The endothelial ERK1/2 pathway plays a crucial role in the maintenance of endothelial homeostasis. Its suppression results in activation of TGFβ signaling, leading to hypertension, renal failure, fibrosis, and sudden death, findings similar to those observed in patients treated with anti-VEGF agents.
Abstract
To define the role of ERK1/2 signaling in the quiescent endothelium, we induced endothelial Erk2 knockout in adult Erk1−/− mice. This resulted in a rapid onset of hypertension, a decrease in eNOS expression, and an increase in endothelin-1 plasma levels, with all mice dying within 5 wk. Immunostaining and endothelial fate mapping showed a robust increase in TGFβ signaling leading to widespread endothelial-to-mesenchymal transition (EndMT). Fibrosis affecting the cardiac conduction system was responsible for the universal lethality in these mice. Other findings included renal endotheliosis, loss of fenestrated endothelia in endocrine organs, and hemorrhages. An ensemble computational intelligence strategy, comprising deep learning and probabilistic programing of RNA-seq data, causally linked the loss of ERK1/2 in HUVECs in vitro to activation of TGFβ signaling, EndMT, suppression of eNOS, and induction of endothelin-1 expression. All in silico predictions were verified in vitro and in vivo. In summary, these data establish the key role played by ERK1/2 signaling in the maintenance of vascular normalcy.
Introduction
Endothelial cells are key players in a number of biological processes, including angiogenesis, control of permeability and vascular integrity, and regulation of blood flow, among many others (Aird, 2007). They receive numerous signals ranging from mechanical to cell–cell contact to paracrine and endocrine inputs. Despite the diverse nature of these signals, most trigger activation of MAPK pathways. The MAPK family of cytoplasmic serine/threonine kinases includes p38, JNK, ERK5, and ERK1-ERK2 (Cargnello and Roux, 2011). While the MAPK family was one of the first signaling pathways discovered, surprisingly little is known about MAPK signaling in endothelial biology. In particular, there is paucity of information regarding the role of ERK signaling in the quiescent endothelium.
Vascular endothelial growth factor A (VEGFA) is the prototypical growth factor acting on the endothelium. Its activation of VEGF receptor 2 (VEGFR2) leads to a robust activation of the ERK1/2 pathway, while inhibition of VEGFR2 signaling results in almost complete suppression of ERK activation and early embryonic mortality (Simons et al., 2016). ERK1 and ERK2 isoforms share >80% of homology at the amino acid sequence level; both are expressed in endothelial cells. Mice with a global deletion of Erk1 are viable, fertile, and have no known vascular phenotype (Pagès et al., 1999). A global deletion of Erk2 results in embryonic lethality with a variety of developmental defects (Hatano et al., 2003). An endothelial-specific deletion of Erk2 on an Erk1−/− background is also embryonic lethal due to impaired endothelial proliferation and migration leading to defective angiogenesis (Srinivasan et al., 2009). Postnatal effects of endothelial Erk2 deletion have never been studied, especially in combination with the Erk1 knockout.
While in embryonic and early postnatal life growth factors induce differentiation, migration, and proliferation of endothelial cells in order to form functional hierarchical vascular networks, in healthy adult vasculature, endothelium remains quiescent. Yet it continues to play important physiological roles ranging from regulation of permeability to maintenance of vascular integrity to generation of paracrine and endocrine signals that control blood flow and blood pressure. In vivo studies in mice have demonstrated that a disruption of certain growth factors inputs, including those of VEGF and fibroblast growth factors (FGFs), results in reduction of ERK signaling and the loss of vascular integrity and vessel involution (Lee et al., 2007; Murakami et al., 2008). At the same time, clinical uses of drugs targeting VEGF, such as bevacizumab, or the downstream signaling pathway, such as the mitogen-activated kinase (MEK) inhibitor trametinib, are associated with extensive and poorly understood cardiovascular side effects, including hemorrhage and hypertension (Porta et al., 2015; Banks et al., 2017; Rubin, 2017). These observations notwithstanding, the role of ERK signaling and specific ERK isoforms in these events is unknown.
To address these issues, we induced endothelial-specific knockout of Erk2 in adult Erk1−/− mice. This resulted in universal lethality (sudden death) 4–5 wk later. Serial studies demonstrated rising blood pressure, progressive myocardial fibrosis, renal failure, and pulmonary hemorrhage. Molecular studies showed the presence of widespread endothelial-to-mesenchymal transition (EndMT) and activation of the endothelial TGFβ pathway. A causal computational intelligence strategy deployed to analyze RNA sequencing (RNA-seq) data from HUVECs treated with siRNA targeting ERK1 and ERK2 confirmed ERK1/2 loss–induced activation of the TGFβ signaling pathway and the central role of the latter in the observed pathologies. Interestingly, many pathological features in these mice are similar to those seen in patients receiving anti-VEGFA therapies and in preeclampsia. These data demonstrate a critical role of endothelial ERK1/2 signaling in the maintenance of vascular integrity and shed light on the molecular nature of complications associated with anti-VEGF therapies.
Results
Deletion of ERK1/2 in the adult quiescent endothelium leads to cardiovascular morbidity and mortality
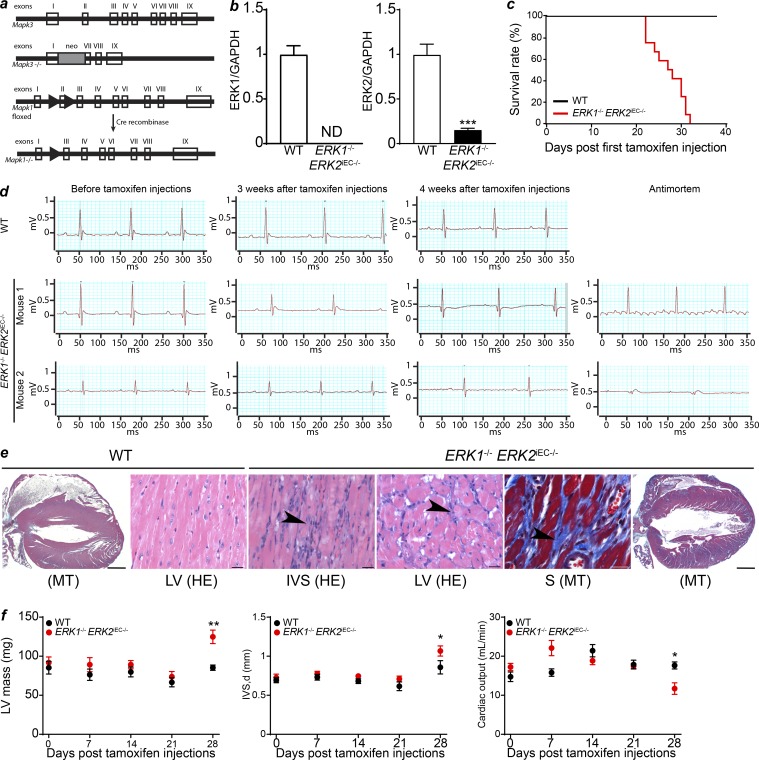
To evaluate the function of the endothelial ERK1/2 pathway in vivo, we studied mice with a global Erk1 deletion (Erk1−/−) and an inducible, endothelial-specific Erk2 deletion (Cdh5CreERT2 Erk2fl/fl, hereafter referred to as Erk2iEC−/−; Selcher et al., 2001; Samuels et al., 2008; Sörensen et al., 2009; Fig. 1 a). Detailed analysis of representative sections of the heart, lung, thymus, liver, and kidneys from Erk1−/− (13 wk) and Erk2iEC−/− (21 and 33 wk of age) mice was unremarkable and within normal limits for all tissues (Fig. S1, a and b). No vascular abnormalities were seen in the retina of Erk1−/− mice (Fig. S1 c). Postnatal endothelial deletion (starting at postnatal day 1) of Erk2 induced a mild delay in retinal angiogenesis; however, all mice survived, and vascular defects were not observed in the adult mice (Fig. S1 c). The two strains were then crossed, generating the Erk1−/− Erk2iEC−/− mouse line. Evaluation of Erk1 and Erk2 expression by quantitative PCR (qPCR) in hepatic endothelial cells showed a total loss of Erk1 and decreased (80%) expression of Erk2 in Erk1−/− Erk2iEC−/− compared with WT mice, confirming the knockout efficiency (Fig. 1 b).
Figure 1.
Erk1−/− Erk2iEC−/− mice genotype, survival, and cardiac ECG and pathology. (a) Generation of Mapk3 (Erk1) knockout mice and Mapk1 (Erk2) conditional knockout mice. Clear boxes represent the exons of the genes. The gray box represents the neomycin cassette. Arrowheads represent LoxP sites. (b) qPCR of ERK1 and ERK2 on hepatic endothelial cells isolated from Erk1−/− Erk2iEC−/− and WT mice to assess the knockout efficiency (n = 4 mice per genotype). We found a total loss of ERK1 and an 80% decrease of ERK2 expression in cells from Erk1−/− Erk2iEC−/− mice compared with the ones from WT mice. ND, not detected. ***, P < 0.001, unpaired t test. (c) 40-d survival curves after tamoxifen injection (n = 12 mice per genotype). (d) ECG recordings of WT and Erk1−/− Erk2iEC−/− mice before tamoxifen injections, 3 and 4 wk after tamoxifen injections, and within a few hours antemortem (n = 4 mice at each time point and n = 7 for antemortem ECG. ECGs from the same mouse are presented at each time point. Two mice are presented for the Erk1−/− Erk2iEC−/− mice). (e) Representative Erk1−/− Erk2iEC−/− mice cardiac photomicrographs show suspect fibrosis (arrowheads) with the left ventricle (LV) and interventricular septum (IVS) by HE, confirmed (blue stain) by MT. Scale bars, 20 µm (n = 4 mice per genotype were analyzed). (f) Left ventricular mass, interventricular septum thickness, and calculated cardiac output evaluated by cardiac echography in Erk1−/− Erk2iEC−/− mice before tamoxifen injections and weekly after tamoxifen injections until 4 wk after injections (n = 4 mice per genotype). *, P < 0.05; **, P < 0.01; two-way ANOVA followed by Sidak’s multiple comparisons. Error bars represent SEM.
To explore the role of ERK signaling in the mature, normal vasculature, endothelial Erk2 deletion was induced in 12–16-wk-old Erk1−/− mice. This resulted in 100% lethality 4–5 wk later (Fig. 1 c). All deaths were sudden, with mice deteriorating rapidly several hours before dying. Electrocardiographic (ECG) monitoring demonstrated a gradual progression from intraventricular conduction delays to atrial flutter with high degree atrial-ventricular block to disappearance of P-waves to complete heart block with an idioventricular escape rhythm followed by death shortly thereafter (Fig. 1 d). Complete cross section of the entire heart and detailed histopathologic examination demonstrated widespread myocardial fibrosis within the intraventricular septum and the left ventricle (Fig. 1 e).
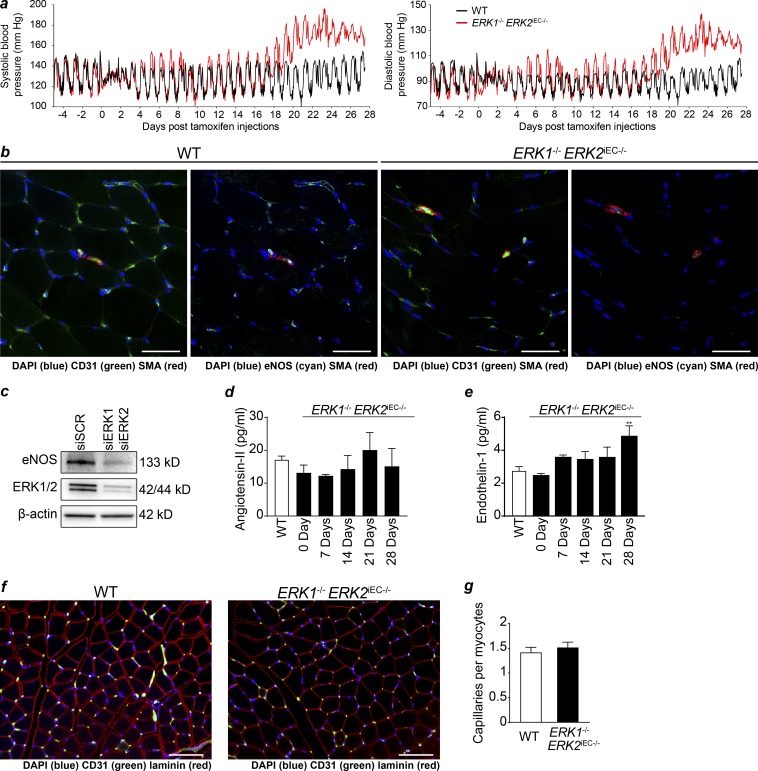
To evaluate changes in cardiac function over time, echocardiography was performed weekly after the initiation of tamoxifen injections. 4 wk after the start of injections, there was a significant increase in the intraventricular septum thickness and left ventricular mass and a significant decline in calculated cardiac output in Erk1−/− Erk2iEC−/− mice (Fig. 1 f). Since one potential explanation for these finding is the development of systemic hypertension, mice were implanted with indwelling pressure transducers 5 d before the start of tamoxifen injections. No difference in systemic arterial blood pressure between WT and Cdh5CreERT2 Erk1−/− Erk2fl/fl mice was detected at this time point (Fig. 2 a). 3 d after the last tamoxifen dose, there was a mild increase in systemic arterial blood pressure (Fig. 2 a) that was followed by a much larger increase 3 wk later (Fig. 2 a and Table S1).
Figure 2.
Systemic hypertension in Erk1−/− Erk2iEC−/− mice. (a) Systemic systolic (left) and diastolic (right) blood pressure in WT and Erk1−/− Erk2iEC−/− mice measured with ambulatory blood pressure transducer (n = 5 mice per genotype). (b) eNOS expression (cyan) in quadriceps with CD31 (green) and SMA (red) costaining in Erk1−/− Erk2iEC−/− mice 1 wk after tamoxifen injections (representative images from n = 4 mice per genotype). Scale bars, 50 µm. (c) eNOS expression by Western blot in HUVECs after knockdown of ERK1 and ERK2 (representative blot from n = 3). (d and e) Circulating angiotensin-II (d) and endothelin-1 (e) in plasma of Erk1−/− Erk2iEC−/− mice before tamoxifen injections and weekly after tamoxifen injections until 4 wk after injections (n = 5 mice per genotype). **, P < 0.01 using two-way ANOVA followed by Sidak’s multiple comparisons. (f and g) Capillary staining (CD31 in green) in quadriceps in Erk1−/− Erk2iEC−/− mice (f) and quantification (g; n = 6 mice per genotype). Scale bars, 100 µm. Error bars represent SEM.
We speculated that the initial rise in blood pressure could be due to decreased nitric oxide production. Indeed, immunocytochemistry of quadriceps muscle sections demonstrated a reduction in endothelial nitric oxide synthase (eNOS) expression 1 wk after tamoxifen injection (Fig. 2 b). To confirm this link, we carried an ERK1/2 knockdown in HUVECs. In agreement with the in vivo data, this led to a nearly complete disappearance of eNOS expression (Fig. 2 c). To explain the late rise in blood pressure, we performed serial measurements of plasma levels of two potent vasoconstrictors, angiotensin-II and endothelin-1. While angiotensin-II remained unchanged, there was a significant rise in endothelin-1 levels in Erk1−/− Erk2iEC−/− mice that temporally corresponded to the late increase in blood pressure (Fig. 2, d and e). Finally, immunocytochemical analysis of the skeletal muscle vascular density revealed no abnormalities in the knockout mice suggesting that vascular rarefication is not responsible for hypertension in these mice (Fig. 2, f and g).
Endothelial ERK1/2 knockout induces EndMT
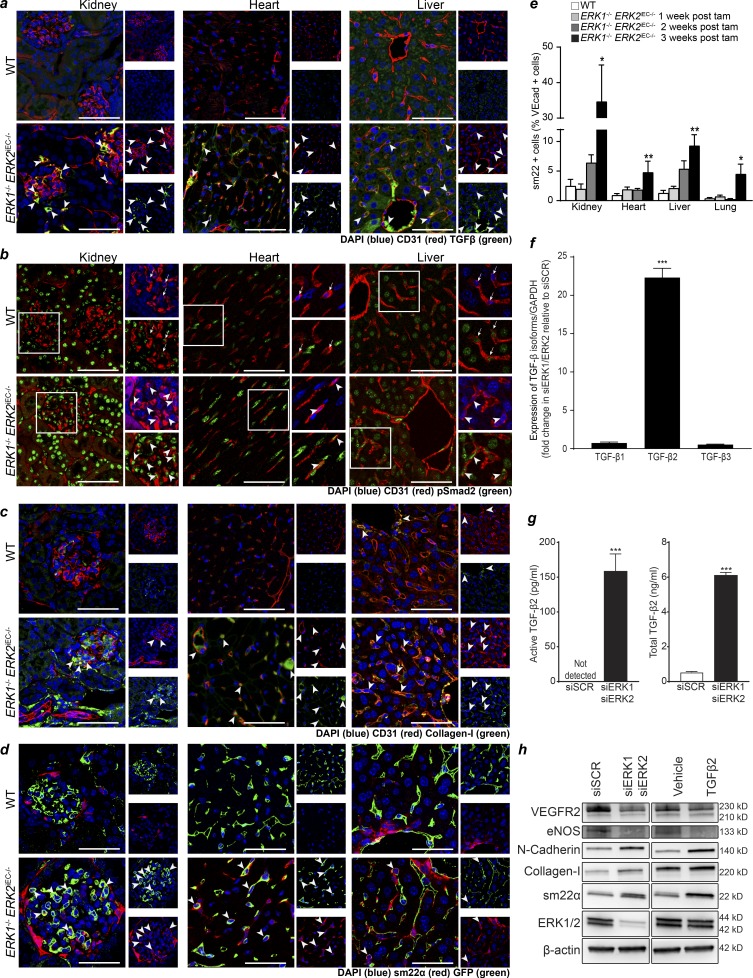
Given extensive cardiac fibrosis in Erk1−/− Erk2iEC−/− mice, we investigated whether ERK gene deletion resulted in activation of TGFβ signaling, a well-established inducer of fibrosis (Meng et al., 2016). Analysis of kidney, heart, and liver tissues demonstrated markedly increased expression of TGFβ in endothelial cells in Erk1−/− Erk2iEC−/− mice, whereas no endothelial TGFβ expression was seen in WT mice (Fig. 3 a). This increase in endothelial expression of TGFβ was detected as early as 1 wk after tamoxifen injections in the kidney and 2 wk in the other organs (Fig. S2 a). In agreement with these findings, staining with an anti-phospho-Smad2 (pSmad2) antibody showed a strong increase in pSmad2 levels in cardiac, renal, and liver tissues (Fig. 3 b). Interestingly, while the increase in pSmad2-positive endothelial cells is significant in the three studied tissues, the increase is particularly important in the renal endothelium (Fig. S2 b), starting 2 wk after tamoxifen injections (Fig. S2 c). Finally, there also was an increase in expression of collagen-I, a known TGFβ-induced gene (Fig. 3 c). In contrast, there was no increase in pSmad2 levels in tissues from ERK1−/− mice and ERK2iEC−/− mice (Fig. S2 d).
Figure 3.
Widespread EndMT in Erk1−/− Erk2iEC−/− mice. (a) TGFβ staining in the kidney, heart, and liver of Erk1−/− Erk2iEC−/− mice 3 wk after tamoxifen injections (representative of n = 4 mice per genotype). Scale bars, 50 µm. Arrowheads point to endothelial cells expressing TGFβ. The small picture represents the single channel of the large picture. (b) pSmad2 staining in kidney, heart, and liver from Erk1−/− Erk2iEC−/− mice 4 wk after tamoxifen injections (representative of n = 4 mice per genotype). Scale bars, 50 µm. Arrows point to endothelial cells with no or low pSmad2 staining. Arrowheads point to endothelial cells with intense pSmad2 staining. Small pictures are a higher magnification of the field in the white square in the large pictures. (c) Collagen-I staining in kidney, heart, and liver from Erk1−/− Erk2iEC−/− mice 3 wk after tamoxifen injections (representative of n = 4 mice per genotype). Scale bars, 50 µm. Arrowheads point to endothelial cells expressing collagen-I. The small picture represents the single channel of the large picture. (d) GFP and sm22α staining in the kidney, heart, and liver of Cdh5Cre-mTmG mice (denoted WT) and Erk1−/− Erk2iEC−/− mTmG mice 4 wk after tamoxifen injections (representative of n = 4 mice per genotype). Scale bars, 50 µm. Arrowheads point to endothelial cells expressing sm22α. The small picture represents the single channel of the large picture. (e) Quantification of the proportion of VE-cadherin+ cells expressing sm22α by FACS (n = 4 mice per genotype and per time point). *, P < 0.05; **, P < 0.01; Kruskal–Wallis test followed by Dunn’s multiple comparisons to WT mice. (f) Evaluation of Tgfb1, Tgfb2, and Tgfb3 mRNA expression by qPCR in HUVECs after Erk1 and Erk2 knockdown (n = 6). ***, P < 0.001; Mann–Whitney U test. (g) Active and latent TGFβ2 concentration in conditioned media from HUVECs knocked down for ERK1 and ERK2 (n = 9). ***, P < 0.001; Mann–Whitney U test. (h) Expressions of endothelial markers (VEGFR2 and NOS3) and mesenchymal markers (N-cadherin, collagen-I, and sm22α) by Western blot in HUVECs knocked down for ERK1 and ERK2 and HUVECs treated with TGF-β2 at 10 ng/ml for 4 d (representative blot from n = 3). Error bars represent SEM.
These data point to the development of EndMT induced by endothelial ERK1/2 deletion. To confirm this and track endothelial cells fate in vivo, we crossed the Erk1−/− Erk2iEC−/− mice with a mouse line carrying a floxed mTmG allele, generating a Cdh5CreERT2 mTmG Erk2fl/fl Erk1−/− strain. Activation of Cdh5-Cre in this strain results in fate mapping of all endothelial cells with GFP in addition to the excision of Erk2.
Immunocytochemical examination of the heart, liver, and kidney tissues 4 wk after tamoxifen treatment showed widespread expression of mesenchymal markers Sm22α and smooth muscle actin (SMA) in GFP+ cells (Figs. 3 d and S3 a), confirming the occurrence of EndMT. To quantify the proportion of endothelial cells undergoing EndMT in different organs, we used FACS (Fig. S3, b and c). While there was an increase in endothelial cells (VE-cadherin+) expressing Sm22α in all organs examined 3 wk after tamoxifen injection, the proportion of endothelial cells undergoing EndMT was the highest in the kidney (Fig. 3 e).
To validate these results in vitro, we performed qPCR analysis of TGFβ1-3 gene expression in HUVECs treated with ERK1 and ERK2 siRNAs. As expected, there was a highly significant increase in TGFβ2 levels (Fig. 3 f). ELISA measurement of secreted latent and active TGFβ in the media confirmed a highly significant increase in TGFβ2 production (Fig. 3 g). The let-7 family of miRNAs is a known regulator of TGFβ expression (Chen et al., 2012). We found a decrease in most let-7 family members expression in HUVECs treated with siRNAs targeting ERK1 and ERK2 (Fig. S4 a). Furthermore, Western blot analysis showed an increase in endothelial expression of a number of mesenchymal markers including N-cadherin, collagen-1, Sm22α, and a decrease in expression of endothelial fate marker genes VEGFR2 and eNOS (Fig. 3 h). A prolonged (4 d) treatment of HUVECs with TGFβ2 induced similar changes in the expression of endothelial and mesenchymal markers (Fig. 3 h). No changes in the expression of inhibitory Smads (Smad6 and Smad7) or Smads activated by the TGFβ pathway (Smad2 and Smad3) were detected (Fig. S4 b).
Organ pathology in endothelial ERK1/2 knockout mice
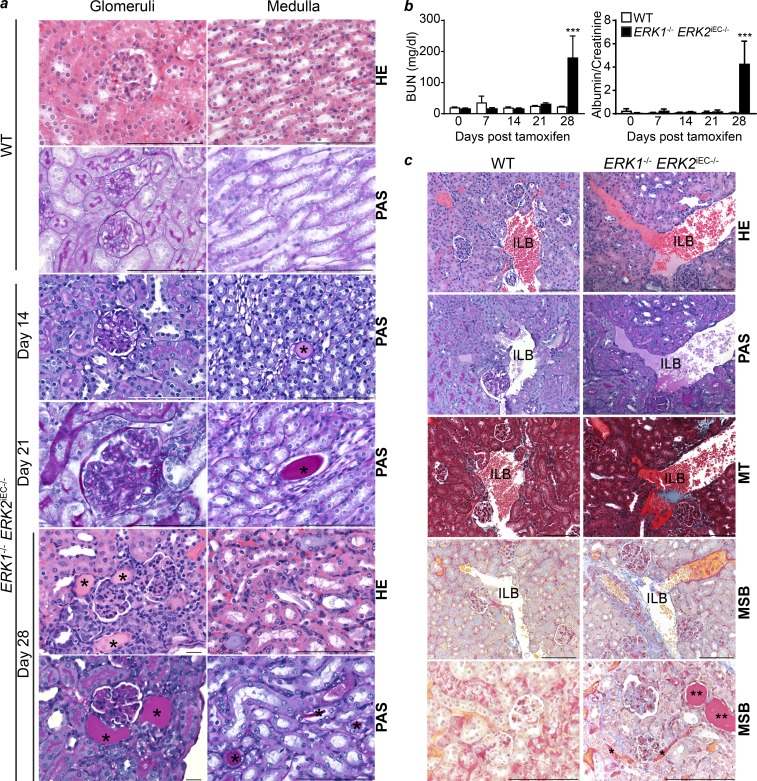
Given a particularly strong increase in renal endothelial pSmad2 staining in Erk1−/− Erk2iEC−/− mice, we performed a detailed examination of the kidney in these animals. Both Erk1−/− Erk2iEC−/− and C57Bl6 WT mice were injected with tamoxifen daily for 4 d, and four mice from each genotype were sacrificed at days 7, 14, 21, and 28 following the first tamoxifen injection. In each case, serum was collected for blood urea nitrogen (BUN) determination, urine was collected to measure the albumin/creatinine ratio, and tissues were harvested for histopathology. No abnormalities at any time point were seen in WT mice. While kidneys from Erk1−/− Erk2iEC−/− mice showed no pathological abnormalities at day 7, scattered glomerular basement membrane thickening and tubular proteinosis were both visible at day 14, with the severity and extent of the lesions increasing with time (Fig. 4 a). In agreement with these findings, BUN in plasma and the urine albumin/creatinine ratio in Erk1−/− Erk2iEC−/− mice remained normal until day 28, at which point there was a sharp rise in both measurements (Fig. 4 b).
Figure 4.
Renal pathology in Erk1−/− Erk2iEC−/− mice. (a) Representative renal pathology HE and PAS photomicrographs from Erk1−/− Erk2iEC−/− mice on days 14, 21, and 28 after tamoxifen injections revel scattered glomerular basement membrane thinking and protein (*) within medullary tubules positive by PAS stain starting at day 14 and increasing in severity by day 21, with enlarged glomeruli compared with WT mice, who lacked pathological changes at all time points. By day 28, Erk1−/− Erk2iEC−/− mice show more severe changes in tubular proteinosis compared with WT mice. Scale bars, 100 µm. (b) BUN in plasma and urine albumin/creatinine ratio from Erk1−/− Erk2iEC−/− mice before tamoxifen injection and weekly after tamoxifen injections until 4 wk after injections are both markedly elevated at day 28 compared with WT mice (n = 4). ***, P < 0.001; two-way ANOVA followed by Sidak’s multiple comparisons. (c) Representative HE, PAS, MT, and MSB photomicrographs of kidney from Erk1−/− Erk2iEC−/− mice at day 28 after tamoxifen injection show scattered antemortem blood clots in glomerular (arrowhead) and peritubular capillaries (*), and increased connective tissue (MT and MSB blue) compared with WT mice. Both Erk1−/− Erk2iEC−/− and WT mice have visible postmortem blood clots within peritubular (arrows) and interlobular blood vessels (ILB). Representative images from n = 4 mice per genotype and per time point. MT, collagen (blue). MSB, new fibrin/erythrocytes (yellow), old fibrin in vessels (red), connective tissue (blue), and protein in tubules stain less intense red (**). Scale bars, 100 µm. Error bars represent SEM.
To evaluate the presence of intravascular thrombosis, we used a Martius scarlet blue (MSB) staining to distinguish antemortem and perimortem thrombosis. This showed scattered antemortem blood clots in Erk1−/− Erk2iEC−/− mice at day 28 in glomerular capillary lumina and peritubular capillaries (Masson trichrome [MT] and MSB red), while no antemortem clots were detected in WT mice. Both Erk1−/− Erk2iEC−/− and WT mice had scattered visible postmortem blood clots within peritubular (arrows) and interlobular blood vessels (Fig. 4 c).
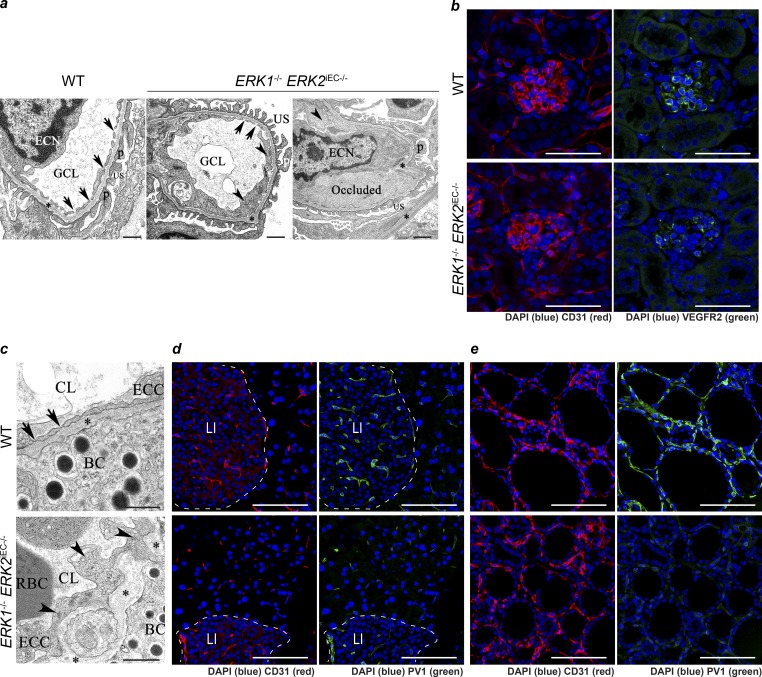
Transmission electron microscopic (TEM) evaluation of kidney sections revealed multifocal loss of glomerular capillary endothelial cell (GEC) fenestration, an irregular and thickened glomerular basement membrane, and multifocal podocyte effacement consistent with the onset of proteinuria in Erk1−/− Erk2iEC−/− mice, but not WT mice (Fig. 5 a). There was widespread GEC cytoplasmic swelling, and some GEC lumens were partially to completely occluded by electron-opaque material suggestive of fibrin (Fig. 5 a). The VEGF pathway is well known to maintain fenestration in glomeruli endothelial cells (Eremina et al., 2007). To verify the loss of GEC fenestration mediated through VEGF, which is expressed by podocytes and binds the receptor VEGFR2 in glomeruli endothelial cells, we stained kidney sections with an anti-VEGFR2 antibody. We found a decrease in VEGFR2 expression in glomeruli that is consistent with the decrease in endothelial fenestration (Fig. 5 b). We also observed a widespread loss of fenestration in renal peritubular and pancreatic islet capillaries (Fig. 5 c). PV1 is a protein expressed in endocrine endothelial cells and is critical to diaphragm fenestra formation in these endothelia (Stan et al., 2012). Immunocytochemical analysis of pancreas and thyroid sections with an anti-PV1 antibody showed a marked decrease in its expression, suggesting that ERK1/2 signaling is involved in endothelial fenestration in endocrine glands in part by controlling PV1 expression.
Figure 5.
Loss of endothelial fenestration in glomeruli endothelium and endocrine gland endothelium in Erk1−/− Erk2iEC−/− mice. (a) Transmission electron photomicrographs of kidney Erk1−/− Erk2iEC−/− mice reveal swollen endothelial cells (arrowheads) with loss of glomerular capillary cell fenestrations (arrows) and basement membrane (*) thickening and distortion with localized effacement of podocyte foot processes (p) and scattered occlusion of the glomerular capillary lumen (GCL) by fibrin (presumptive). ECN, endothelial cell nucleus; US, urinary space. Representative images from n = 4 mice per genotype. Scale bars, 500 nm. (b) Immunofluorescent staining of VEGFR2 (green) in glomeruli from Erk1−/− Erk2iEC−/− and WT mice 2 wk after tamoxifen injections. Representative images from n = 4 mice per genotype. Scale bars, 50 µm. (c) Transmission electron photomicrograph of pancreas from Erk1−/− Erk2iEC−/− mice reveal swollen endothelial cells (arrowheads) with loss of capillary endothelial cell fenestrations (arrows) and basement membrane (*) thickening and distortion compared with WT mice. BC, β cells; CL, capillary lumen; ECC, endothelial cell cytoplasm. Representative images from n = 4 mice per genotype. Scale bars, 500 nm. (d and e) Immunofluorescent staining of PV1 (green) in pancreas (d) and thyroid (e) in Erk1−/− Erk2iEC−/− and WT mice. Representative images from n = 4 mice per genotype. Scale bars, 50 µm. LI, Langerhans islet.
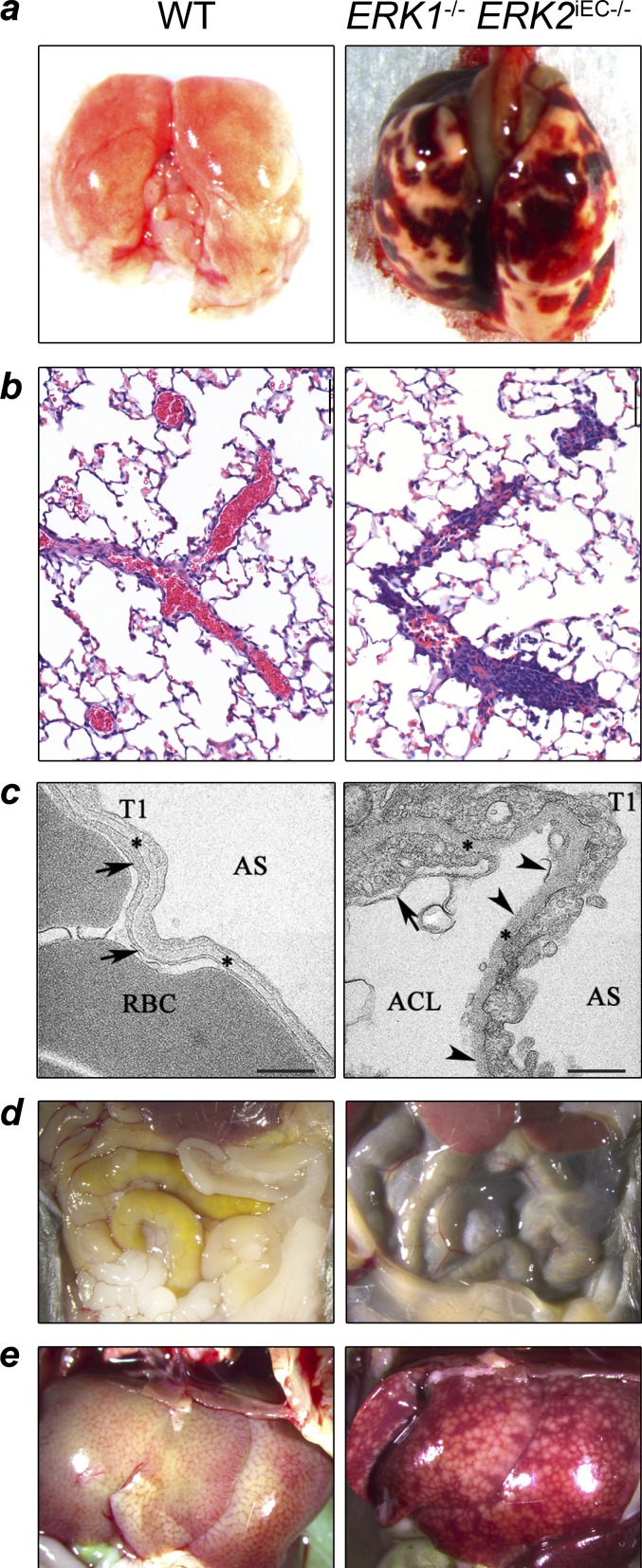
Other prominent findings in Erk1−/− Erk2iEC−/− mice were macroscopic pulmonary hemorrhages (Fig. 6 a) and microscopic evidence of intravascular pulmonary thrombosis (Fig. 6 b). Pulmonary TEM demonstrated the presence of the same morphological abnormalities in alveolar capillary endothelial cells that were seen in GECs, including cytoplasmic thickening and irregular and thickened basement membrane in Erk1−/− Erk2iEC−/− mice that were absent in the WT mice (Fig. 6 c). Additional pulmonary changes in Erk1−/− Erk2iEC−/− mice include alveolar capillary endothelial cell delamination and cytoplasmic changes in type 1 alveolar pneumocytes. Hemorrhage was also observed in other organs in Erk1−/− Erk2iEC−/− mice, including the intestine (dark loops in the small bowel) and liver (Fig. 6, d and e).
Figure 6.
Pulmonary, hepatic, and intestinal hemorrhage in Erk1−/− Erk2iEC−/− mice. (a) Erk1−/− Erk2iEC−/− mice 4 wk after tamoxifen injection show macroscopic pulmonary and pleural hemorrhage compared with WT. (b) Erk1−/− Erk2iEC−/− mice necropsied 28 d after tamoxifen injections have partially or completely occluded pulmonary vessels compared with WT mice (n = 4 mice per genotype; a thrombus was found in three out of four mice). (c) Lung electron photomicrographs from Erk1−/− Erk2iEC−/− mice show capillary endothelial cell membrane (arrows) loss and delamination (arrowheads) compared with WT mice. Arrowheads show the capillary endothelial cell membrane, and asterisks show the basement membrane. RBCs within the alveolar capillary lumen (ACL) are shown. AS, alveolar space; T1, alveolar type 1 epithelial cell. Scale bars, 500 nm. (d and e) Erk1−/− Erk2iEC−/− mice 5 wk after tamoxifen injection show macroscopic dark loops of intestine (d) and hepatic sinusoidal congestion (e) consistent with intraluminal and hepatic hemorrhage, respectively, compared with WT mice.
TGFβ2 is the main driver of endothelial ERK1/2 knockout phenotypes
To gain insights into the links among the loss of endothelial ERK1/2 expression, activation of TGFβ signaling, and the observed phenotypes, we used a causal statistical computing framework designed to generate working biological hypotheses from high-dimensional “omics” datasets. To this end, we analyzed RNA-seq data from HUVECs treated with siRNA against ERK1 and ERK2 (siERK1/2, n = 19) and scrambled controls (siSCR, n = 18; Fig. S5 a). The normalized overall dataset was split into training (80% of total samples) and test (20% of total samples) datasets, and all subsequent processing and modeling steps were performed on the training dataset only (the test dataset was only used to evaluate model generalizability and performance). As deep learning becomes computationally difficult with >10,000 input features (13,985 genes were measured by RNA-seq), we first applied a simple feature learning technique to reduce the dimensionality of the input by filtering out differentially expressed genes (edgeR, negative binomial model) of the training dataset with a q-value ≥ 10−35 (Fig. S5 b, i–iii; and Table S2; Robinson et al., 2010). We then trained a deep artificial neural network (DANN), using only the training dataset, to classify siSCR versus siERK1/2 cell populations with the 983 resultant gene expression profiles as inputs (Fig. S5 b, iv; and Table S3; tuning model hyperparameters with an inner cross-validation loop using the training datasets; LeCun et al., 2015). Although our dataset was significantly smaller than dataset sizes typically employed for deep learning (n = 37 compared with n >> 500), our model generalized a robust test dataset (20% of the total dataset) with area under the curve and classification accuracy of 1.0 (Fig. S5 b, v).
To determine the most informative of the 983 genes in classifying samples as siSCR or siERK1/2, we applied a local gradient-based sensitivity analysis technique, termed sensitivity mapping (Fig. S5 b, iv; see Materials and methods), which resulted in a ranked list of the 983 genes (Table S4; Smilkov et al., 2017 Preprint). While useful, maximizing class separation with statistical learning approaches does not provide biological context or predict relationships among the most informative molecular features (Bennett et al., 2003; Mahieu et al., 2016). Therefore, we added an additional level of biological context by hypothesizing the causal dependency structure between the most informative genes of the deep learning model by building a Bayesian belief network (BBN; Fig. S5 b′, vi). Genes closer to the top of the directed acyclic graph (DAG) are predicted to have causal influence over genes lower in the DAG (i.e., changes in upstream genes are more likely to lead to altered states of downstream genes).
Using the top 100 most informative genes of the deep learning model, we generated a BBN that allowed us to better understand the conditional dependency between these genes (Fig. S5 c). A significant portion of the top 100 genes were related to TGFβ signaling; however, TGFβ ligands were not included in this BBN (TGFβ2, the highest ranked TGFβ ligand, fell within the top 200 genes by sensitivity mapping; see Table S4). Thus, we expanded the total number of genes in our BBN analysis to the top 600 genes and discovered that TGFβ2 was one of the most upstream drivers of this predicted network (Fig. S5 c′). However, due to the expanded number of genes, the BBN was significantly less connected between vertices (decreased average degree per vertex).
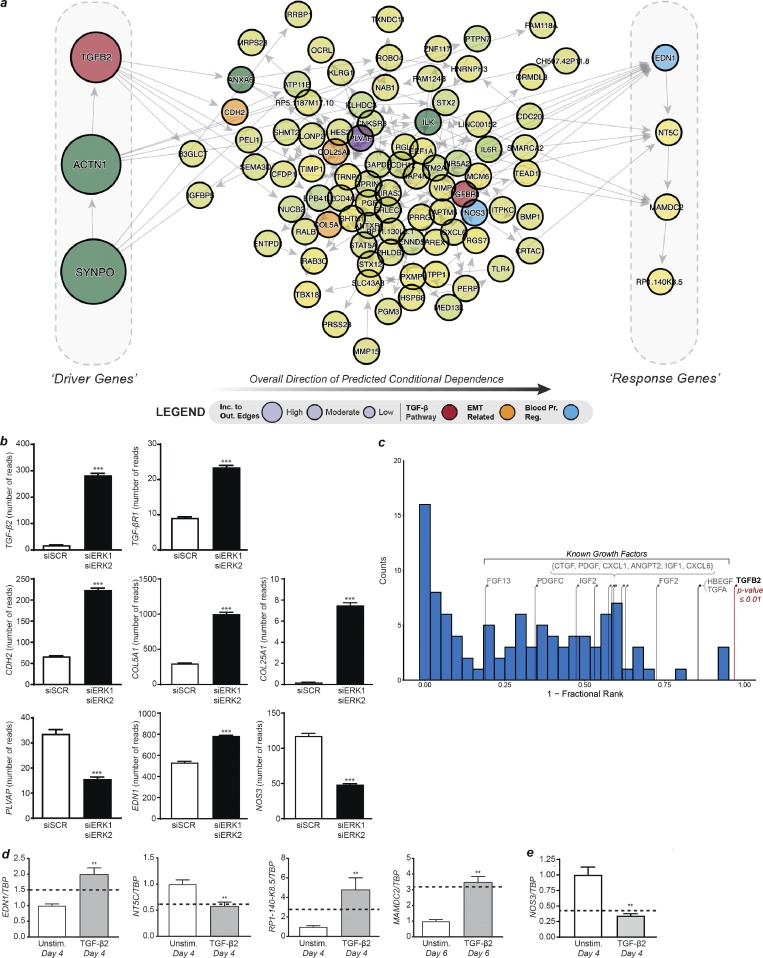
Therefore, to determine a core causal structure with respect to TGFβ signaling, we predicted a BBN for the top 100 most informative genes with the addition of TGFβ2 and any additional TGFB2 related genes within the 983 input genes (Laumonnier et al., 2000; Saura et al., 2002). We again found that TGFβ2 was one of the most upstream drivers of the core network, thus indicating a robust hypothesis of involvement of TGFβ signaling (Fig. 7 a). Within the core network, predicted to be driven by TGFβ2, several genes encoded proteins involved in the process of EndMT (Fig. 7, a and b, orange vertices) such as CDH2, COL25A1, and COL5A1 (Dejana et al., 2017). In addition, PVLAP encodes a protein involved in endothelial fenestration (Fig. 7, a and b, purple vertices), EDN1 encodes endothelin-1, a vasoconstrictor hormone, and NOS3 encodes eNOS, an enzyme producing the vasodilator nitric oxide (Fig. 7, a and b, blue vertices; Marasciulo et al., 2006; Stan et al., 2012).
Figure 7.
Hypothesizing and validating the causal structure between the most informative genes. (a) DAG derived from the most informative genes with the addition of TGFβ2, TGFβR1, and NOS3. Genes on the left side of the DAG are the most likely upstream causal drivers, while genes on the right are the most likely downstream targets (determined based on incoming and outgoing edges). (b) Expression levels determined by RNA-seq of genes involved in the TGFβ pathway (TGFβ2 and TGFβR1), EndMT (CDH2, COL5A1, and COL25A1), endothelial fenestration (PVLAP), and blood pressure regulation (NOS3 and EDN1) in HUVECs knocked down for ERK1 and ERK2 (n = 18 for siSCR and n = 19 for siERK1 siERK2). (c) Permutation analysis by randomly selecting genes to replace TGFβ2 (relative position shown with the red stem) and determining the relative position within the predicted BBN (Westfall–Young permutation analysis). The blue histogram shows the null distribution, while the stems indicate the relative position in the DAG of other known signaling ligands that were highly ranked by sensitivity analysis. (d) Expression levels of predicted downstream genes by qPCR after 4 or 6 d of TGFβ2 stimulation in HUVECs compared with unstimulated HUVECs. The dotted line represents the fold change in expression level of these genes in HUVECs knocked down for ERK1 and ERK2 versus control (n = 6). **, P < 0.01; Mann–Whitney U test. (e) Expression levels of NOS3 by qPCR after 4 or 6 d of TGFβ2 stimulation in HUVECs compared with unstimulated HUVECs. The dotted line represents the fold change in expression level of these genes in HUVECs knocked down for ERK1 and ERK2 versus control (n = 6). **, P < 0.01; ***, P < 0.001; Mann–Whitney U test. Error bars represent SEM.
To verify that the predicted involvement of TGFβ2 was not an artifact, we performed permutation analysis by randomly selecting genes to replace TGFβ2 and determining the relative position within the predicted BBN (Fig. 7 c). We observed that one would not find the position of TGFβ2 within the predicted network by chance (Fig. 7 c, Westfall–Young permutation analysis, P ≤ 0.01). In addition, we tested other known signaling ligands acting on the endothelium that were highly ranked by sensitivity analysis (see Table S4) and found that none of the relative positions of these ligands within the predicted causal structures were significant. These results indicated that our TGFβ2 hypothesis was robust and strengthened the prediction that it was a likely an upstream driver within the network.
Next, we experimentally validated our BBN in vitro. We stimulated HUVECs with TGFβ2 and measured expression of the predicted downstream genes (EDN1, NT5C, MAMDC2, and RP1-140-K8.5) 4–6 d later. There was a statistically significant difference (P ≤ 0.05, Mann–Whitney U test) in the expression level of all of the predicted downstream genes in TGFβ2 stimulated HUVECs compared with unstimulated cells (Fig. 7 d). TGFβ2 was also predicted to be a regulator of NOS3 expression (Fig. 7 a). HUVECs stimulated with TGFβ2 for 4 d have significantly reduced expression of NOS3 compared with unstimulated cells (Fig. 7 e).
Altogether, our computational analysis demonstrated that overexpression of TGFβ2 in ERK1/2 knockdown HUVECs is the main driver of the EndMT, the increase in EDN1 expression, and the decrease in NOS3 expression.
Discussion
While much effort has been focused on understanding signaling pathways controlling endothelial behavior in development, little is known about regulation of vascular quiescence. The phenotypes of Erk1−/− Erk2iEC−/− mice are particularly instructive in this regard. The most prominent feature was a widespread EndMT that was responsible, at least in part, for the progressive renal failure, hemorrhages, and fibrosis in multiple organs, including fibrosis of the cardiac conduction system. While EndMT is not the sole cause of cardiac fibrosis, the development of advanced conduction defects due to fibrosis in the interventricular septum was the dominant cause of after ablation of endothelial ERK expression. We also observed a two-phase hypertensive response. The early increase in the blood pressure was likely due to a decrease in endothelial eNOS expression, whereas the late increase correlated with rising blood levels of endothelin-1.
Taken together, these observations implicate endothelial ERK1/2 signaling in regulation of endothelial integrity and cell fate maintenance. Previous studies have suggested a possible role of ERK1/2 in cell fate determination during development (Xu et al., 2008; Deng et al., 2013a,b; Shin et al., 2016a,b), but its involvement in continuous maintenance of endothelial fate (and prevention of EndMT) in the adult vasculature has not been appreciated. The link of endothelial ERK1/2 expression to hypertension, the loss of NOS3 expression, and increased endothelin-1 levels are also new and unexpected findings.
While a number of receptor tyrosine kinases can activate the ERK1/2 pathway in quiescent endothelial cells, VEGF and FGFs are likely the key mediators. The loss of FGF signaling was previously shown to result in the loss of endothelial integrity, increased permeability and, ultimately, frank hemorrhage previously (Murakami et al., 2008, 2011). Similarly, inhibition of autocrine VEGF signaling was also reported to lead to endothelial degeneration, hemorrhages, and sudden death (Lee et al., 2007). Renal failure with proteinuria, endotheliosis, and loss of endothelial fenestration was described in mice in which Vegfa knockout in podocytes disrupted paracrine VEGF signaling in glomeruli (Eremina et al., 2003). Emphysema has also been observed in mice treated with the VEGFR2 inhibitor SU5416 (Kasahara et al., 2000), and pulmonary hemorrhages were observed in mice heterozygous for Dll4 (Rostama et al., 2015), a well-established VEGF target gene (Fish et al., 2017).
Many of the pathological findings observed in Erk1−/− Erk2iEC−/− mice have also been reported in patients treated with anti-VEGF neutralizing agents. In particular, systemic hypertension has been observed in patients treated with anti-VEGF neutralizing agents or a tyrosine kinase inhibitor targeting VEGFR2 signaling (Touyz and Herrmann, 2018; Touyz et al., 2018). Similar to findings in our mice, these patients exhibited decreased nitric oxide production and increased endothelin-1 plasma levels (Facemire et al., 2009; Kappers et al., 2010; de Jesus-Gonzalez et al., 2012). Loss of endothelial fenestration in endocrine glands was also reported in mice treated with an anti-VEGF therapy (Zhang et al., 2016). Hemorrhages were described in patients treated with sunitinib and sorafenib, two tyrosine kinase inhibitors targeting VEGFR2 signaling (Je et al., 2009). Finally, renal failure, including proteinuria and glomerulosclerosis, is a common side effect found in patients treated with anti-VEGF therapies (Eremina et al., 2008).
Preeclampsia is a pregnancy-associated disease thought to be due to elevated levels of circulating VEGFR1 leading to sequestration of VEGFA and a reduction in VEGFR2 signaling (Maynard et al., 2003; Shibuya, 2013; Armaly et al., 2018). It is characterized by progressive hypertension with decreased nitric oxide production and increased endothelin-1 plasma levels (Nova et al., 1991; Bernardi et al., 2008) as well as myocardial dysfunction (Patten et al., 2012). In severe cases, this leads to renal failure, seizures, and death. Interestingly, changes in renal histology seen in these patients (Phipps et al., 2016) are similar to that seen in Erk1−/− Erk2iEC−/− mice, as is the observed elevation in TGFβ2 plasma levels (Shaarawy et al., 2001).
In addition, clinical uses of inhibitors of Braf (vemurafenib and dabrafenib) and an inhibitor of MEK (trametinib), two upstream regulators of the ERK1/2 pathway, have reported side effects similar to phenotypes observed in Erk1−/− Erk2iEC−/− mice. In particular, hypertension is the most common severe side effect seen in patients treated with trametinib (Welsh and Corrie, 2015; Banks et al., 2017), while many patients on these three drugs also develop renal toxicity (Porta et al., 2015; Wanchoo et al., 2016a,b; Rubin, 2017). Lastly, abnormal bleeding was seen in patients treated with trametinib alone or in combination with Braf inhibitors, particularly cerebral and gastrointestinal hemorrhages (Rubin, 2017). Taken together, these clinical observations are in line with the critical role of endothelial ERK1/2 signaling in the maintenance of vascular normalcy.
One unexpected finding in this study is the activation of TGFβ signaling in ERK1/2 null endothelial cells leading to EndMT. There is by now an extensive literature dealing with TGFβ as the driver of EndMT and fibrosis (reviewed in Dejana et al., 2017). Normal endothelial cells express very low levels of TGFβ receptors and thus are not particularly sensitive to TGFβ stimulation. In contrast, endothelial cells subjected to inflammatory stress activate TGFβ receptor expression and become TGFβ sensitive. Stimulation of these activated endothelial cells with TGFβ induces EndMT and further promotes and amplifies inflammation, leading to permanent tissue organ damage, including fibrosis (Schwartz et al., 2018). Remarkably, the activation of TGFβ signaling in ERK1/2 null endothelial cells was predicted by our ensemble computational intelligence strategy, comprising both deep learning and probabilistic programing of RNA-seq data. The model, based on analysis of RNA-seq data from HUVECs treated with either scrambled and ERK1/2 siRNAs, predicted an increase in TGFβ2 expression and activation of TGFβ signaling leading to EndMT. In addition, this statistical computing approach correctly predicted a TGFβ-driven increase in endothelin-1 and a reduction in eNOS expression in addition to changes in expression of a long non-coding RNA of unknown function. Importantly, these in silico predictions were validated both in vivo and in vitro. These findings provide evidence that intelligently engineered causal statistical-learning strategies can glean meaningful insights into more complex disease etiology from relatively small experimental designs, and by coupling deep learning with probabilistic programming, such approaches can robustly infer informative gene network dependency structures, thus generating further insight into the molecular underpinnings of cellular behavior.
TGFβ can signal through the canonical Smad2/3 pathway and noncanonical pathways, including the ERK1/2 pathway (Massagué, 2012). Interestingly, in epithelial cells, the activation of the ERK1/2 pathway by TGFβ has been shown to be involved in the epithelial-to-mesenchymal transition (EMT; Xie et al., 2004; Buonato and Lazzara, 2014), whereas other studies showed that the Smad2/3 pathway is involved in EMT (Thuault et al., 2008; Tan et al., 2012). Here, we show that loss of the ERK1/2 pathway in endothelial cells activates the TGFβ pathway, leading to EndMT. While we did not identify the signaling pathway activated downstream of TGFβ and involved in the EndMT phenotype, the ERK1/2 pathway is not the pathway involved, as we knocked down both ERK isoforms. These findings show that the downstream signaling pathways activated by TGFβ and involved in EMT and EndMT may be different.
The results of this study indicate that in the quiescent endothelium, the paracrine continuous ERK1/2 signaling input is required for the maintenance of endothelial normalcy. In large put this is mediated by ERK-dependent suppression of TGFβ signaling. When this protective input is lost, activation of TGFβ signaling results in induction of EndMT, decreased nitric oxide production, increased endothelin-1 expression, and the loss of endothelial fenestration. The end result is a spectrum of pathologies that closely resembles findings in patients treated with VEGFA neutralizing agents or inhibitors of ERK signaling.
Materials and methods
Cell culture, siRNA transfection, and qPCR
HUVECs were purchased at Lonza and cultivated in EBM-2 MV medium (Lonza). siRNA transfections were done in OptiMem medium (ThermoFisher) with the lipofectamine RNAimax transfection reagent (ThermoFisher). siRNA was purchased from Qiagen (siRNA MAPK1_10 and MAPK3_6). TGFβ2 stimulation (R&D Systems) was done at 10 ng/ml in EBM-2 MV medium changed daily. RNA extractions were done using RNeasy mini kit (Qiagen) with DNase digestion (Qiagen). cDNA was synthetized with iScript Reverse Transcription Supermix (Bio-Rad), and qPCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). For miRNA, RNA was isolated with miRNease Mini Kit (Qiagen), and cDNA and qPCR were realized using miScript PCR System and primers (Qiagen).
RNA-seq processing and differential analysis
Next-generation sequencing was performed at the Shanghai Sequencing Lab of WuXi NextCODE (Illumina; Gene Expression Omnibus accession no. GSE130737). RNA-seq reads were aligned to human genome build 38 (GRCh38) using the short reads aligner STAR, and expression quantification was performed using RSEM with GENCODE annotation (release 24: http://www.gencodegenes.org; Li and Dewey, 2011; Dobin et al., 2013). Lowly expressed genes were filtered out by requiring one count per million in ≥10% of samples. The edgeR R package was used to normalize the mRNA read counts (using the trimmed mean of M-values method) and determine differential expression between experimental groups (siERK1/2 versus siSCR; Robinson et al., 2010). The top 983 differentially expressed genes (adjusted P value ≤ 1E-35) were used for further downstream analysis.
Deep learning: DANNs
We adapted common deep learning methodologies to analyze genomic datasets (Alipanahi et al., 2015). Typical deep neural networks use a series of nonlinear transformations (termed layers), with the final output considered a prediction of class or regression variable. Each layer consists of a set of weights (W) and biases (b) that are tuned during a training phase to learn which nonlinear combinations of input features are most important for the prediction task. These types of models “automatically” learn patterns in the data and combine them, in some abstract nonlinear fashion, to gain an ability to make predictions about the dataset.
The basic formulation of a fully connected DANN is given as
where the dimensions of W and b are determined by the number of neurons in each layer (d1, d2, … dm). Each layer used rectified linear units as activation functions:
The final layer used a softmax function, with the number of neurons equal to the number of class (K), to convert the logits to probabilities:
In addition, we used the concept of “dropout,” which randomly sets a portion of input values (η) to the layer to zero during the training phase (Srivastava et al., 2014). This has a strong regularization effect (essentially by injecting random noise) that helps prevent models from overfitting. Layers that included dropout were formulated as
where ml ~ Bernoulli(η).
When evaluating models on test datasets, the dropout mask is not used. We used the categorical cross-entropy loss function to train DANNs, where (Bn) is the minibatch size, ti is the correct class index, and pi is the class probability from the softmax layer:
We used minibatch stochastic gradient descent with Nesterov momentum to update the DANN parameters based on the loss function above (Sutskever, I., J. Martens, G. Dahl, and G. Hinton. 2013. Proceedings of the 30th International Conference on International Conference on Machine Learning). We initialized weights and biases with a Golrot initialization. We used the lasagna (Dieleman et al., 2015), nolearn (Nouri, 2014), and Theano (Theano Development Team, 2016 Preprint) python packages to construct DANNs.
DANN hyperparameter calibration
The following hyperparameters for DANNs were optimized using a cross-validated grid search over the parameter grid using only the training dataset: learning rate, layer size, dropout rate, and maximum epoch number. A detailed list of model hyperparameters and ranges used for evaluating hyperparameters (using only the training dataset with cross-validation) is given in Table S2.
Sensitivity mapping
Sensitivity mapping was originally developed for visualizing the sensitivities of image classification to input pixels from deep neural networks (Smilkov et al., 2017 Preprint). It is a gradient-based local sensitivity analysis technique that evaluates the relative importance of input variables with respect to output classes. Given an input sample x, the final layer of a neural network is traditionally a class activation function FK, which gives an associated score to each of the K classes. The final class is determined by taking the maximum of the class activation function. A very simplistic way of estimating the importance of each feature in a sample, known as the sensitivity map, Gk, is to differentiate the class activation function with respect to the input sample x:
Therefore, Gk indicates how sensitive the class activation function, for a specific class k, is to perturbations in the features of sample x and thus gives a metric of feature importance. This has shown to be a useful strategy for attempting to deconstruct the black box of deep learning techniques (Smilkov et al., 2017 Preprint). Herein, we use this simple raw-gradient approach to calculate the importance rankings of the input genes to our DANN. Contrary to image analysis, where each image is unique in the location, shape, morphology, etc. of the object to classify, there is no spatial information/diversity encoded in genomic datasets. Thus, an individual feature measurement at a position in the input data (e.g., feature d across n samples, x1,d, …, xn,d) is always the same feature (representing the same genomic measurement). Therefore, we aggregated sensitivity maps, calculated using only the training dataset across all samples of a specific predicted class k using the mean statistic.
BBNs
BBNs were used to assess the conditional dependence and probabilistic relationships between the most informative genes. Briefly, a BBN is a graphical model where nodes represent random variables and the directed edges represent conditional dependence between nodes (Koller and Friedman, 2009). BBNs are DAGs, given by G = (V, E), where V is the node set and E is the edge set, that satisfies the local directed Markov condition for G. We rely on a set of common assumptions to determine the causal structure: (1) causal sufficiency assumption, where there are no unobserved cofounders; (2) causal Markov assumption, where all d-separations in the graph (G) imply conditional independence in the observed probability distribution; and (3) causal faithfulness assumption, where all of the conditional independences in the observed probability distribution imply d-separations in the graph (G). We acknowledge that our data may not strictly meet all of these assumptions, however the generated BBNs provide useful biological hypothesis that could be experimentally validated.
We determined BBNs using the bnlearn R package with the score-based hill-climbing algorithm that heuristically searched the optimality space of all possible DAGs (Scutari, 2010). During the hill-climbing process, each candidate BBN was assessed with the Bayesian information criterion (BIC) score (Lam and Bacchus, 1994; Scutari, 2010):
where X1, …, Xv is the node set, d is the number of free parameters, n is the sample size of the dataset, and L is the likelihood. The penalty term was used to prevent overly complicated structures and overfitting. Each run of the hill-climbing algorithm returns a structure that maximizes the BIC score (including evaluating the directions of edges). A caveat is that these structures may be partially oriented graphs (i.e., situations where the directionality of some edges cannot be effectively determined). We use the cextend function from the bnlearn package to construct a DAG that is a consistent extension of X. As the hill-climbing algorithm can get stuck in local minima, we generated BBN consensus networks for each run by constructing 100 BBNs starting from different random network seeds. Any residual undirected edges contained in the consensus network were discarded (typically <<1% of edges). We assessed statistical significance of edges within the imposed consensus network by randomly permuting the dataset 10,000 times and constructing BBNs based on these scrambled datasets (thus providing an estimate of the null distribution). BBN edges with a false discovery rate of 5% (i.e., the edge occurred in ≥500 of the random BBNs) or greater were removed from the final network.
We generated BBNs for the top 100 and top 600 most informative genes as defined by sensitivity mapping. To determine the core causal structure, we added the genes TGFβ2, TGFβR1, and NOS3 to the top 100 most informative genes and determined the BBN. We performed permutation analysis by randomly selecting genes to replace TGFβ2 and determined the relative position within the predicted BBN (Westfall–Young permutation analysis). In addition, we tested known signaling ligands that were highly ranked by sensitivity analysis by replacing TGFβ2 with these ligands and determined their P values based on the above null distribution.
Western blot
Cells were lysed in radioimmunoprecipitation assay buffer (Boston BioProducts). Proteins were titrated using Bio-Rad Protein Assay Dye Reagent (Bio-Rad). 20 ng of proteins was loaded on a 4–12% acrylamide gel (Bio-Rad) and then transferred on a polyvinylidene fluoride membrane (Millipore). Primary antibodies used were Collagen-I, sm22 (Abcam), N-cadherin (Santa Cruz Biotechnology), VEGFR2, eNOS, ERK1/2 (Cell Signaling), and β-actin (Sigma-Aldrich).
Mouse models
Mapk3−/− mice (denominated ERK1−/−), Mapk1tm1Gela/J mice (denominated ERK2fl/fl), and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J mice (denominated mTmG) were purchased from the Jackson Laboratory. Cdh5CreERT2 mice were a generous gift from Ralf Adams (Max Planck Institute, Münster, Germany). All mice, including the WT mice, are Mus musculus on a pure C57Bl6 genetic background. Cardiac echography was performed under isoflurane anesthesia using PSLAX AM-Mode. Animals were housed and used in accordance with protocols and policies approved by the Yale Institutional Animal Care and Use Committee.
Blood collection, necropsy, and histopathology
Serial blood samples were collected by retro-orbital sampling with the mouse anesthetized by isoflurane. Plasma and urine samples were analyzed using BUN (plasma samples), creatine, and albumin (urine samples) assays. At necropsy, blood was collected as a terminal procedure from mice rendered unconscious by CO2 narcosis by cardiac puncture. Tissues were immersion-fixed in 10% neutral buffered formalin or Bouins fixative (Ricca Chemical), processed, sectioned and stained by hematoxylin and eosin (HE), periodic acid–Schiff (PAS), MT, and MSB by routine methods. Slides were examined blind to experimental manipulation and genotype and evaluated for pathological change by a veterinarian trained and experienced in mouse pathology. Digital light microscopic images were recorded with a Zeiss Axio Imager.A1 microscope, AxioCam MRc5 camera, and AxioVision 4.9.1 imaging software (Carl Zeiss Microimaging) and optimized using Adobe Photoshop CC 2015.1.2.
ELISA
TGFβ2 ELISA (R&D Systems) was performed on conditioned medium collected 4 d after transfection. Endothelin-1 ELISA (R&D Systems) and Angiotensin-II enzyme immunoassay (Sigma-Aldrich) were performed on mouse plasma collected by retro-orbital bleeding.
Ambulatory blood pressure transducer implantation
A blood pressure transducer (TA11-PA-C10; Data Sciences International) was surgically implanted under isoflurane anesthesia (1–3% in oxygen) into the carotid artery of mice using sterile surgical technique. Postoperative analgesics (Meloxicam) were provided for 48 h. The skin was closed with surgical staples and the mouse allowed to recover for ∼7 d. Surgical staples were removed, and the mice were transferred to a fresh cage (singly housed for data collection) and allowed to acclimate for several days before initiation of the study. All test substances were administered under brief isoflurane anesthesia. A 10-s segment was collected every minute for the duration of the experiment.
Immunofluorescent staining
Tissues were collected and fixed in paraformaldehyde 4%. Paraffin sections were treated with xylene, ethanol 100%, ethanol 90%, and ethanol 70% baths. Permeabilization was done in Triton 0.1%. Antigen retrieval was done in citrate buffer (ThermoFisher). Primary antibodies were Acta2 (Sigma-Aldrich), CD31 (R&D Systems), collagen-I (Abcam), eNOS (BD Biosciences), GFP (Abcam), laminin (Sigma-Aldrich), pSmad2 (Millipore), TGFβ (R&D Systems), VEGFR2 (Cell Signaling), and PV1 (Novus Biologicals). Pictures were taken using an SP5 confocal microscope (Leica).
Ultrastructure analysis
Dissected kidneys, lungs, and pancreas were fixed in 0.1 M cacodylate buffer, pH 7.5, containing 2% glutaraldehyde and 4% formaldehyde and postfixed in 1% OsO4. Fixed specimens were dehydrated and embedded in epoxy resin for sectioning. Ultrathin resin sections were stained with a solution containing uranyl acetate and lead acetate and viewed in FEI Tecnai G2 transmission electron microscope. Ultrastructural photomicrographs were evaluated independently for morphological changes by two investigators trained in ultrastructure pathology.
FACS
Tissues were collected and digested in a solution of collagenase and dispase (Roche) with DNase. The suspensions were then washed and filtered. RBCs were lysed using an RBC lysis buffer (Invitrogen). Cells were then fixed (paraformaldehyde 4%), permeabilized (Triton 0.5%), and stained (anti–VE-cadherin and anti-Sm22 were from Novus Biologicals). Cells were sorting on an LSR-II (BD Biosciences), and analysis was done using FlowJo software.
Cell isolation
Tissues were collected and digested in a solution of collagenase and dispase (Roche) with DNase. The suspensions were then washed and filtered. Endothelial cells were isolated using magnetic beads anti-rat IgG (Invitrogen) previously coated with rat anti-mouse CD31 antibody (BD Biosciences). After extensive washing, cells were lysed, and RNA was isolated using the PicoPure RNA isolation kit (ThermoFisher).
Statistical analyses
Statistical tests were performed using the software GraphPad Prism 7. For two-group comparisons, a Mann–Whitney U test was performed. For two-group and time course comparisons, a two-way ANOVA followed by Sidak’s multiple comparisons was used. Results were considered significant when the P value was <0.05.
Online supplemental material
Fig. S1 shows histology from Erk1−/− and Erk2iEC−/− mice. Fig. S2 shows the time course of TGFβ pathway activation in Erk1−/− Erk2iEC−/− mice. Fig. S3 shows additional staining demonstrating EndMT in Erk1−/− Erk2iEC−/− mice and presents the FACS strategy. Fig. S4 shows the regulation of the let-7 miRNA family by ERK1/2. Fig. S5 shows the knockdown efficiency of the siRNA against ERK1 and ERK2 and presents the bioinformatics strategy used to analyze RNA-seq data. Table S1 shows the systemic blood pressure data from Erk1−/− Erk2iEC−/− mice with statistical significance. Table S2 shows the differential analysis of RNA-seq from siSCR- and siERK1/2-treated HUVECs. Table S3 shows the DANN hyperparameter search ranges. Table S4 shows the sensitivity mapping ranges of the 983 input genes to the DANN.
Supplementary Material
Acknowledgments
We would like to thank Pengwei Yang and Zehua Chen for their assistance in RNA-seq data analysis, Kris Holton for the Gene Expression Omnibus submission, Rita Webber for mouse colony maintenance, and Nicole Mikush for cardiac echography. We would like to thank Gordon Terwilliger and Michael Schadt for help with necropsy and histology (HE, PAS, and MT) respectively (Mouse Research Pathology, Department of Comparative Medicine, Yale University School of Medicine, New Haven, CT).
We would like to thank Lonnette Diggs and the George M. O’Brien Kidney Center at Yale University School of Medicine (grant P30 DK079310). This work is supported in part by National Institutes of Health grants 1R01HL135582 (M. Simons), 1R01HL124120-01 (S.E. Quaggin), and 1P30DK114857-01A1 (S.E. Quaggin).
N.A. Cilfone, J.L. Baylon, J.R. Gulcher, and T.W. Chittenden were employed by WuXi NextCODE during the research project. The remaining authors declare no competing financial interests.
Author contributions: N. Ricard and M. Simons designed the experiments and oversaw the entire project. N. Ricard bred the mice, generated samples for RNA-seq, validated bioinformatics findings and performed the immunofluorescence and confocal microscopy, ECG studies, ELISA assays, macroscopic photography, qPCR, cell isolation from mice, FACS, Western blotting, retina dissection, and images. H. Velazquez realized the telemetric blood pressure measurement. C.J. Booth and N. Ricard mouse necropsy. C.J. Booth histopathologic analysis, characterization, and photomicroscopy. R.P. Scott and S.E. Quaggin performed MSB stain and electron microscopy. R.P. Scott, S.E. Quaggin, and C.J. Booth evaluated and characterized the TEM photomicrographs. N.A. Cilfone, J.L. Baylon, J.R. Gulcher, and T.W. Chittenden developed and applied causal statistical learning strategies. N. Ricard, N.A. Cilfone, R.P. Scott, S.E. Quaggin, T.W. Chittenden, C.J. Booth, and M. Simons wrote the manuscript. All authors discussed the results.
References
- Aird W.C. 2007. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 100:158–173. 10.1161/01.RES.0000255691.76142.4a [DOI] [PubMed] [Google Scholar]
- Alipanahi B., Delong A., Weirauch M.T., and Frey B.J.. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33:831–838. 10.1038/nbt.3300 [DOI] [PubMed] [Google Scholar]
- Armaly Z., Jadaon J.E., Jabbour A., and Abassi Z.A.. 2018. Preeclampsia: Novel Mechanisms and Potential Therapeutic Approaches. Front. Physiol. 9:973 10.3389/fphys.2018.00973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks M., Crowell K., Proctor A., and Jensen B.C.. 2017. Cardiovascular Effects of the MEK Inhibitor, Trametinib: A Case Report, Literature Review, and Consideration of Mechanism. Cardiovasc. Toxicol. 17:487–493. 10.1007/s12012-017-9425-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L., Palucka A.K., Arce E., Cantrell V., Borvak J., Banchereau J., and Pascual V.. 2003. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197:711–723. 10.1084/jem.20021553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F., Constantino L., Machado R., Petronilho F., and Dal-Pizzol F.. 2008. Plasma nitric oxide, endothelin-1, arginase and superoxide dismutase in pre-eclamptic women. J. Obstet. Gynaecol. Res. 34:957–963. [DOI] [PubMed] [Google Scholar]
- Buonato J.M., and Lazzara M.J.. 2014. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 74:309–319. 10.1158/0008-5472.CAN-12-4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M., and Roux P.P.. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75:50–83. 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Y., Qin L., Barnes C., Charisse K., Yi T., Zhang X., Ali R., Medina P.P., Yu J., Slack F.J., et al. . 2012. FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Reports. 2:1684–1696. 10.1016/j.celrep.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana E., Hirschi K.K., and Simons M.. 2017. The molecular basis of endothelial cell plasticity. Nat. Commun. 8:14361 10.1038/ncomms14361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus-Gonzalez N., Robinson E., Penchev R., von Mehren M., Heinrich M.C., Tap W., Wang Q., Demetri G., George S., and Humphreys B.D.. 2012. Regorafenib induces rapid and reversible changes in plasma nitric oxide and endothelin-1. Am. J. Hypertens. 25:1118–1123. 10.1038/ajh.2012.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Atri D., Eichmann A., and Simons M.. 2013a Endothelial ERK signaling controls lymphatic fate specification. J. Clin. Invest. 123:1202–1215. 10.1172/JCI63034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Larrivée B., Zhuang Z.W., Atri D., Moraes F., Prahst C., Eichmann A., and Simons M.. 2013b Endothelial RAF1/ERK activation regulates arterial morphogenesis. Blood. 121:3988–3996: S1–S9. 10.1182/blood-2012-12-474601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieleman S., Schlüter J., Raffel C., Olson E., Sønderby S.K., Nouri D., Maturana D., Thoma M., Battenberg E., Kelly J., et al. . 2015. Lasagne: First release. https://zenodo.org/record/27878
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina V., Sood M., Haigh J., Nagy A., Lajoie G., Ferrara N., Gerber H.P., Kikkawa Y., Miner J.H., and Quaggin S.E.. 2003. Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J. Clin. Invest. 111:707–716. 10.1172/JCI17423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina V., Baelde H.J., and Quaggin S.E.. 2007. Role of the VEGF--a signaling pathway in the glomerulus: evidence for crosstalk between components of the glomerular filtration barrier. Nephron, Physiol. 106:32–37. 10.1159/000101798 [DOI] [PubMed] [Google Scholar]
- Eremina V., Jefferson J.A., Kowalewska J., Hochster H., Haas M., Weisstuch J., Richardson C., Kopp J.B., Kabir M.G., Backx P.H., et al. . 2008. VEGF inhibition and renal thrombotic microangiopathy. N. Engl. J. Med. 358:1129–1136. 10.1056/NEJMoa0707330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facemire C.S., Nixon A.B., Griffiths R., Hurwitz H., and Coffman T.M.. 2009. Vascular endothelial growth factor receptor 2 controls blood pressure by regulating nitric oxide synthase expression. Hypertension. 54:652–658. 10.1161/HYPERTENSIONAHA.109.129973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J.E., Cantu Gutierrez M., Dang L.T., Khyzha N., Chen Z., Veitch S., Cheng H.S., Khor M., Antounians L., Njock M.S., et al. . 2017. Dynamic regulation of VEGF-inducible genes by an ERK/ERG/p300 transcriptional network. Development. 144:2428–2444. 10.1242/dev.146050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano N., Mori Y., Oh-hora M., Kosugi A., Fujikawa T., Nakai N., Niwa H., Miyazaki J., Hamaoka T., and Ogata M.. 2003. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells. 8:847–856. 10.1046/j.1365-2443.2003.00680.x [DOI] [PubMed] [Google Scholar]
- Je Y., Schutz F.A., and Choueiri T.K.. 2009. Risk of bleeding with vascular endothelial growth factor receptor tyrosine-kinase inhibitors sunitinib and sorafenib: a systematic review and meta-analysis of clinical trials. Lancet Oncol. 10:967–974. 10.1016/S1470-2045(09)70222-0 [DOI] [PubMed] [Google Scholar]
- Kappers M.H., van Esch J.H., Sluiter W., Sleijfer S., Danser A.H., and van den Meiracker A.H.. 2010. Hypertension induced by the tyrosine kinase inhibitor sunitinib is associated with increased circulating endothelin-1 levels. Hypertension. 56:675–681. 10.1161/HYPERTENSIONAHA.109.149690 [DOI] [PubMed] [Google Scholar]
- Kasahara Y., Tuder R.M., Taraseviciene-Stewart L., Le Cras T.D., Abman S., Hirth P.K., Waltenberger J., and Voelkel N.F.. 2000. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J. Clin. Invest. 106:1311–1319. 10.1172/JCI10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller D., and Friedman N.. 2009. Probabilistic graphical models: principles and techniques. MIT Press, Cambridge, MA. 1231 pp. [Google Scholar]
- Lam W., and Bacchus F.. 1994. Learning Bayesian Belief Networks: An Approach Based on The MMDL Principle. Comput. Intell. 10:269–293. 10.1111/j.1467-8640.1994.tb00166.x [DOI] [Google Scholar]
- Laumonnier Y., Nadaud S., Agrapart M., and Soubrier F.. 2000. Characterization of an upstream enhancer region in the promoter of the human endothelial nitric-oxide synthase gene. J. Biol. Chem. 275:40732–40741. 10.1074/jbc.M004696200 [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., and Hinton G.. 2015. Deep learning. Nature. 521:436–444. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- Lee S., Chen T.T., Barber C.L., Jordan M.C., Murdock J., Desai S., Ferrara N., Nagy A., Roos K.P., and Iruela-Arispe M.L.. 2007. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 130:691–703. 10.1016/j.cell.2007.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu M.A., Strand V., Simon L.S., Lipsky P.E., and Ramsey-Goldman R.. 2016. A critical review of clinical trials in systemic lupus erythematosus. Lupus. 25:1122–1140. 10.1177/0961203316652492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasciulo F.L., Montagnani M., and Potenza M.A.. 2006. Endothelin-1: the yin and yang on vascular function. Curr. Med. Chem. 13:1655–1665. 10.2174/092986706777441968 [DOI] [PubMed] [Google Scholar]
- Massagué J. 2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13:616–630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S.E., Min J.Y., Merchan J., Lim K.H., Li J., Mondal S., Libermann T.A., Morgan J.P., Sellke F.W., Stillman I.E., et al. . 2003. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Invest. 111:649–658. 10.1172/JCI17189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.M., Nikolic-Paterson D.J., and Lan H.Y.. 2016. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12:325–338. 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- Murakami M., Nguyen L.T., Zhuang Z.W., Moodie K.L., Carmeliet P., Stan R.V., and Simons M.. 2008. The FGF system has a key role in regulating vascular integrity. J. Clin. Invest. 118:3355–3366. 10.1172/JCI35298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Nguyen L.T., Hatanaka K., Schachterle W., Chen P.Y., Zhuang Z.W., Black B.L., and Simons M.. 2011. FGF-dependent regulation of VEGF receptor 2 expression in mice. J. Clin. Invest. 121:2668–2678. 10.1172/JCI44762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri D.2014. nolearn: scikit-learn compatible neural network library. https://github.com/dnouri/nolearn (accessed January 12, 2018).
- Nova A., Sibai B.M., Barton J.R., Mercer B.M., and Mitchell M.D.. 1991. Maternal plasma level of endothelin is increased in preeclampsia. Am. J. Obstet. Gynecol. 165:724–727. 10.1016/0002-9378(91)90317-K [DOI] [PubMed] [Google Scholar]
- Pagès G., Guérin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., and Pouysségur J.. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 286:1374–1377. 10.1126/science.286.5443.1374 [DOI] [PubMed] [Google Scholar]
- Patten I.S., Rana S., Shahul S., Rowe G.C., Jang C., Liu L., Hacker M.R., Rhee J.S., Mitchell J., Mahmood F., et al. . 2012. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature. 485:333–338. 10.1038/nature11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps E., Prasanna D., Brima W., and Jim B.. 2016. Preeclampsia: Updates in Pathogenesis, Definitions, and Guidelines. Clin. J. Am. Soc. Nephrol. 11:1102–1113. 10.2215/CJN.12081115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta C., Cosmai L., Gallieni M., Pedrazzoli P., and Malberti F.. 2015. Renal effects of targeted anticancer therapies. Nat. Rev. Nephrol. 11:354–370. 10.1038/nrneph.2015.15 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostama B., Turner J.E., Seavey G.T., Norton C.R., Gridley T., Vary C.P., and Liaw L.. 2015. DLL4/Notch1 and BMP9 Interdependent Signaling Induces Human Endothelial Cell Quiescence via P27KIP1 and Thrombospondin-1. Arterioscler. Thromb. Vasc. Biol. 35:2626–2637. 10.1161/ATVBAHA.115.306541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin K.M. 2017. MAPK Pathway-Targeted Therapies: Care and Management of Unique Toxicities in Patients With Advanced Melanoma. Clin. J. Oncol. Nurs. 21:699–709. 10.1188/17.CJON.699-709 [DOI] [PubMed] [Google Scholar]
- Samuels I.S., Karlo J.C., Faruzzi A.N., Pickering K., Herrup K., Sweatt J.D., Saitta S.C., and Landreth G.E.. 2008. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J. Neurosci. 28:6983–6995. 10.1523/JNEUROSCI.0679-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura M., Zaragoza C., Cao W., Bao C., Rodríguez-Puyol M., Rodríguez-Puyol D., and Lowenstein C.J.. 2002. Smad2 mediates transforming growth factor-beta induction of endothelial nitric oxide synthase expression. Circ. Res. 91:806–813. 10.1161/01.RES.0000040397.23817.E5 [DOI] [PubMed] [Google Scholar]
- Schwartz M.A., Vestweber D., and Simons M.. 2018. A unifying concept in vascular health and disease. Science. 360:270–271. 10.1126/science.aat3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutari M. 2010. Learning Bayesian Networks with the bnlearn R Package. J. Stat. Softw. 35:1–22. 10.18637/jss.v035.i0321603108 [DOI] [Google Scholar]
- Selcher J.C., Nekrasova T., Paylor R., Landreth G.E., and Sweatt J.D.. 2001. Mice lacking the ERK1 isoform of MAP kinase are unimpaired in emotional learning. Learn. Mem. 8:11–19. 10.1101/lm.37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaarawy M., El Meleigy M., and Rasheed K.. 2001. Maternal serum transforming growth factor beta-2 in preeclampsia and eclampsia, a potential biomarker for the assessment of disease severity and fetal outcome. J. Soc. Gynecol. Investig. 8:27–31. 10.1177/107155760100800105 [DOI] [PubMed] [Google Scholar]
- Shibuya M. 2013. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J. Biochem. 153:13–19. 10.1093/jb/mvs136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M., Beane T.J., Quillien A., Male I., Zhu L.J., and Lawson N.D.. 2016a Vegfa signals through ERK to promote angiogenesis, but not artery differentiation. Development. 143:3796–3805. 10.1242/dev.137919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M., Male I., Beane T.J., Villefranc J.A., Kok F.O., Zhu L.J., and Lawson N.D.. 2016b Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development. 143:3785–3795. 10.1242/dev.137901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Gordon E., and Claesson-Welsh L.. 2016. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17:611–625. 10.1038/nrm.2016.87 [DOI] [PubMed] [Google Scholar]
- Smilkov D., Thorat N., Kim B., Viégas F., and Wattenberg M.. 2017. SmoothGrad: removing noise by adding noise. arXiv. https://arxiv.org/abs/1706.03825 (Preprint posted June 12, 2017).
- Sörensen I., Adams R.H., and Gossler A.. 2009. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 113:5680–5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Zabuawala T., Huang H., Zhang J., Gulati P., Fernandez S., Karlo J.C., Landreth G.E., Leone G., and Ostrowski M.C.. 2009. Erk1 and Erk2 regulate endothelial cell proliferation and migration during mouse embryonic angiogenesis. PLoS One. 4:e8283 10.1371/journal.pone.0008283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., and Salakhutdinov R.. 2014. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 15:1929–1958. [Google Scholar]
- Stan R.V., Tse D., Deharvengt S.J., Smits N.C., Xu Y., Luciano M.R., McGarry C.L., Buitendijk M., Nemani K.V., Elgueta R., et al. . 2012. The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev. Cell. 23:1203–1218. 10.1016/j.devcel.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.J., Thuault S., Caja L., Carletti T., Heldin C.H., and Moustakas A.. 2012. Regulation of transcription factor Twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J. Biol. Chem. 287:7134–7145. 10.1074/jbc.M111.291385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theano Development Team 2016. Theano: A Python framework for fast computation of mathematical expressions. arXiv. https://arxiv.org/abs/1605.02688 (Preprint posted May 9, 2016).
- Thuault S., Tan E.J., Peinado H., Cano A., Heldin C.H., and Moustakas A.. 2008. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 283:33437–33446. 10.1074/jbc.M802016200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touyz R.M., and Herrmann J.. 2018. Cardiotoxicity with vascular endothelial growth factor inhibitor therapy. NPJ Precis Oncol. 2:13 10.1038/s41698-018-0056-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touyz R.M., Herrmann S.M.S., and Herrmann J.. 2018. Vascular toxicities with VEGF inhibitor therapies-focus on hypertension and arterial thrombotic events. J. Am. Soc. Hypertens. 12:409–425. 10.1016/j.jash.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanchoo R., Devoe C., and Jhaveri K.D.. 2016a BRAF inhibitors - do we need to worry about kidney injury? Expert Opin. Drug Saf. 15:579–581. 10.1517/14740338.2016.1164139 [DOI] [PubMed] [Google Scholar]
- Wanchoo R., Jhaveri K.D., Deray G., and Launay-Vacher V.. 2016b Renal effects of BRAF inhibitors: a systematic review by the Cancer and the Kidney International Network. Clin. Kidney J. 9:245–251. 10.1093/ckj/sfv149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh S.J., and Corrie P.G.. 2015. Management of BRAF and MEK inhibitor toxicities in patients with metastatic melanoma. Ther. Adv. Med. Oncol. 7:122–136. 10.1177/1758834014566428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Law B.K., Chytil A.M., Brown K.A., Aakre M.E., and Moses H.L.. 2004. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 6:603–610. 10.1593/neo.04241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Liu X., Jiang Y., Chu L., Hao H., Liua Z., Verfaillie C., Zweier J., Gupta K., and Liu Z.. 2008. MAPK/ERK signalling mediates VEGF-induced bone marrow stem cell differentiation into endothelial cell. J. Cell. Mol. Med. 12(6A, 6A):2395–2406. 10.1111/j.1582-4934.2008.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang Y., Hosaka K., Huang G., Zang J., Chen F., Zhang Y., Samani N.J., and Cao Y.. 2016. Endocrine vasculatures are preferable targets of an antitumor ineffective low dose of anti-VEGF therapy. Proc. Natl. Acad. Sci. USA. 113:4158–4163. 10.1073/pnas.1601649113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.