Abstract
Purpose: The present study aims to investigate the involvement of lncRNA GAPLINC in non-small lung cancer (NSCLC).
Patients and methods: The study included 70 patients with NSCLC (39 males and 31 females, 33 to 68 years, 49.3 ± 6.4 years). RT-qPCR, transient cell transfections, measurement of in vitro cell migration and invasion abilities and western blot were carrying out during the research.
Results: We showed that GAPLINC was up-regulated in NSCLC tissues and positively correlated with TGF-β1. In vitro cell experiment showed that over-expression of TGF-β1 significantly up-regulated the expression of GAPLINC, while over-expression of GAPLINC failed to affect TGF-β1. Follow-up study showed that high GAPLINC level in NSCLC tissue was closely correlated with poor survival rate of NSCLC patients. Over-expressions of TGF-β1 and GAPLINC resulted to accelerated migration and invasion of NSCLC cells. In addition, the silencing of GAPLINC siRNA attenuated the effect of TGF-β1 treatment.
Conclusion: TGF-β1 may mediate lncRNA GAPLINC expression to promote NSCLC cell invasion and migration.
Keywords: non-small cell lung cancer, lncRNA GAPLINC, TGF-β1, prognosis
Introduction
A large population of the cancer patients are diagnosed with inoperable conditions. Therefore, the accurate prognosis is important to clinicians to design individualized treatment strategies, which are beneficial to improve the patients’ survival rate.1,2 Non-small cell lung cancer (NSCLC) is the major subtype of lung cancer and possesses more than 80% of all lung cancer cases.3 NSCLC is the most common type of malignancies as well as the leading cause of deaths in cancer patients in many regions of the world, including China and USA.4,5 After the initial diagnosis, the 5-year survival rate of NSCLC patients is only less than 20%.6 Therefore, novel effective prediction markers and therapeutic targets are urgently needed.
Long non-coding RNAs (lncRNAs, >200 nt) play critical roles in gene expression regulation. They have attracted more and more attention from researchers in cancer research field.7,8 The altered expression of certain key regulative lncRNAs in cancer development and progression is closely related with dysregulated expression of some oncogenes or tumor suppressors. Therefore, regulation of the expression of certain lncRNAs may indirectly modulate cancer progression.9,10 However, the functions of most lncRNAs remain to be elusive. LncRNA GAPLINC has been characterized as an oncogene in gastric cancer and colorectal cancer,11,12 while its role in lung cancer is rarely known. The present study was carried out to investigate the roles of GAPLINC in NSCLC.
Materials and methods
Patients
The Ethics Committee of Jinan Central Hospital approved this study. It included 70 patients with NSCLC (39 males and 31 females, 33–68 years, 49.3±6.4 years). All the patients were enrolled in Jinan Central Hospital between June 2011 and June 2013. The inclusion criteria were: 1) new NSCLC cases were diagnosed by histopathological examinations; 2) patients were willing to donate biopsies and participate in a 5-year follow-up. The exclusion criteria were: 1) patients received any drug or other types of treatment before admission; 2) patients were complicated with other clinical disorders. Based on the AJCC staging system, there were 15, 20, 21, and 14 cases that were at stage I–IV, respectively. All the 70 patients signed the informed consent.
Follow-up
All the 70 patients were followed up for 5 years (or until their death) to observe their survival conditions. Patients who were lost, died of other clinical disorders, or traffic accidents were not included in this study.
Specimens and cells
The biopsy was performed on all patients before therapies. NSCLC cancer tissues and their adjacent (within 2 cm around the tumor) non-cancer tissues (0.1–0.3 g) were collected from each patient during biopsy.
H23 and H522 cell lines were used in this study, which were purchased from ATCC (USA). The cells were cultured in RPMI-1640 medium (10% FBS) at 37°C and 5% CO2.
RT-qPCR
Total RNA extraction was conducted using Ribozol reagent (Thermo Fisher Scientific). cDNA synthesis was performed using Reverse Transcriptase AMV (Sigma-Aldrich, USA). qPCR mixtures were prepared using SYBR Green Master Mix (Bio-Rad, USA) to detect the expression of GAPLINC and TGF-β1 with 18s rRNA or GAPDH, respectively. All PCR reactions were repeated 3 times. All data were normalized using 2−ΔΔCT method.
Transient cell transfections
GAPLINC and TGF-β1 expression vectors were constructed using the pcDNA3.1 vector (Sangon, Shanghai, China). GAPLINC siRNA (TGATAGCCCGAGGATGTGGATG) and Scrambled negative control (NC) siRNA (5ʹ-UUCUCCGAACGUGUCACGUdTdT-3ʹ) were purchased from Sangon (Shanghai, China). H23 and H522 cells were cultured overnight to reach a confluence of 70–80%. All transient transfections were then performed with 35 nM siRNA (NC siRNA as NC group) or 12 nM vector (empty vector as NC group). All subsequent experiments were performed at 24 hrs after transfections. Control (C) cells for all groups were untransfected cells.
Measurement of in vitro cell migration and invasion abilities
H23 and H522 cells were harvested at 24 hrs after transfections. Cell suspensions were prepared by mixing 1 mL RPMI-1640 medium (1% FBS) with 3×104 cells. The upper Transwell chamber was filled with cell suspension, while the lower Transwell chamber was filled with a mixture of 80% RPMI-1640 medium and 20% FBS. Before invasion assay, Matrigel (356234, Millipore, USA) was used to coat the upper chamber at 37°C for 6 hrs to mimic in vivo invasion conditions. Cells were cultured at 37°C and 5% CO2 for 3 hrs to allow cell migration and invasion. After that, the upper chamber was stained for 15 mins at room temperature with 0.5% crystal violet (Sigma-Aldrich, USA). The upper chamber membrane was then cleaned. The stained cells were observed under a light microscope.
Western blot
H23 and H522 cells were subjected to total protein extraction using Total Protein Extraction Kit (NBP2-37853, Novus Biologicals). After denaturing, protein samples were subjected to 10% SDS-PAGE gel electrophoresis. After gel transferred to PVDF membranes, blocking was performed in 5% non-fat milk at room temperature for 2 hrs. Membranes were then subjected to incubation with rabbit anti-human GAPDH (1:1300, ab8245, Abcam) and TGF‑β1 (1:1300, ab92486, Abcam) primary antibodies at 4°C overnight. In the next day, the membrane was incubated with IgG-HRP goat anti-rabbit secondary antibody (1:1000, MBS435036, MyBioSource) at room temperature for 2 hrs. Signals were detected using ECL™ Western Blotting Analysis System (Sigma-Aldrich) and analyzed using Image J v1.46 software.
Statistical process
Data were presented as the mean values, based on three biological replicates of each experiment. Differences between NSCLC and non-cancer tissues were analyzed by paired t-test. Differences among cell transfection groups or clinical stages were analyzed by one-way ANOVA and Tukey test. The correlation between GAPLINC and TGF-β1 was analyzed by linear regression analysis. Survival analysis was performed by dividing the 70 patients into high GAPLINC expression (n=33) and low GAPLINC expression (n=37) groups based on Youden’s index. Based on survival data, survival curves were plotted and analyzed using K-M method and log-rank test, respectively. The statistically significant cutoff value was p<0.05.
Results
GAPLINC and TGF-β1 were upregulated in NSCLC tissues
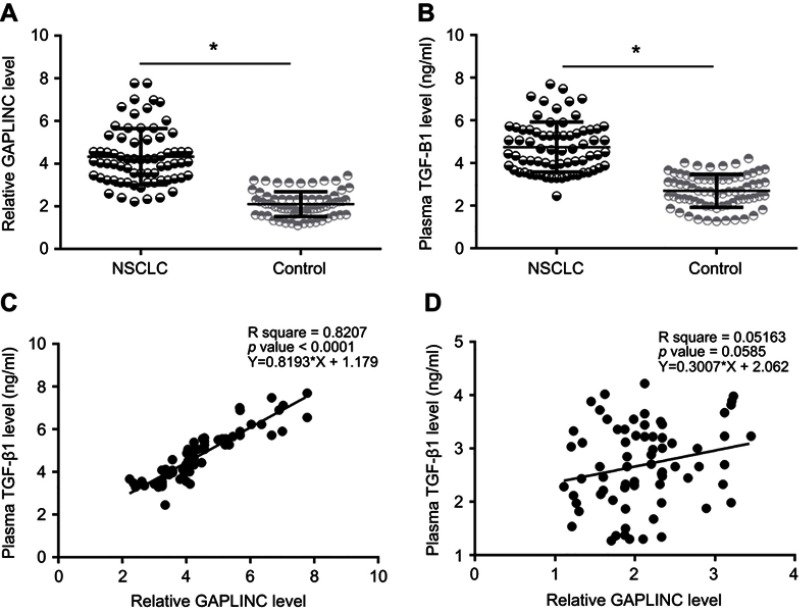
GAPLINC and TGF-β1 mRNA were detected by performing RT-qPCR experiments and analyzed by paired t-test. Comparing to adjacent non-cancer tissues, expression levels of GAPLINC (Figure 1A) and TGF-β1 mRNA (Figure 1B) were significantly increased in NSCLC tissues (p<0.05). The correlation between GAPLINC and TGF-β1 was analyzed by performing linear regression analysis. It was observed that GAPLINC and TGF-β1 were positively correlated in NSCLC tissues (Figure 1C). However, the correlation between them was not significant in non-cancer tissues (Figure 1D).
Figure 1.
GAPLINC and TGF-β1 were upregulated in non-small cell lung cancer (NSCLC) tissues. Analysis of GAPLINC and TGF-β1 mRNA expression data by one-way ANOVA and Tukey test. (A) and TGF-β1 mRNA (B) was significantly upregulated in NSCLC tissues than in non-cancer tissues (*p<0.05). Linear regression analysis showed that GAPLINC and TGF-β1 were positively correlated in NSCLC tissues (C), but not in adjacent non-cancer tissues (D).
TGF-β1 overexpression resulted in the upregulation of GAPLINC
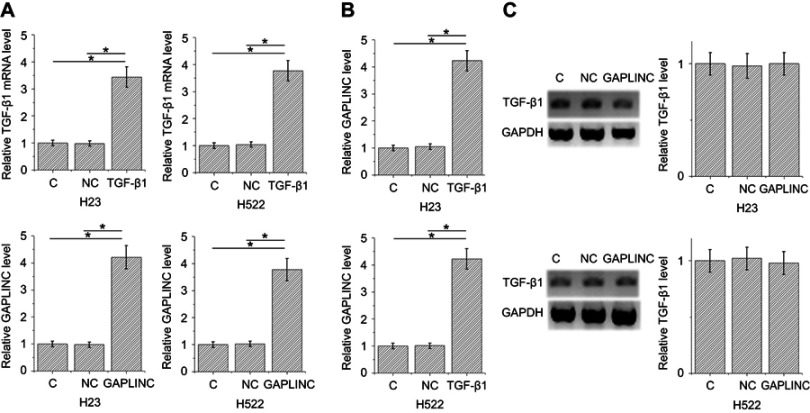
TGF-β1 and GAPLINC expression vectors were transfected into H23 and H522 cells to further investigate the relationship between TGF-β1 and GAPLINC. Expression levels of TGF-β1 and GAPLINC were significantly increased in both cell lines after 24-hr transfections, comparing to NC and C. It indicates the successful overexpression (Figure 2A, p<0.05). Moreover, TGF-β1 overexpression resulted in the upregulated expression of GAPLINC (Figure 2B, p<0.05), while GAPLINC overexpression failed to affect TGF-β1 (Figure 2C).
Figure 2.
TGF-β1 overexpression resulted in the upregulation of GAPLINC. Expression levels of TGF-β1 and GAPLINC were significantly increased in both cell lines after 24-hr transfections, comparing to negative control and control, indicating the successful overexpression (A). TGF-β1 overexpression resulted in upregulated expression of GAPLINC (B) (p<0.05), but GAPLINC overexpression failed to affect TGF-β1 (C); (*p<0.05).
High GAPLINC level in NSCLC tissue predicted poor survival
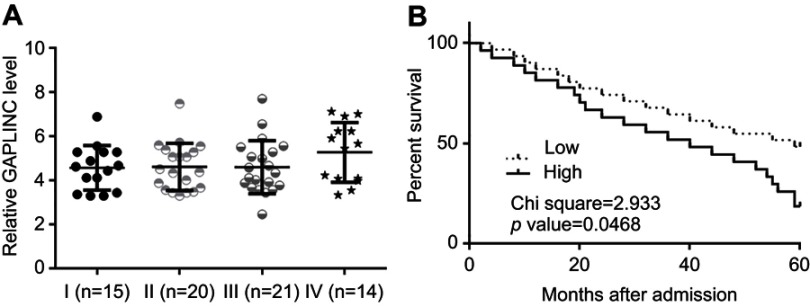
Differences in expression levels of GAPLINC in NSCLC tissues among patients with different clinical stages were analyzed by one-way ANOVA and Tukey test. No significant differences in expression levels of GAPLINC were found (Figure 3A). Through the aforementioned methods, survival curves were plotted and compared. It was observed that patients with high expression levels of GAPLINC showed significantly worse overall survival (Figure 3B). It is noted that patients with high levels of TGF-β1 also showed a lower overall survival rate, compared to patients with low levels of TGF-β1.
Figure 3.
High GAPLINC level in non-small cell lung cancer (NSCLC) tissue predicted poor survival. Analysis of expression levels of GAPLINC in NSCLC tissues by one-way ANOVA and Tukey test (A). Survival curve analysis showed that patients with high expression levels of GAPLINC had significantly worse overall survival (B).
TGF-β1 promoted GAPLINC expression to accelerate NSCLC cell migration and invasion
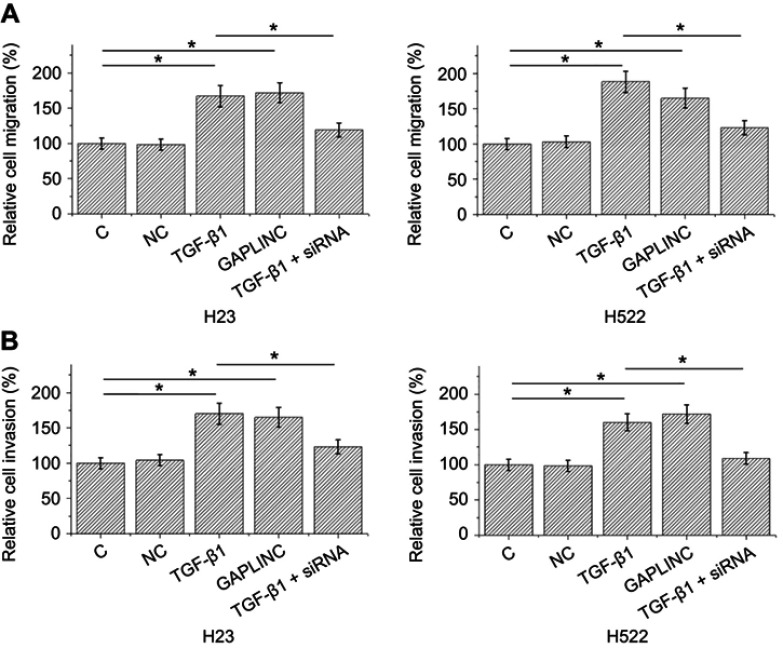
Transwell migration and invasion data were analyzed by one-way ANOVA and Tukey test. Compared to NC and C, TGF-β1, and GAPLINC overexpressions resulted in accelerated migration (Figure 4A) and invasion (Figure 4B) of H23 and H522 cells (p<0.05). In addition, silencing of GAPLINC siRNA attenuated the effect of TGF-β1 treatment.
Figure 4.
TGF-β1 promoted GAPLINC expression to accelerate non-small cell lung cancer (NSCLC) cell migration and invasion. Analysis of transwell migration and invasion showed that TGF-β1 and GAPLINC overexpression caused the accelerated migration (A) and invasion (B) of H23 and H522 cells. In addition, silencing of GAPLINC siRNA attenuated the effect of TGF-β1 treatment (*p<0.05).
Discussion
Our study mainly investigated the functionality of GAPLINC in NSCLC. We found GAPLINC was upregulated in NSCLC and had prognostic values. In addition, we found that GAPLINC was upregulated by TGF-β1 to promote NSCLC cell migration and invasion.
Accurate prediction of cancer development is critical for the survival of cancer patients but is also challenged by the lack of sensitive markers. Several biomarkers, such as serum SELDI proteomic patterns13 and glycans14 have been developed to predict NSCLC. But the sensitivity and specificity are not satisfactory and the combined application of multiple biomarkers is limited. With the advantages with non-invasiveness, circulating biomarkers, such as tumor cells, miRNA, and lncRNAs in blood, have been widely used in clinical practices.14–16 LncRNA GAPLINC was discovered to be upregulated in gastric cancer and colorectal cancer,11,12 indicating oncogenic roles. Our data also showed the upregulating of GAPLINC in NSCLC. It confirmed that high level of GAPLINC in NSCLC tissues was a potential prognostic biomarker for NSCLC. Besides these efforts, more clinical studies are needed to further test the accuracy of plasma circulating GAPLINC in the prognosis of NSCLC.
TGF-β signaling participates in cancer biology through many aspects.17 In most types of cancers, TGF-β exerts tumor suppressive function during the early cancer development stage by inhibiting cancer cell proliferation. However, it promotes cancer metastasis during late stages by elevating the ability of cancer cell migration and invasion.18 Due to the fact that most NSCLC are diagnosed at advanced stages, the inhibiting of TGF-β1 is a promising method to inhibit cancer cell migration and invasion.17 TGF-β may participate in cancer biology by regulating the expression of certain lncRNAs.19,20 In the present study, we proved that GAPLINC was at the downstream of TGF-β signaling, during the regulation of NSCLC cell migration and invasion. It is widely known that GAPLINC regulates SNAI2, PSF, and NONO in cancer biology.12 Therefore, GAPLINC may be a mediator between TGF-β and those downstream effectors.
Conclusion
GAPLINC was overexpressed in NSCLC and regulated by TGF-β1. The regulation of GAPLINC by TGF-β1 is involved in the regulation of NSCLC cell migration and invasion.
Ethical approval and informed consent
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration. Informed consent was obtained from all individual participants included in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hogdall E. Cancer antigen 125 and prognosis. Curr Opin Obstet Gynecol. 2008;20(1):4–8. doi: 10.1097/GCO.0b013e3282f2b124 [DOI] [PubMed] [Google Scholar]
- 2.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–S15. doi: 10.1016/S0959-8049(01)00231-3 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 4.Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. doi: 10.1016/S0140-6736(10)62101-0 [DOI] [PubMed] [Google Scholar]
- 5.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastorino U. Lung cancer screening. Br J Cancer. 2010;102(12):1681–1686. doi: 10.1038/sj.bjc.6605660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 8.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi: 10.1038/modpathol.2012.160 [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74(23):6890–6902. doi: 10.1158/0008-5472.CAN-14-0686 [DOI] [PubMed] [Google Scholar]
- 12.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7(27):42183–42194. doi: 10.18632/oncotarget.9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SY, Xiao XY, Zhang WG, et al. Application of serum SELDI proteomic patterns in diagnosis of lung cancer. BMC Cancer. 2005;5:83. doi: 10.1186/1471-2407-5-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820(9):1347–1353. doi: 10.1016/j.bbagen.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 15.Punnoose EA, Atwal SK, Spoerke JM, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5(9):e12517. doi: 10.1371/journal.pone.0012517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colak S, Ten Dijke P. Targeting TGF-beta Signaling in Cancer. Trends Cancer. 2017;3(1):56–71. doi: 10.1016/j.trecan.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 18.Akhurst RJ, Derynck R. TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol. 2001;11(11):S44–S51. [DOI] [PubMed] [Google Scholar]
- 19.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 20.Fan Y, Shen B, Tan M, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–1541. doi: 10.1158/1078-0432.CCR-13-1455 [DOI] [PubMed] [Google Scholar]