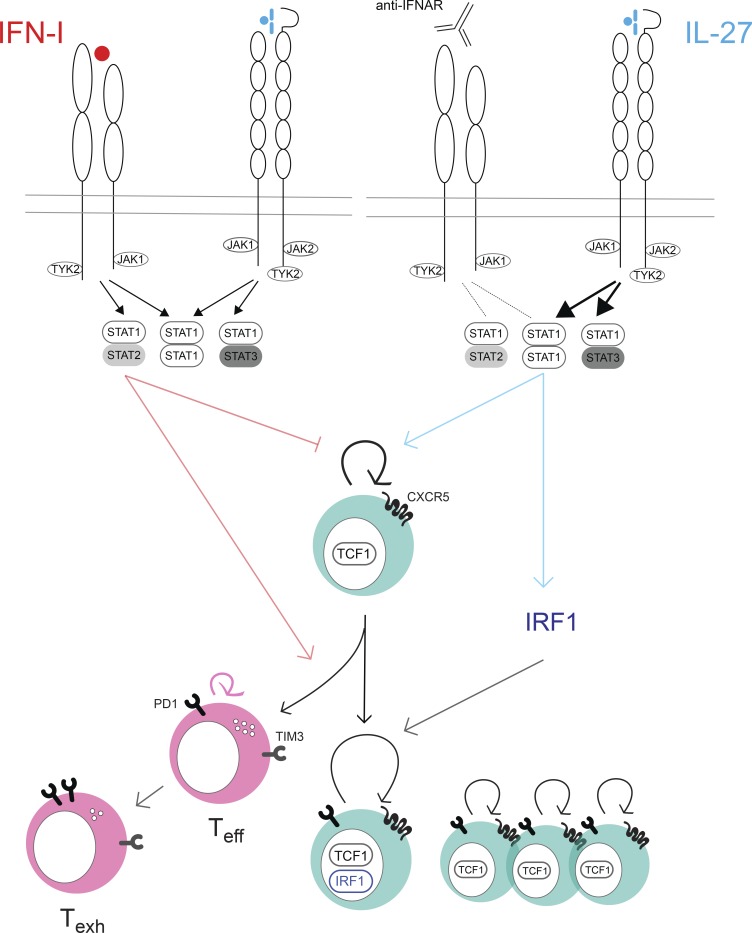
CXCR5+ TCF1+ CD8+ T cells sustain responses during persistent viral infection and mediate the proliferative burst following anti-PD1 treatment. Huang et al. show that IL-27 supports rapid division of these cells by competing with type 1 interferon for STAT1, driving IRF1 expression and preventing cell death.
Abstract
Chronic infection and cancer are associated with suppressed T cell responses in the presence of cognate antigen. Recent work identified memory-like CXCR5+ TCF1+ CD8+ T cells that sustain T cell responses during persistent infection and proliferate upon anti-PD1 treatment. Approaches to expand these cells are sought. We show that blockade of interferon type 1 (IFN-I) receptor leads to CXCR5+ CD8+ T cell expansion in an IL-27– and STAT1-dependent manner. IFNAR1 blockade promoted accelerated cell division and retention of TCF1 in virus-specific CD8+ T cells. We found that CD8+ T cell–intrinsic IL-27 signaling safeguards the ability of TCF1hi cells to maintain proliferation and avoid terminal differentiation or programmed cell death. Mechanistically, IL-27 endowed rapidly dividing cells with IRF1, a transcription factor that was required for sustained division in a cell-intrinsic manner. These findings reveal that IL-27 opposes IFN-I to uncouple effector differentiation from cell division and suggest that IL-27 signaling could be exploited to augment self-renewing T cells in chronic infections and cancer.
Graphical Abstract
Introduction
During persistent viral infections and cancer, high antigen levels and the inflammatory environment promote functional exhaustion of antigen-specific T cells (Zajac et al., 1998; Wherry and Kurachi, 2015). Exhausted T cells display poor effector functions and increased surface expression of multiple inhibitory receptors which maintain the hyporesponsive state (Barber et al., 2006; Blackburn et al., 2009). Blockade of inhibitory receptors can rescue specific functions and enhance their ability to control persistent viral infections and cancer (Barber et al., 2006; Blackburn et al., 2009; Pardoll, 2012). Recently, a subpopulation of exhausted, memory-like, self-renewing CD8+ T cells expressing CXCR5 and the transcription factor TCF1 was identified in mouse persistent lymphocytic choriomeningitis mammarenavirus (LCMV) clone 13 (Cl13) infection and cancer models and patients infected with hepatitis C virus and HIV (He et al., 2016; Im et al., 2016; Leong et al., 2016; Lin et al., 2016; Utzschneider et al., 2016; Wu et al., 2016; Petrovas et al., 2017; Wieland et al., 2017). These CXCR5+ CD8+ T cells, also termed T follicular cytotoxic cells (Leong et al., 2016), were required to sustain antiviral T cell responses to chronic infections (Utzschneider et al., 2016) and provide the proliferative burst and effector function following anti–PD-1/PD-L1 therapy (Im et al., 2016). Given the efficacy of anti–PD-1 in cancer therapy coupled to the fact that only a finite number of PD-1–responsive CD8+ T cells exist in patients (Huang et al., 2017), understanding the cellular and molecular mechanisms that regulate CXCR5+ CD8+ T cell differentiation and expansion will provide crucial knowledge for improving the efficacy of immunotherapy in both persistent infections and cancer.
The characterization of the CXCR5+ CD8+ T cell phenotype in persistent LCMV Cl13 infection catalyzed in-depth cellular and molecular analyses of this subpopulation. Multiple transcription factors controlling CXCR5+ CD8+ T cell generation have been identified: in addition to TCF1, which is required for CXCR5+ CD8+ T cell formation, there are CXCR5+ CD8+ T cell–promoting transcription factors BCL6 and FOXO1 and CXCR5+ CD8+ T cell–restraining factors BLIMP1, ID2, IRF4, RUNX3, and STAT4, which favor effector CD8+ differentiation (Leong et al., 2016; Utzschneider et al., 2016, 2018; Man et al., 2017; Shan et al., 2017; Danilo et al., 2018). Despite recent understanding of the transcription factors required for CXCR5+ CD8+ T cell generation, less is known about how specific cytokine inputs regulate CXCR5+ CD8+ T cell differentiation and expansion. To date, only type 1 IFN (IFN-I) and IL-12 have been demonstrated to directly regulate the CXCR5+ CD8+ T cell population, with both cytokines reported to negatively control CXCR5+ CD8+ T cell formation (Wu et al., 2016; Danilo et al., 2018). During clonal expansion, effector differentiation is coupled to cell division (Lin et al., 2016). In the course of viral infection, TCF1hi cells can enter a rapid-amplifying state that is characterized by a higher expression of effector-associated molecules such as T-bet and IFNγ compared with quiescent TCF1hi cells (Lin et al., 2016). Whether IFN-I restrains CXCR5+ CD8+ T cells by promoting loss of TCF1 in all CD8+ T cells, or by inhibiting division of TCF1hi cells, is unknown. Moreover, cytokines that positively regulate CXCR5+ CD8+ T cell differentiation or expansion are yet to be identified (Hashimoto et al., 2019).
IL-27 is a heterodimeric cytokine in the IL-6 and IL-12 family composed of the p28 and Epstein-Barr virus–inducible protein 3 (EBI3) subunits. IL-27 signals through a heterodimeric receptor consisting of WSX-1 and gp130, which is shared with the IL-6 receptor complex (Kastelein et al., 2007). Stimulation of IL-6 or IL-27 can elicit STAT1/STAT1, STAT1/STAT3, or STAT3/STAT3 complexes and thus can induce overlapping gene signatures (Hirahara et al., 2015). IL-27 has been demonstrated to regulate diverse functions in lymphocytes based on its ability to activate both STAT1 and STAT3 transcription factors (Yoshida and Hunter, 2015). During persistent viral infection, IL-27R is required for viral control, which correlates with significantly impaired anti-viral CD4+ T cell responses in IL-27R–deficient mice (Harker et al., 2013, 2018).
Most studies focusing on how IL-27 signaling regulates T cell responses center on modulation of CD4+ T cell subsets (Hunter and Kastelein, 2012). Effector CD8+ T cell responses seem to be attenuated by IL-27 during coronavirus and influenza infections (de Aquino et al., 2014; Liu et al., 2014), but not LCMV Cl13 infection (Harker et al., 2013). IL-27 enhances the expansion of memory CD8+ T cells following subunit vaccination or tumor challenge (Liu et al., 2013; Pennock et al., 2014) but not vaccinia virus vaccination, and Sendai and influenza virus–specific memory CD8+ T cells seem to lose responsiveness to IL-27 (Perona-Wright et al., 2012). IL-27 also promotes CD8+ T cell function in vitro (Morishima et al., 2005), whereas others reported direct suppression of IFNγ production by IL-27 (Liu et al., 2014). Even in cancer, some have observed enhancement of CD8+ responses by IL-27 (Salcedo et al., 2004; Liu et al., 2013), while others attribute induction of an inhibitory gene program in CD8+ T cells to IL-27 (Chihara et al., 2018). Thus, the regulation of CD8+ T cell responses by IL-27 is complex, and IL-27’s effect on CXCR5+ CD8+ T cells is unknown. Here we uncover a role for IL-27 in promoting amplification of CXCR5+ CD8+ T cells, preventing death of rapidly proliferating nonterminally differentiated virus-specific cells, and show that this effect is dependent on STAT1 and IRF1 and is unique to IL-27 among IL-12 family cytokines.
Results
IFNα/β receptor 1 (IFNAR1) blockade results in expansion of TCF1hi virus-specific CD8+ T cells
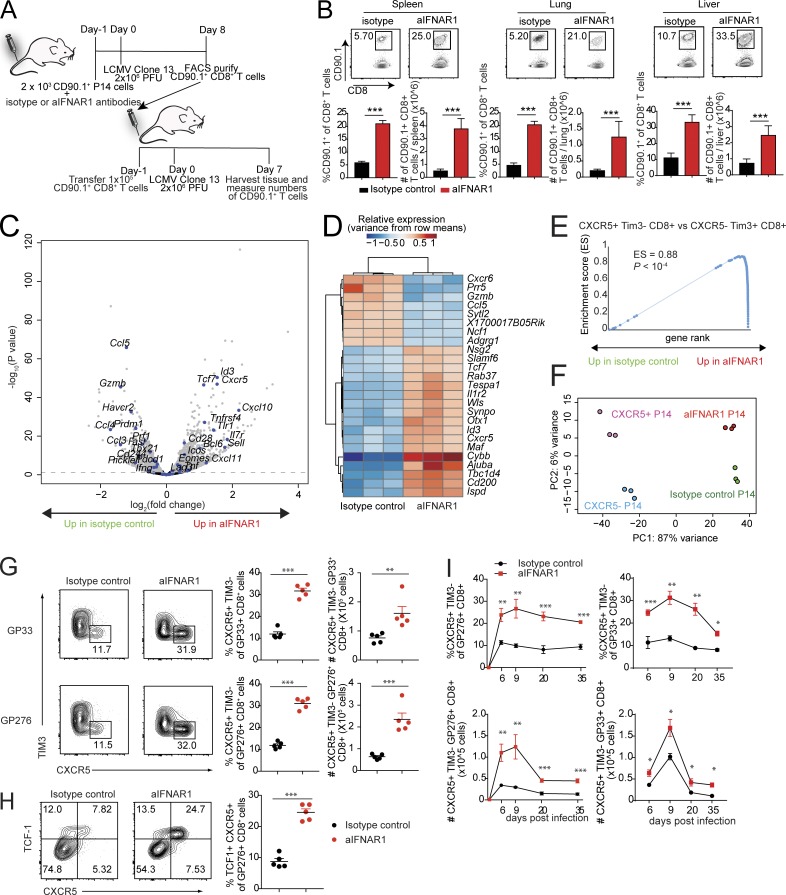
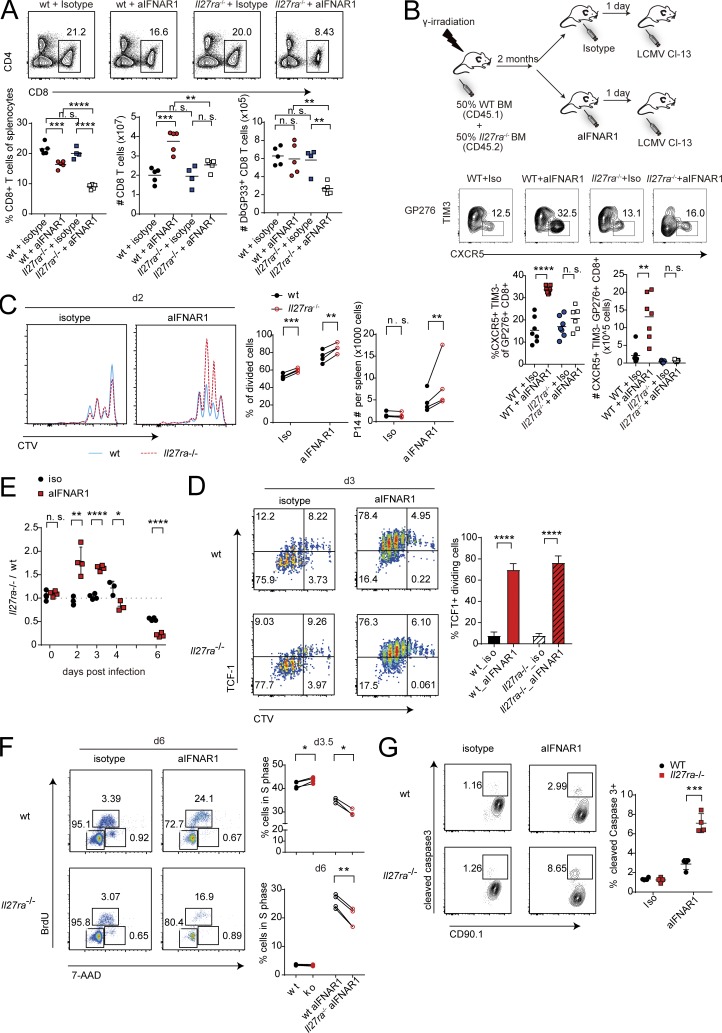
Blockade of IFN-I signaling during persistent LCMV infection enhances T cell–mediated control of persistent virus infection (Teijaro et al., 2013; Wilson et al., 2013; Ng et al., 2015). Here we asked whether virus-specific CD8+ T cells generated in an IFN-I–deficient environment would respond differently than CD8+ T cells generated in an IFN-I–replete environment. To address this question, we used CD8+ T cells from mice with a transgenic T cell antigen receptor specific for the glycoprotein 33–41 antigen (GP33–41) of LCMV (Pircher et al., 1987, 1989). CD90.1+ P14 cells were transferred into CD90.2+ congenic mice, which were then treated with isotype control or aIFNAR1 antibody and infected with a persistent dose of LCMV Cl13 (Fig. 1 A). Virus-specific CD90.1+ effector CD8+ T cells were purified 8 d post infection (dpi) and adoptively transferred into CD90.2+ congenic hosts, followed by infection with Cl13 (Fig. 1 A). At 7 dpi, we observed significant increases in the percentages and total numbers of virus-specific CD8+ T cells isolated from aIFNAR1-treated mice in spleen, lung, and liver compared with virus-specific CD8+ T cells isolated from the isotype controls (Fig. 1 B).
Figure 1.
Blockade of IFN-I promotes the expansion of virus-specific CXCR5+ TIM3− CD8+ T cells. (A and B) Experimental scheme for adoptive transfer experiments. C57BL/6J host mice received 2 × 103 CD90.1+ P14 cells and isotype control or aIFNAR1 antibodies 1 d before infection with Cl13 (2 × 106 focus-forming units). At 8 dpi, 105 P14 cells were sorted from these mice and then adoptively transferred into another two groups of CD90.2+ mice, followed by LCMV Cl13 infection the next day. P14 cells in spleen, lung, and liver of the recipient mice were quantified at 7 dpi. (C–F) P14 cells were transferred into isotype- or aIFNAR1-treated mice, followed by LCMV Cl13 infection the next day. P14 cells were then sorted at day 10 after infection and subjected to RNA-seq analysis. (C) Volcano plot showing statistical significance (P value) versus fold increase of gene expression comparing aIFNAR1-treated P14 cells to isotype-treated control. Left half: Down-regulated with aIFNAR1 treatment. Right half: Up-regulated with aIFNAR1 treatment. Genes with important function in T cells are highlighted. (D) Heat map of hierarchically clustered top differentially expressed genes (by P value) between isotype- and aIFNAR1-treated mice. (E) GSEA demonstrating a significant enrichment of CXCR5+ CD8+ T cell gene signature within P14 of the aIFNAR1-treated group but not the isotype-treated controls. The CXCR5+ CD8+ T cell gene set was constructed from a publicly available dataset (GEO accession no. GSE84105) by taking the top 100 genes most significantly up-regulated in CXCR5+ Tim3− CD8+ cells versus CXCR5− Tim3+ CD8+ T cells. (F) Principal component analysis of transcriptomes of purified CXCR5+ and CXCR5− P14 cells using reanalyzed RNA-seq reads from Leong et al. (2016) and P14 cells from mice treated with isotype control or aIFNAR1 antibody. (G–I) Isotype- or aIFNAR1-treated mice were infected with LCMV Cl13, and endogenous virus-specific CXCR5+ CD8+ T cells were analyzed. (G) Frequency and total number of DbGP33+ and DbGP276+ CXCR5+ CD8+ T cells in the spleens of isotype- or aIFNAR1-treated mice at 9 dpi. (H) Frequency and total number of DbGP276+ TCF1+ CXCR5+ cells in the spleens of isotype- or aIFNAR1-treated mice at 9 dpi. (I) Frequency and total number of DbGP276+ CXCR5+ CD8+ T cells in the spleen of isotype- or aIFNAR1-treated, Cl13-infected mice at different time points after infection. For panels A, B, G, and H, data show n = 5 mice per group and are representative of two independent experiments. Numbers on flow plots indicate the frequency of each gated population. In G and H, axes indicate log10 fluorescence. Statistical analyses of differential expression and gene set enrichment were performed using DESeq2 and GSEA, respectively; statistical comparison of experimental groups in B, G, and I were performed using Student’s two-tailed t test; bars represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Fig. S1.
We next asked whether virus-specific CD8+ T cells from Cl13-infected aIFNAR1-treated mice could promote control of persistent virus infection. As done above, virus-specific CD8+ T cells from Cl13-infected isotype- or aIFNAR1-treated control mice were purified and adoptively transferred into Cl13-infected mice depleted of CD4+ T cells, which are unable to clear Cl13 (Fig. S1 A). We observed substantial reductions in viral loads in mice that received T cells from aIFNAR1-treated mice infected with Cl13, and 50% and 100% of mice had completely cleared virus below the limit of detection in serum and liver, respectively, compared with mice receiving CD8+ T cells from isotype control–treated animals (Fig. S1 B). These experiments demonstrate that IFNAR1 signaling limits the proliferative potential of virus-specific CD8+ T cells in a cell-intrinsic manner.
To understand the mechanism by which IFNAR1 prevents CD8+ T cells from proliferating, we compared transcriptomes of CD90.1+ P14 cells from aIFNAR1- versus isotype-treated mice by RNA sequencing (RNA-seq) 10 dpi. Strikingly, among the top 15 genes most significantly up-regulated in aIFNAR1- versus isotype-treated mice were Cxcr5, Tcf7, and Id3 (Fig. 1 C). These genes mark the recently identified population of CXCR5+ CD8+ T cells, characterized by a lower degree of exhaustion and higher proliferative potential. Moreover, genes associated with effector function such as Gzmb were significantly down-regulated in the aIFNAR1 group (Fig. 1, C and D). Gene set enrichment analysis (GSEA) of genes that distinguish CXCR5+ TIM3− CD8+ T cells from their CXCR5− TIM3+ counterparts (Im et al., 2016) showed a significant up-regulation of the CXCR5+ TIM3− gene signature in aIFNAR1-treated mice (Fig. 1 E). Indeed, when we examined specifically CXCR5+ P14 and CXCR5− P14 cells using RNA-seq data (Leong et al., 2016), the transcriptional variance separating CXCR5+ from CXCR5− P14 cells was described by the same principal component as variance separating P14 cells from aIFNAR1- versus isotype-treated mice (Fig. 1 F). These results suggest that IFNAR activity skews CD8+ T cells toward the CXCR5− phenotype. Accordingly, in WT mice treated with aIFNAR1 versus isotype antibodies, we observed a significant increase in the percentage and total number of virus-specific CXCR5+ CD8+ T cells (Fig. 1 G). We also observed commensurate increases in the expression of BCL6 and TCF1, transcription factors required for CXCR5+ CD8+ T cell differentiation, following IFNAR1 blockade (Figs. 1 H and S1 C). Moreover, the increases in CXCR5+ CD8+ T cell numbers following aIFNAR1 treatment were sustained for over a month (Fig. 1 I). Interestingly, treatment with aIFNAR1 at 30 dpi still caused a significant increase in the frequency and number of CXCR5+ CD8+ T cells, demonstrating that IFN-I signaling limits their expansion even in the persistent phase of infection (Fig. S1 D). Our results confirm and extend a recent study by Wu et al. (2016) and collectively demonstrate that with aIFNAR1 treatment, mice infected with LCMV Cl13 exhibit increased and sustained CXCR5+ CD8+ T cell generation compared with isotype-treated controls.
Expansion of CXCR5+ CD8+ T cells is restrained by STAT2 but requires STAT1
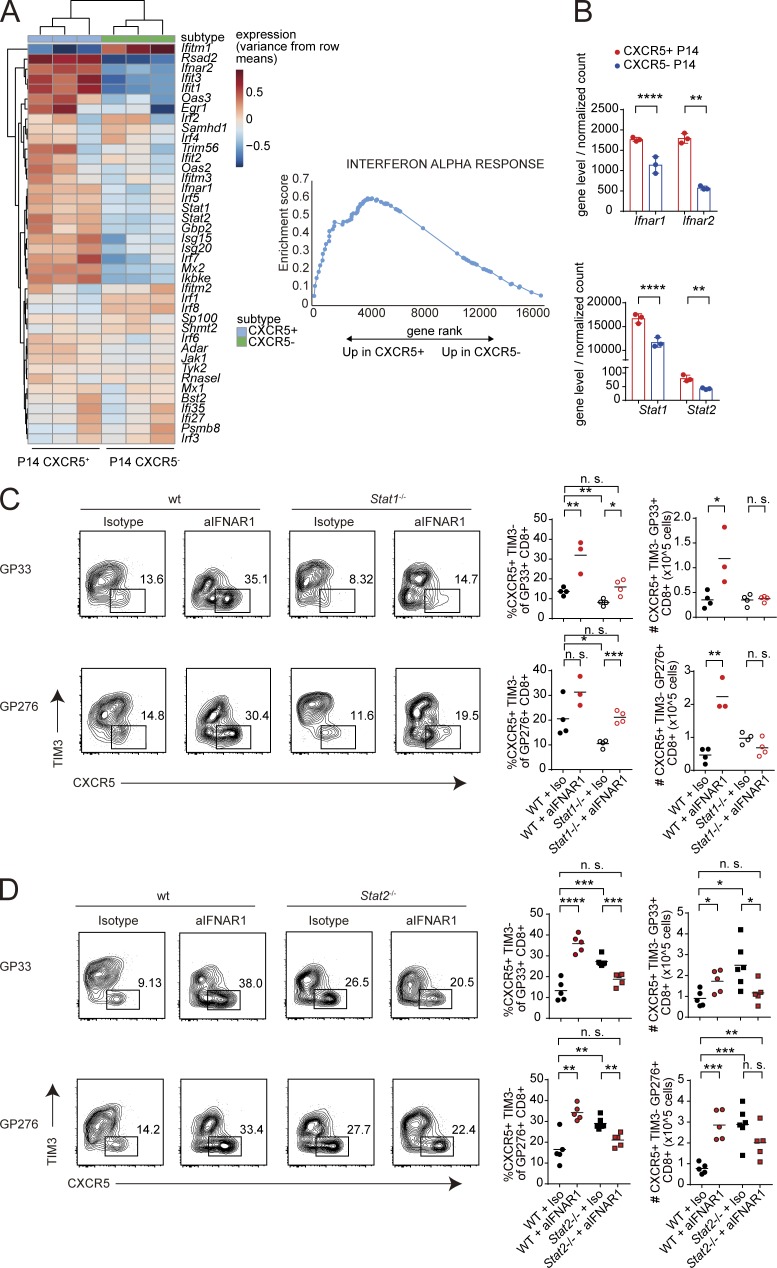
We next examined the expression of IFN-stimulated genes (ISGs) in CXCR5+ CD8+ T cells. Publicly available datasets revealed that purified CXCR5+ CD8+ T cells express higher levels of ISGs and IFN-I receptor genes compared with CXCR5− effector cells, suggesting that CXCR5+ CD8+ T cells are more sensitive to IFN-I than CXCR5− CD8+ T cells (Fig. 2, A and B; and Fig. S2 A). Paradoxically, ISGs were also significantly up-regulated in virus-specific CD8+ T cells from aIFNAR1-treated mice compared with isotype-treated controls (Fig. S2 B). Given that mice were treated with aIFNAR1 antibody 24 h before infection, the observed up-regulation of ISGs at 9 dpi could potentially be explained by a rebound in IFNAR signaling in aIFNAR1-treated mice. To determine whether latent up-regulation of ISGs was required for CXCR5+ CD8+ T cell expansion, we administered aIFNAR1 continuously throughout Cl13 infection and examined virus-specific CD8+ T cells by RNA-seq and CXCR5+ CD8+ T cells by flow cytometry. Continuous aIFNAR1 treatment still caused a significant expansion of percentage and number of virus-specific CXCR5+ CD8+ T cells (Fig. S2 C). However, ISGs were significantly down-regulated overall in virus-specific CD8+ T cells from the continuously treated mice (Fig. S2 D), apart from a limited number of individual ISGs such as Irf1. These results suggest that global up-regulation of ISGs is not required for CXCR5+ CD8+ T cell expansion.
Figure 2.
Expansion of CXCR5+ CD8+ T cells is restrained by STAT2 but requires STAT1. (A) Up-regulation of type I ISGs in purified CXCR5+ versus CXCR5− P14 cells. RNA-seq data were reanalyzed from GEO series GSE76279, in which mice receiving adoptively transferred P14 cells were infected with LCMV Docile; RNA-seq reads were normalized using DESeq2 and analyzed by GSEA. (B) Normalized mRNA levels of Ifnar1, Ifnar2, Stat1, and Stat2 in purified CXCR5+ versus CXCR5− P14 cells (raw data from GSE76729). Bars represent SD. (C) Percentages and total numbers of GP33-41–specific CXCR5+ TIM3− CD8+ T cells in WT and congenic STAT1-deficient mice (Stat1−/−) treated with IgG1 isotype control or aIFNAR1 antibodies and infected with Cl13. (D) Percentages and total numbers of GP33-41–specific CXCR5+ TIM3− follicular cytotoxic CD8+ T cells in WT and congenic STAT2-deficient (Stat2−/−) mice treated with IgG1 isotype control or aIFNAR1 antibodies and infected with Cl13. Splenic CXCR5+ CD8+ T cells were analyzed at 9 dpi. Experiments in C and D were performed with three to five mice per group, and data are representative of two to three independent experiments. Numbers on flow plots in C and D indicate the frequency of each gated population; axes indicate log10 fluorescence. Statistical comparison of experimental groups was performed using two-tailed Student’s t test: not significant (n.s.), P > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001. wt, wild type.
IFN-I is known to activate STAT1 and STAT2, which can signal either as homodimers or as part of the STAT1–STAT2–IRF9 heterotrimeric complex (IGSF3). To determine how IFN-I prevents CXCR5+ CD8+ T cell expansion, we treated STAT1-deficient and STAT2-deficient mice with aIFNAR1 and isotype control antibody and analyzed CXCR5+ CD8+ T cells at 9 dpi. Although aIFNAR1-treated STAT1-deficient mice exhibited a slight increase in the frequency of CXCR5+ CD8+ T cells among virus-specific CD8+ T cells, the total number of CXCR5+ CD8+ T cells was unaffected by aIFNAR1 treatment (Fig. 2 C). In contrast, STAT2-deficient mice experienced an inversion of the effect that aIFNAR1 has on CXCR5+ CD8+ T cells in WT mice: percentages and numbers of CXCR5+ CD8+ T cells were high in isotype-treated STAT2-deficient mice compared with WT controls and significantly decreased upon aIFNAR1 treatment (Fig. 2 D). These data suggest that IFN-I signaling through STAT2 restrains CXCR5+ CD8+ T cell expansion, whereas STAT1 is required for aIFNAR1-mediated CXCR5+ CD8+ T cell expansion.
IL-27 and IL-27R drive CXCR5+ CD8+ T cell expansion following IFNAR1 blockade
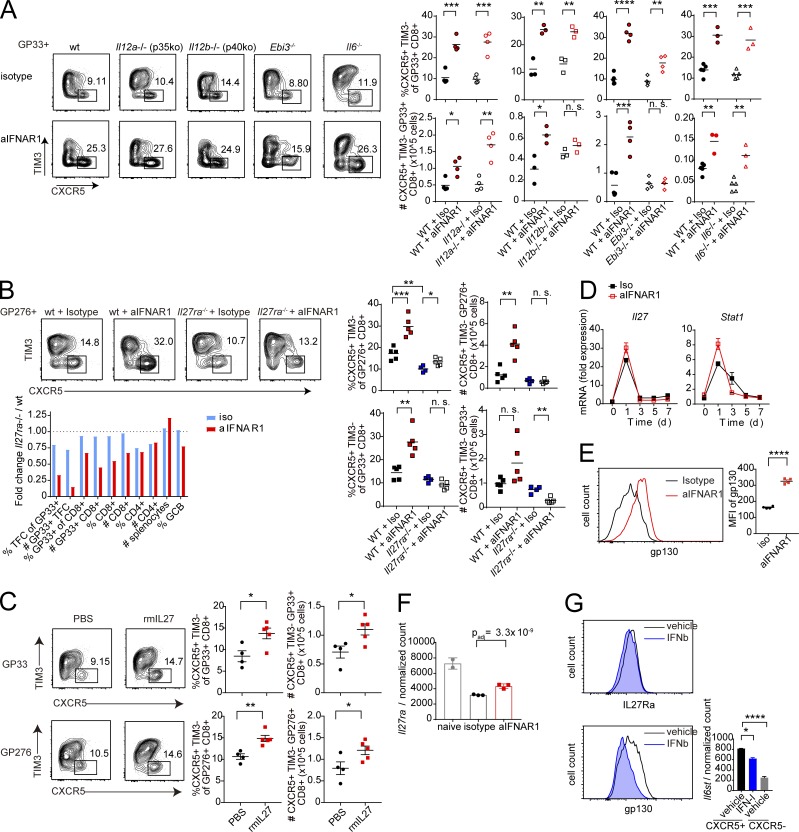
The observation that STAT1 is required for the expansion of CXCR5+ CD8+ T cells following IFNAR1 blockade suggests that a distinct STAT1-dependent pathway may positively regulate CXCR5+ CD8+ T cell expansion. Among STAT1 activators are two cytokines of the IL-12 family: IL-27 and IL-35 (Vignali and Kuchroo, 2012). We tested whether cytokine subunits of the IL-12 family played a role in the expansion of CXCR5+ CD8+ T cells after IFNAR1 blockade. Mice deficient in p40, a component of IL-12 and IL-23 cytokines, responded similarly to WT mice upon aIFNAR1 treatment (Figs. 3 A and S3 A). In contrast, CXCR5+ CD8+ T cell expansion was completely abolished in mice deficient in EBI3 (a component of IL-27 and IL-35 cytokines; Figs. 3 A and S3 A). Mice treated with aIFNAR1 also showed a significant increase in percentage and total number of CXCR5+ CD8+ T cells in inguinal lymph nodes compared with isotype-treated controls, and this effect was fully dependent on EBI3 (Fig. S3 B). Further, deficiency of p35 (an IL-35–specific subunit) or deficiency of IL-6 allowed normal CXCR5+ CD8+ T cell expansion following aIFNAR1 treatment (Figs. 3 A and S3 A). These data suggest that IL-27, but not IL-12, IL-23, IL-35, or IL-6, is required for aIFNAR1-induced CXCR5+ CD8+ T cell expansion.
Figure 3.
IL-27 and IL-27R drive CXCR5+ CD8+ T cell expansion following IFNAR1 blockade. (A) Percentages and total numbers of GP33-41–specific CXCR5+ TIM3− follicular cytotoxic CD8+ T cells in WT and congenic IL-12 p35-deficient (Il12a−/−), IL-12 p40-deficient (Il12b−/−), IL-27 EBI3-deficient (Ebi3−/−), and IL-6–deficient (Il6−/− mice treated with IgG1 isotype control or aIFNAR1 antibodies and infected with Cl13. Splenic CXCR5+ CD8+ T cells were analyzed at 9 dpi. (B) Percentages and total numbers of virus-specific CXCR5+ TIM3− follicular cytotoxic CD8+ T cells in WT and congenic IL-27R–deficient (Il27ra−/−) mice treated with IgG1 isotype control or aIFNAR1 antibodies and infected with Cl13. Splenic CXCR5+ CD8+ T cells were analyzed at 9 dpi. Bottom: Ratio of individual splenocyte populations in IL-27R–deficient versus WT mice treated with isotype or aIFNAR1. (C) Percentages and total numbers of virus-specific CXCR5+ TIM3− CD8+ T cells in WT mice that were treated with recombinant IL-27 protein on days 0–8 of Cl13 infection by intraperitoneal injection; data shown for 9 dpi. (D) Dynamics of mRNA expression of IL-27 p28 (Il27) and Stat1 in splenocytes from mice treated with aIFNAR1 or isotype control through 0–7 dpi with Cl13. (E) Expression of GP130 protein on P14 cells from aIFNAR1- or isotype-treated mice infected with Cl13, measured by MFI. (F) Normalized mRNA expression of IL-27R (Il27ra) on virus-specific P14 cells from naive mice or mice treated with isotype or aIFNAR1 and infected with Cl13. Bars represent mean ± SD. (G) Level of IL-27R subunits IL-27RA and GP130 on virus-specific CD8+ T cells treated with IFNβ or vehicle. Right: mRNA level of Il6st (encoding GP130) on WT CXCR5+ CD8+ T cells from Cl13-infected mice treated with vehicle or IFN-I and vehicle-treated CXCR5− CD8+ T cells. Numbers on flow plots (A–C) indicate the frequency of each gated population; axes in A–C and horizontal axes in E and G indicate log10 fluorescence. Experiments were performed with three to five mice per group, and data are representative of three independent experiments. Statistical comparisons of experimental groups were performed using Student’s two-tailed t test: not significant (n.s.), P > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001. rmIL27, recombinant mouse IL-27 protein; wt, wild type. See also Fig. S3.
To substantiate the requirement of IL-27 in this process, CXCR5+ CD8+ T cell expansion was examined in IL-27 receptor (Il27ra)-deficient mice. Similar to Ebi3−/− mice, Il27ra−/− mice also failed to expand virus-specific CXCR5+ CD8+ T cells following aIFNAR1 treatment (Fig. 3 B). Moreover, virus-specific CD8+ T cells numbers were also reduced in Il27ra−/− aIFNAR1-treated mice compared with WT mice treated with aIFNAR1, while other splenocyte populations were not significantly different (Fig. 3 B). CXCR5+ CD8+ T cell numbers in Il27ra−/− mice treated with aIFNAR1 did not increase even later after infection, although some CXCR5+ CD8+ T cells survived, indicating that IL-27R is not absolutely required for CXCR5+ CD8+ T cell survival (Fig. S3 C). Therefore, virus-specific CXCR5+ CD8+ T cells fail to expand following IFNAR blockade in IL-27R–deficient mice, and this is associated with a reduced splenic CD8+ T cell pool.
Given the ability of CXCR5+ CD8+ T cells to home inside B cell follicles, we asked whether the importance of IL-27 for CXCR5+ CD8+ T cell expansion could be related to impaired expansion of other follicular cell populations following aIFNAR1 treatment. Interestingly, there was a relative reduction in the number of T follicular helper (TFH) cells in both Il27ra−/− and Il6−/− mice compared with WT mice in response to aIFNAR1 (Fig. S3, D and E). The number of splenic germinal center B (GCB) cells increased upon aIFNAR1 treatment, and this increase in GCB number was unimpaired in both Il27ra−/− and Il6−/− mice (Fig. S3 F). Therefore, although IL-6 and IL-27 both affect aIFNAR1-induced expansion of the follicular cell populations TFH and GCB, IL-27 is specifically required for the expansion of CXCR5+ CD8+ T cells. This also suggests that differences in other follicular cell populations are not the main driver of the observed IL-27 dependency of CXCR5+ CD8+ T cells.
We next asked whether IL-27 was able to affect the CXCR5+ CD8+ T cell population in the absence of aIFNAR1. Treatment of mice with recombinant IL-27 on days 0–8 of Cl13 infection resulted in a significant increase in frequency and total number of both GP33-specific and GP276-specific CXCR5+ CD8+ T cells (Fig. 3 C). IL-27 treatment led to an approximately twofold reduction of viral titers in the brain and a plasma titer reduction in some mice that did not reach statistical significance (Fig. S3 G). In contrast, mice treated with IL-35 showed no increase in the CXCR5+ CD8+ T cell population at any dose tested (Fig. S3 H). Therefore, IL-27 can drive CXCR5+ CD8+ T cell expansion even in the presence of uninhibited IFNAR signaling.
We hypothesized that IFNAR blockade might promote STAT1 activation through IL-27 signaling, through either an increase in IL-27 levels and/or up-regulation of IL-27 receptor following aIFNAR1 treatment. RT-PCR revealed a slight increase in the abundance of IL-27 (p28) transcript in spleens of aIFNAR1-treated mice (Fig. 3 D). However, there was a more than twofold increase in the expression of GP130 subunit of the IL-27 receptor on virus-specific CD8+ T cells following aIFNAR1 treatment compared with isotype control treatment (Fig. 3 E). The mRNA level of Il27ra was also increased in P14 from aIFNAR1-treated hosts (Fig. 3 F). Moreover, treatment of cells with recombinant IFN-I led to a significant reduction in the expression of GP130 at both mRNA and protein levels (Fig. 3 G). These observations suggest that IFN-I directly modulates IL-27 signaling by reducing the expression of IL-27R subunit GP130. In summary, these results show that IL-27 signaling is required for the expansion of CXCR5+ CD8+ T cells upon aIFNAR1 treatment, but that IL-27 treatment is sufficient to drive an expansion of CXCR5+ CD8+ T cells even in the presence of IFN-I signals.
Cell-intrinsic requirement for IL-27R in sustained proliferation of TCF1+ CD8+ T cells
To provide further detail on how IL-27 signaling enables CXCR5+ CD8+ T cell expansion, we assessed the numbers of splenic CD8+ T cells in WT and Il27ra−/− mice from the experiment described in Fig. 3 B. Interestingly, the proportion of CD8+ T cells among splenocytes and the total number of GP33-specific CD8+ T cells at 9 dpi were the lowest in Il27ra−/− mice treated with aIFNAR1 of all experimental groups (Fig. 4 A). We therefore asked whether CD8+ T cell–intrinsic IL-27 signaling was required for clonal expansion following aIFNAR1 treatment. To address this, WT CD45.1+ mice were subjected to lethal irradiation followed by bone marrow transplantation of 50% CD45.1+ Il27ra+/+ and 50% CD45.2+ Il27ra−/− bone marrow, treated with aIFNAR1 or isotype antibodies, and infected with Cl13 (Fig. 4 B). There was a slight but significant excess of CD45.2+ (Il27ra−/−) versus CD45.1+ (WT) cells in the spleens of the infected chimeric mice (Fig. S4 A). Although we saw a modest increase in the percentage of GP33-specific CXCR5+ CD8+ T cells in CD8+ T cells originating from IL-27R–deficient CD45.2+ bone marrow, the GP276-specific CXCR5+ CD8+ T cells did not increase in percentage or total number in aIFNAR1- versus isotype-treated mice (Figs. 4 B and S4 B). In comparison, CD45.1+ bone marrow gave rise to T cells that produced CXCR5+ CD8+ T cell increases after aIFNAR1 treatment similar to those of WT mice. Moreover, the percentage of GP33-specific T cells of CD8+ T cells were similar across all groups except for Il27ra−/− CD8+ T cells in aIFNAR1-treated mice, in which the GP33-specific proportion was dramatically reduced (Fig. S4 C). Collectively, these data show that cell-intrinsic IL-27R is required for the aIFNAR1-induced expansion of CXCR5+ CD8+ T cells and suggest that IL-27R compensates for the loss of CD8+ T cell–intrinsic IFNAR signaling.
Figure 4.
Cell-intrinsic requirement for IL-27R in sustained proliferation of TCF1+ CD8+ T cells. (A) Percent CD8+ T cells of all splenocytes, total number of splenic CD8+ T cells, and number of GP33-specific CD8+ T cells in mice from experiment described in Fig. 3 C. (B) WT mice were subjected to lethal irradiation (2× 450 rad) followed by transplantation of an equal mixture of bone marrows from CD45.1+ IL27R-WT mice and CD45.2+ IL27R-deficient mice. 2 mo later, mice were treated with IgG1 isotype control or aIFNAR1 antibody 1 d before infection with Cl13. Shown are percentages and total numbers of splenic CXCR5+ TIM3− follicular cytotoxic T cells derived from CD45.1 WT and CD45.2+ IL27R-deficient bone marrow in mice treated with isotype control or aIFNAR1 antibody as described above. (C–E) WT CD90.1+ and Il27ra−/− CD90.1+ CD90.2+ P14 cells were labeled with CTV and adoptively transferred to WT CD90.2+ recipients that were treated with aIFNAR1 or isotype. The mice were infected with Cl13 the next day, and splenic P14 was analyzed at day 2 (C), day 3 (D), or days 2–6 (E) after infection. Ratio of Il27ra−/− to WT P14 cells detected in host mice is shown in E. (F) BrdU and 7-AAD staining of Il27ra−/− and WT P14 treated with aIFNAR1 or isotype and analyzed at 3.5 and 6 dpi. (G) Cell death in Il27ra−/− versus WT P14 assayed by cleaved caspase-3 staining. Splenocytes were cultured in vitro for 4 h and then analyzed by FACS. Experiments were performed with three to five mice per group, and data are representative of two to three independent experiments. Numbers on flow plots (A, B, and D–G) indicate the frequency of each gated population; axes in A, B, and D–G and horizontal axes in C indicate log10 fluorescence. Bars represent mean ± SD; statistical comparison of experimental groups was performed using Student’s two-tailed t test: not significant (n.s.), P > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001. wt, wild type.
The cell-intrinsic requirement for IL-27R in CD8+ T cells prompted us to ask how IL-27R controls expansion of virus-specific CD8+ T cells. IL-27 and IFN-I have been shown to induce MHC class I expression in T cells, and IFNAR is required to prevent T cell killing by natural killer (NK) cells (Crouse et al., 2014; Xu et al., 2014). Therefore, we hypothesized that IL-27 might be required to compensate for IFN-I by maintaining MHC-I expression to prevent NK-mediated killing. However, depleting NK cells with anti-NK1.1 did not rescue the CXCR5+ CD8+ T cell expansion in aIFNAR1-treated Il27ra−/− mice, arguing against NK-mediated killing as the factor limiting CXCR5+ CD8+ T cell expansion in IL-27R–deficient mice (Fig. S4 D).
Next, we asked if IL-27R affected CXCR5+ CD8+ T cell proliferation. Mice carrying the P14 transgene were crossed with Il27ra−/− mice to generate a source of CD90.1+/CD90.2+ Il27ra−/− P14 cells, which were then labeled with cell tracer dye (CellTrace Violet [CTV]) and cotransferred with WT CD90.1+/CD90.1+ P14 cells into CD90.2+/CD90.2+ hosts (Fig. S4 E). Recipient mice treated with isotype control showed similar proliferation profiles for Il27ra−/− and WT P14 cells at 2 and 3 dpi (Fig. 4, C and D). In contrast, aIFNAR1-treated mice exhibited a significant increase in divided P14 cells, and most of these proliferating cells were TCF1hi compared with isotype-treated mice (Fig. 4, C and D). Although IL-27R deficiency seemed to have no effect on the proliferation of TCF1hi P14 cells, there was an excess of Il27ra−/− P14 cells that had undergone at least one division compared with their WT counterparts (Fig. 4 C), and this was associated with a larger Il27ra−/− P14 versus WT P14 cell pool at 2–3 dpi in each recipient mouse (Fig. 4 E). However, Il27ra−/− P14 cells were unable to maintain the accelerated division and were outnumbered by WT P14 cells from day 4 onward (Fig. 4 E). In accordance with the reduced abundance of Il27ra−/− P14, we observed a decreased proportion of Il27ra−/− versus WT P14 in S phase of the cell cycle from day 3.5, and this difference became more dramatic at 6 dpi (Figs. 4 F and S4 F). In addition, Il27ra−/− P14 cells were more susceptible to caspase-3–dependent cell death (Fig. 4 G). Previously, rapidly dividing TCF1+ CD8+ T cells were found to express high levels of T-bet, a known target induced by IL-27 (Lin et al., 2016). We observed a small but statistically significant reduction of T-bethigh divided Il27ra−/− versus WT P14 cells (Fig. S4 G). These results suggest that IL-27R is required to maintain CD8+ T cell proliferation and survival in IFNAR-attenuated conditions.
We also generated Stat1−/− P14 cells to determine the role of CD8+ T cell–intrinsic STAT1 in CXCR5+ CD8+ T cell expansion. In a mixed adoptive transfer experiment analogous to the experiment above, significantly fewer Stat1−/− P14 cells were recovered from recipient mice following infection, which could be due to activation-induced cell death as previously reported in vaccinia infection (Quigley et al., 2008). In aIFNAR1-treated mice, we observed a reduction in the frequency of Stat1−/− P14 cells in S phase compared with WT P14, and increased cell death in Stat1−/− versus WT P14 cells comparable to the increase observed between Il27ra−/− and WT P14 (Fig. S4, H and I; and Fig. 4 G). In summary, IFNAR blockade enables the proliferation of TCF1hi CD8+ T cells, and cell-intrinsic IL-27R and STAT1 are required to sustain their proliferation.
IL-27 and IFN-I compete to induce partially overlapping transcriptomes
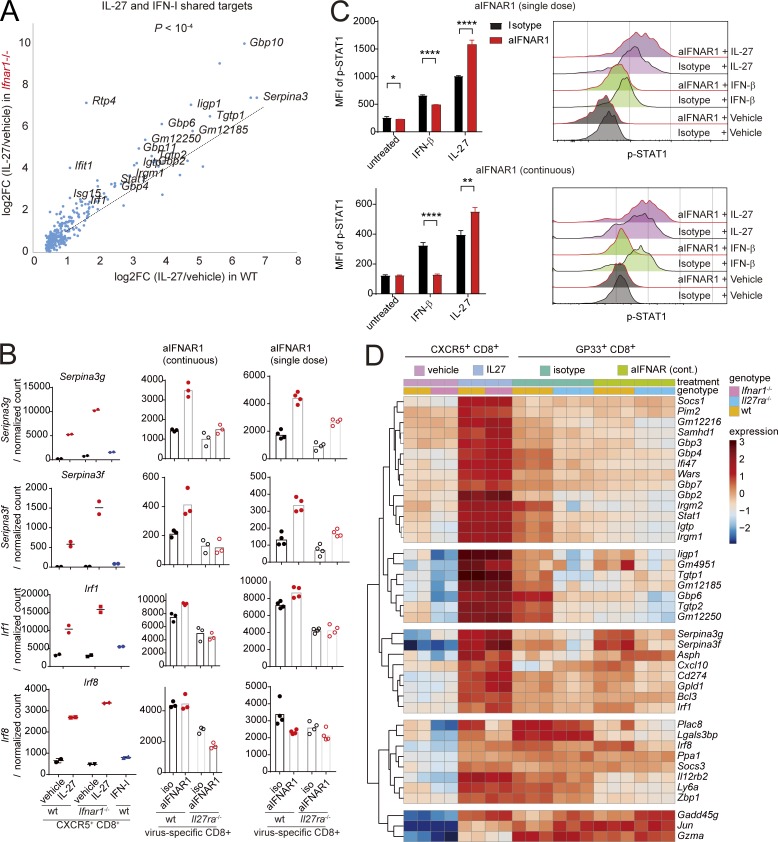
Given that IFN-I and IL-27 exert opposing effects on CXCR5+ CD8+ T cell expansion, we sought to understand their downstream genes. Purified CXCR5+ CD8+ T cells were isolated using flow cytometry and incubated in the presence of IFN-I, IL-27, or growth medium, and their transcriptomes were analyzed by RNA-seq. The transcriptomes induced by each cytokine showed a considerable overlap, and the overlapping target genes were highly enriched in annotated ISGs (Fig. S5 A). Given that STAT1 is activated by both cytokines, we hypothesized that IFNAR might compete with IL-27R to activate the downstream genes. To test this, we purified CXCR5+ CD8+ T cells from IFNAR1-deficient and WT mice, treated them with IL-27, and analyzed them by RNA-seq. Interestingly, target genes shared by IL-27 and IFN-I were induced significantly more strongly by IL-27 in Ifnar1−/− compared with WT CXCR5+ CD8+ T cells (Fig. 5, A and B). These genes feature canonical ISGs as well as transcription factors Stat1 and Irf1 (Fig. 5, A and B). In addition, P14 cells isolated from aIFNAR1-treated mice infected with Cl13 showed a stronger phosphorylation of STAT1 in response to IL-27 than isotype-treated mice (Fig. 5 C), in accordance with the previously observed increase in GP130 expression (Fig. 3 E). Conversely, P14 cells from aIFNAR1-treated mice exhibited a weaker induction of pSTAT1 in response to IFN-I than cells from isotype-treated mice (Fig. 5 C). These results suggest that CXCR5+ CD8+ T cells are sensitized to IL-27–mediated STAT1 activation by IFNAR1 deficiency.
Figure 5.
IL-27 and IFN-I compete to induce partially overlapping transcriptomes. (A) Sensitization of IFNAR1-deficient CXCR5+ CD8+ T cells to IL-27: log2 of fold-change in IL-27 versus vehicle-treated CXCR5+ CD8+ T cells from Ifnar1−/− (ordinate) and WT (abscissa) mice. (B) Normalized expression levels of selected genes induced by IL-27 to a higher level in IFNAR1-deficient versus WT CXCR5+ CD8+ T cells. These genes also require IL-27R for their physiological expression in aIFNAR1-treated mice in both single-dose and continuous treatment experiments (right panels). (C) The response of CD90.1+ P14 cells to IFN-I and IL-27 in aIFNAR1 versus isotype control treated mice, measured by pSTAT1 expression following stimulation. CD90.1+ P14 cells were adoptively transferred to mice that were treated with isotype or aIFNAR1 1 d before Cl13 infection or continuously treated with aIFNAR1. Splenocytes were harvested 6 dpi (single dose) or 9 dpi (continuous treatment); stimulated with vehicle, IFN-I, or IL-27 in vitro for 30 min; and then analyzed for pSTAT1 levels by FACS. Bars represent mean ± SEM. Horizontal axes indicate log10 fluorescence. (D) Hierarchical clustering of expression of genes induced by IL-27 in purified CXCR5+ CD8+ T cells and endogenous GP33-specific CD8+ T cells. CXCR5+ GP33+ cells were sorted at 9 dpi and treated with medium or medium containing recombinant IL-27 (20 ng/ml) for 12 h; RNA was then extracted from the cells for RNA-seq; endogenous GP33-specific CD8+ T cells were purified by flow cytometry from WT or Il27ra−/− mice treated with aIFNAR1 or isotype (days −1, 2, 5, and 8) and then infected with Cl13; spleens were harvested 9 dpi. Statistical comparison of experimental groups was performed using the paired two-tailed Student’s t test (A), unpaired two-tailed t test (C), and R package DESeq2 (D): *, P < 0.05; **, P < 0.005; ****, P < 0.0001. wt, wild type.
To understand how IL-27–induced genes (IL-27SGs) might control CXCR5+ CD8+ T cell expansion, we compared the in vitro IL-27 targets in CXCR5+ CD8+ T cells to the transcriptomes of endogenous virus-specific CD8+ T cells isolated from aIFNAR1 (continuous) or isotype-treated WT and Il27ra−/− mice described at 10 dpi. Hierarchical clustering of IL-27SGs by expression revealed distinct groups of genes (Fig. 5 D). A large cluster of shared targets of IFN-I and IL-27 was attenuated by aIFNAR1 treatment (Fig. 5 D, top cluster), suggesting these genes are unlikely to mediate the CXCR5+ CD8+ T cell–expanding effects of aIFNAR1. However, a separate cluster of IL-27SGs comprising genes with unattenuated or increased expression following aIFNAR1 treatment emerged (Fig. 5 D, middle cluster). Notably, this cluster featured Serpina3g and Bcl3, important prosurvival factors of memory CD8+ T cells (Mitchell et al., 2001; Liu et al., 2004), and Irf1, which regulates clonal expansion of CD8+ T cells during viral infection (Brien et al., 2011). IL-27R is required for the expression of Serpina3g and Irf1 in aIFNAR1-treated mice, because IL-27R–deficient mice showed significantly reduced expression in both the single-dose and continuous treatment experiments (Fig. 5 B). In addition, the transcription factor Irf8, previously shown to regulate proliferation of CD8+ T cells, was induced by IL-27 in CXCR5+ CD8+ T cells and significantly reduced in Il27ra−/− mice (Fig. 5 B). Taken together, these results suggest that IL-27 might be required for the expression of prosurvival and cell cycle–controlling genes in CXCR5+ CD8+ T cells under IFNAR-attenuated conditions.
CD8+ T cell–intrinsic IL-27R is required to sustain IRF1 expression in rapidly dividing cells
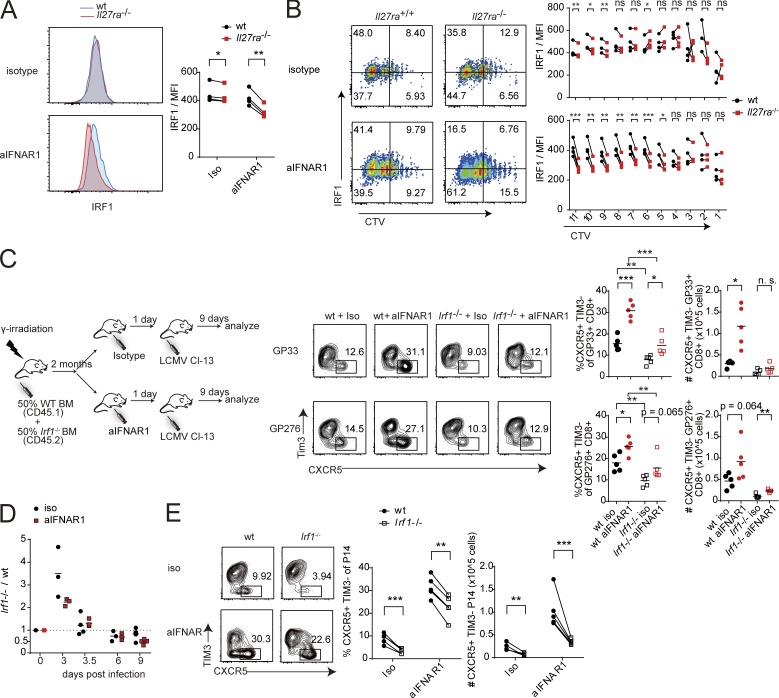
We asked if CD8+ T cell–intrinsic IL-27R was required for IRF1 expression. Performing a mixed transfer experiment with Il27ra−/− and WT P14 cells as described in Fig. 4, at 3 dpi we observed comparable levels of IRF1 in Il27ra−/− and WT P14 cells in isotype control–treated hosts, but significantly reduced levels of IRF1 in Il27ra−/− P14 compared with WT P14 in hosts receiving aIFNAR1 (Fig. 6 A). Strikingly, CTV labeling of WT and Il27ra−/− P14 cells before infection followed by analysis of IRF1 expression at 3 dpi revealed that the loss of IRF1 expression in Il27ra−/− versus WT P14 cells was specific to cells that have undergone a higher number of divisions (Fig. 6 B). The deficit of IRF1 in Il27ra−/− versus WT P14 cells was most dramatic in aIFNAR1-treated mice, and the magnitude of the loss progressively increased with the number of divisions, reaching a mean fluorescence intensity (MFI) ratio (Il27ra−/−/WT) of <0.7 in the most highly divided population of cells (Figs. 6 B and S5 D). Dividing Stat1−/− P14 cells also showed a markedly reduced expression of IRF1 compared with WT P14 cells in the same host (Fig. S5 E).
Figure 6.
CXCR5+ CD8+ T cell expansion following IFNAR blockade requires IL-27–induced transcription factor IRF1. (A and B) WT CD90.1+ P14 and Il27ra−/− CD90.1+ CD90.2+ P14 were labeled with CTV and adoptively transferred to WT CD90.2+ recipients treated with isotype or aIFNAR1 and infected with Cl13. Cells were analyzed at 3 dpi for IRF1 expression and CTV level by flow cytometry. (C) CXCR5+ CD8+ T cell expansion in mixed Irf1−/− and WT bone marrow chimera at 9 dpi. Mixed bone marrow chimeras were generated using a 1:1 ratio of WT CD45.1+ and Irf1−/− CD45.2+ bone marrow as depicted in the experimental diagram. (D and E) CD45.1+ WT host mice received equal numbers of WT CD45.2+ CD90.1+ P14 and Irf1−/− CD45.2+ CD90.2+ P14 cells, then aIFNAR1 or isotype control antibodies, and were infected with Cl13 the following day. (D) The ratio of splenic Irf1−/− versus WT P14 cells at selected time points is shown. (E) The frequency and total number of CXCR5+ cells of Irf1−/− and WT P14 cells in spleens of mice at 9 dpi. Experiments are representative of two to three independent experiments; individual points represent individual mouse readings. Numbers on flow plots (B, C, and E) indicate the frequency of each gated population; axes in B, C, and E and horizontal axes in A indicate log10 fluorescence. Statistical comparisons of experimental groups were performed using the paired two-tailed Student’s t test (A and B), unpaired two-tailed t test (C), and ratio paired two-tailed t test (E): not significant (n.s.), P > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005. wt, wild type. See also Fig. S5.
Transcription factor IRF1 is required for CXCR5+ CD8+ T cell expansion
We next investigated whether IRF1 plays a role in the expansion of CXCR5+ CD8+ T cells. A mixed bone marrow chimera using WT and Irf1−/− bone marrow was generated (Fig. 6 C). IRF1-deficient bone marrow gave rise to significantly fewer splenocytes than WT bone marrow, possibly due to a defect in thymic selection as previously reported (Fig. S5 F; Penninger et al., 1997; Brien et al., 2011). The chimeras were treated with isotype or aIFNAR1 and infected with Cl13 (Fig. 6 C). Interestingly, isotype-treated mice showed a reduced frequency of CXCR5+ CD8+ T cells among Irf1−/− bone marrow–derived virus-specific CD8+ T cells compared with WT bone marrow–derived T cells (Fig. 6 C). Moreover, whereas the total number of Irf1+/+ GP33+ CXCR5+ CD8+ cells increased, the number of Irf1−/− GP33+ CXCR5+ CD8+ cells did not increase in aIFNAR1-treated mice compared with isotype-treated mice (Fig. 6 C). These data suggest that IRF1 promotes CXCR5+ CD8+ T cell expansion.
To separate the effect of IRF1 on CXCR5+ CD8+ T cell expansion from its role in T cell thymic selection, Irf1-deficient P14 mice (Irf1−/− Thy1.1+ P14) were generated as a source of IRF1-deficient P14 cells. In a mixed transfer experiment, WT host mice received equal numbers of WT Thy1.1+ P14 and Irf1−/− Thy1.1+/Thy1.2+ P14 cells, then aIFNAR or isotype control, and were infected with cl13 the next day. At 3 dpi, Irf1−/− P14 cells outnumbered WT P14 cells in the spleen, but the ratio gradually reversed, with WT P14 cells approximately twofold more abundant than Irf1−/− P14 cells by 9 dpi in the anti-IFNAR–treated group (Fig. 6 D). Isotype-treated mice experienced a milder loss of Irf1−/− P14, but the knockout/WT ratio was also <1 from 6 dpi onward (Fig. 6 D). Interestingly, the frequency and total number of CXCR5+ Irf1−/− P14 cells was significantly lower compared with WT CXCR5+ P14 cells even in isotype control–treated hosts, suggesting IRF1 is required for the general expansion of CXCR5+ CD8+ T cells (Fig. 6 E).
Discussion
Recently, work by multiple groups uncovered the importance of CXCR5+ CD8+ T cells in the immunological response to PD1 blockade (He et al., 2016; Im et al., 2016), supporting IgG production and class switching by B cells (Quigley et al., 2007; Valentine et al., 2018) and sustaining the immune response to chronic viral infection (Leong et al., 2016; Utzschneider et al., 2016; Wieland et al., 2017). The emergent knowledge about the superior self-renewal properties of CXCR5+ CD8+ T cells in multiple models of viral infection and cancer underscores the importance of deciphering the upstream regulation of CXCR5+ CD8+ T cell formation and proliferation.
In viral infections, IL-27 appears to limit CD8+ T cell responses, in accordance with its established regulatory function (de Aquino et al., 2014; Liu et al., 2014; Harker et al., 2018). Consistently, T cell–intrinsic IL-27 signaling is required for the expression of negative regulatory molecules on tumor-infiltrating CD8+ T cells in a mouse melanoma model (Chihara et al., 2018), IL-27 was dispensable for memory CD8+ expansion following Listeria and vaccinia virus vaccinations (Pennock et al., 2014), and virus-specific memory CD8+ T cells seem to eventually lose IL-27 responsiveness (Perona-Wright et al., 2012). However, IL-27 also elicits powerful antitumor effects directly dependent on CD8+ T cells (Hisada et al., 2004; Salcedo et al., 2004, 2009; Liu et al., 2013), drives CD8+ T cell proliferation and IFNγ production (Mayer et al., 2008; Schneider et al., 2011; Liu et al., 2013), and augments memory CD8+ expansion following subunit vaccination (Pennock et al., 2014). Here, we showed that IL-27 promotes CXCR5+ CD8+ T cell expansion. Treatment of LCMV-infected mice with recombinant IL-27 was sufficient to drive an increase in frequency and number of virus-specific CXCR5+ CD8+ T cells. IL-27 and IL-27R were also required for the expansion of CXCR5+ CD8+ T cells induced by IFNAR blockade. Our observations of IL-27–driven CXCR5+ CD8+ T cell expansion in a persistent infection further support the stimulatory function of IL-27. Which factors dictate whether IL-27 elicits suppressive or activating effects on CD8+ T cells is unknown; one possibility is that IL-27 up-regulates prosurvival factors in CD8+ T cells under limited inflammation, such as during subunit vaccination, with localized tumors, or in a persistent phase of viral infection. If these genes can be induced redundantly by other cytokines, the suppressive effects of IL-27 might dominate.
Multiple factors that oppose CXCR5+ CD8+ T cell formation and drive effector CD8+ T cell differentiation have been identified: IFN-I, IL-12, BLIMP1, ID2, IRF4, RUNX3, and STAT4. RUNX3 cooperates with CBP-β to repress Tcf7 transcription (Shan et al., 2017). Similarly, IL-12 drives loss of Tcf7 transcription in dividing CD8+ T cells (Danilo et al., 2018). In CD4+ T cells, IL-27 can induce Tcf7 (encoding TCF1) expression, suggesting that IL-27 might promote CXCR5+ CD8+ T cell differentiation by sustaining TCF1 levels. Our results suggest that IL-27 is dispensable for TCF1 expression but required for the survival of proliferating TCF1+ CD8+ T cells, thus serving as an amplification factor of the nonterminally differentiated virus-specific CD8+ T cell pool.
Interestingly, IFN-I and IL-27 both activate STAT1 yet exerted opposing effects on CXCR5+ CD8+ T cell expansion. IFN-I, acting in concert with TCR activation (termed “in-sequence”), acts as a costimulatory cytokine and enhances effector function through STAT4; however out-of-sequence action (where IFN-I signal precedes TCR stimulation) has potent antiproliferative and apoptotic effects mediated by STAT1 (Platanias, 2005; Crouse et al., 2015). Both IFN-I and IL-27 enhance effector CD8+ T cell generation and function and induce the expression of T-bet (Hibbert et al., 2003; Morishima et al., 2005, 2010; Mayer et al., 2008; Schneider et al., 2011). However, whereas IFN-I favors short-lived effector cell formation, IL-27 also supports memory precursor-like effector cell formation (Liu et al., 2013). Moreover, whereas STAT1 activation by IFN-I has antiproliferative effects in CD8+ T cells, IL-27 activation of STAT1 is not known to inhibit proliferation in this cell type. On the contrary, IL-27 simultaneously increased STAT1 protein levels, STAT1 phosphorylation, and proliferation in human CD8+ T cells (Schneider et al., 2011). According to our competitive cell transfer studies, IFN-I couples cell division to loss of TCF1, particularly in rapidly dividing cells. In contrast, IL-27 allowed fast-dividing TCF1+ cells to sustain proliferation and avoid cell death, thereby uncoupling rapid division from TCF1 loss. IL-27 induced prosurvival genes Serpina3g, encoding cathepsin B inhibitor Spi2A, which is required for the maintenance of central memory CD8+ T cells (Liu et al., 2004; Byrne et al., 2012), and Bcl3, another inhibitor of cell death in activated CD8+ T cells (Mitchell et al., 2001; Valenzuela et al., 2005). Given the increased cell death in IL-27R–deficient P14 cells compared with WT P14 cells in mice treated with anti-IFNAR, it is likely that IL-27–dependent induction of prosurvival genes prevents attrition of TCF1hi cells undergoing division.
The notion that IFN-I and IL-27 compete to restrain or promote rapid proliferation of TCF1hi CD8+ T cells, respectively, is further supported by the observations that virus-specific CD8+ T cells from anti-IFNAR–treated mice expressed significantly higher levels of the IL-27R subunit GP130, and that IFN-I treatment caused a significant reduction in the mRNA and protein levels of GP130. Consistently, IL-27 induced STAT1 phosphorylation more strongly in anti-IFNAR– versus isotype-treated mice, and IL-27 achieved a stronger fold induction of target genes in Ifnar1−/− compared with WT CXCR5+ CD8+ T cells. Moreover, deletion of STAT2, which acts downstream of IFN-I but not IL-27, led to increased frequency and number of virus-specific CXCR5+ TIM3− CD8+ T cells. STAT2 is also required for the antiproliferative effect of IFN-I on IL-2–induced thymocyte proliferation (Gimeno et al., 2005), suggesting type 1 interferon’s antiproliferative effect on TCF1+ CD8+ T cells could be mediated by the STAT1–STAT2–IRF9 complex, whereas IL-27’s protective effect is mediated by STAT1 homodimers. These results, along with the requirement of STAT1 for CXCR5+ CD8+ T cell expansion following anti-IFNAR treatment, suggest that the relative magnitude of IFN-I and IL-27 signaling on TCF1hi cells influences CXCR5+ CD8+ T cell expansion via differential control of STAT1 and STAT2.
IL-27 was also required for IRF1 expression, a transcription factor we found to be essential in a cell-autonomous manner for the expansion of CXCR5+ CD8+ T cells following IFNAR blockade. IRF1 is a known regulator of CD8+ T cell differentiation and was recently shown to signal downstream of IL-27 in inducing type 1 regulatory CD4+ T cells in a STAT1-dependent manner (Brien et al., 2011; Karwacz et al., 2017). Irf1 is also an ISG, but IL-27 induced Irf1 more strongly than IFN-I, and Ifnar1 deletion further increased the magnitude of its induction by IL-27. We observed that IL-27 endows rapidly dividing CD8+ T cells with high IRF1 levels. IL-27R–deficient virus-specific (P14) CD8+ T cells underwent rapid division shortly after infection but were unable to sustain this, gradually being outcompeted by WT cells. In competitive transfer experiments, IRF1-deficient P14 cells showed highly similar kinetics to Il27ra−/− P14 cells and exhibited reduced frequency of CXCR5+ cells compared with WT P14 cells in the same host. This suggests that IL-27R supports CXCR5+ CD8+ T cell expansion in part by inducing sustained IRF1 expression. Irf1−/− P14 cells retain some ability to expand following anti-IFNAR treatment, suggesting that other IL-27 target genes play a role, possibly by decreasing the rate of cell death in proliferating Irf1−/− CXCR5+ P14 cells.
Our findings center on the ability of IL-27 to promote and sustain rapid division of memory-like CXCR5+ cells during viral infection, but might also have implications for other diseases. For example, CXCR5+ CD8+ T cells are essential for the proliferative burst of CD8+ T cells following anti-PD1 treatment (Im et al., 2016). Anti-PD1, which belongs to the class of checkpoint inhibitors, has become standard of care in the treatment of certain cancers. IL-27 has shown beneficial effects on antitumor CD8+ T cell responses (Murugaiyan and Saha, 2013). It would be interesting to test whether IL-27 can enhance the ability of anti-PD1 treatment to drive the proliferation of CD8+ T cells in cancer patients.
Materials and methods
Mice and virus strains
C57BL/6, C57BL/6 CD45.1+, C57BL/6 CD90.1+DbGP33–41TCR tg (P14), C57BL/6 CD90.1+ GP66-77 TCR tg (SMARTA), Ebi3−/− (Jackson Laboratory; stock 008701), Il12a−/− (Jackson Laboratory; stock 2692), Il12b−/− (Jackson Laboratory; stock 2693), Il27ra−/− (Jackson Laboratory; stock 018078; Yoshida et al., 2001), Il6−/− (Jackson Laboratory; stock 002650), Irf1−/− (Jackson Laboratory; stock 002762), Stat1−/− (Jackson Laboratory stock; 012606), and Stat2−/− (Jackson Laboratory stock; 023309) mice were from the Jackson Laboratory. Il27ra−/− P14 CD90.1+, Irf1−/− P14 CD45.2+, and Stat1−/− CD90.1+ P14 mice were generated by mating the single-allele mice described above. All mice were bred and housed under specific pathogen–free conditions. For virus infection, LCMV Cl13 was injected by the intravenous route at a dose of 2 × 106 PFU per mouse. For quantification of viremia, blood was drawn from the retroorbital sinus under isoflurane anesthesia. Serum was isolated by low-speed centrifugation to remove red and white blood cells. 10 µl of serum was used to perform 10-fold serial dilutions and quantified by focus forming assay on VeroE6 cells as previously described (Battegay et al., 1991). Tissues (spleen, lung, liver, and kidney) were harvested from euthanized mice and frozen at −80°C. At the time of the assay, tissues were thawed, weighed, homogenized in medium to 10% wt/vol, and clarified by low-speed centrifugation. 10 µl of the supernatant was used to quantify viral titers in a focus-forming assay. All mice were used in accordance with guidelines from the Institutional Animal Care and Use Committee of the Scripps Research Institute.
Mixed bone marrow chimeras
CD45.1+ mice were irradiated with two rounds of 450 cGy with a 3-h interval. Bone marrow from Il27ra+/+ (CD45.1+) and Il27ra −/− (CD45.2+) was subjected to lysis of red blood cells and was depleted of T cells by biotin-CD4 (clone GK1.5; BioLegend), biotin-CD8a (clone 53-6.7; BioLegend) antibodies, and Anti-Biotin MicroBeads (Miltenyi Biotec). Bone marrow cells were mixed at a ratio of 1:1 (2 × 106 cells for each genotype) and were transferred by injection intravenously. 2 mo later, recipient mice were treated with isotype control or aIFNAR1 antibody, then infected with LCMV Cl13. An equivalent procedure was followed to generate mixed chimeras using Irf1−/− CD45.2+ and WT CD45.2+ bone marrow.
Antibody treatments
Mice were treated with 1 mg of aIFNAR1 antibody intraperitoneally (clone MAR1-5A3; Leinco Technologies) or a mouse IgG1 isotype control (clone MOPC21; Leinco Technologies) 1 d before virus infection. To deplete NK cells, mice received a single intraperitoneal injection of 25 µg anti-NK1.1 monoclonal antibodies (PK136) or a control mouse IgG2a (Bio-X-Cell) 1 d before virus infection.
Cytokine in vivo treatments
C57BL/6 mice were infected with Cl13 (2 × 106 PFU) and recombinant mouse IL-27 comprising both functional subunits (R&D Systems) or recombinant mouse IL-35 comprising both functional subunits (Adipogen), each at the specified concentration, administered daily by intraperitoneal injection on 0–8 dpi.
Cytokine in vitro treatments
For a flow cytometric analysis of STAT1 phosphorylation, cells were treated with IFN-I (5,000 U/ml) or IL-27 (50 ng/ml; R&D Systems) for 30 min and then fixed with Lyse/Fix Buffer (BD Biosciences) for 12 min at 37°C. Surface markers were stained after fixation. Cells were then permeabilized with Perm Buffer III (BD Biosciences) for 30 min on ice, washed, and stained for phosphorylated STAT1 (BD Biosciences). For analysis of IFN-I effect on IL-27R expression, mouse CD8+ T cells from spleen were purified with CD8+ T cell negative selection kit (Stemcell Technologies) and stimulated with plate-coated anti-CD3 and anti-CD28 (BioLegend) in the absence or presence of 5,000 U/ml IFNβ (PBL Assay Science) for 2 d. Il27RA and GP130 expression were examined by flow cytometry. For RNA-seq analysis, splenic CXCR5+ CD8+ and CXCR5− CD8+ T cells were purified by flow cytometry, plated at a concentration of 106 cell/ml, and treated with growth medium (RPMI 1640 containing 10% FBS and supplemented with nonessential amino acids, sodium pyruvate, Hepes, l-glutamine, 2-mercaptoethanol, and penicillin-streptomycin [Gibco]) with or without recombinant IL-27 (20 ng/ml; R&D Systems) or recombinant IFNβ (1,000 U/ml, carrier-free; PBL Assay Science) for 12 h at 37°C.
Flow cytometry, sorting, and intracellular staining
Single-cell suspensions were prepared from spleen. Antibodies for cell surface markers were added, followed by incubation for 30 min at 4°C. For CXCR5 staining of CXCR5+ CD8+ T cells, single-cell suspensions were stained with tetramer and purified antibody to mouse CXCR5, followed by 20 µM d-biotin (Avidity Biosciences) and biotin-conjugated antibody to rat IgG2a (MRG2a-83; BioLegend). Samples were then stained with streptavidin-PE (BioLegend) and other antibodies to surface markers. For intracellular cytokine staining, splenocytes were stimulated with 2 µg/ml of MHC class I restricted LCMV-GP33–41 (AnaSpec) or 10 µg/ml of MHC class II restricted LCMV-GP61–80 peptide (AnaSpec) for 1 h in the absence of brefeldin A and then 5 h in the presence of 4 µg/ml brefeldin A (Sigma-Aldrich). Cells were fixed and permeabilized with 2% saponin, and intracellular staining was performed with antibodies to IFNγ (XMG1.2; BioLegend), TNF-α (MP6-XT22; BioLegend), and IL-2 (JES6-5H4; BioLegend). Flow cytometric analysis was performed using a BD LSR II (BD Biosciences), and data were analyzed using FlowJo (TreeStar). Absolute number of cells was determined by multiplying the frequency of specific cell populations by the total number of viable cells. CD90.1+ P14 CD8+ T cells were sorted from Cl13-infected mice treated with either isotype or aIFNAR1 antibody using a MoFlo Astrios flow cytometer (Beckman Coulter).
Cells for RNA analysis were prepared in the following experiments. (1) CD90.1+ P14 cells were isolated from naive CD90.1+ Tg(P14) mice; 2,000 cells were adoptively transferred to a congenic CD90.2+ host; and mice were treated with aIFNAR1 or IgG1 isotype control antibodies by intraperitoneal injection and then infected with Cl13 (2 × 106 PFU) by intravenous injection. Splenocytes were harvested at 10 dpi, enriched for mouse T cells using a kit (Stemcell Technologies), and stained, and CD90.1+ CD8+ cells were sorted from spleens by flow cytometry. (2) WT, Il27ra−/−, and Ifnar1−/− mice were treated with aIFNAR1 or IgG1 isotype control antibodies 1 d before infection with Cl13 (2 × 106 PFU). Splenocytes were harvested at 10 dpi and stained, and GP33- and GP276-tetramer+ CD8+ B220− cells were sorted from the spleen by flow cytometry. Cells from individual mice were used as samples for RNA preparation and sequencing. (3) CXCR5+ CD8+ and CXCR5− CD8+ cells were isolated using flow cytometry from spleens of WT, Il27ra−/−, and Ifnar1−/− Cl13-infected mice at 9 dpi. The cells were plated at 106 cells/ml and treated with growth medium or cytokines as described above.
Cell division tracking
P14 cells were isolated from donor DbGP33–41TCR tg (P14), Il27ra−/− P14, or Irf1−/− P14 mouse spleens using the EasySep Mouse CD8+ T cell Isolation Kit (Stemcell Technologies), and 50% WT and 50% Il27ra−/− P14 or 50% Irf1−/− P14 cells were mixed and labeled with CTV (Thermo Fisher Scientific) according to the manufacturer’s instructions. 0.2–2 × 106 CTV-labeled cells were adoptively transferred to congenic hosts by intravenous injection 1 d before infection with 2 × 106 PFU Cl13. At the selected time after infection, splenocytes were harvested, and CTV levels in donor CD8+ T cells were analyzed by flow cytometry.
Cell cycle analysis
Mice were euthanized after priming with 1 mg BrdU (BD Biosciences) intraperitoneally for 1 h. Splenocytes were then stained for surface markers, fixed, permeabilized, treated with DNase at 37°C for 1 h, and stained for BrdU and 7-AAD (BD Biosciences; cat. no. 559619) according to the manufacturer’s instructions.
Caspase-3 staining
Splenocytes were cultured ex vivo for 4 h in RPMI 1640. Cells were then stained for surface markers, fixed, permeabilized, and stained with PE Active Caspase-3 Apoptosis Kit (BD PharMingen).
RT-PCR
RNA was isolated from splenocytes or sorted cells with TRIzol (Thermo Fisher Scientific) as described by manufacturer’s protocol. The RNA was reverse-transcribed into cDNA with the Quantitect Reverse Transcription Kit (Qiagen). Gene expression analysis was performed with GAPDH (QT01658692; Qiagen), Stat1 (QT00162183), and IL-27 (QT00143017; Qiagen). Relative quantities (RQs) were determined with the equation RQ = 2–ΔΔCt.
RNA-seq
RNA isolation from sorted cells was performed using the PicoPure RNA Isolation Kit (Thermo Fisher Scientific). RNA integrity was analyzed using the RNA Nano Kit (Agilent). Amplified cDNA was prepared using SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) and sheared using Covaris tubes, and Illumina libraries were constructed using the NEBNext Ultra DNA Library Prep Kit (NEB) and sequenced using the NextSeq 500 system (Illumina) at a depth of 20–25 million single-end 75-bp reads per sample. Illumina’s bcl2fastq Conversion Software (version 2.17.1.14) was used to demultiplex reads and convert BCL files generated by the Illumina NextSeq 500 to FASTQ. Raw data were deposited in Gene Expression Omnibus under the accession no. GSE97139.
Bioinformatic analysis of RNA-seq data
Quality of raw RNA-seq reads was checked using FastQC version 0.11.3 (Barbaham Institute; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the mouse genome (GRCm38 assembly with Ensembl v87 transcriptome annotations), and gene counts were calculated using STAR version 2.5.2a (Dobin et al., 2013) using the following parameters: --quantMode GeneCounts, --outSAMtype None, --sjdbGTfile /Ensembl/Mus_musculus.GRCm38.87.gtf, and --sjdbOverhang 74. Statistical normalization and differential expression analysis were performed using the R package DESeq2 version 1.14.1 (Love et al., 2014). Principal component analysis was performed using R function prcomp, and the top 500 genes with the highest variance between samples were used. R version 3.4.3 was used for statistical analyses (R Core Development Team, 2008). Heatmaps were generated using R package pheatmap version 1.0.8 (Kolde, 2013) and using expression values transformed by the rld function from the DESeq2 package. GSEA version 2.2.3 was used (Subramanian et al., 2005) with default settings. Overrepresentation analysis of gene ontology (GO) gene sets was performed using AmiGO 2 version 2.4.26 (Ashburner et al., 2000; The Gene Ontology Consortium, 2017) and PANTHER (Mi et al., 2017); Benjamini–Hochberg multiple-hypothesis correction was used to assess false discovery rate. Basic expression level plots were produced using Prism version 7.03 (GraphPad Software), and other graphics were generated using R programming language (R Core Development Team, 2008).
Analysis of publicly available expression data
Gene expression analysis of publicly available RNA-seq studies was performed using the pipeline described above on raw SRA files obtained from the Sequence Read Archive. Gene expression analysis of publicly available microarrays was performed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) and R package limma version 3.30.13. An adjusted P value threshold of 0.05 was used to derive differentially expressed genes. Expression data of CXCR5+ Tim3− CD8+ versus CXCR5− Tim3+ CD8+ T cells (used in Figs. 1 E and S2 A) were retrieved from GSE84105 by comparing CXCR5+ Tim3− to CXCR5− Tim3+ samples with GEO2R and calculating top 100 CXCR5+-specific genes by statistical significance (Barrett et al., 2013). Data for CXCR5+ and CXCR5− P14 cells (used in Fig. 1 F; Fig. 2, A and B; and Fig. S4 D) were analyzed using the uniform RNA-seq analysis pipeline starting with SRA data for GSE76279 (Leong et al., 2016). Expression data for T cells treated with medium, IL-27, or IL-6 (used in Fig. S4 F) were derived from GSE65621 (Hirahara et al., 2015). All heatmaps were generated using rld-transformed data using R packages DESeq2 and pheatmap. Fold-change/fold-change plots were generated using log2-transformed expression data using R package DESeq2 and function results.
Analysis of IL-27 target genes
Genes induced by IL-27 and IFN-I plotted in Fig. 5 A were empirically selected as genes significantly up-regulated in CXCR5+ CD8+ T cells in both IL-27 and IFN-I treatment conditions compared with growth medium–treated cells (DESeq2, adjusted P < 0.05). Statistical comparison of fold-change induction by IL-27 in Ifnar1−/− versus WT cells was performed using paired t test. Genes plotted in Fig. 5 D were selected as the top 40 genes induced by IL-27 in CXCR5+ CD8+ T cells, and expression levels from both in vitro experiments and in vivo continuous aIFNAR1- versus isotype-treatment experiments were subjected to hierarchical clustering using R functions pheatmap and hclust.
Selection of ISG sets
The Gene Ontology Resource website (http://geneontology.org/), AmiGO2 version 2.4.26, was used as source of annotated ISGs. Search criteria were as follows: search “ontology,” species “Mus musculus,” and GO terms response to type I IFN (GO:0034340), cellular response to type I IFN (GO:0071357), response to IFNβ (GO:0035456); and cellular response to IFNβ (GO:0035458; Table S1). The downloaded gene sets were filtered for duplicated entries to obtain the numbers of genes listed below. As an additional ISG gene set, the GO set “response to type I IFN” was downloaded from MSigDB. This MSigDB gene set contains mouse orthologues of human genes of the Homo sapiens GO gene set.
Statistical analysis
Statistical analyses were performed as described in the figure legends. R version 3.4.3 was used for computational analysis and plotting (R Core Development Team, 2008); Prism was used for plotting selected panels.
Online supplemental material
Fig. S1 shows the viral titers of mice following adoptive transfer of CD8+ T cells from aIFNAR1- versus isotype-treated donors, and the frequency of virus-specific CXCR5+ CD8+ T cells following aIFNAR1 treatment in persistent infection phase. Fig. S2 shows the expression of ISGs, Ifnar1, Ifnar2, Stat1, and Stat1 in CXCR5+ versus CXCR5− CD8+ T cells; frequency and number of CXCR5+ TIM3− CD8+ T cells; and expression of ISGs in virus-specific CD8+ T cells following continuous treatment with aIFNAR1. Fig. S3 shows the frequency and number of virus-specific CXCR5+ TIM3− CD8+ T cells in spleens of WT, Il12a−/−, Il12b−/−, Ebi3−/−, and Il6−/− mice and in lymph nodes of Ebi3−/− mice following aIFNAR1 treatment; the expansion of TFH cells in Il27ra−/− and Il6−/− mice following aIFNAR1 treatment; expansion of GCB cells in Il27ra−/− mice following aIFNAR1 treatment; and the frequency of virus-specific CXCR5+ TIM3− CD8+ T cells in mice treated with recombinant IL-35, all during Cl13 infection. Fig. S4 shows the frequency of cells derived from mixed WT and Il27ra−/− bone marrow and the expansion of GP33-specific CXCR5+ TIM3− CD8+ T cells in these mixed bone marrow chimeras following aIFNAR1 treatment; the number of virus-specific CXCR5+ TIM3− CD8+ T cells in Il27ra−/− mice following aIFNAR1 treatment with or without NK cell depletion; the cell cycle profile and T-bet levels of WT and Il27ra−/− P14 cells in aIFNAR1- or isotype-treated hosts on day 2 after infection; and the cell cycle profile and caspase-3 activation of WT and Stat1−/− P14 cells in aIFNAR1-treated hosts. Fig. S5 shows the enrichment of ISGs and G1-S genes in IL-27–treated CXCR5+ CD8+ T cells, the expression profile of selected marker genes and of IRF1 across cell divisions in virus-specific CD8+ T cells in WT and Il27ra−/− mice following aIFNAR1 treatment, expression of IRF1 in WT and Stat1−/− P14 cells in host mice following aIFNAR1 treatment, and the ratio of WT versus Irf1−/− cells in a mixed bone marrow chimera. Table S1 provides annotated sets of ISGs.
Supplementary Material
Acknowledgments
The authors are grateful to Pamela Schwartzberg (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD) for sharing the P14 Stat1−/− mouse strain. We thank the Scripps Research Institute Flow Cytometry and Genomics cores for technical assistance. Reagents for tetramer generation were kindly provided by the National Institutes of Health Tetramer Core. We also thank Donna Farber, Ari Theofilopoulos, and Michael Oldstone for critical reading of the manuscript.
This work was supported by National Institutes of Health grants R01 AI118862 and R01 AI123210 (to J.R. Teijaro) and R01 AI121155 and R21 AI143529 (to C. Xiao). J.R. Teijaro is partly supported by Array BioPharma and Pfizer. Array Biopharma and Pfizer had no involvement in this study.
The authors declare no competing financial interests.
Author contributions: Z. Huang, J. Zak, I. Pratumchai, and J.R. Teijaro conceived and designed the project; Z. Huang, J. Zak, I. Pratumchai, T. Wu, and J.R. Teijaro designed experiments; Z. Huang, J. Zak, I. Pratumchai, N. Shaabani, V.F. Vartabedian, N. Nguyen, and T. Wu performed experiments; Z. Huang, J. Zak, and I. Pratumchai analyzed data; J. Zak and J.R. Teijaro wrote the manuscript; C. Xiao cosupervised the study; J.R. Teijaro supervised the study; and all authors reviewed the manuscript.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium . 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., and Ahmed R.. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439:682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. 2013. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41(D1):D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battegay M., Cooper S., Althage A., Bänziger J., Hengartner H., and Zinkernagel R.M.. 1991. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods. 33:191–198. 10.1016/0166-0934(91)90018-U [DOI] [PubMed] [Google Scholar]
- Blackburn S.D., Shin H., Haining W.N., Zou T., Workman C.J., Polley A., Betts M.R., Freeman G.J., Vignali D.A., and Wherry E.J.. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10:29–37. 10.1038/ni.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien J.D., Daffis S., Lazear H.M., Cho H., Suthar M.S., Gale M. Jr., and Diamond M.S.. 2011. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8(+) T cell immune responses against West Nile virus infection. PLoS Pathog. 7:e1002230 10.1371/journal.ppat.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S.M., Aucher A., Alyahya S., Elder M., Olson S.T., Davis D.M., and Ashton-Rickardt P.G.. 2012. Cathepsin B controls the persistence of memory CD8+ T lymphocytes. J. Immunol. 189:1133–1143. 10.4049/jimmunol.1003406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara N., Madi A., Kondo T., Zhang H., Acharya N., Singer M., Nyman J., Marjanovic N.D., Kowalczyk M.S., Wang C., et al. 2018. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 558:454–459. 10.1038/s41586-018-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J., Bedenikovic G., Wiesel M., Ibberson M., Xenarios I., Von Laer D., Kalinke U., Vivier E., Jonjic S., and Oxenius A.. 2014. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 40:961–973. 10.1016/j.immuni.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Crouse J., Kalinke U., and Oxenius A.. 2015. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 15:231–242. 10.1038/nri3806 [DOI] [PubMed] [Google Scholar]
- Danilo M., Chennupati V., Silva J.G., Siegert S., and Held W.. 2018. Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation. Cell Reports. 22:2107–2117. 10.1016/j.celrep.2018.01.072 [DOI] [PubMed] [Google Scholar]
- de Aquino M.T.P., Kapil P., Hinton D.R., Phares T.W., Puntambekar S.S., Savarin C., Bergmann C.C., and Stohlman S.A.. 2014. IL-27 limits central nervous system viral clearance by promoting IL-10 and enhances demyelination. J. Immunol. 193:285–294. 10.4049/jimmunol.1400058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R., Lee C.-K., Schindler C., and Levy D.E.. 2005. Stat1 and Stat2 but not Stat3 arbitrate contradictory growth signals elicited by alpha/beta interferon in T lymphocytes. Mol. Cell. Biol. 25:5456–5465. 10.1128/MCB.25.13.5456-5465.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Dolgoter A., and Zuniga E.I.. 2013. Cell-intrinsic IL-27 and gp130 cytokine receptor signaling regulates virus-specific CD4+ T cell responses and viral control during chronic infection. Immunity. 39:548–559. 10.1016/j.immuni.2013.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Wong K.A., Dallari S., Bao P., Dolgoter A., Jo Y., Wehrens E.J., Macal M., and Zuniga E.I.. 2018. Interleukin-27R Signaling Mediates Early Viral Containment and Impacts Innate and Adaptive Immunity after Chronic Lymphocytic Choriomeningitis Virus Infection. J. Virol. 92:e02196-17 10.1128/JVI.02196-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Im S.J., Araki K., and Ahmed R.. 2019. Cytokine-Mediated Regulation of CD8 T-Cell Responses During Acute and Chronic Viral Infection. Cold Spring Harb. Perspect. Biol. 1:pii:a028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Hou S., Liu C., Zhang A., Bai Q., Han M., Yang Y., Wei G., Shen T., Yang X., et al. 2016. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 537:412–428. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Hibbert L., Pflanz S., De Waal Malefyt R., and Kastelein R.A.. 2003. IL-27 and IFN-alpha signal via Stat1 and Stat3 and induce T-Bet and IL-12Rbeta2 in naive T cells. J. Interferon Cytokine Res. 23:513–522. 10.1089/10799900360708632 [DOI] [PubMed] [Google Scholar]
- Hirahara K., Onodera A., Villarino A.V., Bonelli M., Sciumè G., Laurence A., Sun H.W., Brooks S.R., Vahedi G., Shih H.Y., et al. 2015. Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity. Immunity. 42:877–889. 10.1016/j.immuni.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada M., Kamiya S., Fujita K., Belladonna M.L., Aoki T., Koyanagi Y., Mizuguchi J., and Yoshimoto T.. 2004. Potent antitumor activity of interleukin-27. Cancer Res. 64:1152–1156. 10.1158/0008-5472.CAN-03-2084 [DOI] [PubMed] [Google Scholar]
- Huang A.C., Postow M.A., Orlowski R.J., Mick R., Bengsch B., Manne S., Xu W., Harmon S., Giles J.R., Wenz B., et al. 2017. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 545:60–65. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., and Kastelein R.. 2012. Interleukin-27: balancing protective and pathological immunity. Immunity. 37:960–969. 10.1016/j.immuni.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacz K., Miraldi E.R., Pokrovskii M., Madi A., Yosef N., Wortman I., Chen X., Watters A., Carriero N., Awasthi A., et al. 2017. Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat. Immunol. 18:412–421. 10.1038/ni.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein R.A., Hunter C.A., and Cua D.J.. 2007. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 25:221–242. 10.1146/annurev.immunol.22.012703.104758 [DOI] [PubMed] [Google Scholar]
- Kolde R.2013. pheatmap: Pretty Heatmaps (version 1.0.8). https://rdrr.io/cran/pheatmap
- Leong Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., Minnich M., Meckiff B.J., Wei Y., Hou Z., et al. 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17:1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Lin W.W., Nish S.A., Yen B., Chen Y.H., Adams W.C., Kratchmarov R., Rothman N.J., Bhandoola A., Xue H.H., and Reiner S.L.. 2016. CD8+ T Lymphocyte Self-Renewal during Effector Cell Determination. Cell Reports. 17:1773–1782. 10.1016/j.celrep.2016.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.D.M., Kenngott E.E., Schröter M.F., Kühl A., Jennrich S., Watzlawick R., Hoffmann U., Wolff T., Norley S., Scheffold A., et al. 2014. Timed action of IL-27 protects from immunopathology while preserving defense in influenza. PLoS Pathog. 10:e1004110 10.1371/journal.ppat.1004110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Phillips T., Zhang M., Wang Y., Opferman J.T., Shah R., and Ashton-Rickardt P.G.. 2004. Serine protease inhibitor 2A is a protective factor for memory T cell development. Nat. Immunol. 5:919–926. 10.1038/ni1107 [DOI] [PubMed] [Google Scholar]
- Liu Z., Liu J.-Q., Talebian F., Wu L.-C., Li S., and Bai X.-F.. 2013. IL-27 enhances the survival of tumor antigen-specific CD8+ T cells and programs them into IL-10-producing, memory precursor-like effector cells. Eur. J. Immunol. 43:468–479. 10.1002/eji.201242930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. 2017. Transcription Factor IRF4 Promotes CD8+ T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity. 47:1129–1141.e5. 10.1016/j.immuni.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Mayer K.D., Mohrs K., Reiley W., Wittmer S., Kohlmeier J.E., Pearl J.E., Cooper A.M., Johnson L.L., Woodland D.L., and Mohrs M.. 2008. Cutting edge: T-bet and IL-27R are critical for in vivo IFN-gamma production by CD8 T cells during infection. J. Immunol. 180:693–697. 10.4049/jimmunol.180.2.693 [DOI] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., and Thomas P.D.. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T.C., Hildeman D., Kedl R.M., Teague T.K., Schaefer B.C., White J., Zhu Y., Kappler J., and Marrack P.. 2001. Immunological adjuvants promote activated T cell survival via induction of Bcl-3. Nat. Immunol. 2:397–402. 10.1038/87692 [DOI] [PubMed] [Google Scholar]
- Morishima N., Owaki T., Asakawa M., Kamiya S., Mizuguchi J., and Yoshimoto T.. 2005. Augmentation of effector CD8+ T cell generation with enhanced granzyme B expression by IL-27. J. Immunol. 175:1686–1693. 10.4049/jimmunol.175.3.1686 [DOI] [PubMed] [Google Scholar]
- Morishima N., Mizoguchi I., Okumura M., Chiba Y., Xu M., Shimizu M., Matsui M., Mizuguchi J., and Yoshimoto T.. 2010. A pivotal role for interleukin-27 in CD8+ T cell functions and generation of cytotoxic T lymphocytes. J. Biomed. Biotechnol. 2010:605483 10.1155/2010/605483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan G., and Saha B.. 2013. IL-27 in tumor immunity and immunotherapy. Trends Mol. Med. 19:108–116. 10.1016/j.molmed.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Ng C.T., Sullivan B.M., Teijaro J.R., Lee A.M., Welch M., Rice S., Sheehan K.C., Schreiber R.D., and Oldstone M.B.. 2015. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. 17:653–661. 10.1016/j.chom.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D.M. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 12:252–264. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninger J.M., Sirard C., Mittrücker H.-W., Chidgey A., Kozieradzki I., Nghiem M., Hakem A., Kimura T., Timms E., Boyd R., et al. 1997. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity. 7:243–254. 10.1016/S1074-7613(00)80527-0 [DOI] [PubMed] [Google Scholar]
- Pennock N.D., Gapin L., and Kedl R.M.. 2014. IL-27 is required for shaping the magnitude, affinity distribution, and memory of T cells responding to subunit immunization. Proc. Natl. Acad. Sci. USA. 111:16472–16477. 10.1073/pnas.1407393111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona-Wright G., Kohlmeier J.E., Bassity E., Freitas T.C., Mohrs K., Cookenham T., Situ H., Pearce E.J., Woodland D.L., and Mohrs M.. 2012. Persistent loss of IL-27 responsiveness in CD8+ memory T cells abrogates IL-10 expression in a recall response. Proc. Natl. Acad. Sci. USA. 109:18535–18540. 10.1073/pnas.1119133109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C., Ferrando-Martinez S., Gerner M.Y., Casazza J.P., Pegu A., Deleage C., Cooper A., Hataye J., Andrews S., Ambrozak D., et al. 2017. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med. 9:eaag2285 10.1126/scitranslmed.aag2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H., Michalopoulos E.E., Iwamoto A., Ohashi P.S., Baenziger J., Hengartner H., Zinkernagel R.M., and Mak T.W.. 1987. Molecular analysis of the antigen receptor of virus-specific cytotoxic T cells and identification of a new V alpha family. Eur. J. Immunol. 17:1843–1846. 10.1002/eji.1830171226 [DOI] [PubMed] [Google Scholar]
- Pircher H., Mak T.W., Lang R., Ballhausen W., Rüedi E., Hengartner H., Zinkernagel R.M., and Bürki K.. 1989. T cell tolerance to Mlsa encoded antigens in T cell receptor V beta 8.1 chain transgenic mice. EMBO J. 8:719–727. 10.1002/j.1460-2075.1989.tb03431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias L.C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375–386. 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- Quigley M.F., Gonzalez V.D., Granath A., Andersson J., and Sandberg J.K.. 2007. CXCR5+ CCR7- CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur. J. Immunol. 37:3352–3362. 10.1002/eji.200636746 [DOI] [PubMed] [Google Scholar]
- Quigley M., Huang X., and Yang Y.. 2008. STAT1 signaling in CD8 T cells is required for their clonal expansion and memory formation following viral infection in vivo. J. Immunol. 180:2158–2164. 10.4049/jimmunol.180.4.2158 [DOI] [PubMed] [Google Scholar]
- R Core Development Team 2008. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Salcedo R., Stauffer J.K., Lincoln E., Back T.C., Hixon J.A., Hahn C., Shafer-Weaver K., Malyguine A., Kastelein R., and Wigginton J.M.. 2004. IL-27 mediates complete regression of orthotopic primary and metastatic murine neuroblastoma tumors: role for CD8+ T cells. J. Immunol. 173:7170–7182. 10.4049/jimmunol.173.12.7170 [DOI] [PubMed] [Google Scholar]
- Salcedo R., Hixon J.A., Stauffer J.K., Jalah R., Brooks A.D., Khan T., Dai R.-M., Scheetz L., Lincoln E., Back T.C., et al. 2009. Immunologic and therapeutic synergy of IL-27 and IL-2: enhancement of T cell sensitization, tumor-specific CTL reactivity and complete regression of disseminated neuroblastoma metastases in the liver and bone marrow. J. Immunol. 182:4328–4338. 10.4049/jimmunol.0800471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Yaneva T., Beauseigle D., El-Khoury L., and Arbour N.. 2011. IL-27 increases the proliferation and effector functions of human naïve CD8+ T lymphocytes and promotes their development into Tc1 cells. Eur. J. Immunol. 41:47–59. 10.1002/eji.201040804 [DOI] [PubMed] [Google Scholar]
- Shan Q., Zeng Z., Xing S., Li F., Hartwig S.M., Gullicksrud J.A., Kurup S.P., Van Braeckel-Budimir N., Su Y., Martin M.D., et al. 2017. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 18:931–939. 10.1038/ni.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C.F., Welch M., Schreiber R.D., de la Torre J.C., and Oldstone M.B.A.. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 340:207–211. 10.1126/science.1235214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider D.T., Charmoy M., Chennupati V., Pousse L., Ferreira D.P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D., et al. 2016. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 45:415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Utzschneider D.T., Delpoux A., Wieland D., Huang X., Lai C.-Y., Hofmann M., Thimme R., and Hedrick S.M.. 2018. Active Maintenance of T Cell Memory in Acute and Chronic Viral Infection Depends on Continuous Expression of FOXO1. Cell Reports. 22:3454–3467. 10.1016/j.celrep.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine K.M., Davini D., Lawrence T.J., Mullins G.N., Manansala M., Al-Kuhlani M., Pinney J.M., Davis J.K., Beaudin A.E., Sindi S.S., Gravano D.M., and Hoyer K.K.. 2018. CD8 Follicular T Cells Promote B Cell Antibody Class Switch in Autoimmune Disease. J. Immunol. 201:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J.O., Hammerbeck C.D., and Mescher M.F.. 2005. Cutting edge: Bcl-3 up-regulation by signal 3 cytokine (IL-12) prolongs survival of antigen-activated CD8 T cells. J. Immunol. 174:600–604. 10.4049/jimmunol.174.2.600 [DOI] [PubMed] [Google Scholar]
- Vignali D.A.A., and Kuchroo V.K.. 2012. IL-12 family cytokines: immunological playmakers. Nat. Immunol. 13:722–728. 10.1038/ni.2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., and Kurachi M.. 2015. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15:486–499. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland D., Kemming J., Schuch A., Emmerich F., Knolle P., Neumann-Haefelin C., Held W., Zehn D., Hofmann M., and Thimme R.. 2017. TCF1+ hepatitis C virus-specific CD8+ T cells are maintained after cessation of chronic antigen stimulation. Nat. Commun. 8:15050 10.1038/ncomms15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., and Brooks D.G.. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 340:202–207. 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Ji Y., Moseman E.A., Xu H.C., Manglani M., Kirby M., Anderson S.M., Handon R., Kenyon E., Elkahloun A., Wu W., Lang P.A., Gattinoni L., McGavern D.B., and Schwartzberg P.L.. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1:eaai8593. [DOI] [PMC free article] [PubMed]
- Xu H.C., Grusdat M., Pandyra A.A., Polz R., Huang J., Sharma P., Deenen R., Köhrer K., Rahbar R., Diefenbach A., et al. 2014. Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity. Immunity. 40:949–960. 10.1016/j.immuni.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Yoshida H., and Hunter C.A.. 2015. The immunobiology of interleukin-27. Annu. Rev. Immunol. 33:417–443. 10.1146/annurev-immunol-032414-112134 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Hamano S., Senaldi G., Covey T., Faggioni R., Mu S., Xia M., Wakeham A.C., Nishina H., Potter J., et al. 2001. WSX-1 is required for the initiation of Th1 responses and resistance to L. major infection. Immunity. 15:569–578. 10.1016/S1074-7613(01)00206-0 [DOI] [PubMed] [Google Scholar]
- Zajac A.J., Blattman J.N., Murali-Krishna K., Sourdive D.J., Suresh M., Altman J.D., and Ahmed R.. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.