the search strategy, 290 publications were initially retrieved. After 47 duplicates were excluded, 213 unrelated articles and 25 publications with insufficient data or non-MDR1 polymorphisms and HCC risk were excluded by reading the title, abstract, and full text. A total of 5 qualified publications were screened and included in the meta-analysis8–12 (Figure 1).
Figure 1.
Flow diagram of study selection process.
Basic characteristics and quality evaluation of the included studies
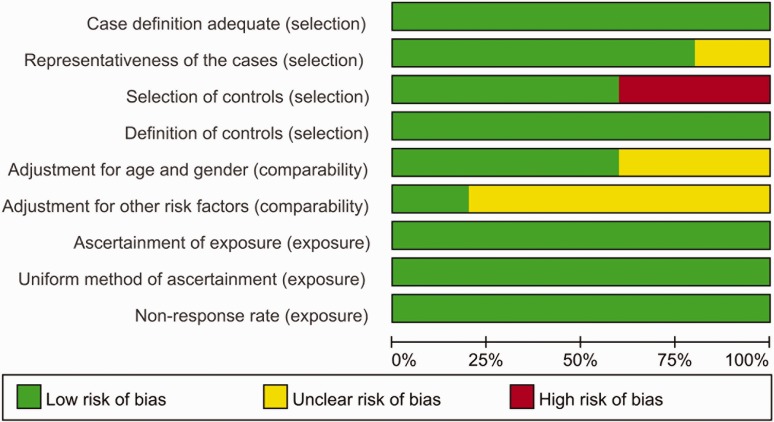
After the studies included in the literature were integrated and differentiated, 7 studies were available from 5 publications for analysis in this study. Of these, 4 studies were on the rs1128503 polymorphism and HCC susceptibility, and 3 studies were on the rs1045642 locus. The quality of the studies was scored using the NOS, and the results ranged from 5 to 9 points, indicating that the included studies were of medium to high quality (Table 1, Figure 2).
Table 1.
Characteristics of studies on the associations between rs1128503 (T > C) and rs1045642 (T > C) polymorphisms in MDR1 and hepatocellular cancer.
| Author | Year | Country | Ethnicity | MDR1 type | Source of controls | Control type | Cases | Controls | Cases |
Controls |
HWE | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WH | WM | MH | WH | WM | MH | |||||||||||
| De | 2017 | Italy | Caucasian | rs1128503 | HB | HBV/HCV | 192 | 167 | 46 | 92 | 54 | 22 | 93 | 52 | 0.15 | 7 |
| Italy | Caucasian | rs1128503 | PB | Healthy | 192 | 192 | 46 | 92 | 54 | 39 | 95 | 58 | 1.00 | 7 | ||
| Dong | 2013 | China | Asian | rs1045642 | PB | Healthy | 109 | 109 | 16 | 45 | 48 | 22 | 47 | 40 | 0.50 | 8 |
| Jing | 2013 | China | Asian | rs1128503 | PB | Healthy | 109 | 109 | 36 | 54 | 19 | 39 | 48 | 22 | 0.60 | 9 |
| Chen | 2011 | China | Asian | rs1128503 | HB | Cholecyst | 36 | 50 | 15 | 6 | 15 | 24 | 14 | 12 | 0.02 | 6 |
| China | Asian | rs1045642 | HB | Cholecyst | 36 | 50 | 8 | 10 | 18 | 11 | 12 | 27 | <0.01 | 6 | ||
| Fukuda | 2010 | Japan | Asian | rs1045642 | PB | Healthy | 58 | 61 | 13 | 29 | 16 | 8 | 39 | 14 | 0.08 | 6 |
HBV, hepatitis B virus; HCV, hepatitis C virus; HB, hospital-based; PB, population-based; WH, wild homozygous genotype; WM, wild/mutant heterozygous genotype; MH, mutant homozygous genotype; HWE, Hardy-Weinberg equilibrium; NOS, Newcastle-Ottawa scale.
Figure 2.
Quality assessment scale of eligible studies.
Meta-analysis
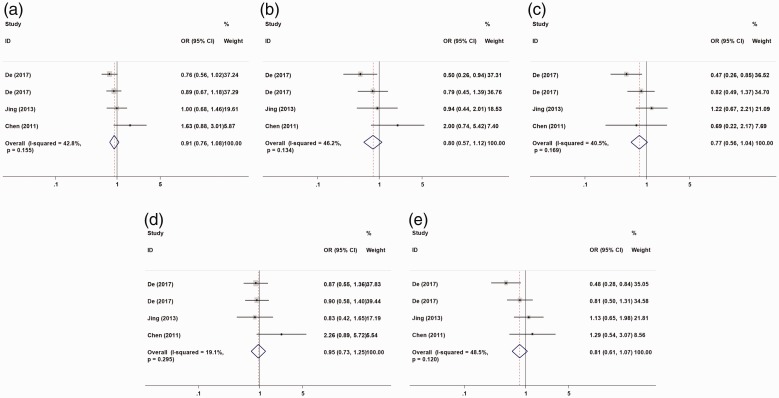
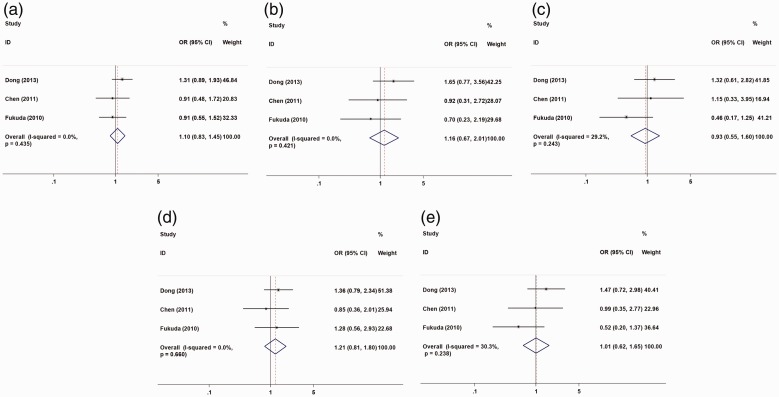
The association between susceptibility to HCC and two MDR1 polymorphism sites (rs1128503 and rs1045642) was analyzed in an allele model (C vs. T), a homozygous model (CC vs. TT), a heterozygous model (CT vs. TT), a recessive model (CC vs. CT + TT), and a dominant model (CC + CT vs. TT), respectively. Four studies were included and we found no significant relation between MDR1 rs1128503 polymorphism and susceptibility to HCC under the five genetic models (Table 2, Figure 3). Meta-analysis of MDR1 rs1045642 polymorphism and susceptibility to HCC from 3 studies also showed no significant relation (Table 2, Figure 4). Racial subgroup analysis showed a relation between MDR1 rs1128503 polymorphism and HCC risk in Caucasians (CC vs. TT: OR = 0.64, 95% CI = 0.42–0.98, P = 0.039, I2 = 12.4%; CT vs. TT: OR = 0.64, 95% CI = 0.44–0.94, P = 0.024, I2 = 48.2%; CC + CT vs. TT: OR = 0.64, 95% CI =0.45–0.93, P = 0.017, I2 = 47.4%). A subgroup analysis of control species showed a relation between MDR1 rs1128503 polymorphism and HCC risk in patients with hepatitis virus infection and gallstones (CT vs. TT: OR = 0.51, 95% CI = 0.30–0.86, P = 0.011, I2 = 0.0%) (Table 3).
Table 2.
OR and 95% CI for hepatocellular cancer and rs1128503 or rs1045642 polymorphism in MDR1 under different genetic models.
| Genetic models | n | OR (95% CI) | P (OR) | Analysis model | I2(%) | P(H) | P (Egger) | P (Begg) |
|---|---|---|---|---|---|---|---|---|
| rs1128503 T>C | ||||||||
| Allele (C vs. T) | 4 | 0.91 (0.76, 1.08) | 0.260 | F (M-H) | 42.8 | 0.155 | 0.082 | 0.308 |
| Homozygous model (CC vs. TT) | 4 | 0.80 (0.57, 1.12) | 0.193 | F (M-H) | 46.2 | 0.134 | 0.225 | 0.308 |
| Heterozygous model (CT vs. TT) | 4 | 0.77 (0.56, 1.04) | 0.093 | F (M-H) | 40.5 | 0.169 | 0.914 | 1.000 |
| Recessive model (CC vs. CT+TT) | 4 | 0.95 (0.73, 1.25) | 0.729 | F (M-H) | 19.1 | 0.295 | 0.230 | 0.734 |
| Dominant model (CC+CT vs. TT) | 4 | 0.81 (0.61, 1.07) | 0.137 | F (M-H) | 48.5 | 0.120 | 0.592 | 0.734 |
| rs1045642 T>C | ||||||||
| Allele (C vs. T) | 3 | 1.10 (0.83, 1.45) | 0.505 | F (M-H) | 0 | 0.435 | 0.293 | 1.000 |
| Homozygous model (CC vs. TT) | 3 | 1.16 (0.67, 2.01) | 0.587 | F (M-H) | 0 | 0.421 | 0.100 | 0.296 |
| Heterozygous model (CT vs. TT) | 3 | 0.93 (0.55, 1.60) | 0.800 | F (M-H) | 29.2 | 0.243 | 0.755 | 1.000 |
| Recessive model (CC vs. CT+TT) | 3 | 1.21 (0.81, 1.80) | 0.354 | F (M-H) | 0 | 0.660 | 0.481 | 0.296 |
| Dominant model (CC+CT vs. TT) | 3 | 1.01 (0.62, 1.65) | 0.962 | F (M-H) | 30.3 | 0.238 | 0.450 | 1.000 |
OR, odds ratio; CI, confidence interval; P(OR), probability for odds ratio; P(H), P for heterogeneity; n, number of the included studies; F, fixed-effect model; M-H, Mantel-Haenszel method.
Figure 3.
Forest plot of hepatocellular cancer risk associated with rs1128503 (C>T) models. (a) allele model; (b) homozygous model; (c) heterozygous model; (d) recessive model; (e) dominant model. The horizontal line indicates the lower and upper limits of the 95% CI; the square indicates the OR, with the size of the square indicating the weight of the study and the dotted red line indicating the combined OR value. The diamond represents the combined effect size, and the larger the diamond, the larger the confidence interval. A cross between the diamond and the ineffective line indicates no statistical correlation between the factors studied and the outcome; if the diamond falls on the left side of the invalid vertical line, it indicates a protective factor; if the diamond falls on the right side of the line, it indicates a risk factor. OR, odds ratio; CI, confidence interval.
Figure 4.
Forest plot of hepatocellular cancer risk associated with rs1045642 (C>T) models. (a) allele model; (b) homozygous model; (c) heterozygous model; (d) recessive model; (e) dominant model. The horizontal line indicates the lower and upper limits of the 95% CI; the square indicates the OR, with the size of the square indicating the weight of the study and the dotted red line indicating the combined OR value. The diamond represents the combined effect size, and the larger the diamond, the larger the confidence interval. A cross between the diamond and the ineffective line indicates no statistical correlation between the factors studied and the outcome; if the diamond falls on the left side of the invalid vertical line, it indicates a protective factor; if the diamond falls on the right side of the line, it indicates a risk factor. OR, odds ratio; CI, confidence interval.
Table 3.
Stratified analyses of relation between hepatocellular cancer and rs1128503 or rs1045642 polymorphism in MDR1 under different genetic models.
| Subgroup | N | Allele (C vs. T) |
Homozygous model (CC vs. TT) |
Heterozygous model (CT vs. TT) |
Recessive model (CC vs. CT + TT) |
Dominant model (CC + CT vs. TT) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P (OR) | I2 (%) | OR (95% CI) | P (OR) | I2 (%) | OR (95% CI) | P (OR) | I2 (%) | OR (95% CI) | P (OR) | I2 (%) | OR (95% CI) | P (OR) | I2 (%) | ||
| rs1128503 T>C | ||||||||||||||||
| Ethnicity | ||||||||||||||||
| Asian | 2 | 1.15 (0.83, 1.58) |
0.409 | 43.4 | 1.24 (0.68, 2.26) |
0.484 | 29.0 | 1.06 (0.64, 1.82) |
0.784 | 0.0 | 1.18 (0.69, 2.03) |
0.543 | 65.3 | 1.18 (0.74, 1.88) |
0.499 | 0.0 |
| Caucasian | 2 | 0.82 (0.67, 1.01) |
0.063 | 0.0 | 0.64 (0.42, 0.98) |
0.039 | 12.4 | 0.64 (0.44, 0.94) |
0.024 | 48.2 | 0.89 (0.65, 1.21) |
0.449 | 0.0 | 0.64 (0.45, 0.93) |
0.017 | 47.4 |
| Control type | ||||||||||||||||
| Healthy | 2 | 0.93 (0.74, 1.17) |
0.524 | 0.0 | 0.84 (0.53, 1.32) |
0.447 | 0.0 | 0.97 (0.66,1.43) |
0.883 | 0.0 | 0.88 (0.61, 1.28) |
0.510 | 0.0 | 0.93 (0.65, 1.34) |
0.710 | 0.0 |
| Others | 2 | 0.88 (0.67, 1.14) |
0.324 | 79.6 | 0.75 (0.44, 1.26) |
0.269 | 81.3 | 0.51 (0.30, 0.86) |
0.011 | 0.0 | 1.04 (0.70, 1.57) |
0.835 | 69.9 | 0.64 (0.40, 1.01) |
0.058 | 71.7 |
| rs1045642 T>C | ||||||||||||||||
| Controls type | ||||||||||||||||
| Healthy | 2 | 1.15 (0.84, 1.56) |
0.379 | 20.2 | 1.26 (0.67, 2.36) |
0.473 | 32.7 | 0.89 (0.49, 1.61) |
0.702 | 63.0 | 1.33 (0.85, 2.10) |
0.214 | 0.0 | 1.02 (0.58, 1.78) |
0.946 | 65.1 |
| others | 1 | 0.91 (0.48, 1.72) |
0.774 | NU | 0.92 (0.31, 2.72) |
0.876 | NU | 1.15 (0.33, 3.95) |
0.829 | NU | 0.85 (0.36, 2.01) |
0.714 | NU | 0.99 (0.35, 2.77) |
0.980 | NA |
N, number of comparisons; OR, odds ratio; CI, confidence interval; NU, null, NA, not available.
Publication bias and sensitivity analysis
On the basis of the results of Egger’s test, there was no publication bias in this study (Table 2). To evaluate the stability of this meta-analysis, we excluded the included studies one by one and compared the differences between the effect values before and after each elimination. This analysis showed that the results were stable.
Discussion
The MDR1 rs1128503 and rs1045642 polymorphisms are synonymous mutations in exon 26 and exon 12,13 respectively, and the CC genotype in the mutation site is considered the wild type.14 The wild-type P-glycoprotein not only pumps drugs out of cells but, synergistically with immune function, also inhibits tumorigenesis and development.15 Some studies have found that P-glycoprotein can delay the apoptosis cascade of tumor cells by inhibiting caspase.16 Five relevant studies were included in this meta-analysis, and the results showed that the rs1128503 polymorphism may be related to HCC risk in Caucasian individuals and in patients with hepatitis virus infection or gallstones.
A previous meta-analysis showed that mutations in the MDR1 gene are associated with susceptibility to HCC and are risk factors for HCC.17 The different results between that meta-analysis and the current meta-analysis may be explained by two factors. First, in the early study, the association analysis between MDR1 polymorphisms and susceptibility to HCC was based on pooled results from 11 mutation sites in MDR1. In the current meta-analysis, the relation analysis between MDR1 polymorphism and susceptibility to HCC was conducted for only two polymorphic sites. Second, the subjects included in the previous study were Asian (Japanese and Chinese), whereas those in the current analysis were Asian (Japanese and Chinese) and Caucasian (Italian). Compared with the previous study, a more appropriate detailed analysis of the relation between MDR1 mutation and hepatocarcinogenesis, involving different populations, was performed in this meta-analysis, and the results were shown to be reliable.
The occurrence of HCC is a complex pathological process, involving multiple genes and environmental factors.18–20 Hepatitis virus infection,21,22 smoking,23,24 drinking,25,26 and genetic factors are non-negligible causes of HCC. These risk factors may cause chronic inflammation and accumulation of toxic products in the liver, leading to HCC.27,28 The protein encoded by MDR1 is involved in the elimination of endogenous and exogenous harmful substances.29 Polymorphisms in MDR1 will alter the structure or expression of the encoded protein, thereby affecting its efflux effect on carcinogens and the normal physiological functions of hepatocytes.30 However, abnormal protein expression of MDR1 also affects the sensitivity of cancer cells to drugs, thereby affecting the development of HCC.31 In this meta-analysis, we analyzed the relationship between two mutations of MDR1 and susceptibility to HCC without considering the influence of other factors. However, the number of subjects included in the current meta-analysis was limited, and the control group was included with different standards. Therefore, further stringent analyses with larger sample sizes are necessary to confirm the results of this meta-analysis.
In conclusion, this meta-analysis showed that MDR1 polymorphism rs1128503 may be related to HCC risk in Caucasians and in patients with hepatitis viral infection or gallstones. A better understanding of the effect of MDR1 gene polymorphisms on HCC risk by analyzing the relation between rs1128503 or rs1045642 and HCC might improve our understanding of the role of genetic factors in HCC risk.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was supported by a project of applied Development Program in Beibei of Chongqing (Grant No.: 2016-34).
References
- 1.Wang JB, Abnet CC, Chen W, et al. Association between serum 25(OH) vitamin D, incident liver cancer and chronic liver disease mortality in the Linxian Nutrition Intervention Trials: a nested case-control study. Br J Cancer 2013; 109: 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 3.McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis 2015; 19: 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mann JF, Schmieder RE, McQueen M, et al. Renal outcomes with telmisartan, ramipril, or both, in people at high vascular risk (the ONTARGET study): a multicentre, randomised, double-blind, controlled trial. Lancet 2008; 372: 547–553. [DOI] [PubMed] [Google Scholar]
- 5.Jamroziak K, Mlynarski W, Balcerczak E, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol 2004; 72: 314–321. [DOI] [PubMed] [Google Scholar]
- 6.He H, Yin J, Li X, et al. Association of ABCB1 polymorphisms with prognostic outcomes of anthracycline and cytarabine in Chinese patients with acute myeloid leukemia. Eur J Clin Pharmacol 2015; 71: 293–302. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Huang Z, Zheng K, et al. Two SNPs of ATP-binding cassette B1 gene on the risk and prognosis of colorectal cancer. Int J Clin Exp Pathol 2015; 8: 3083–3089. [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda M, Kawahara Y, Hirota T, et al. Genetic polymorphisms of hepatic ABC-transporter in patients with hepatocellular carcinoma. J Cancer Ther 2010; 1: 114–123. [Google Scholar]
- 9.Chen XJ, Wang XG, Shen YJ, et al. Correlation of MDR1 single nucleotide polymorphism with prognosis of hepatocellular carcinoma. J Chin Oncol 2011; 17: 209–211. [Google Scholar]
- 10.De Mattia E, Cecchin E, Polesel J, et al. Genetic biomarkers for hepatocellular cancer risk in a caucasian population. World J Gastroenterol 2017; 23: 6674–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jing R, Dong X, Deng W, et al. Correlation between MDR1 polymorphism and primary liver cancer in Guangxi. Chin J Oncol Prev Treat 2013; 5: 122–126. [Google Scholar]
- 12.Dong X. Association of MDR1 gene polymorphisms with susceptibility and external exposure factors to primary liver cancer in a case-control study. Guangxi Med Univ (Master's thesis) 2013.
- 13.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta 2009; 1794: 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA 2000; 97: 3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnstone RW, Ruefli AA, Smyth MJ. Multiple physiological functions for multidrug transporter P-glycoprotein? Trends Biochem Sci 2000; 25: 1–6. [DOI] [PubMed] [Google Scholar]
- 16.Ruefli AA, Tainton KM, Darcy PK, et al. P-glycoprotein inhibits caspase-8 activation but not formation of the death inducing signal complex (disc) following Fas ligation. Cell Death Differ 2002; 9: 1266–1272. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZC, Liu LZ, Liu XY, et al. Genetic polymorphisms of the multidrug resistance 1 gene MDR1 and the risk of hepatocellular carcinoma. Tumour Biol 2015; 36: 7007–7015. [DOI] [PubMed] [Google Scholar]
- 18.Liu ZM, Li LQ, Peng MH, et al. Hepatitis B virus infection contributes to oxidative stress in a population exposed to aflatoxin B1 and high-risk for hepatocellular carcinoma. Cancer Lett 2008; 263: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang ZY. Hepatocellular carcinoma–cause, treatment and metastasis. World J Gastroenterol 2001; 7: 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Nie Y, Zhao J, et al. Genetic polymorphism at miR-181a binding site contributes to gastric cancer susceptibility. Carcinogenesis 2012; 33: 2377–2383. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Chang CC, Marsh GM, et al. Population attributable risk of aflatoxin-related liver cancer: systematic review and meta-analysis. Eur J Cancer 2012; 48: 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirk GD, Lesi OA, Mendy M, et al. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene 2005; 24: 5858–5867. [DOI] [PubMed] [Google Scholar]
- 23.Wozniak A, Kulza M, Senczuk-Przybylowska M, et al. Selected biochemical parameters of oxidative stress as a result of exposure to tobacco smoke in animals addicted to ethyl alcohol. Przegl Lek 2012; 69: 824–832. [PubMed] [Google Scholar]
- 24.Miah S, Dudziec E, Drayton RM, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer 2012; 107: 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology 2004; 127: S87–S96. [DOI] [PubMed] [Google Scholar]
- 26.Purohit V, Rapaka R, Kwon OS, et al. Roles of alcohol and tobacco exposure in the development of hepatocellular carcinoma. Life Sci 2013; 92: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 2005; 5: 558–567. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka M, Katayama F, Kato H, et al. Hepatitis B and C virus infection and hepatocellular carcinoma in China: a review of epidemiology and control measures. J Epidemiol 2011; 21: 401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szakács G, Váradi A, Ozvegy-Laczka C, et al. The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME-Tox). Drug Discov Today 2008; 13: 379–393. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Xie H, Wei B, et al. Association of MDR1 gene SNPs and haplotypes with the tacrolimus dose requirements in Han Chinese liver transplant recipients. PLoS One 2011; 6: e25933. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Lasagna N, Fantappiè O, Solazzo M, et al. Hepatocyte growth factor and inducible nitric oxide synthase are involved in multidrug resistance-induced angiogenesis in hepatocellular carcinoma cell lines. Cancer Res 2006; 66: 2673–2682. [DOI] [PubMed] [Google Scholar]