The authors report a mutation in the long noncoding RNA Reg1cp that induces osteogenesis via vascular induction in humans. This mutation affects angiogenesis by blocking Klf3’s repressing activity. The Klf3 antagonist Ophiopogonin D could promote CD31hiEmcnhi vessel formation and osteogenesis in osteoporosis mice.
Abstract
High bone mass (HBM) is usually caused by gene mutations, and its mechanism remains unclear. In the present study, we identified a novel mutation in the long noncoding RNA Reg1cp that is associated with HBM. Subsequent analysis in 1,465 Chinese subjects revealed that heterozygous Reg1cp individuals had higher bone density compared with subjects with WT Reg1cp. Mutant Reg1cp increased the formation of the CD31hiEmcnhi endothelium in the bone marrow, which stimulated angiogenesis during osteogenesis. Mechanistically, mutant Reg1cp directly binds to Krüppel-like factor 3 (KLF3) to inhibit its activity. Mice depleted of Klf3 in endothelial cells showed a high abundance of CD31hiEmcnhi vessels and increased bone mass. Notably, we identified a natural compound, Ophiopogonin D, which functions as a KLF3 inhibitor. Administration of Ophiopogonin D increased the abundance of CD31hiEmcnhi vessels and bone formation. Our findings revealed a specific mutation in lncRNA Reg1cp that is involved in the pathogenesis of HBM and provides a new target to treat osteoporosis.
Introduction
The skeleton is one of the most complex tissues in mammals and undergoes continuous shaping, remodeling, and repair throughout adulthood to provide protection of the vital organs and rigid support for the whole body (Long and Ornitz, 2013; Riddle and Clemens, 2017). The structure and function of bone is maintained by the balance between bone resorption and formation (Long, 2011; Niedźwiedzki and Filipowska, 2015; Croucher et al., 2016). Osteoporosis, characterized by increased fragility in skeletal tissue, typically reflects an imbalance of bone remodeling in which bone resorption exceeds bone formation (Rachner et al., 2011). With the progressive aging of the general population, osteoporosis has emerged as a medical and socioeconomic problem. However, most current treatment options for osteoporosis have limitations and side effects that affect their long-term administration and patient adherence (Rachner et al., 2011; Jaleel et al., 2018). Disturbance of osteoblast or osteoclast regulation would also lead to abnormal accumulation of bone, such as osteosclerosis, which is classified according to its causative factor as either acquired or hereditary. Hereditary osteosclerosis includes osteopetrosis and high bone mass (HBM). Osteopetrosis is a rare inherited disorder involving decreased bone resorption. HBM results in increased bone formation that leads to an abnormal elevation in bone density (Boyden et al., 2002; Bonewald, 2011). Numerous studies have shown that mutations in the regulators of bone metabolism are the genetic determinants of HBM (Johnson et al., 1997; Boyden et al., 2002; Leupin et al., 2011). Boyden et al. (2002) performed genetic and biochemical analyses in a kindred with HBM and detected gain-of-function mutations in the gene encoding low-density lipoprotein receptor–related protein 5 (LRP5). Identifying the pathway(s) that affect the imbalance in bone remodeling during HBM pathogenesis might lead to the identification of therapeutic targets for osteoporosis.
Specialized vessels formed in tissues participate in the formation of a specific microenvironment that decides the fate of progenitor cells (Jabalee and Franz-Odendaal, 2015; Ramasamy et al., 2015, 2016; Rafii et al., 2016). CD31hiEMCNhi vessels (CD31, also known as PECAM1 [platelet and endothelial cell adhesion molecule 1]; EMCN, endomucin), which are located in the metaphysis and endosteum of postnatal long bones and stain strongly for CD31 and EMCN, were identified as specific vessels in the skeletal system that couple angiogenesis and osteogenesis (Kusumbe et al., 2014; Ramasamy et al., 2014). However, the abundance of CD31hiEMCNhi vessels declines markedly during aging (Kusumbe et al., 2014; Wang et al., 2017; Yang et al., 2017). In our previous study, we demonstrated that inducing CD31hiEMCNhi vessels could prevent bone loss in osteoporosis (Xie et al., 2014; Yang et al., 2017). Angiogenesis coupled with osteogenesis plays an important role in bone metabolism and could be a new target to treat low bone mass diseases.
In the present study, we identified a novel mutation in a long noncoding RNA (lncRNA) gene, Reg1cp, which is associated with HBM. The homozygous mutant of Reg1cp binds directly to Krüppel-like factor 3 (KLF3), a transcription factor, to regulate angiogenesis. Endothelial-specific klf3 knockout mice showed increased CD31hiEMCNhi vessels and bone formation. Notably, we identified a natural compound as a KLF3 inhibitor, which could increase the CD31hiEMCNhi endothelium and promote bone formation in aged mice. Taken together, our study identified a potential therapeutic target to treat osteoporosis.
Results
Reg1cp is a new HBM-associated gene
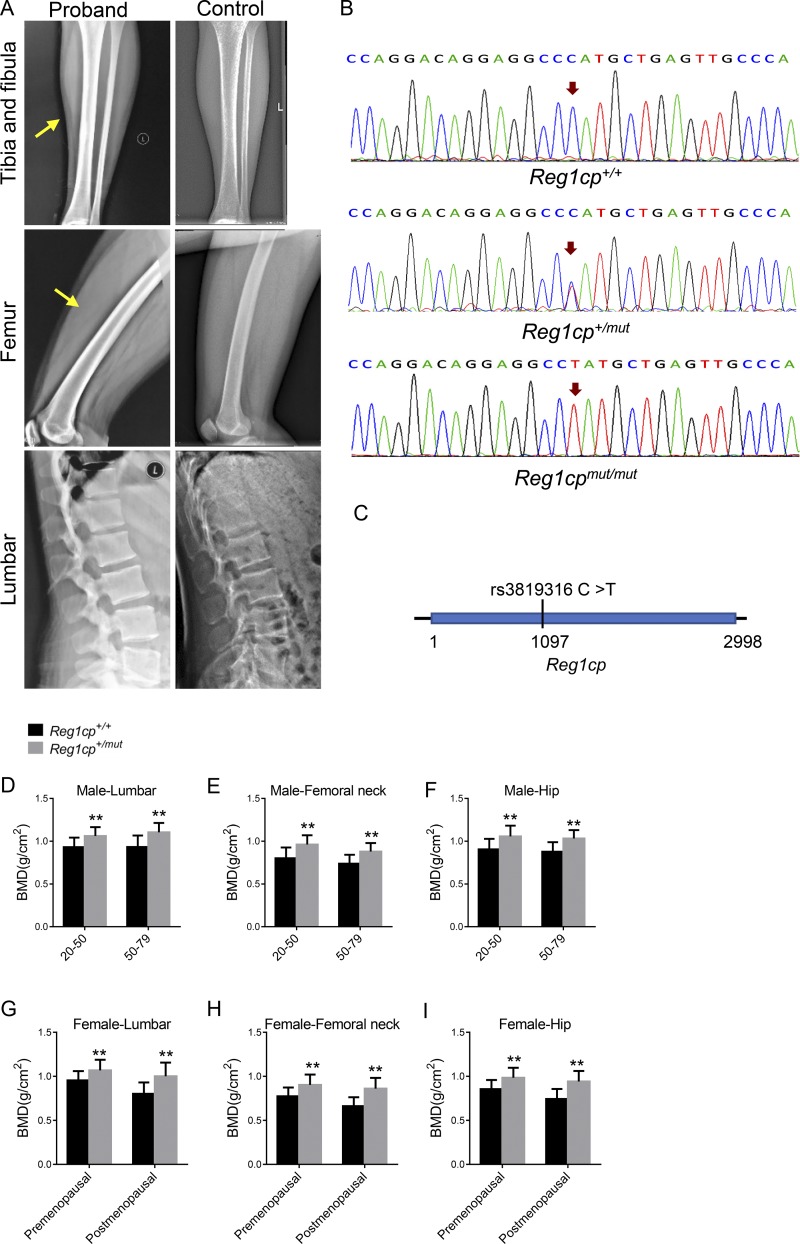
During clinical screening, we identified one patient who had extremely high bone density. This patient was female, 20 yr old, with a date of birth of April 22, 1997. She was of normal height (163.5 cm) and weight (60 kg). Clinical examination was not remarkable, except for HBM, a bone mineral density (BMD) of 1.266 g/cm2 in the hip (Z-score = +4.0), 1.169 g/cm2 in the femoral neck (Z-score = +3.9), and 1.191 g/cm2 in the lumbar spine (Z-score = +2.9). Radiographical examination at several skeletal sites showed normal skeletal morphology, except for a significant thickening of the cortical bones (Fig. 1 A). The circulating levels of type I procollagen amino-terminal propeptide (PINP), indicated that the level of bone formation was slightly increased; however, the circulating levels of β-isomerized type I collagen (COL 1) C-telopeptide breakdown products (β-CTX), which reflect the level of bone resorption, were normal (Table 1). This patient also had normal levels of bone-related hormones, such as parathyroid hormone (PTH), human chorionic thyrotropin (hCT), and 25-hydroxy vitamin D (25-OHVD; Table 1). After excluding a unique dietary lifestyle or conditions affecting bone metabolism, we hypothesized that the HBM phenotype had a genetic cause. To investigate the potential genetic variation related to HBM, samples from the patient and her parents were analyzed using whole-exome sequencing. We searched for the potential mutation site using several steps. First, we filtered variants in which allele frequency was >5% in the Exome Variant Server and 1000 Genomes databases. Then, nonsynonymous mutations, multiallelic variants, and frameshift mutations were removed. Ultimately, six variants in the RYR3, MUC7, END2, REG1CP, SERINC4, and RDH16 genes remained. No mutation was found among the genes known to cause hyperostosis. Bioinformatic analysis allowed us to select the variant in the Reg1cp gene, which validated the allele frequency in control individuals and the relation between the phenotype and the variant. The patient had a homozygous mutation in the lncRNA Reg1cp gene on chromosome 2 (rs3819316 C >T), and the heterozygous mutation at the same site was found in her parents (Fig. 1, B and C; and Fig. S1 A).
Figure 1.
Reg1cp is a new HBM-associated gene. (A) Radiographical features of the proband with a homozygous mutation in the Reg1cp gene and age-related normal controls. Yellow arrows indicate thickening of the cortical bones in tibia and femur. (B) DNA sequencing data of an unaffected man (Reg1cp+/+), the proband with homozygous mutation (Reg1cpmut/mut), and one subject with heterozygous mutation (Reg1cp+/mut). Representative of three independent experiments. (C) Genomic structure of the human Reg1cp gene. The mutation identified is marked. (D–F) BMD in lumbar (D), femoral neck (E), and hip (F) of male Reg1cp+/mut subjects and age-matched Reg1cp+/+ controls. (G–I) BMD in lumbar (G), femoral neck (H), and hip (I) of the female Reg1cp+/mut subjects and age-matched Reg1cp+/+ controls. The number of human samples is indicated in Tables S2 and S3. Data shown as mean ± SD. **, P < 0.01; Student’s t test.
Table 1. Biochemical survey for bone remodeling parameters in the proband.
| Parameter | Value | Reference value |
|---|---|---|
| PINP | 78.53 ng/ml | 57.84–78.31 ng/ml |
| β-CTX | 496.78 pg/ml | 350.52–573.44 pg/ml |
| ALP | 67.1 U/liter | 32–120 U/liter |
| PTH | 36.2 pg/ml | 15–65 pg/ml |
| hCT | 17.2 mg/ml | 6–24.66 mg/ml |
| 25-OHVD | 35.6 ng/ml | 30–100 ng/ml |
| GH | 0.18 ng/ml | <7.5 ng/ml |
GH, growth hormone.
We extended this analysis to a larger cohort (n = 1,465, including 616 males and 849 females). Among them, 67 heterozygous Reg1cp individuals (Reg1cp+/mut) were identified (Table S1). These Reg1cp+/mut individuals had higher bone densities and higher serum PINP levels compared with the subjects with the WT gene (Reg1cp+/+; Fig. 1, D–I; and Tables S2, S3, S4, and S5). There is no consensus about the definition of HBM, and Z-score cutoffs of greater than +2.5 or greater than or equal to +4 have been suggested (Paccou et al., 2018). In the present study, we used the Z-score cutoff score of greater than +2.5 at the lumbar as the definition of HBM. The proportion of HBM was 0.715% (10 of 1,398 subjects) among the Reg1cp+/+ samples and 5.970% (4 of 67 subjects) among the Reg1cp+/mut samples. We also extended the family line investigation among the subjects with heterozygous mutation of Reg1cp randomly. Two families agreed to be investigated. The family line investigation showed hereditary genetic correlations of the Reg1cp mutation (Fig. S1, B and C). These results suggested that Reg1cp might be a new HBM-associated gene.
Reg1cp is involved in the regulation of CD31hiEMCNhi endothelium formation
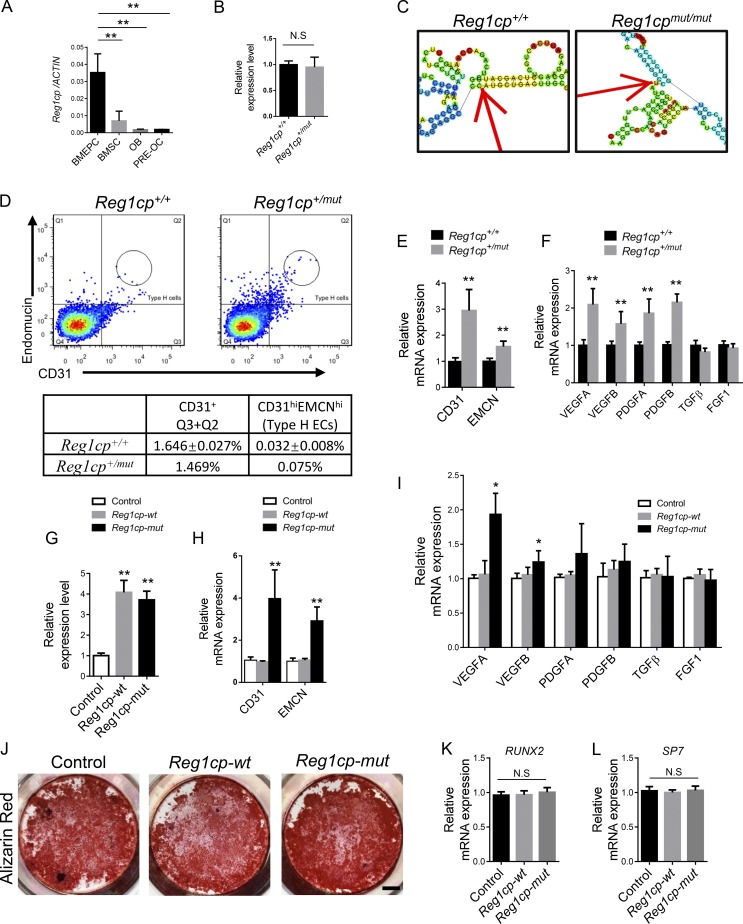
LncRNA Reg1cp belongs to the Reg family and is encoded as a pseudogene that is mainly expressed in the pancreas (Gharib et al., 1993). Reg1cp has little reported association with bone metabolism regulation. To determine the role of Reg1cp and its mutant in bone metabolism, we examined the expression of Reg1cp in isolated bone marrow endothelial progenitor cells, bone marrow mesenchymal stem cells (BMSCs), osteoblasts, and osteoclast precursor cells from human bone samples. Reg1cp was mainly expressed in bone marrow endothelial progenitor cells in skeletal tissue (Fig. 2 A). The expression level of Reg1cp was similar in Reg1cp+/mut individuals and age-matched Reg1cp+/+ controls (Fig. 2 B). However, structure prediction suggested that the mutation led to a large change in the structure of Reg1cp (Fig. 2 C and Fig. S2).
Figure 2.
Reg1cpis involved in the regulation of CD31hiEMCNhi endothelium formation. (A) Reg1cp expression levels in different cells isolated from human bone marrow. (B) Reg1cp expression levels in ECs. (C) The predicted secondary structure of Reg1cp and mutant Reg1cp around the mutant site (RNAfold Webserver, http://rna.tbi.univie.ac.at/cgibin/RNAfold.cgi). (D) FACS analysis dot plot and quantitation of CD31hiEMCNhi ECs (Type H ECs) of bone samples from one 37-yr-old male Reg1cp+/mut subject and five age-matched Reg1cp+/+ controls. (E) qRT-PCR analysis of CD31 and EMCN expression levels in ECs. (F) qRT-PCR analysis of VEGFA, VEGFB, PDGFA, PDFB, TGFβ, and FGF1 expression level in ECs. (G) Reg1cp expression level in human ECs transfected with Reg1cp-mut or Reg1cp-wt plasmid. (H and I) CD31 and EMCN expression levels (H) and VEGFA, VEGFB, PDGFA, PDFB, TGFβ, and FGF1 expression levels (I) in ECs. (J–L) Representative images of Alizarin Red S staining (J) and qRT-PCR analysis of the levels of SP7 and RUNX2 expression (K and L) in human BMSCs transfected with Reg1cp-mut or Reg1cp-wt plasmid with osteogenic induction. Scale bar, 0.5 cm. In A, B, E–I, K, and L, n = 5 in each group from three independent experiments. J is representative of three independent experiments. Data are shown as the mean ± SD. *, P < 0.05; **, P < 0.01; N.S, no significance; Student’s t test (B, E, and F) and ANOVA (A, G–I, K, and L). BMEPC, bone marrow endothelial progenitor cell; OB, osteoblast; PRE-OC, osteoclast precursor cell.
The CD31hiEMCNhi endothelium couples angiogenesis with osteogenesis. We observed that CD31, EMCN, and vessel growth factors (VEGFA, VEGFB, PDGFA, and PDGFB) transcripts were expressed at higher levels in endothelial cells (ECs) isolated from the Reg1cp+/mut individuals than in cells from age-matched Reg1cp+/+ controls (Fig. 2, E and F). We also found one Reg1cp+/mut subject that had pronounced induction of CD31hiEMCNhi ECs in the bone marrow compared with that in aged-matched Reg1cp+/+ controls (Fig. 2 D). To clarify whether the increased levels of growth factors are related to mutation of Reg1cp, we transferred plasmids encoding mutant Reg1cp (Reg1cp-mut) or WT Reg1cp (Reg1cp-wt) into ECs isolated from Reg1cp+/+ subjects. Quantitative real-time RT-PCR (qRT-PCR) confirmed the successful transfection (Fig. 2 G). The delivery of the exogenous Reg1cp-mut plasmid significantly induced the transcription of CD31, EMCN, VEGFA, and VEGFB (Fig. 2, H and I). However, the expression levels of these genes did not change after delivery of the exogenous Reg1cp-wt plasmid (Fig. 2, H and I). Reg1cp was expressed in BMSCs (Fig. 2 A). To investigate whether Reg1cp-mut could affect osteogenic differentiation, we transfected Reg1cp-mut or Reg1cp-wt plasmids into BMSCs undergoing osteogenic induction. Alizarin Red staining showed no difference between the Reg1cp-mut and Reg1cp-wt groups (Fig. 2 J). The expression level of the osteoblast transcription factors SP7 and RUNX2 remained unchanged (Fig. 2, K and L). This result indicated that the mutant Reg1cp was involved in the regulation of angiogenesis in ECs.
Mutant Reg1cp directly binds to KLF3 and affects its binding to downstream genes in ECs
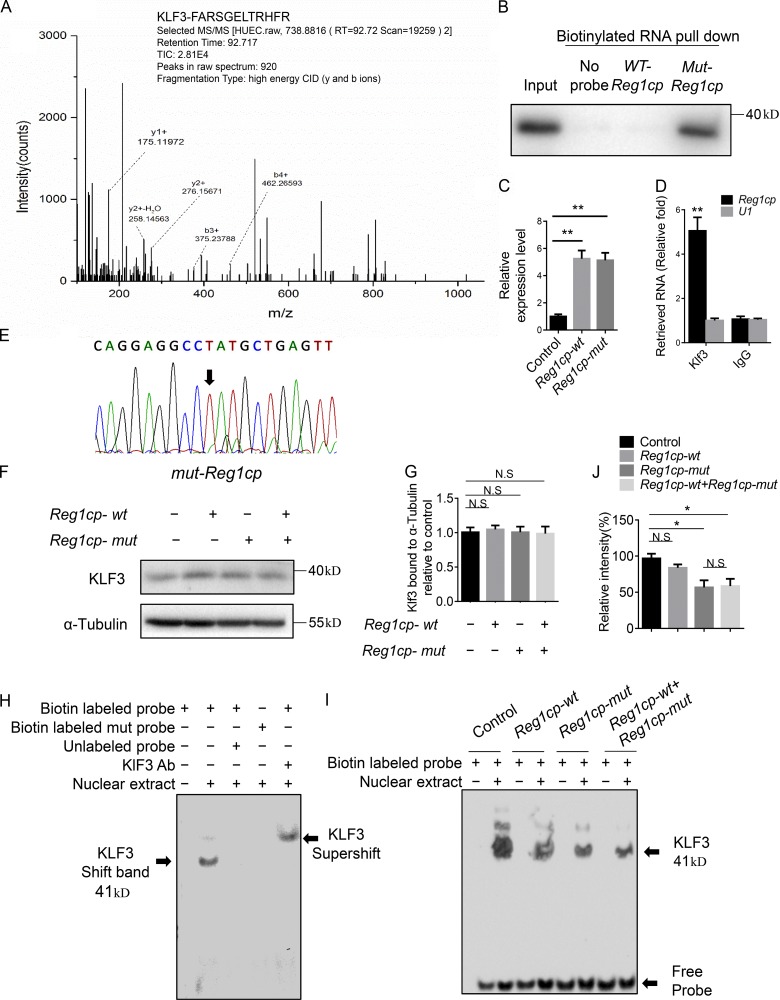
To identify the downstream molecules of Reg1cp that mediate angiogenesis, we conducted mass spectrometry (MS) of proteins after RNA pulldown using WT Reg1cp or mutant Reg1cp in human microvascular ECs (HMECs). Among the identified proteins, KLF3 showed the largest difference (Fig. 3 A). Mutant Reg1cp (Mut-Reg1cp), but not WT Reg1cp (WT-Reg1cp), specifically retrieved KLF3 (Fig. 3 B). Phylogenetic analysis of the C-terminal DNA-binding domains divides the KLF family into three subgroups. KLF3 is a member of group 2, which also contains KLF8 and KLF12 (Pearson et al., 2011). However, neither KLF8 nor KLF12 was retrieved by Mut-Reg1cp (Fig. S3 A). To further validate the interaction between KLF3 and Mut-Reg1cp, we transfected HMECs with the Reg1cp-mut or Reg1cp-wt plasmids; qRT-PCR confirmed the successful transfection (Fig. 3 C). We then conducted RNA immunoprecipitation of HMECs transfected with the Reg1cp plasmids. The anti-KLF3 antibody pulled down significantly more Reg1cp than did the IgG control (Fig. 3 D). To further distinguish which Reg1cp (Mut-Reg1cp or WT-Reg1cp) bound to KLF3, the pulled down transcripts were reverse transcribed into cDNA and subjected to Sanger sequencing. The sequencing result showed that KLF3 pulled down Mut-Reg1cp, but not WT-Reg1cp (Fig. 3 E). Thus, these results indicated that Mut-Reg1cp binds directly to KLF3 in ECs.
Figure 3.
Mutant Reg1cp binds directly to KLF3 and affects its binding to downstream genes in ECs. (A) KLF3’s interaction with Mut-Reg1cp was identified by MS. The b ions and y ions indicate peptide backbone fragment containing the N and C termini, respectively (y1+, y2+, b3+, and b4+ are labeled). CID, collision induced dissociation. (B) Mut-Reg1cp retrieves KLF3, as detected by immunoblotting. (C) Reg1cp expression level in HMECs transfect with Reg1cp-mut or Reg1cp-wt plasmid. (D) KLF3 retrieves Reg1cp RNA specifically, as detected by qRT-PCR. (E) Sanger sequencing results of transcripts pulled down by an anti-KLF3 antibody. (F and G) Western blotting analysis (F) and quantitation (G) of the relative levels of KLF3 protein expression in Hek293T cells transfected with Reg1cp-mut or Reg1cp-wt plasmids. (H) EMSA of the binding of KLF3 with JunB CACCC probe in vitro. Shift band stands for specific KLF3/probe complexes. Ab, antibody. (I and J) Nuclear extracts were prepared from Hek293T cells transfected with Reg1cp-mut or Reg1cp-wt plasmids. Binding of KLF3 to a JunB CACCC probe was assessed using EMSA (I) and quantified using ImageJ software (J). B and E–J are representative of three independent experiments. In C, n = 5 in each group from three independent experiments. In D, n = 3 in each group from three independent experiments. Data are shown as the mean ± SD. *, P < 0.05; **, P < 0.01; N.S, not significant; ANOVA (C, G, and J) and Student’s t test (D).
KLF3 acts as transcriptional repressor that mediates transcriptional silencing via recruiting the corepressor C-terminal binding protein (CTBP; Pearson et al., 2011; Dewi et al., 2015). To investigate whether the binding of Mut-Reg1cp affected the function of KLF3, we transfected Hek293T cells with the Reg1cp-mut plasmid, the Reg1cp-wt plasmid, or both. Transfection of the Reg1cp plasmids did not affect the expression of KLF3 (Fig. 3, F and G). JUNB (encoding JunB proto-oncogene, AP-1 transcription factor subunit) is one of KLF3’s downstream genes. We conducted an electrophoretic mobility shift assay (EMSA) using the nuclear extracts from the Hek293T cells and an oligonucleotide probe designed based on the predicted KLF3 binding site on the promoter of JUNB. The EMSA results demonstrated that KLF3 could specifically bind to the promoter of JUNB (Fig. 3 H). KLF3 binding was reduced after Reg1cp-mut plasmid transfection; however, there was no difference between the group transfected with the Reg1cp-mut plasmid and the group transfected with both plasmids, which suggested that the mutant Reg1cp does not compete with the WT form (Fig. 3, I and J). Transfection with Reg1cp-mut plasmid did not affect the recruitment of corepressors CTBP1 and CTBP2 to KLF3 (Fig. S3, C and D). These results indicated that Mut-Reg1cp binds directly to KLF3 and affects the binding of KLF3 to its downstream genes.
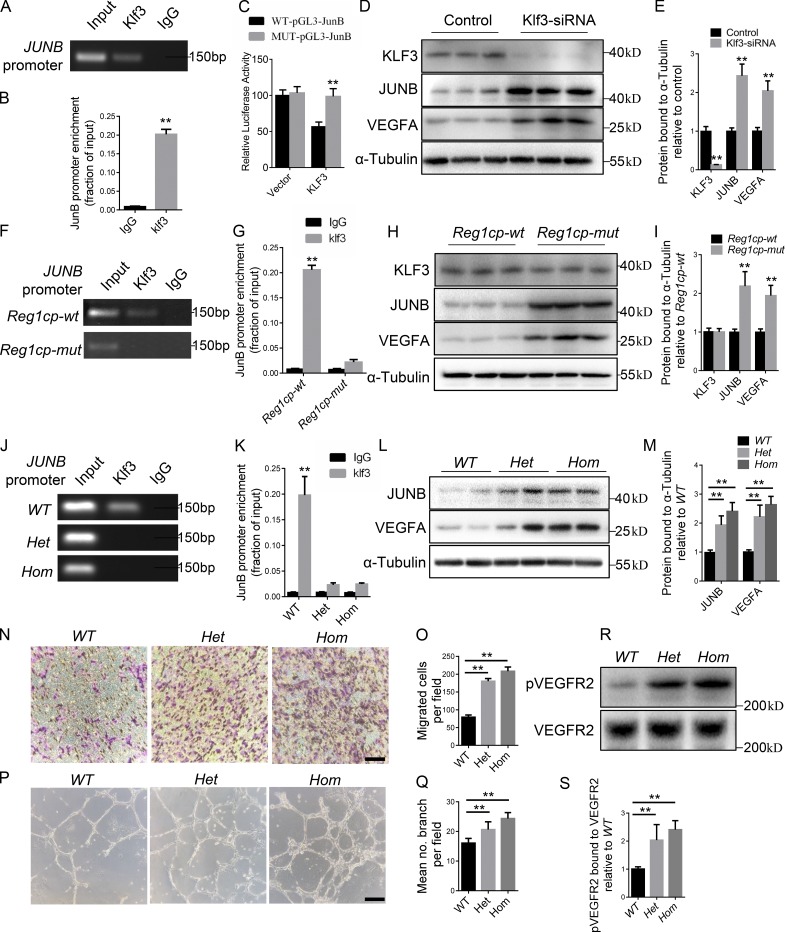
Mutant Reg1cp abolishes the role of KLF3 in CD31hiEMCNhi vessel formation
KLF3 is a potent transcriptional repressor with widespread roles in differentiation (Pearson et al., 2011). Therefore, we investigated how the binding of Mut-Reg1cp to KLF3 affected CD31hiEMCNhi vessel formation. We performed chromatin immunoprecipitation (ChIP)–PCR assays to assess the binding of KLF3 at the reported downstream genes involved in angiogenesis (Ilsley et al., 2017). Among them, JUNB has been reported as a critical independent regulator of VEGFA transcription (Schmidt et al., 2007). ChIP-PCR assays showed strong KLF3 binding to the promoter of JUNB (Fig. 4, A and B). Luciferase reporter constructs containing the WT or mutant predicted KLF3-binding site of JUNB (WT-pGL3-JunB and MUT-pGL3-JunB) were generated. We transfected WT-pGL3-JunB or MUT-pGL3-JunB with the KLF3 plasmid or empty vector into HMECs and measured luciferase fluorescence. KLF3 repressed the luciferase activity of the WT-pGL3-JunB reporter gene, but not that of MUT-pGL3-JunB (Fig. 4 C). We also transfected HMECs with a KLF3 siRNA to silence the KLF3 gene. Western blotting analysis showed significantly lower KLF3 but higher JUNB protein levels after transfection of siRNA-KLF3 (Fig. 4, D and E). The level of VEGFA was elevated along with JUNB (Fig. 4, D and E). These results confirmed the direct repression of JUNB transcription by KLF3.
Figure 4.
Mutant Reg1cp abolishes the role of KLF3 in CD31hiEMCNhi vessel formation. (A) ChIP-PCR assays with anti-KLF3 antibodies or anti-IgG antibodies using specific primers targeting the promoter regions of JUNB. (B) qRT-PCR analysis of the JUNB expression level after anti-KLF3 or anti-IgG ChIP. (C) HMECs were transfected with luciferase reporter carrying WT-pGL3-JunB or MUT-pGL3-JunB, respectively, and cotransfected with the Klf3 plasmid or vector. Firefly luciferase values, normalized for renilla luciferase, are presented. (D and E) Western blotting analysis (D) and quantitation (E) of the relative levels of KLF3, JUNB, and VEGFA protein expression. (F) ChIP-PCR assays with anti-KLF3 antibodies or anti-IgG antibodies in HMECs transfected with Reg1cp-mut or Reg1cp-wt plasmids. (G) JUNB expression level of anti-KLF3 or anti-IgG ChIP. (H and I) Western blotting analysis (H) and quantitation (I) of the relative levels of KLF3, JUNB, and VEGFA protein expression. (J) ChIP-PCR assays with anti-KLF3 antibodies or anti-IgG antibodies in HMECs with different Reg1cp genotypes. Het, heterozygous mutation; Hom, homozygous mutation. (K) JUNB expression level of anti-KLF3 or anti-IgG ChIP. (L and M) Western blotting analysis (L) and quantitation (M) of the relative levels of JUNB and VEGFA protein expression. (N and O) Representative images (N) and relative quantification (O) of a transwell migration assay. Scale bar, 150 µm. (P and Q) Representative images (P) and relative quantification (Q) of tube branch numbers of a Matrigel tube formation assay. Scale bar, 750 µm. (R and S) HMECs were cultured under hypoxia for 24 h and analyzed (R) and quantified (S) for VEGFR2 phosphorylation (pVEGFR2, top) and VEGFR2 total levels (bottom). All panels were representative of three independent experiments. Data are shown as the mean ± SD. **, P < 0.01; Student’s t test (B, C, E, G, I, and K) and ANOVA (M, O, Q, and S).
To examine whether Mut-Reg1cp regulates angiogenesis by directly affecting the function of KLF3 in the transcription of JUNB, we performed the same ChIP-PCR assays in HMECs transfected with the Reg1cp-mut or Reg1cp-wt plasmids. Mutant Reg1cp abolished the binding of KLF3 to the promoter region of JUNB (Fig. 4, F and G). In addition, the HMECs transfected with the Reg1cp-mut plasmid showed significantly increased expression of JUNB and VEGFA, without affecting KLF3 expression (Fig. 4, H and I). To further investigate how the heterozygous mutation could have a dominant effect, we obtained an HMEC cell line with a heterozygous (Het) or homozygous (Hom) mutation of the Reg1cp gene using CRISPR/Cas9-mediated genome engineering. Sanger sequencing confirmed the successful construction (Fig. S3 B). The HMECs with the heterozygous mutation affected the binding of KLF3 to the JUNB promoter (Fig. 4, J and K), and further increased the protein level of JUNB and VEGFA in HMECs (Fig. 4, L and M), similar to the homozygous mutation. Both the heterozygous and homozygous mutation could increase the migration and tube formation ability of HMECs, and the homozygous mutation was more effective (Fig. 4, N–Q). The HMECs with the Reg1cp mutation showed increased VEGF signaling compared with that in the WT controls under hypoxic conditions (Fig. 4, R and S).
These results indicated that KLF3 represses the expression of JUNB in ECs; however, the mutated Reg1cp directly binds to KLF3 and abolished this process.
Endothelial-specific Klf3 knockout mice show increased CD31hiEMCNhi vessels and bone formation
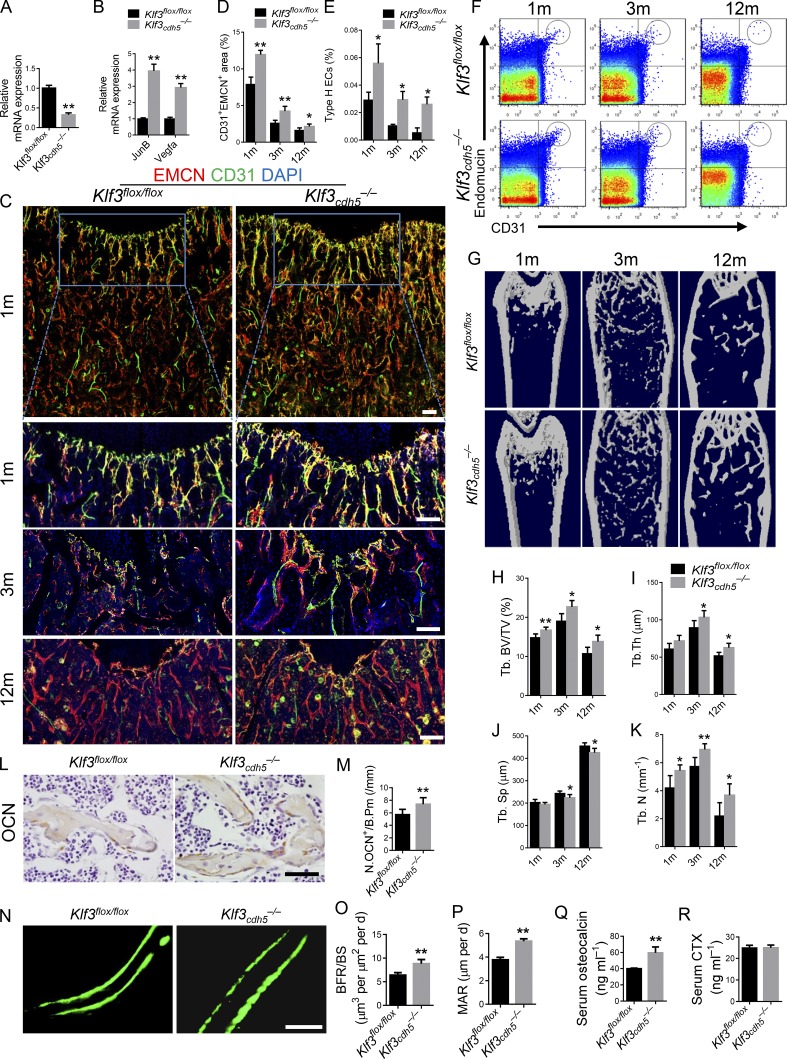
KLF3 was broadly expressed in bone marrow, and there were no regional differences among the diaphysis, metaphysis, or endosteum in KLF3 expression in the bone vasculature (Fig. S4, A and B). To investigate the role of Klf3 in ECs in vivo, we crossed Cdh5 (PAC)-Cre transgenic mice with Klf3flox/flox mice to specifically knock out Klf3 in ECs (Klf3cdh5−/−). As expected, the expression level of Klf3 was significant decreased in the ECs of Klf3cdh5−/− mice (Fig. 5 A). Specific knockout of Klf3 in ECs did not affect the body weight of the mice (Fig. S4 J). The expression levels of JunB and Vegfa increased significantly in the ECs of Klf3cdh5−/− mice (Fig. 5 B). Co-immunostaining for CD31 and EMCN showed higher levels of the CD31hiEMCNhi endothelium in the Klf3cdh5−/− mice compared with that in their Klf3flox/flox littermates (Fig. 5, C and D). Flow cytometry confirmed a significant increase in CD31hiEMCNhi ECs in the bone marrow of the Klf3cdh5−/− mice (Fig. 5, E and F). Microcomputed tomography (µCT) identified a significantly increased trabecular bone volume, number, and thickness, and decreased trabecular separation in Klf3cdh5−/− mice (Fig. 5, G–K). The number of bone surface osteocalcin+ (OCN+) osteoblasts was increased in the Klf3cdh5−/− mice (Fig. 5, L and M). However, the number and surface of osteoclasts in the bone surface were unchanged (Fig. S4, C–E). Calcein double labeling confirmed that the Klf3cdh5−/− mice had increased bone formation rates (BFRs) and mineral apposition rates (MARs; Fig. 5, N–P). The Klf3cdh5−/− mice had increased circulating OCN levels; however, the serum type I collagen C-telopeptide breakdown product (CTX) levels were comparable to those of the controls (Fig. 5, Q and R). There were no differences in the expression level of Wnt target genes in the Klf3cdh5−/− mice (Fig. S4 F). Yadav et al. (2008) reported that LRP5 regulates bone metabolism by regulating serotonin synthesis, which is reflected by Tph1 expression. We detected the expression level of Tph1 in the duodenum. The result showed no differences between Klf3cdh5−/− mice and their Klf3flox/flox controls (Fig. S4 G). Aggrecan and SOX9 staining showed that specific knockout of Klf3 in ECs did not affect the morphology of the growth plate (Fig. S4, H and I).
Figure 5.
Endothelial-specific Klf3 knockout mice show increased CD31hiEMCNhi vessels and bone formation. (A) Expression level of Klf3 in ECs. (B) qRT-PCR analysis of JunB and Vegfa levels in ECs. (C and D) Representative images (C) and quantitation (D) of CD31 (green) and EMCN (red) immunostaining in femora from endothelial-specific Klf3 knockout mice (Klf3cdh5−/−) and their littermate controls (Klf3flox/flox). Scale bars, 100 µm. (E and F) Quantitation (E) and FACS analysis dot plot (F) of CD31hiEMCNhi ECs (Type H ECs) from long bones of 1-, 3-, and 12-mo-old Klf3cdh5−/− mice and their littermate controls. (G–K) Representative μCT images (G) and quantitative μCT analysis (H–K) of trabecular bone microarchitecture in femora. (L and M) Immunohistochemical staining (L) and quantification (M) of OCN+ cells (brown) in femora. Scale bar, 50 µm. (N–P) Representative images of calcein double labeling of trabecular bone (N) with quantification of BFR per bone surface (BFR/BS; O) and MAR (P). Scale bar, 25 µm. (Q and R) Serum levels of OCN (Q) and CTX (R) at the time of harvest. n = 6 mice in each group from three independent experiments. Data are shown as the mean ± SD. *, P < 0.05; **, P < 0.01. Student’s t test.
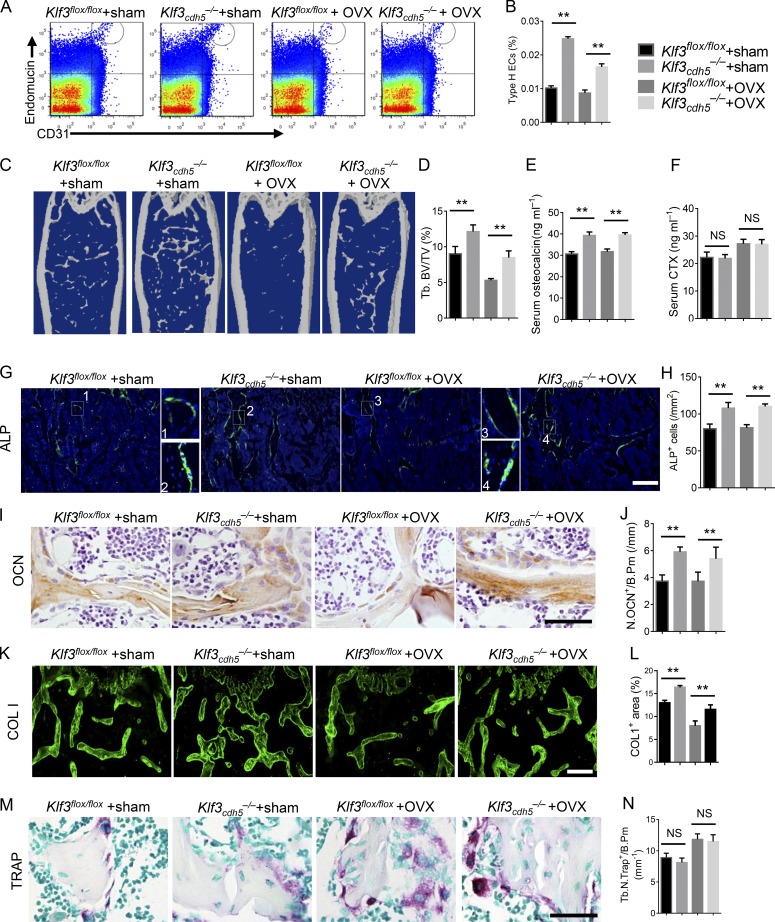
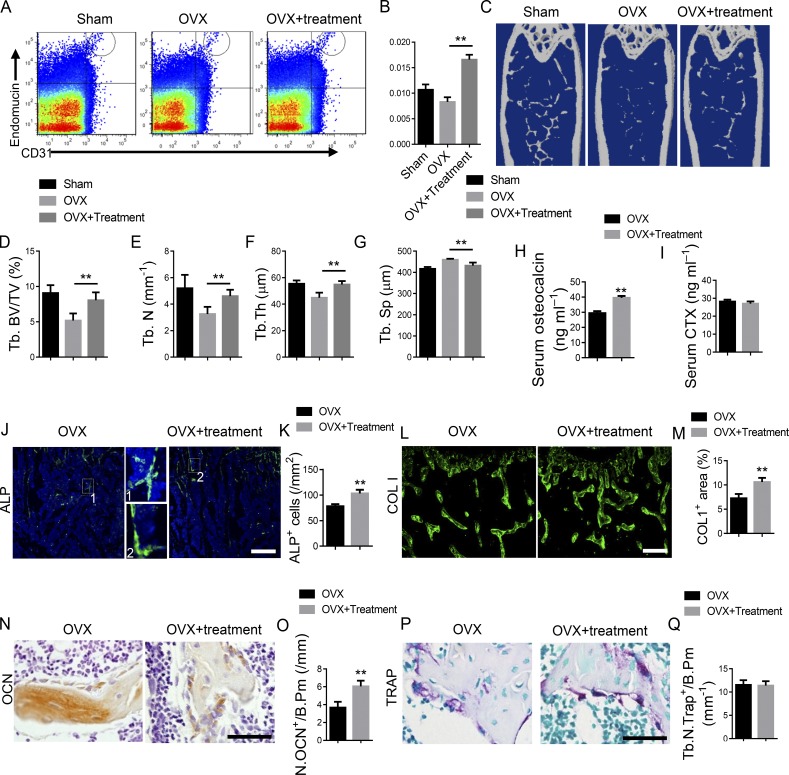
To investigate the role of KLF3 in ECs in an ovariectomized (OVX) model of osteoporosis, OVX surgery was conducted in Klf3cdh5−/− mice and their Klf3flox/flox littermates at 2 mo old. The bone phenotype was detected 3 mo later. Specific knockout of Klf3 in ECs increased the amount of CD31hiEMCNhi endothelium, the bone volume, the number of alkaline phosphatase–positive (ALP+) osteoprogenitors, bone surface OCN+ osteoblasts, and the serum OCN level and bone marrow COL 1–positive area in mice, with or without OVX surgery (Fig. 6, A–E, and G–L). However, endothelium-specific knockout of Klf3 did not affect the number of osteoclasts and or their function, as indicated by tartrate-resistant acid phosphatase (TRAP) staining (Fig. 6, M and N) and serum CTX levels (Fig. 6 F). Taken together, these results suggested that the Klf3cdh5−/− mice had increased numbers CD31hiEMCNhi vessels and increased bone formation.
Figure 6.
Endothelial-specific Klf3 knockout in OVX mice show increased CD31hiEMCNhi vessels and bone formation. (A and B) FACS analysis dot plot (A) and quantification (B) of CD31hiEMCNhi ECs (Type H ECs). (C and D) Representative μCT images (C) and quantitative μCT analysis (D) of trabecular bone microarchitecture in femora. (E and F) Serum levels of OCN (E) and CTX (F) at the time of harvest. (G and H) Representative images (G) and quantification (H) of ALP (green) immunostaining in femora. Scale bar, 200 µm. (I and J) Immunohistochemical staining (I) and quantification (J) of OCN+ cells (brown) in femora. Scale bar, 50 µm. (K and L) Immunohistochemical staining (K) and quantification (L) of COL 1 (green) in femora. Scale bar, 200 µm. (M) Representative images of TRAP staining of femora. Scale bar, 50 µm. (N) Quantification data of TRAP+ cells in trabecular bone surface. Number of TRAP+ cells per bone perimeter (Tb.N.Trap+/B.Pm) was measured. n = 6 mice in each group from three independent experiments. Data are shown as the mean ± SD. **, P < 0.01; ANOVA.
Molecular docking identified a natural compound as a KLF3 inhibitor
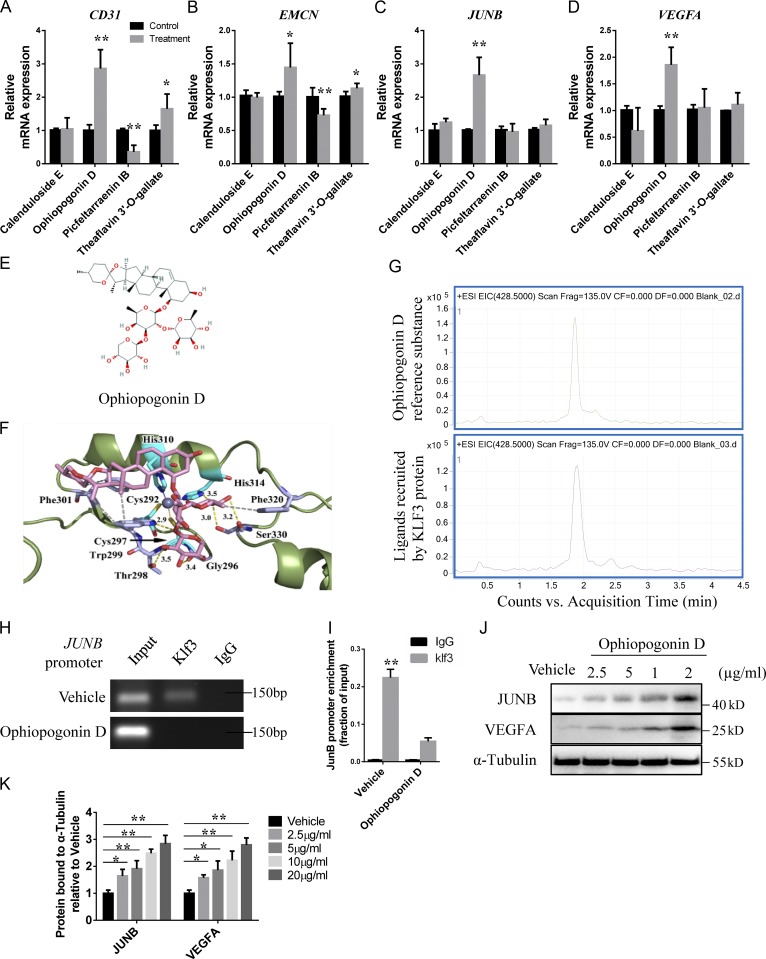
To identify novel small molecule inhibitors of KLF3, we performed virtual screening based on compounds docking to the binding pocket of KLF3 protein and obtained a small number of diverse compounds (Table S6). The top four compounds, Calenduloside E, Theaflavin 3′-O-gallate, Ophiopogonin D, and Picfeltarraenin IB, were selected for further experimentation. We treated HMECs with the four compounds for 48 h, and then detected the expression levels of CD31, EMCN, JUNB, and VEGFA. Ophiopogonin D treatment significantly increased the CD31, EMCN, JUNB, and VEGFA expression levels (Fig. 7, A–D). The structure and binding mode of Ophiopogonin D and KLF3 showed that Ophiopogonin D covered the zinc finger structure of KLF3 through a hydrophobic skein ring and three glycoside structures (Fig. 6, E and F).
Figure 7.
A natural compound is identified as a KLF3 inhibitor by molecular docking. (A–D) qRT-PCR analysis of the relative levels of CD31 (A), EMCN (B), JUNB (C), and VEGFA (D) expression in HMECs treated with four different compounds. n = 3 in each group from three independent experiments. (E) The structure of Ophiopogonin D selected by molecular docking. (F) Crystal structure of Ophiopogonin D bound to Klf3. (G) HPLC-MS chromatograms of Ophiopogonin D reference substance (upper panel) and KLF3 recruit ligand (lower panel). Representative of two independent experiments. (H) ChIP-PCR assays with anti-Klf3 antibodies or anti-IgG antibodies in HMECs treated with Ophiopogonin D and control groups. Representative of three independent experiments. (I) qRT-PCR analysis of JUNB expression after anti-KLF3 or anti-IgG ChIP. n = 3 in each group from three independent experiments. (J and K) Western blotting analysis (J) and the quantification (K) of the levels of JUNB and VEGFA in HMECs treated with vehicle or different doses of Ophiopogonin D. Representative of three independent experiments. Data are shown as the mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test (A–D and I) and ANOVA (K).
To further demonstrate the direct binding of Ophiopogonin D to KLF3, Ophiopogonin D in a mixture was allowed to bind to the His-tag KLF3 protein. HPLC with tandem MS chromatograms of Ophiopogonin D reference substance and the KLF3 recruited ligand showed high consistency, indicating that the KLF3 protein could directly recruit Ophiopogonin D (Fig. 6 G). To further confirm the effects of Ophiopogonin D on KLF3 activity, we treated HMECs with Ophiopogonin D for 48 h. ChIP-PCR assays indicated that Ophiopogonin D treatment affected the binding of KLF3 to the promoter of JUNB (Fig. 6, H and I). Western blotting also showed increased protein levels of JUNB and VEGFA in HMECs treated with Ophiopogonin D (Fig. 6, J and K). These data indicated that Ophiopogonin D acts as a KLF3 inhibitor and has a positive effect on the formation of the CD31hiEMCNhi endothelium.
The KLF3 inhibitor promotes CD31hiEMCNhi vessels and bone formation in aged mice and in an OVX model of osteoporosis
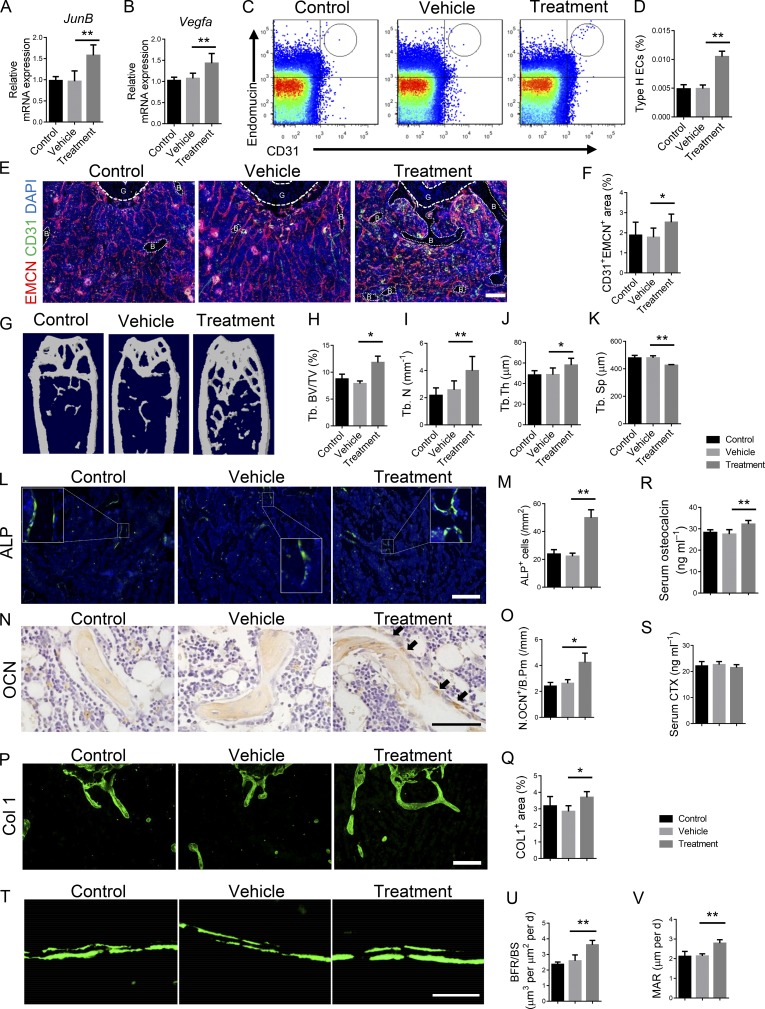
To investigate the therapeutic effects of Ophiopogonin D on osteoporosis, 2-mo-old C57/B6 mice after OVX surgery and 12-mo-old C57/B6 mice were treated intraperitoneally with Ophiopogonin D at 20 mg/kg every other day for 3 mo. Ophiopogonin D treatment increased the expression of JunB and Vegfa in ECs in the aged mice (Fig. 8, A and B), and increased the amount of CD31hiEMCNhi endothelium, the bone volume, the number of ALP+ osteoprogenitors, bone surface OCN+ osteoblasts, and bone marrow COL 1–positive area in the aged mice without disturbing the number of TRAP+ osteoclasts (Fig. 8, C–Q; and Fig. S5, A–C). Aged mice treated with Ophiopogonin D had increased circulating OCN levels compared with those in the controls; however, the serum CTX levels were comparable (Fig. 8, R and S). Calcein double labeling confirmed that trabecular BFRs and MARs were increased in mice treated with Ophiopogonin D (Fig. 8, T–V). Aggrecan staining indicated that Ophiopogonin D treatment did not affect the morphology of the growth plate (Fig. S5 D). Ophiopogonin D–treated OVX mice also showed increased numbers of CD31hiEMCNhi ECs, bone volume, serum OCN levels, numbers of osteoblasts and ALP+ osteoprogenitors on the bone surfaces, and the COL 1+ area in the bone marrow (Fig. 9, A–H, and J–O), without changing the number of TRAP+ osteoclasts or the serum CTX levels (Fig. 9, I, P, and Q).
Figure 8.
Ophiopogonin D treatment promotes CD31hiEMCNhi vessels and bone formation in aged mice. 12-mo-old C57/B6 mice were intraperitoneally treated with Ophiopogonin D at 20 mg/kg every other day for 3 mo. (A) qRT-PCR analysis of JunB level in ECs. (B) qRT-PCR analysis of Vegfa level in ECs. (C and D) FACS analysis dot plot (C) and quantification (D) of CD31hiEmcnhi ECs (Type H ECs). (E and F) Representative images (E) and quantification (F) of CD31 (green) and EMCN (red) immunostaining in femora from Ophiopogonin D–treated mice and the vehicle control group. G, growth plate. B, bone. Scale bar, 100 µm. (G–K) Representative μCT images (G) and quantitative μCT analysis (H–K) of trabecular bone microarchitecture in femora. (L and M) Representative images (L) and quantification (M) of ALP (green) immunostaining in femora. Scale bar, 200 µm. (N and O) Immunohistochemical staining (N) and quantification (O) of OCN+ cells (brown) in femora. Scale bar, 50 µm. (P and Q) Immunohistochemical staining (P) and quantification (Q) of COL 1 (green) in femora. Scale bar, 200 µm. (R and S) Serum levels of OCN (R) and CTX (S) at the time of harvest. (T–V) Representative images of calcein double labeling of trabecular bone (T) with quantification of BFR/BS (U) and MAR (V). Scale bar, 25 µm. n = 6 mice in each group from three independent experiments. Data are shown as the mean ± SD. *, P < 0.05; **, P < 0.01; ANOVA. Tb. BV/TV, trabecular bone volume per tissue volume; Tb. N, trabecular number; Tb. Sp, trabecular separation; Tb. Th, trabecular thickness.
Figure 9.
Ophiopogonin D treatment promotes CD31hiEMCNhi vessels and bone formation in OVX mice. 2-mo-old C57/B6 mice underwent OVX surgery and were intraperitoneally treated with Ophiopogonin D at 20 mg/kg every other day for 3 mo. (A and B) FACS analysis dot plot (A) and quantification (B) of CD31hiEMCNhi ECs (Type H ECs). (C–G) Representative μCT images (C) and quantitative μCT analysis (D–G) of trabecular bone microarchitecture in femora. (H and I) Serum levels of OCN (H) and CTX (I) at the time of harvest. (J and K) Representative images (J) and quantification (K) of ALP (green) immunostaining in femora. Scale bar, 200 µm. (L and M) Immunohistochemical staining (L) and quantification (M) of COL 1 (green) in femora. Scale bar, 200 µm. (N and O) Immunohistochemical staining (N) and quantification (O) of OCN+ cells (brown) in femora. Scale bar, 50 µm. (P) Representative images of TRAP staining of femora from Ophiopogonin D–treated mice and their controls. (Q) Quantification of TRAP+ cells in trabecular bone surfaces. Number of TRAP+ cells per bone perimeter (Tb.N.Trap+/B.Pm) was measured. Scale bar, 50 µm. (n = 6 mice in each group from three independent experiments. Data are shown as the mean ± SD. **, P < 0.01; ANOVA (B and D–G) and Student’s t test (H, I, K, M, O, and Q). Tb. BV/TV, trabecular bone volume per tissue volume; Tb. N, trabecular number; Tb. Sp, trabecular separation; Tb Th, trabecular thickness.
Taken together, these results suggested that inhibition of KLF3 in ECs by intraperitoneal administration of Ophiopogonin D promoted CD31hiEMCNhi vessel formation and stimulated new bone formation in both aged and OVX osteoporosis mouse models.
Discussion
Angiogenesis and osteogenesis show a close spatial–temporal association during skeletal development (Maes and Clemens, 2014; Jabalee and Franz-Odendaal, 2015; Huang et al., 2016) Specialized CD31hiEMCNhi bone capillaries identified in the skeletal system have a close relationship with perivascular osteoprogenitors and mediate their differentiation (Kusumbe et al., 2014). Xu et al. (2018) demonstrated that osteoblast-derived SLIT3 could increase the formation of the CD31hiEMCNhi endothelium. Our previous studies revealed that PDGF-BB secreted by preosteoclasts could induce the CD31hiEMCNhi vessel subtype, subsequently preventing bone loss in osteoporosis (Xie et al., 2014). The microRNA miR-497∼195 cluster contributes to the increase in CD31hiEMCNhi vessels and bone formation in aged mice (Yang et al., 2017). The decreased abundance of CD31hiEMCNhi vessels is an early marker of bone loss in humans (Wang et al., 2017). Therefore, it is a rational therapeutic strategy to target osteoporosis via the induction of CD31hiEMCNhi vessels. In the present study, we identified a specific point mutation in lncRNA Reg1cp that is involved in HBM pathogenesis. Mutant Reg1cp regulates the coupling of angiogenesis with osteogenesis via promoting CD31hiEMCNhi vessel formation in bone marrow. Furthermore, we provided a practical treatment option for age-related bone loss based on the mechanism of Mut-Reg1cp in regulating bone metabolism.
The major hormonal regulators of bone remodeling are hCT, PTH, 25-OHVD, and estrogen. Growth factors such as insulin-like growth factors, transforming growth factor-β (TGF-β), fibroblast growth factors, epidermal growth factor, WNTs, and bone morphogenetic proteins also play crucial roles in the regulation of bone remodeling (Khosla et al., 2012; Baron and Kneissel, 2013; Siddiqui and Partridge, 2016; Karner and Long, 2017; Lim et al., 2017). Our research began with the clinical screening of a patient with extremely high bone density. After excluding hormonal derangement and other conditions disrupting bone remodeling, we hypothesized the patient’s condition was genetic. The majority of established HBM mutations causing osteosclerosis involve Wnt signaling (Boyden et al., 2002; Balemans et al., 2007; Paccou et al., 2018). Wnt signaling regulated bone resorption as well as bone formation (Kramer et al., 2010; Baron and Kneissel, 2013). In our case, no mutation was found in genes related to Wnt signaling or other known bone remodeling regulating factors. However, whole-exome sequencing identified a homozygous mutation in lncRNA Reg1cp gene in this patient. A larger cohort analysis showed that Reg1cp+/mut subjects had higher bone density compared with that in Reg1cp+/+ subjects. This confirmed that the mutation on Reg1cp was related to the HBM phenotype.
Regenerating genes (Reg) belong to the calcium-dependent lectin gene superfamily (Miyashita et al., 1995; Zhang et al., 2003), and are involved in cell proliferation and differentiation (Kiji et al., 2003; Tohma et al., 2017). However, the function of Reg1cp, which is mainly expressed in pancreas as a lncRNA, has not been reported. In addition to the pancreas, we found that Reg1cp was also expressed in bone samples, especially in bone marrow ECs, suggesting local regulation by Mut-Reg1cp on bone formation. The identified mutation on Reg1cp did not affect its expression but led to a large change in its structure. Our results indicated that Mut-Reg1cp, but not WT-Reg1cp, takes part in bone formation regulation. The mutant Reg1cp does not compete with the WT form. HMECs with a Reg1cp heterozygous mutation also showed increased JUNB and VEGFA expression, and increased migration and tube formation abilities. This indicated that Reg1cp+/mut subjects also expressed the mutant form of Reg1cp, which resulted in increased bone mass.
Mutated Reg1cp binds directly to KLF3 in ECs to abolish its function. KLF3 is a member of Krüppel-like factor family of transcription factors, which consists three C-terminal C2H2 zinc fingers that recognize the CACCC boxes and GC-rich motif in the promoters and enhancers of target genes (Pearson et al., 2011). KLF3 is considered a potent transcriptional repressor that mediates transcriptional silencing via recruiting the corepressor CTBP (Pearson et al., 2011; Dewi et al., 2015). KLF3 is expressed in many tissues, and modulates diverse physiological processes, such as erythropoiesis, B lymphopoiesis, adipogenesis, myogenesis, and even cardiac development (Sue et al., 2008; Himeda et al., 2010; Kelsey et al., 2013). JunB, one of the KLF3 downstream genes, is a critical regulator of Vegfa transcription (Schmidt et al., 2007; Ilsley et al., 2017). In the present study, we demonstrated that KLF3 could inhibit the expression of JunB and Vegfa, thus repressing angiogenesis. The specific knockout of Klf3 in ECs increased the amount of CD31hiEMCNhi vessel formation and increased bone formation, without affecting body weight. Administration of a KLF3 inhibitor to osteoporosis mouse models produced similar results to knockout of Klf3. Thus, KLF3 might be a novel therapeutic target to treat osteoporosis.
Numerous factors, such as hormones, growth factors, and neurotransmitters, participate in bone metabolism regulation (Oury et al., 2010; He et al., 2017; Lee et al., 2018). It was possible that in addition to angiogenesis, the mutant Reg1cp might also affect other signaling pathways that regulate bone mass. In this project, we focused on the influence of Mut-Reg1cp on angiogenesis because Reg1cp is mainly expressed in ECs in bone marrow, and there was no sign of systemic hormone disarrangement in the patient with the homozygous Reg1cp mutation. Bone resorption was not affected in Klf3cdh5−/− mice. The osteogenic differentiation of BMSCs and the levels of osteoblast transcription factor SP7 and RUNX2 in BMSCs showed no difference after transfection with the Reg1cp-mut plasmid, indicating that Reg1cp has no direct influence on osteoblast differentiation. LRP5 gain-of-function mutation is the well-known cause of HBM; however, LRP5 acting as Wnt-coreceptor was challenged by the study of Yadav et al. (2008). They demonstrated Lrp5 controlling bone formation by inhibiting serotonin synthesis in the duodenum. Wnt signaling was not altered in Klf3cdh5−/− mice; neither was serotonin synthesis–related gene Tph1. We did not investigate whether Mut-Reg1cp is involved in other signaling pathways; however, this should be addressed in a future study.
With the progressive aging of the general population, the medical and socioeconomic burden of osteoporosis will increase further. One of the major requirements for novel treatments of bone loss is to develop anabolic agents that can stimulate bone formation. Ophiopogonin D is a natural compound isolated from the traditional Chinese herbal agent Radix Ophiopogon japonicus. It was reported to have anti-osteoporotic effects in a murine OVX model (Huang et al., 2015). In the present study, we identified that Ophiopogonin D functions as a mimic of mutant Reg1cp. Ophiopogonin D could bind to KLF3 and suppress its function, thus further promoting angiogenesis. Aged and OVX osteoporosis mice treated with Ophiopogonin D showed increased numbers of CD31hiEMCNhi vessels and new bone formation.
In summary, we revealed a specific mutation in the lncRNA Reg1cp that is involved in HBM pathogenesis by promoting angiogenesis during coupling with osteogenesis. We identified a natural compound that could act as a mimic of mutant Reg1cp, which increased the number of CD31hiEMCNhi vessels and promoted bone formation in aged and OVX osteoporosis mice. Our findings provide a potentially novel strategy to treat osteoporosis.
Materials and methods
Whole exome sequencing
Genomic DNA was collected and extracted from peripheral whole blood samples using a FlexiGene DNA kit (51206; Qiagen). Whole exome sequencing was conducted by Sangon Biotech by using the Aglient SureSelect Human All Exon V5+UTR and HiSeq X Ten Reagent Kit v2.5 for capture and Illumina sequencing platforms for data generation (paired end, 300 cycles). Sequencing was performed with 150-bp paired-end reads on the Illumina HiSeq XTen sequencing platform. The whole exome sequencing data were deposited on the Sequence Read Archive website with the accession no. PRJNA542422.
Bone density measurements
We measured BMD in the lumbar spine, femoral neck, and hip by dual-energy x-ray absorptiometry using a Hologic device (Hologic Discovery). The results are expressed as BMD and Z-scores (the number of SDs from the mean value for persons in the general population matched for age, sex, and race).
Massive sequencing and biochemistry
Human peripheral whole blood samples were obtained from 1,465 healthy subjects (616 males and 849 females) with ages ranging 20–80 yr. These volunteers were all residents of Changsha and surrounding areas. (The whole blood samples collection was conducted by the Physical Examination Center,Endocrinology Department of Xiangya Hospital of Central South University, Changsha, China and Department of Metabolic Endocrinology of the Second People’s Hospital of Xiangxiang, Xiangxiang, China.) All subjects were screened using a detailed questionnaire, disease history, and physical examination. Subjects were excluded from the study if they had conditions affecting bone metabolism, including diseases of the kidney, liver, parathyroid, or thyroid, hyperprolactinemia, oophorectomy, rheumatoid arthritis, ankylosing spondylitis, malabsorption syndromes, malignant tumors, hematologic diseases, menopause before age 40 yr, or previous pathological fractures within 1 yr. If the subjects had received treatment with glucocorticoids, estrogens, thyroid hormone, PTH, fluoride, bisphosphonate, calcitonin, thiazide diuretics, barbiturates and anti-seizure medication, they were also excluded.
Genomic DNA was collected and extracted from peripheral whole blood samples using a FlexiGene DNA kit (51206; Qiagen). The genomic DNA met the sequencing requirements of the purity (OD 260/280 > 1.8) and concentration (50 ng/ml) of each sample. Reg1cp gene was amplified in 1,465 subjects by using DNA polymerase (MIX [GREEN]), and the primers of forward: 5′-GGGAGCTTTGTGTGTAGAGAAACTG-3′; reverse: 5′-CTCCTCACTCACATCTGCATAAACC-3′. Forward and reverse sequencing reactions were performed with the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems), and the products were analyzed on an ABI 3730XL automated sequencer (PE Applied Biosystems). The clinical study was approved by the Ethics Committee of Xiangya Hospital of Central South University, and written informed consent was obtained from all participants before whole blood collection. Serum PINP and β-CTX were measured with the use of commercial kits (FC-007; Human PINP ELISA Kits, Phicon; FC-008; Human β-CTX ELISA Kits, Phicon).
Human samples
Human trabecular bone and bone marrow samples were obtained from 23 patients (13 male and 10 female) based on the inclusion and exclusion criteria. 20 patients with aseptic necrosis of femoral head or fracture undergoing hip arthroplasty replacement or open reduction internal fixation (ORIF), with ages ranging from 34 to 79 yr; one patient with ulna fracture, female, 27 yr old; and two patients with tibia fracture undergoing ORIF, female, 43 and 52 yr old respectively. We also performed Reg1cp sequencing of genomic DNA collected and extracted from bone marrow cells from those surgery patients. Among them, one male patient, 37 yr old, with femur shaft fracture undergoing ORIF was detected with Reg1cp heterozygous mutation (human trabecular bone and bone marrow collection were conducted by the Department of Orthopedics, Xiangya Hospital of Central South University, Changsha, China). All subjects were screened using a detailed questionnaire, disease history, and physical examination. Subjects were excluded from the study if they had conditions affecting bone metabolism, including diseases of the kidney, liver, parathyroid, or thyroid, hyperprolactinemia, oophorectomy, rheumatoid arthritis, ankylosing spondylitis, malabsorption syndromes, malignant tumors, hematologic diseases, or menopause before age 40 yr. If the subjects had received treatment with glucocorticoids, estrogens, thyroid hormone, PTH, fluoride, bisphosphonate, calcitonin, thiazide diuretics, barbiturates, or anti-seizure medication, they were also excluded. These participants had trabecular bone and bone marrow collection performed during bone fracture surgery or hip arthroplasty replacement. The clinical study was approved by the Ethics Committee of Xiangya Hospital of Central South University, and written informed consent was obtained from all participants before bone marrow collection.
Mice
To generate endothelium-specific Klf3 knockout mice, mice carrying loxP-flanked Klf3 alleles (Klf3flox/flox) and Cdh5-Cre transgenics were interbred. The Klf3flox/flox littermates were used as controls. The Cdh5-Cre transgenic mice (017968) were purchased from Jackson Laboratory; loxP-flanked Klf3 mice were purchased from Cyagen Biosciences. For endothelium-specific Klf3 knockout experiments, six male mice were used for each group at each observed time point (1, 3, and 12 mo) for each independent experiment. All mice we used were on a C57/B6 background. 2-mo-old female mice were used to perform OVX surgery and were sacrificed at 5 mo old. For Ophiopogonin D treatment experiments, 12-mo-old C57/B6 mice were intraperitoneally treated with Ophiopogonin D at a dosage of 20 mg/kg every other day for 3 mo. 2-mo-old C57/B6 mice after undergoing OVX surgery were intraperitoneally treated with Ophiopogonin D at dosage of 20 mg/kg 1 wk after surgery every other day for 3 mo. Six mice were used for each group for each independent experiment. Mice treated with vehicle treatment were used as controls. All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committees of the Laboratory Animal Research Center at Xiangya Medical School of Central South University. All mice were maintained in the specific pathogen–free facility of the Department of Laboratory Animals, Central South University.
Primary cell isolation and cell culture
For mouse bone marrow EC isolation, we collected bone marrow from tibiae and femurs of mice in sterile Ca2+- and Mg2+-free PBS. For human bone marrow EC isolation, we collected bone marrow from the patients with aseptic necrosis of femoral head or fracture undergoing hip arthroplasty replacement or ORIF. After the bone was crushed, the mixture was digested with type IA collagenase (Sigma-Aldrich) to obtain a single-cell suspension. Mice ECs were then sorted using EMCN antibody (SC-65495) and Dynabeads sheep anti-Rat IgG (Invitrogen). Human ECs were sorted using EMCN antibody (ab45772; Abcam). Sorted ECs were then plated on dishes coated with fibronectin and cultured in EC growth medium (EBM-2; Clonetics; Lonza) supplemented with EGM-2 Single Quots (CC-4176; Clonetics; Lonza). At first passage, cells were again magnetic cell separation sorted with EMCN antibody and plated for culture. Cells were fed every other day and passaged upon confluency. Cultures were maintained at 37°C with 5% CO2 in a humidified atmosphere.
For human endothelial progenitor cell isolation, peripheral whole blood samples were collected from Reg1cp+/mut and age-matched Reg1cp+/+ subjects recruited by the Endocrinology Department of Xiangya Hospital of Central South University, as described earlier in Human samples and Massive sequencing and biochemistry. Total mononuclear cells were isolated from blood by density gradient centrifugation with Histopaque-1077 (10771; Sigma-Aldrich). Mononuclear cells were plated in 0.5 ml EC growth medium (EBM-2; Clonetics; Lonza) supplemented with EGM-2 Single Quots (CC-4176; Clonetics; Lonza) on fibronectin/gelatin–coated 24-well plates. After 7 d of culture, the attached endothelia cells were collected for RNA extraction.
For human BMSCs isolation, bone marrow cells were collected and incubated with FITC-, APC-, and PE- conjugated antibodies that recognized human Stro-1 (340106; BioLegend), CD45 (304012; BioLegend), and CD146 (361008; BioLegend) at 4°C for 30 min. The acquisition was performed on a FACS Aria model (BD Biosciences), and the analysis was performed using FACS DIVE software version 6.1.3 (BD Biosciences). The sorted human CD146+Stro-1+CD45− BMSCs were cultured with α-MEM (Gibco-BRL) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
For human osteoblasts isolation, the bone samples were collected from patients with aseptic necrosis of femoral head or fracture undergoing hip arthroplasty replacement or ORIF and were cut into 1- to 2-mm lengths. Then we used warmed collagenase solution (4 mg/ml type IA collagenase in α-MEM) to incubate bone pieces three times at 25°C for 25 min. After that the bone samples were incubated with warmed collagenase solution (4 mg/ml type IA collagenase in α-MEM) and EDTA solution (5 mM EDTA solution PBS with 1% BSA) to incubate bone pieces at 25°C alternately; we aspirated the solution and retained it for cell plating. The isolated primary osteoblasts were cultured with α-MEM (Gibco-BRL) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
For preosteoclasts, we harvested monocytes and macrophages from bone marrow samples of patients with aseptic necrosis of femoral head or fracture undergoing hip arthroplasty replacement or ORIF. Bone marrow cells were cultured overnight on Petri dishes in α-MEM containing 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. After discarding the adherent cells, we incubated floating cells with M-CSF (30 ng/ml; R&D Systems) to obtain pure monocytes and macrophages. Upon incubation of monocytes and macrophages with 30 ng/ml M-CSF and 60 ng/ml receptor activator of nuclear factor-κ B ligand (PeproTech), all cells became preosteoclasts after a 3-d culture.
Osteogenic differentiation assay
Isolated BMSCs were cultured in 6-well plates at 2 × 106 cells/well with α-MEM (Gibco-BRL) supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 0.1 mM dexamethasone, 10 mM β-glycerol phosphate, and 50 mM ascorbate-2-phosphate for 21 d. Culture medium was changed every 3 d. Cells were collected for RNA extraction or stained with 2% Alizarin Red S (Sigma-Aldrich) at pH 4.2 to evaluate the cell matrix mineralization.
EC culture and functional assays
HMECs were cultured in MCDB131 medium (Gibco) containing 10% FBS (Gibco), 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were fed every other day and passaged upon confluency. Cells were maintained at 37°C with 5% CO2 in a humidified atmosphere. EC migration assay was set up in transwell 24-well plates with 8-μm pore filters. In brief, we seeded 105 cells per well in the upper chamber after 1 h serum starvation. After 12 h incubation, we removed the cells in the upper surface of each filter using cotton swabs. The cells that migrated into the lower surface were fixed with 4% paraformaldehyde for 30 min and then stained with crystal violet. We counted the cell number in four random microscope visual fields in each well (three wells for each condition). EC tube formation assay was conducted in 48-well plates precoated with Matrigel (BD Biosciences). In brief, we seeded 105 cells per well after 1 h serum starvation. After 12 h incubation at 37°C, we observed the tube formation of HMECs and quantified the number of tube branches by counting four random microscope visual fields in each well (three wells for each condition).
Construction of mutant Reg1cp HMEC cell line
The mutation Reg1cp HMEC cell line was constructed using a LentiCRISPRv2 system. At first, the editing efficiency of predicted small guide RNAs was detected in Hek293T cells. The one that had the highest efficiency was chosen to construct lentiviruses. The sequence of small guide RNA used in this study is 5′-GGCCCATGCTGAGTTGCCCA-3′. Then, the HMEC cells were infected with lentiviruses. 2 d after infection, 5 μg/ml puromycin was added to the culture medium to kill off cells not transduced by virus until all cells in the no virus infection control group died out. Afterwards, the lentivirus-infected HMEC cells were transfected with Mut-Reg1cp expressing plasmids or empty plasmids as the editing template. The single subclone of transfected HMECs was selected and validated for editing efficiency. The genomic DNA was isolated with a FlexiGene DNA kit (51206; Qiagen) according to the protocol of the manufacturer. The genomic DNA was analyzed by further Sanger sequencing. The single subclones with heterozygous or homozygous mutation on the identified site of Reg1cp were used for further experiments.
RNA immunoprecipitation and RNA pulldown
RNA immunoprecipitation was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (17–700; Millipore) according to the manufacturer’s instructions. RNA for in vitro experiments was transcribed using TranscriptAid T7 High Yield Transcription Kit (K0441; Thermo Fisher Scientific) according to the manufacturer’s instructions. 5 pmol of 30-biotinylated RNA was used in each pulldown assay. The RNA pulldown experiment was performed using a Ribocluster profiler RiboTrap Kit (RN1011/RN1012; MBL) according to the manufacturer’s instructions.
Luciferase reporter assay
For functional analysis of klf3, the segments of the human JUNB promoter, including the predicted klf3 binding site, were PCR-amplified. The PCR products were purified and inserted into the XbaI-FseI site immediately downstream of the stop codon in the pGL3 control luciferase reporter vector (Promega), resulting in human WT-pGL3-JunB. The JUNB mutants for the klf3 seed regions were prepared using the Quik Change Site-Directed Mutagenesis Kit (Stratagene) to get human MUT-pGL3-JunB. Human HMECs were transfected with either WT or mutant pGL3 construct, the pRL-TK renilla luciferase plasmid (Promega), and klf3 plasmid or vector for 48 h using Lipofectamine 2000 (Invitrogen). The dual luciferase reporter assay system (Promega) was used to quantify luminescent signal using a luminometer (Glomax; Promega).
RNA extraction and qRT-PCR
Total RNA from cells was extracted using Trizol reagent (Invitrogen). RT was performed using 1 µg total RNA by using the PrimeScript RT reagent Kit (Takara). Amplification reactions were set up in 25-µl reaction volumes containing SYBR Green PCR Master Mix (PE Applied Biosystems), 1 µl volume of cDNA, and amplification primers. Relative quantification was calculated by normalizing the test crossing thresholds (Ct) with the β-actin amplified control Ct. The results were normalized to β-actin. Primer sequences are listed in Table S7.
ChIP assay
ChIP assay was performed using SimpleChip Kit (9003; Magnetic Beads; Cell Signaling Technology) according to manufacturer’s instructions. Briefly, chromatin was cross-linked with 1% formaldehyde and sheared to 100- to 500-bp fragments by sonication (Sonics VCX130; 30% amplitude, 15 s on and 45 s off for four cycles). Klf3 antibody (1:100; PA5-18030; Invitrogen Antibodies) and normal rabbit IgG (2729; belongs to SimpleChip Kit) were used to immunoprecipitated the relevant protein–DNA complex. The cross-linked DNA and protein were reversed by addition of 5 M NaCl; DNA was extracted by phenol/chloroform and precipitated with ethanol and glycogen. Purified DNA was used to perform normal PCR or was quantified using real-time PCR. The primer sequences are listed in Table S8.
EMSAs
The EMSA was operated using a LightShift chemiluminescent EMSA kit (GS009; Beyotime Biotechnology) according to the manufacturer’s instructions. 3 μl nuclear extract was used in each binding reaction in a total volume of 20 µl. Unlabeled WT and mutant probes were added to the binding reaction as competition experiments. 2 µg of anti-Klf3 antibodies (PA5-18030; Invitrogen Antibodies) was added to the binding reaction as supershift experiments. 20 min later, biotin-labeled WT probe was added. The probes used for EMSA are listed in Table S9.
Co-immunoprecipitation assays
HMECs were transfected with Reg1cp-mut plasmid, His-Klf3, Myc-CTBP1, and HA-CTBP2. Following transfection, cells were lysed on ice in 1 ml of cell lysis buffer for 30 min. Supernatants were collected by centrifugation; two 50-µl aliquots were kept for input, whereas the remaining whole cell extracts were used for immunoprecipitation. Extracts were incubated with 5 µg of antibody (ab9110; anti-HA tag antibody; Abcam; ab9106; anti-Myc tag antibody; Abcam; ab9108; anti-His tag antibody; Abcam) overnight at 4°C, and the products were collected on Dynabeads Protein G.
Western blot
Total cell lysates were separated by SDS-PAGE and blotted on polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% milk (170–6404; Bio-Rad) and incubated with specific antibodies to klf3 (1:1,000; PA5-18030; Invitrogen Antibodies), JunB (1:1,000; 3753; Cell Signaling Techology), VEGFa (1:1,000; 19003–1-AP; Proteintech), α-Tubulin (1:2,000; 11224–1-AP; Proteintech), VEGFR2 (1:1,000; 2479; Cell Signaling Techology), and pVEGFR2 (1:1,000; 2478; Cell Signaling Techology), then reprobed with appropriate HRP-conjugated secondary antibodies. Blots were visualized using SuperSignal West Pico PLUS Chemiluminescent Substrate (SD251210; Thermo Fisher Scientific). Phosphorylation of VEGFR2 was performed as previously described (Lee et al., 2007). Phosphorylation of VEGFR2 under hypoxia was measured after exposure to CoCl2 (100 mM) for 24 h.
ELISA analysis
We performed OCN or CTX ELISA analysis of serum using a mouse OCN Enzyme Immunoassay kit (Biomedical Technologies Inc.) or a RatLaps Enzyme Immunoassay kit (Immunodiagnostic Systems). We did all ELISA assays according to the manufacturers’ instructions.
Molecular docking
The first run of virtual screening was performed using Autodock Vina. First, receptor structure was processed in MGLTools 1.5.6 by addition of hydrogen atoms and Gasteiger charges. Openbabel was then employed to convert compounds into PDBQT files. The box centers of site1∼3 were set (15.34, −0.81, 11.06; −0.56, 2.93, −6.55; and −5.65, −9.39, −23.06, respectively). The box sizes in all dimensions were set at 30 Å. Only one pose was output for each compound. The screening results were filtered based on the Affinity threshold of −7.0 kcal/mol. The candidate compounds were further docked using UCSF Dock 6.7. The receptor was added hydrogens and assigned an AMBER ff14SB force field by UCSF Chimera. Compounds were added hydrogens and MMFF94 charges by Openbabel, and energy minimized before saving to MOL2 files. The box centers for the three sites were set (14.652, −0.931, 11.141; −1.089, 3.818, −7.619; and −5.209, −11.407, −22.547, respectively). The box sizes were set (27.792, 31.437, 27.393; 28.947, 29.056, 29.650; and 27.107, 26.406, 28.328, respectively). Semi-flexible dockings were performed in Dock 6.7, and 10,000 different orientations were produced. A root mean square deviation threshold of 2.0 Å was applied in clustering for docking conformers, and one pose was output for each compound. Grid score = −50 kcal/mol was used as the threshold in filtration. Results for all sites were combined, and duplicates were removed. Finally, clustering analysis was performed to obtain a small number of diverse compounds.
HPLC-MS assays
To demonstrate the direct binding of Ophiopogonin D to KLF3, 2 µg of Ophiopogonin D was incubated with 10 µg of human His-tag KLF3 protein (NBP2-23175; Novus Biologicals) in 150 µl of a buffer (pH 7.5) consisting of 100 mM Tris, 10% glycerol, 50 mM KCl, and 1 mM EDTA at room temperature for 2 h. Ni-NTA Agarose (30210; Qiagen) was used to separate the protein–ligand complexes, and the ligands were dissociated from KLF3 protein using 400 µl of methanol. Finally, we analyzed the recruited ligands by HPLC-MS assays.
Flow cytometry
For the analysis or sorting of mice CD31hiEMCNhi cells, femora and tibia were dissected from mice after euthanization. Then the epiphysis, muscles, and periosteum were removed. The metaphysis and diaphysis regions of the bone were crushed in ice-cold PBS to get the bone marrow. For the analysis of human CD31hiEMCNhi cells in bone, human trabecular bone and bone marrow samples were obtained from people undergoing bone fracture surgery or hip arthroplasty replacement. Bone pieces and whole bone marrow samples were digested using 1 mg/ml type IA collagenase at 37°C for 20 min to obtain single-cell suspensions. For mice samples, after filtration and washing, the cells were counted and incubated for 45 min at 4°C with EMCN antibody (1:100; SC-65495; Santa Cruz Biotechnology), then washed and further incubated with APC-conjugated CD31 antibody (1:100; FAB3628A; R&D Systems) for 45 min at 4°C. For human samples, the cells were incubated with EMCN antibody (1:100; ab45772; Abcam), then washed and further incubated with CD31 antibody (1:100; 561654; BD Biosciences). We performed acquisition on a FACScan cytometer (BD Immunocytometry Systems). For demarcating and sorting CD31hiEMCNhi cells, first standard quadrant gates were set. Subsequently, CD31hiEMCNhi cells were distinguished from the total double-positive cells in quadrant 2, and the gates of quadrant 2 were set at >103 log Fl-4 (CD31-APC) fluorescence and >103 log Fl-2 (EMCN-PE) fluorescence.
µCT analysis
Femora were dissected from mice, carefully removing the attached muscle, and fixed overnight with 10% formalin at 4°C. The µCT analyses were performed using high-resolution μCT (Skyscan 1172; Bruker microCT). The scanner was set at a voltage of 65 kV, a current of 153 μA, and a resolution of 15 µm per pixel. The image reconstruction software (NRecon, version 1.6; Bioz), data analysis software (CT Analyser, version 1.9; Bruker microCT) and three-dimensional model visualization software (μCT Volume, version 2.0; Bruker microCT) were used to analyze the parameters of the distal femoral metaphyseal trabecular bone. The region of interest for analysis was 5% of femoral length below the growth plate. Trabecular bone volume per tissue volume, trabecular number, trabecular separation, and trabecular thickness were measured.
Histochemistry and histomorphometry
Femora were dissected from mice, carefully removing the attached muscle, and fixed overnight with 10% formalin at 4°C. After washing three times with ice-cold PBS, the samples were decalcified at 4°C using 10% EDTA (pH 7.4) for 21 d and then embedded in paraffin. 4-μm-thick sagittal-oriented sections of the knee joint medial compartment were used for staining. The slides were processed for TRAP staining, which was performed using a standard protocol (Sigma-Aldrich). Histomorphometric analysis of two-dimensional parameters of the trabecular bones was performed using OsteoMeasureXP Software (OsteoMetrics). Osteoclast surface per bone surface and osteoclast number per bone perimeter parameters were used to measure the bone resorption.
Immunocytochemistry and histomorphometry
Femora were dissected from mice, carefully removing the attached muscle, and fixed overnight with 10% formalin at 4°C. After washing three times with ice-cold PBS, the samples were decalcified at 4°C using 10% EDTA (pH 7.4) for 21 d and then embedded in paraffin. 4-μm-thick longitudinally oriented bone sections of femora samples were used for staining. The sections were stained with individual primary antibodies to OCN (1:100; M137; Takara Bio) at 4°C overnight. We used the HRP-streptavidin detection system (Dako) to detect immunoactivity. Then we counterstained the sections with hematoxylin (Sigma-Aldrich). Histomorphometric analysis of two-dimensional parameters of the trabecular bones was performed using OsteoMeasureXP Software (OsteoMetrics). Osteoblast number per bone perimeter parameters were used to measure the bone formation.
Immunofluorescence and histomorphometry
For immunofluorescence staining, freshly dissected femora collected from transgenic mice and their control littermates were first fixed in ice-cold 4% paraformaldehyde solution for 4 h, then decalcified in 0.5 M EDTA (pH 7.4) at 4°C for 24 h (1- and 3-mo-old mice) or for 48 h (12- and 15-mo-old mice). The bone samples were then incubated in 20% sucrose and 2% polyvinylpyrrolidone solution overnight, as described previously (Kusumbe et al., 2015). For type H vessel staining, we embedded the tissues in 8% gelatin (porcine) in the presence of 20% sucrose and 2% polyvinylpyrrolidone. 40-μm-thick longitudinally oriented bone sections were stained with primary antibodies to mouse CD31 (1:50; ab28364; Abcam) and EMCN (1:50; V.7C7; Santa Cruz Biotechnology) overnight at 4°C. For other immunofluorescence staining, we embedded the tissues in optimal cutting temperature compound. 4-μm-thick longitudinally oriented bone sections were stained with individual primary antibodies to mouse COL 1 (1:50; AB765P; Millipore), ALP (1:200; ab108337; Abcam), Aggrecan (1:50; AB1031; Millipore), Sox9 (1:200; ab185230; Abcam), and Klf3 (1:100; ab49221; Abcam) overnight at 4°C. Subsequently, we used secondary antibodies conjugated with fluorescence at room temperature for 1 h while avoiding light. We used isotype-matched controls, such as polyclonal rabbit IgG (AB-105-C; R&D Systems), polyclonal goat IgG (AB-108-C; R&D Systems), and monoclonal rat IgG2A (54447; R&D Systems) under the same concentrations and conditions as negative controls. We counted the numbers of positively stained cells in four random visual fields in the distal metaphysis of the femur in five sequential sections per mouse in each group.
Calcein double-labeling
To examine dynamic bone formation, we injected mice intraperitoneally with 0.08% calcein (20 mg/kg, body weight; Sigma-Aldrich) 8 d and 2 d before euthanasia. Calcein double-labeling in undecalcified bone slices was observed under a fluorescence microscope. Four randomly selected visual fields in the distal metaphysis of the femur were measured to test trabecular bone formation in femora.
Statistical analyses
The data are presented as mean ± SD. For comparisons of two groups, the two-tailed Student’s t test was used. For comparisons of multiple groups, one-way ANOVA was used. Differences were considered significant at P < 0.05. No randomization or blinding was used, and no animals were excluded from analysis. Sample sizes were selected on the basis of previous experiments.
Study approval
The clinical study was approved by the Ethics Committee of Xiangya Hospital of Central South University, and written informed consent was obtained from all participants. All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committees of the Laboratory Animal Research Center at Xiangya Medical School of Central South University.
Online supplemental material
Fig. S1 shows the family trees of affected subjects. Fig. S2 shows the secondary structure prediction of Reg1CP and mutated Reg1cp. Fig. S3 shows that mutant Reg1cp does not retrieve KLF8 and KLF12 and does not affect the recruitment of corepressors CTBP1 and CTBP2 to Klf3. Fig. S4 demostrates that endothelial-specific Klf3 knockout mice show no change in osteoclast number, Wnt signaling, Tph1 expression, and growth plate morphology. Fig. S5 shows that Ophiopogonin D treatment does not affect osteoclast number and growth plate morphology in aged mice. Table S1 shows clinical information of the 67 Reg1cp+/mut individuals. Table S2 shows the index of bone mass in 26 male Reg1cp+/mut individuals and 590 male Reg1cp+/+ individuals. Table S3 shows the index of bone mass in 41 female Reg1cp+/mut individuals and 808 female Reg1cp+/+ individuals. Table S4 shows a biochemical survey for bone remodeling parameters in 26 male Reg1cp+/mut individuals and 226 male Reg1cp+/+. Table S5 shows a biochemical survey for bone remodeling parameters in 41 female Reg1cp+/mut individuals and 213 female Reg1cp+/+ individuals. Table S6 shows the top 21 selected small molecules. Table S7 shows nucleotide sequences of primers used for quantitative RT-PCR detection. Table S8 shows nucleotide sequences of primers used for ChIP-PCR. Table S9 shows information from the biotin-labeled probe used for EMSA.
Supplementary Material
Acknowledgments
We acknowledge Elixigen for help editing and proofreading the English in the final manuscript. We acknowledge the Department of Orthopedics, Xiangya Hospital of Central South University for help collecting human trabecular bone and bone marrow samples.
This work was supported by the Major International (Regional) Joint Research Project of the National Natural Science Foundation of China (81520108008), and the National Natural Science Foundation of China (91749105 and 81700785).
The authors declare no competing financial interests.
Author contributions: X-H. Luo designed the experiments; M. Yang, H. Peng, and Q. Guo carried out most of the experiments; Y. Xiao, Y-Z. Xiao, Y. Huang, C-J. Li, Y-L. Zhang, T. Su, M-X. Lei, H-L. Chen, and T-J. Jiang helped to collect the samples; X-H. Luo, M.Yang, and Q. Guo proofread the manuscript; X-H. Luo supervised the experiments, analyzed results, and wrote the manuscript.
References
- Balemans W., Devogelaer J.P., Cleiren E., Piters E., Caussin E., and Van Hul W.. 2007. Novel LRP5 missense mutation in a patient with a high bone mass phenotype results in decreased DKK1-mediated inhibition of Wnt signaling. J. Bone Miner. Res. 22:708–716. 10.1359/jbmr.070211 [DOI] [PubMed] [Google Scholar]
- Baron R., and Kneissel M.. 2013. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 19:179–192. 10.1038/nm.3074 [DOI] [PubMed] [Google Scholar]
- Bonewald L. 2011. The holy grail of high bone mass. Nat. Med. 17:657–658. 10.1038/nm0611-657 [DOI] [PubMed] [Google Scholar]
- Boyden L.M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M.A., Wu D., Insogna K., and Lifton R.P.. 2002. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 346:1513–1521. 10.1056/NEJMoa013444 [DOI] [PubMed] [Google Scholar]
- Croucher P.I., McDonald M.M., and Martin T.J.. 2016. Bone metastasis: the importance of the neighbourhood. Nat. Rev. Cancer. 16:373–386. 10.1038/nrc.2016.44 [DOI] [PubMed] [Google Scholar]
- Dewi V., Kwok A., Lee S., Lee M.M., Tan Y.M., Nicholas H.R., Isono K., Wienert B., Mak K.S., Knights A.J., et al. 2015. Phosphorylation of Krüppel-like factor 3 (KLF3/BKLF) and C-terminal binding protein 2 (CtBP2) by homeodomain-interacting protein kinase 2 (HIPK2) modulates KLF3 DNA binding and activity. J. Biol. Chem. 290:8591–8605. 10.1074/jbc.M115.638338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib B., Fox M.F., Bartoli C., Giorgi D., Sansonetti A., Swallow D.M., Dagorn J.C., and Berge-lefranc J.L.. 1993. Human regeneration protein/lithostathine genes map to chromosome 2p12. Ann. Hum. Genet. 57:9–16. 10.1111/j.1469-1809.1993.tb00882.x [DOI] [PubMed] [Google Scholar]
- He G., Shi Y., Lim J., Bellido T., Ni J., and Long F.. 2017. Differential involvement of Wnt signaling in Bmp regulation of cancellous versus periosteal bone growth. Bone Res. 5:17016 10.1038/boneres.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda C.L., Ranish J.A., Pearson R.C., Crossley M., and Hauschka S.D.. 2010. KLF3 regulates muscle-specific gene expression and synergizes with serum response factor on KLF binding sites. Mol. Cell. Biol. 30:3430–3443. 10.1128/MCB.00302-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Wang W., Li Q., Wang Z., Yan B., Zhang Z., Wang L., Huang M., Jia C., Lu J., et al. 2016. Osteoblasts secrete Cxcl9 to regulate angiogenesis in bone. Nat. Commun. 7:13885 10.1038/ncomms13885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Gao B., Wang L., Zhang H.Y., Li X.J., Shi J., Wang Z., Zhang J.K., Yang L., Luo Z.J., and Liu J.. 2015. Ophiopogonin D: A new herbal agent against osteoporosis. Bone. 74:18–28. 10.1016/j.bone.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Ilsley M.D., Gillinder K.R., Magor G.W., Huang S., Bailey T.L., Crossley M., and Perkins A.C.. 2017. Krüppel-like factors compete for promoters and enhancers to fine-tune transcription. Nucleic Acids Res. 45:6572–6588. 10.1093/nar/gkx441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabalee J., and Franz-Odendaal T.A.. 2015. Vascular endothelial growth factor signaling affects both angiogenesis and osteogenesis during the development of scleral ossicles. Dev. Biol. 406:52–62. 10.1016/j.ydbio.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Jaleel A., Saag K.G., and Danila M.I.. 2018. Improving drug adherence in osteoporosis: an update on more recent studies. Ther. Adv. Musculoskelet. Dis. 10:141–149. 10.1177/1759720X18785539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.L., Gong G., Kimberling W., Reckér S.M., Kimmel D.B., and Recker R.B.. 1997. Linkage of a gene causing high bone mass to human chromosome 11 (11q12-13). Am. J. Hum. Genet. 60:1326–1332. 10.1086/515470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner C.M., and Long F.. 2017. Wnt signaling and cellular metabolism in osteoblasts. Cell. Mol. Life Sci. 74:1649–1657. 10.1007/s00018-016-2425-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey L., Flenniken A.M., Qu D., Funnell A.P., Pearson R., Zhou Y.Q., Voronina I., Berberovic Z., Wood G., Newbigging S., et al. 2013. ENU-induced mutation in the DNA-binding domain of KLF3 reveals important roles for KLF3 in cardiovascular development and function in mice. PLoS Genet. 9:e1003612 10.1371/journal.pgen.1003612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S., Oursler M.J., and Monroe D.G.. 2012. Estrogen and the skeleton. Trends Endocrinol. Metab. 23:576–581. 10.1016/j.tem.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiji T., Dohi Y., Nishizaki K., Takasawa S., Okamoto H., Nagasaka S., Naito H., Yonemasu K., and Taniguchi S.. 2003. Enhancement of cell viability in cryopreserved rat vascular grafts by administration of regenerating gene (REG) inducers. J. Vasc. Res. 40:132–139. 10.1159/000070710 [DOI] [PubMed] [Google Scholar]
- Kramer I., Halleux C., Keller H., Pegurri M., Gooi J.H., Weber P.B., Feng J.Q., Bonewald L.F., and Kneissel M.. 2010. Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol. Cell. Biol. 30:3071–3085. 10.1128/MCB.01428-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., and Adams R.H.. 2014. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 507:323–328. 10.1038/nature13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Starsichova A., and Adams R.H.. 2015. Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue. Nat. Protoc. 10:1904–1914. 10.1038/nprot.2015.125 [DOI] [PubMed] [Google Scholar]
- Lee S., Chen T.T., Barber C.L., Jordan M.C., Murdock J., Desai S., Ferrara N., Nagy A., Roos K.P., and Iruela-Arispe M.L.. 2007. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 130:691–703. 10.1016/j.cell.2007.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Abel E.D., and Long F.. 2018. Glucose metabolism induced by Bmp signaling is essential for murine skeletal development. Nat. Commun. 9:4831 10.1038/s41467-018-07316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupin O., Piters E., Halleux C., Hu S., Kramer I., Morvan F., Bouwmeester T., Schirle M., Bueno-Lozano M., Fuentes F.J., et al. 2011. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J. Biol. Chem. 286:19489–19500. 10.1074/jbc.M110.190330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Burclaff J., He G., Mills J.C., and Long F.. 2017. Unintended targeting of Dmp1-Cre reveals a critical role for Bmpr1a signaling in the gastrointestinal mesenchyme of adult mice. Bone Res. 5:16049 10.1038/boneres.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. 2011. Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13:27–38. 10.1038/nrm3254 [DOI] [PubMed] [Google Scholar]
- Long F., and Ornitz D.M.. 2013. Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5:a008334 10.1101/cshperspect.a008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C., and Clemens T.L.. 2014. Angiogenic-osteogenic coupling: the endothelial perspective. Bonekey Rep. 3:578 10.1038/bonekey.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita H., Nakagawara K., Mori M., Narushima Y., Noguchi N., Moriizumi S., Takasawa S., Yonekura H., Takeuchi T., and Okamoto H.. 1995. Human REG family genes are tandemly ordered in a 95-kilobase region of chromosome 2p12. FEBS Lett. 377:429–433. 10.1016/0014-5793(95)01381-4 [DOI] [PubMed] [Google Scholar]
- Niedźwiedzki T., and Filipowska J.. 2015. Bone remodeling in the context of cellular and systemic regulation: the role of osteocytes and the nervous system. J. Mol. Endocrinol. 55:R23–R36. 10.1530/JME-15-0067 [DOI] [PubMed] [Google Scholar]
- Oury F., Yadav V.K., Wang Y., Zhou B., Liu X.S., Guo X.E., Tecott L.H., Schutz G., Means A.R., and Karsenty G.. 2010. CREB mediates brain serotonin regulation of bone mass through its expression in ventromedial hypothalamic neurons. Genes Dev. 24:2330–2342. 10.1101/gad.1977210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccou J., Michou L., Kolta S., Debiais F., Cortet B., and Guggenbuhl P.. 2018. High bone mass in adults. Joint Bone Spine. 85:693–699. 10.1016/j.jbspin.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Pearson R.C., Funnell A.P., and Crossley M.. 2011. The mammalian zinc finger transcription factor Krüppel-like factor 3 (KLF3/BKLF). IUBMB Life. 63:86–93. [DOI] [PubMed] [Google Scholar]
- Rachner T.D., Khosla S., and Hofbauer L.C.. 2011. Osteoporosis: now and the future. Lancet. 377:1276–1287. 10.1016/S0140-6736(10)62349-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S., Butler J.M., and Ding B.S.. 2016. Angiocrine functions of organ-specific endothelial cells. Nature. 529:316–325. 10.1038/nature17040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S.K., Kusumbe A.P., Wang L., and Adams R.H.. 2014. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 507:376–380. 10.1038/nature13146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S.K., Kusumbe A.P., and Adams R.H.. 2015. Regulation of tissue morphogenesis by endothelial cell-derived signals. Trends Cell Biol. 25:148–157. 10.1016/j.tcb.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S.K., Kusumbe A.P., Itkin T., Gur-Cohen S., Lapidot T., and Adams R.H.. 2016. Regulation of Hematopoiesis and Osteogenesis by Blood Vessel-Derived Signals. Annu. Rev. Cell Dev. Biol. 32:649–675. 10.1146/annurev-cellbio-111315-124936 [DOI] [PubMed] [Google Scholar]
- Riddle R.C., and Clemens T.L.. 2017. Bone Cell Bioenergetics and Skeletal Energy Homeostasis. Physiol. Rev. 97:667–698. 10.1152/physrev.00022.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Textor B., Pein O.T., Licht A.H., Andrecht S., Sator-Schmitt M., Fusenig N.E., Angel P., and Schorpp-Kistner M.. 2007. Critical role for NF-kappaB-induced JunB in VEGF regulation and tumor angiogenesis. EMBO J. 26:710–719. 10.1038/sj.emboj.7601539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui J.A., and Partridge N.C.. 2016. Physiological Bone Remodeling: Systemic Regulation and Growth Factor Involvement. Physiology (Bethesda). 31:233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue N., Jack B.H., Eaton S.A., Pearson R.C., Funnell A.P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J.C., et al. 2008. Targeted disruption of the basic Krüppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28:3967–3978. 10.1128/MCB.01942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohma Y., Dohi Y., Shobatake R., Uchiyama T., Takeda M., Takasawa S., Tanaka Y., and Ohgushi H.. 2017. Reg Gene Expression in Periosteum after Fracture and Its In Vitro Induction Triggered by IL-6. Int. J. Mol. Sci. 18:2257 10.3390/ijms18112257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou F., Zhang P., Wang H., Qu Z., Jia P., Yao Z., Shen G., Li G., Zhao G., et al. 2017. Human type H vessels are a sensitive biomarker of bone mass. Cell Death Dis. 8:e2760 10.1038/cddis.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Cui Z., Wang L., Xia Z., Hu Y., Xian L., Li C., Xie L., Crane J., Wan M., et al. 2014. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat. Med. 20:1270–1278. 10.1038/nm.3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Yallowitz A., Qin A., Wu Z., Shin D.Y., Kim J.M., Debnath S., Ji G., Bostrom M.P., Yang X., et al. 2018. Targeting skeletal endothelium to ameliorate bone loss. Nat. Med. 24:823–833. 10.1038/s41591-018-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V.K., Ryu J.H., Suda N., Tanaka K.F., Gingrich J.A., Schütz G., Glorieux F.H., Chiang C.Y., Zajac J.D., Insogna K.L., et al. 2008. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 135:825–837. 10.1016/j.cell.2008.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li C.J., Sun X., Guo Q., Xiao Y., Su T., Tu M.L., Peng H., Lu Q., Liu Q., et al. 2017. MiR-497∼195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1α activity. Nat. Commun. 8:16003 10.1038/ncomms16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.W., Ding L.S., and Lai M.D.. 2003. Reg gene family and human diseases. World J. Gastroenterol. 9:2635–2641. 10.3748/wjg.v9.i12.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.