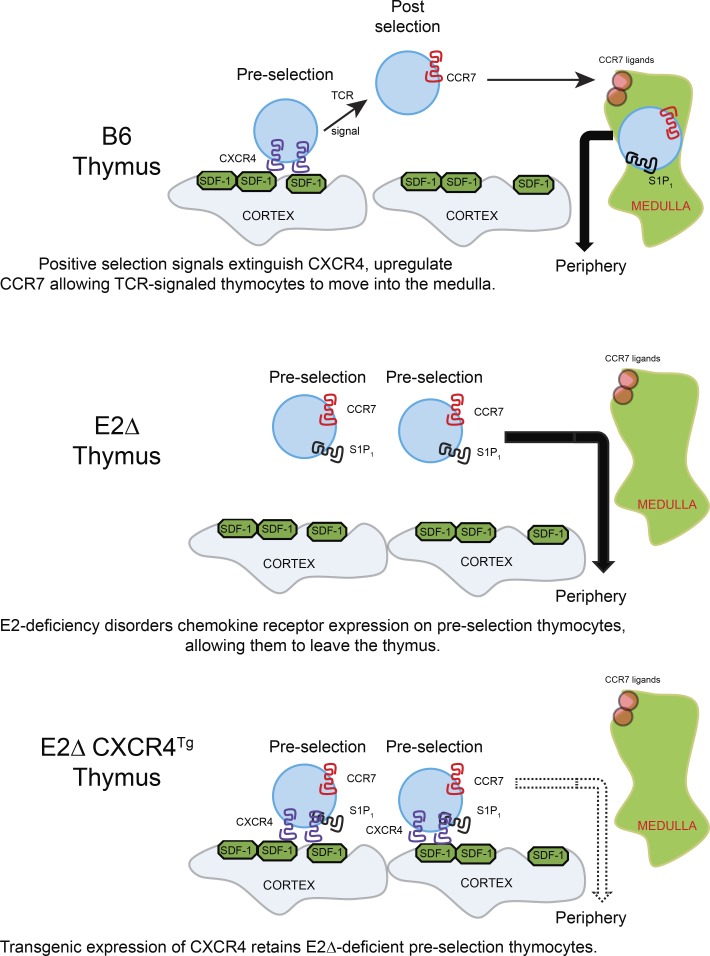
The E-protein transcription factors E2A and HEB regulate thymocyte expression of the chemokine receptor CXCR4 to retain preselection thymocytes in the thymic cortex. TCR-mediated positive selection signals extinguish CXCR4 expression to allow positively selected thymocytes to migrate from the cortex into the thymic medulla.
Abstract
Preselection thymocytes are normally retained in the thymic cortex, but the mechanisms responsible remain incompletely understood. We now report that deletion of genes encoding the E-protein transcription factors E2A and HEB disorders chemokine receptor expression on developing thymocytes to allow escape of preselection TCR−CD8+ thymocytes into the periphery. We document that CXCR4 expression normally anchors preselection thymocytes to the thymic cortex via interaction with its ligand CXCL12 on cortical thymic epithelial cells, and that disruption of CXCR4–CXCL12 engagements release preselection thymocytes from the thymic cortex. We further document that CXCR4 expression must be extinguished by TCR-mediated positive selection signals to allow migration of TCR-signaled thymocytes out of the thymic cortex into the medulla. Thus, E-protein transcription factors regulate the ordered expression pattern of chemokine receptors on developing thymocytes, and the interaction of the chemokine receptor CXCR4 with its ligand adheres TCR-unsignaled preselection thymocytes to the thymic cortex.
Graphical Abstract
Introduction
T cell development is closely coordinated with chemokine receptor–mediated thymocyte migration in the thymus (Petrie and Zúñiga-Pflücker, 2007). Hematopoietic precursor cells from the bone marrow enter the adult thymus via blood vessels in the corticomedullary junction and appear as CD4−CD8− (double-negative [DN]) cells in the thymic cortex (Takahama, 2006; Love and Bhandoola, 2011). Their migration through the cortex is dependent on the chemokine receptor CXCR4, which is expressed on developing DN and double-positive (DP) thymocytes that have not received TCR signals (Suzuki et al., 1999; Plotkin et al., 2003; Petrie and Zúñiga-Pflücker, 2007; Trampont et al., 2010). DP thymocytes that have been signaled by their TCRs to undergo positive selection and to differentiate into mature single-positive (SP) thymocytes extinguish CXCR4 and up-regulate CCR7 expression, which attracts them into the thymic medulla (Campbell et al., 1999; Ueno et al., 2004; Kurobe et al., 2006; Halkias et al., 2013; Lucas et al., 2017). SP thymocytes exit the thymic medulla when they express the chemokine receptor S1P1, which directs their migration into the blood vessels in the corticomedullary junction, taking them into the periphery (Matloubian et al., 2004; McCaughtry et al., 2007).
These carefully choreographed developmental and migratory events in the thymus require thymocyte expression of E2A and HEB, which are members of the E-protein family of basic helix-loop-helix transcription factors (Engel and Murre, 2001; de Pooter and Kee, 2010; Belle and Zhuang, 2014). Thymocyte deletion of Tcf3 and Tcf12, the genes encoding E2A and HEB, dysregulates thymocyte development and results in peripheral T cells that do not express surface TCR complexes (Jones and Zhuang, 2007). As an explanation for the appearance of TCR− peripheral T cells, it has been suggested that E-proteins enforce the TCR signaling requirement for positive selection so that thymocytes lacking E-proteins do not need to be signaled by their TCR to undergo positive selection and leave the thymus (Jones and Zhuang, 2007). Consistent with this perspective, E-protein–deficient TCR− thymocytes that never received TCR signaling nonetheless resembled normal TCR-signaled positively selected thymocytes in their chemokine receptor expression pattern and appeared as CXCR4−CCR7+S1P1+ cells. However, an alternative explanation for TCR− peripheral T cells in E-protein–deficient mice is that they are preselection (not postselection) thymocytes that escaped from the thymus because of their aberrant chemokine receptor expression.
We undertook the present study to specifically determine if E-protein–regulated chemokine receptors were responsible for retaining TCR-unsignaled preselection thymocytes in the thymic cortex. We now report that TCR− peripheral T cells in E-protein–deficient mice are derived from preselection thymocytes that could not be retained in the thymic cortex because of absent CXCR4 expression, and we demonstrate that restoration of CXCR4 expression prevents their appearance in the periphery. Moreover, we document in normal wild-type mice that preselection cells are retained in the thymic cortex by interactions between CXCR4 and its chemokine ligand CXCL12 displayed on cortical thymic epithelial cells (TECs), and that extinction of CXCR4 expression by TCR-mediated positive selection signaling is necessary for TCR-signaled cells to migrate out of the thymic cortex. Thus, CXCR4 adheres preselection thymocytes to the thymic cortex and must be extinguished during positive selection for TCR-signaled thymocytes to freely migrate out of the thymic cortex into the thymic medulla.
Results
E-protein transcription factors regulate thymic retention of preselection thymocytes
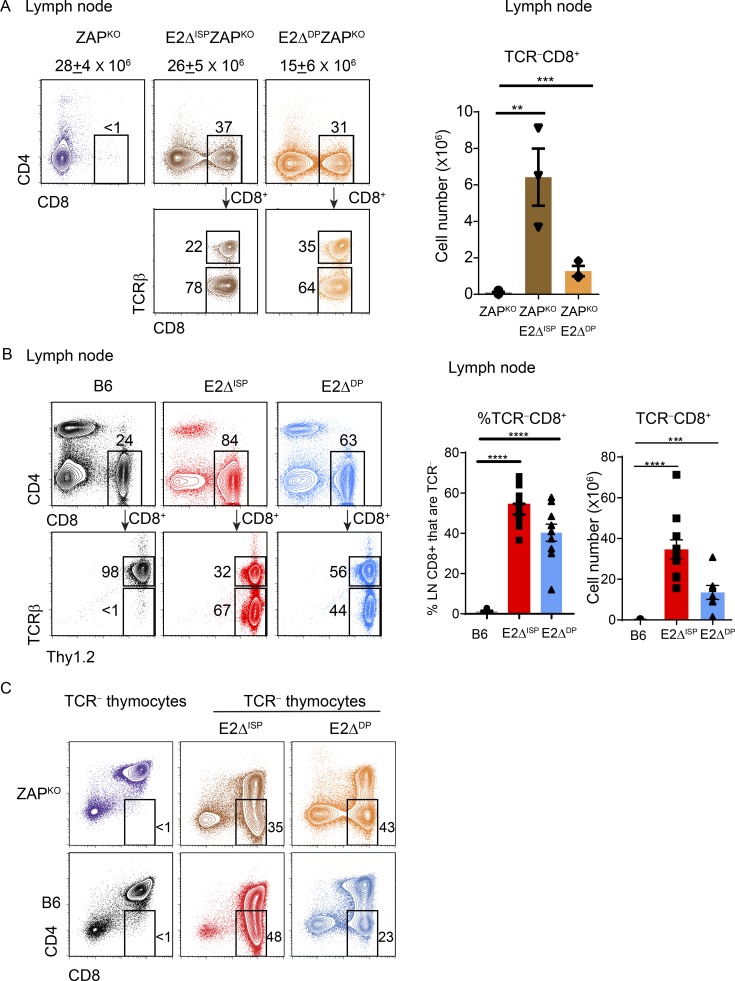
Developing thymocytes acquire the ability to leave the thymus only after receiving TCR-mediated positive selection signals transduced by the protein tyrosine kinase ZAP70 and differentiating into mature T cells (Kadlecek et al., 1998). Consequently, preselection thymocytes that have not received TCR signals are retained in the thymic cortex, but the mechanisms that mediate their retention remain incompletely understood. We began assessing a possible role for E-protein transcription factors in thymic retention of preselection thymocytes by conditionally deleting Tcf3 and Tcf12 genes, which encode E2A and HEB proteins, respectively. Conditional deletion of floxed Tcf3 and Tcf12 genes was accomplished with either CD4-Cre recombinase, which deletes at the late DN/immature SP (ISP) thymocyte stage (E2ΔISP; Lee et al., 2001; Gegonne et al., 2018), or E8III-Cre recombinase, which deletes at the DP thymocyte stage (E2ΔDP; Park et al., 2010). We found that conditional deletion of E-proteins in germline ZAPKO mice caused both TCR−CD8+ and TCR+CD8+ T cells to appear in the periphery even though their precursors could not possibly have been TCR-signaled in the thymus because of absent ZAP70 (Fig. 1 A). We also observed that conditional deletion of E-proteins in ZAP70-sufficient E2ΔISP and E2ΔDP mice resulted in the appearance of peripheral TCR−CD8+ T cells whose thymocyte precursors could not possibly have been TCR-signaled because of absent TCR expression (Figs. 1 B and S1, A and B). Thus, thymic retention of TCR-unsignaled thymocytes requires E-protein transcription factors.
Figure 1.
E2A/HEB double deficiency results in the appearance of TCR− T cells in the periphery. (A) Two-color CD4/CD8 flow cytometry plots of LN cells from the indicated mice. Numbers above histograms indicate total LN numbers. TCRβ staining on gated CD8+ T cells is shown in bottom panels. Boxes within the plots indicate the frequency of cells within that box. Cell numbers of TCRβ−CD8+ LN cells from multiple mice are displayed in the bar graph. (B) LN staining profiles from the indicated mice, with TCRβ versus Thy1.2 staining of gated CD8+ LN cells shown in bottom panels. Bar graphs indicate frequency and numbers of TCRβ−CD8+ LN cells from multiple mice. (C) CD4/CD8 profiles of TCRβ− thymocytes from indicated mouse strains. Mean ± SEM are shown. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 determined by Student’s t test. Data are from 3–12 experiments with 3–6 mice per group (A; mean ± SEM), 10–12 mice per group (B; mean ± SEM), or 3–7 mice per group (C).
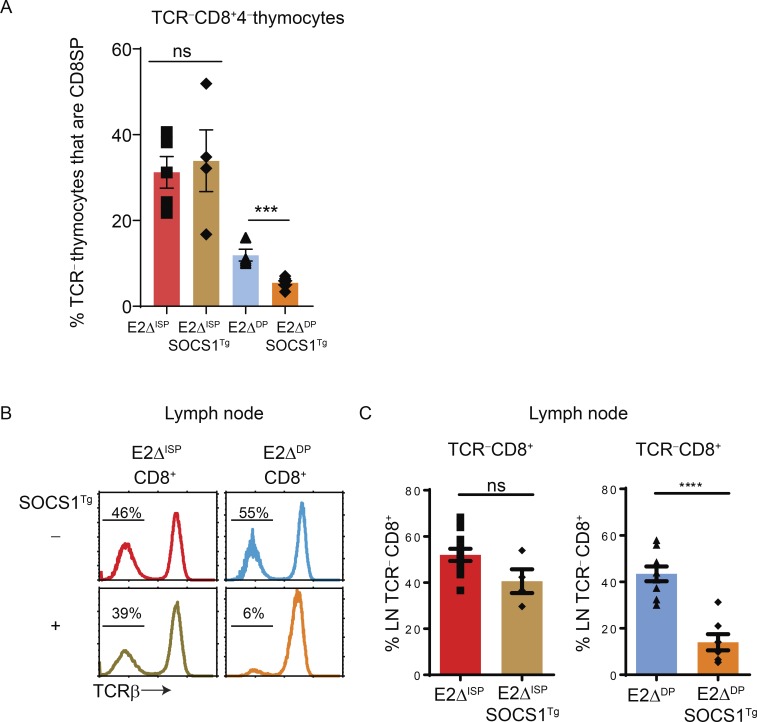
Assessment of thymocyte profiles revealed that TCR−CD8+ thymocytes were markedly increased in both E2ΔISP and E2ΔDP mice compared with E-sufficient mice (both ZAPKO and B6; Fig. 1 C). Because the Cre-recombinase was expressed in DN/ISP thymocytes in E2ΔISP mice (Lee et al., 2001; Gegonne et al., 2018) and was expressed in DP thymocytes in E2ΔDP mice (Park et al., 2010), we considered that the TCR-unsignaled TCR−CD8+ thymocyte populations in these mice might be at different stages of thymocyte development, i.e., before and after DP. If so, generation of TCR−CD8+ thymocytes in E2ΔISP and E2ΔDP mice would have different cytokine requirements, since in vivo cytokine signals induce the Runx-family transcription factor Runx3d, which up-regulates CD8 expression only on post-DP thymocytes (Park et al., 2010; McCaughtry et al., 2012; Etzensperger et al., 2017), but cytokine signals are not required to induce the Runx-family transcription factor Runx1, which up-regulates CD8 expression on pre-DP thymocytes (Egawa et al., 2007; Etzensperger et al., 2017). Consequently, TCR−CD8+ thymocytes in E2ΔISP, which contain intracellular TCRβ (Jones and Zhuang, 2007; Park et al., 2010), might be conventional TCR− ISP cells that are cytokine independent, whereas TCR−CD8+ thymocytes in E2ΔDP mice might be derived from unconventional DP thymocytes that had been signaled by in vivo cytokines, because E-protein deficiency caused DP thymocytes to express IL-7R and quench expression of suppressor of cytokine signaling-1 (SOCS1; Jones and Zhuang, 2007; Park et al., 2010; Fig. S2 A).
To experimentally determine cytokine dependence and cytokine independence, we introduced a SOCS1 transgene (Tg) whose early in vivo expression is driven by the proximal Lck promoter (SOCS1Tg) and inhibits in vivo Jak-Stat–mediated cytokine signaling (Hanada et al., 2001). We observed that the SOCS1Tg did not inhibit generation of TCR−CD8+ cells in the thymus or appearance in the periphery of E2ΔISP mice (Fig. 2, A–C), whereas the SOCS1Tg did significantly inhibit generation of TCR−CD8+ cells in E2ΔDP mice (Fig. 2, A–C). These results are concordant with the concept that TCR−CD8+ cells in E2ΔISP mice are mainly derived from ISP (pre-DP) thymocytes, and that TCR−CD8+ cells in E2ΔDP are derived from cytokine-signaled DP thymocytes. Notably, whereas the SOCS1Tg did not inhibit TCR−CD8+ thymocyte generation in E2ΔISP mice, the SOCS1Tg was functional in E2ΔISP mice, as it significantly inhibited generation in both E2ΔISP and E2ΔDP mice of TCR+CD8+ thymocytes whose surface TCR expression reveals that they are derived from post-DP cells (Fig. S2 B). Thus, we conclude that TCR−CD8+ cells in E2ΔISP mice are mainly derived from pre-DP ISP cells, whereas TCR−CD8+ cells in E2ΔDP mice are derived from cytokine-signaled DP thymocytes, allowing us to assess the impact of E-protein deletion on preselection thymocytes at two different stages (pre-DP and DP) of thymocyte development.
Figure 2.
Distinct cytokine signaling requirements for generation of TCR−CD8+ cells in the thymus and appearance in the periphery of E2ΔISP and E2ΔDP mice. (A) Impact of SOCS1Tg expression on frequencies of TCRβ−CD8+CD4− thymocytes in multiple E2ΔISP and E2ΔDP mice displayed as a bar graph. (B) TCRβ expression on peripheral CD8+ LN cells from E2ΔISP and E2ΔDP mice. Numbers in the histograms indicate frequency of TCRβ−CD8+ T cells. (C) Impact of SOCS1Tg expression on frequency of TCRβ−CD8+ peripheral LN cells in E2ΔISP and E2ΔDP mice displayed as a bar graph. Mean ± SEM are shown. ***, P < 0.001; ****, P < 0.0001; ns, not significant (P > 0.05) determined by Student’s t test. Data are from 3–12 experiments with 4–7 mice per group (A; mean ± SEM) or 4–10 mice per group (B and C; mean ± SEM).
E-proteins coordinate chemokine receptor expression on developing thymocytes
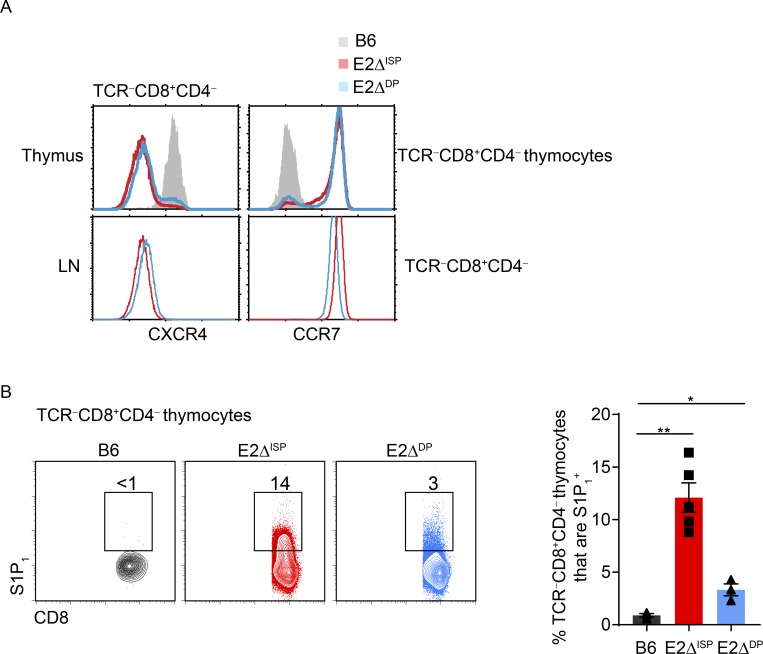
To understand why E-protein deficiency caused TCR-unsignaled thymocytes to escape into the periphery, we examined the impact of E-deficiency on chemokine receptor expression in developing thymocytes (Takahama, 2006; Petrie and Zúñiga-Pflücker, 2007; Bunting et al., 2011; Lancaster et al., 2018). During normal thymocyte development, TCR-unsignaled thymocytes express the chemokine receptor CXCR4 and remain in the thymic cortex until they are TCR-signaled to undergo positive selection (Suzuki et al., 1998, 1999; Lucas et al., 2017). TCR-signaled DP thymocytes then extinguish CXCR4 expression both in vitro and in vivo (Fig. S3, A–C) and up-regulate expression of the chemokine receptor CCR7 (Fig. S3 C), which attracts them to the thymic medulla (Ngo et al., 1998; Ueno et al., 2004). In the medulla, thymocytes then up-regulate expression of the chemokine receptor S1P1 (Matloubian et al., 2004), which promotes migration into the periphery. In accord with this normal sequence of chemokine receptor expression, TCR-unsignaled TCR−CD8+ thymocytes in normal B6 mice are CXCR4+CCR7−S1P1− (Fig. 3, A and B). However, in E2ΔISP and E2ΔDP mice, TCR-unsignaled TCR−CD8+ thymocytes display an aberrant chemokine receptor expression pattern that is CXCR4−CCR7+S1P1+, which resembles peripheral LN T cells (Fig. 3, A and B), suggesting that E-proteins directly or indirectly regulate chemokine receptor expression on developing thymocytes. In this regard, chromatin immunoprecipitation (ChIP) analysis revealed that E2A binds directly to the CXCR4 promoter in DP thymocytes (Fig. S3 D). In contrast, E-protein regulation of CCR7 appears to be indirect but necessary to enforce the TCR signaling requirement for CCR7 expression (Fig. S3 E). Consequently, we suspected that the escape of TCR-unsignaled TCR−CD8+ cells from the thymus into the periphery of E2ΔISP and E2ΔDP mice might be due to their aberrant CXCR4−CCR7+S1P1+ chemokine receptor expression.
Figure 3.
E2A/HEB deficiency results in altered chemokine receptor expression on thymocytes. (A) CXCR4 and CCR7 expression gated on TCRβ−CD8+CD4− thymocytes (top) and peripheral TCRβ−CD8+CD4− LN cells (bottom) from B6, E2ΔISP, and E2ΔDP mice. (B) S1P1 versus CD8 staining of TCRβ−CD8+CD4− thymocytes from the indicated mice. Right panel shows frequencies of TCRβ−CD8+CD4− thymocytes that express S1P1 in the indicated mouse strains displayed as a bar graph. Mean ± SEM are shown. *, P < 0.05; **, P < 0.01 determined by Student’s t test. Data are from three to four experiments with three to five mice per group (mean ± SEM).
We first asked if absent CXCR4 expression on E-deficient thymocytes contributed to escape of TCR-unsignaled thymocytes into the periphery. To do so, we constructed an hCD2-driven CXCR4 Tg (CXCR4Tg) to reconstitute CXCR4 expression on TCR−CD8+ thymocytes in E-deficient mice (Figs. 4 A and S4 A). In fact CXCR4Tg expression markedly reduced the number of TCR−CD8+ cells escaping into the periphery of E2ΔISP and E2ΔDP mice (Figs. 4 B and S4 B), which demonstrated that restoration of CXCR4 expression prevented the escape of TCR-unsignaled E-deficient thymocytes from the thymus, identifying a contribution of CXCR4 to thymic retention. Because the CXCR4Tg was also expressed on TCR-signaled thymocytes (Fig. S3 C), CXCR4Tg expression also markedly reduced the number of TCR+CD8+ T cells escaping into the periphery of E2ΔISP and E2ΔDP mice (Fig. S4 C).
Figure 4.
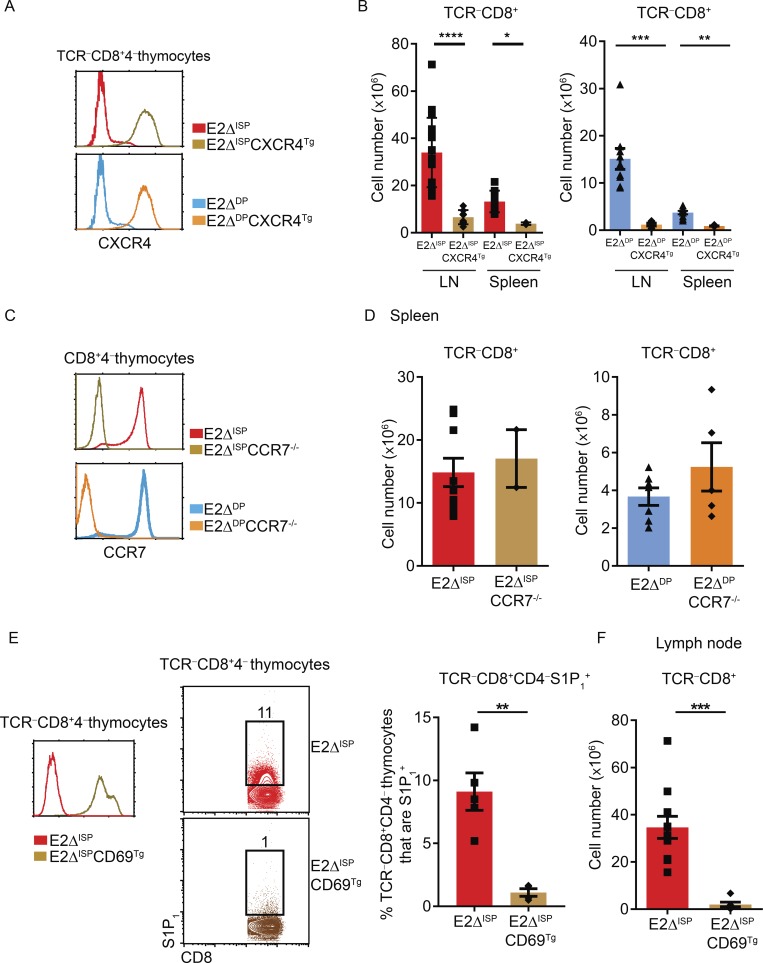
Chemokine receptors responsible for escape of TCR−CD8+ E-deficient thymocytes into the periphery. (A) Profiles of CXCR4 expression on TCRβ−CD8+CD4− thymocytes from E2ΔISP and E2ΔDP mice in the presence and absence of CXCR4Tg. (B) Cell numbers of TCRβ−CD8+ cells in the indicated mouse strains displayed as a bar graph. (C) Profiles of CCR7 expression on CD8+CD4− thymocytes from E2ΔISP and E2ΔDP mice with and without CCR7 deficiency. (D) Cell numbers of TCRβ−CD8+ T cells in the indicated mouse strains displayed as a bar graph. (E) Expression of CD69 (left) and S1P1 (middle) on TCRβ−CD8+CD4− thymocytes from E2ΔISP and E2ΔISPCD69Tg mice. Frequencies of S1P1+ cells among TCRβ−CD8+CD4− thymocytes in E2ΔISP and E2ΔISPCD69Tg mice displayed as a bar graph (right). (F) Numbers of TCRβ−CD8+ LN cells in E2ΔISP and E2ΔISPCD69Tg mice. Mean ± SEM are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 determined by Student’s t test. Data are from ≤12 experiments with 2–16 mice per genotype (A and B; mean ± SEM), 2–9 mice per genotype (C and D; mean + SEM), or 4–11 mice per genotype (E and F).
We next asked if aberrant induction of CCR7 expression on TCR-unsignaled E-deficient thymocytes contributed to their escape into the periphery. We did so by making E2ΔISP and E2ΔDP mice genetically CCR7-deficient by crossing them with CCR7KO mice (Fig. 4 C). However, absent CCR7 expression did not significantly diminish peripheral TCR−CD8+ cell numbers (Figs. 4 D and S4 D), indicating that aberrant CCR7 expression did not contribute to the escape of TCR-unsignaled thymocytes in E-deficient mice.
Finally, S1P1 is a chemokine receptor known to promote thymic egress (Matloubian et al., 2004) and is aberrantly expressed on TCR-unsignaled E-deficient thymocytes. Because S1P1 surface expression is antagonized by CD69 expression (Feng et al., 2002; Bankovich et al., 2010), we introduced a CD69 Tg to eliminate aberrant S1P1 surface expression on TCR-unsignaled thymocytes in E2ΔISP mice, which did not affect their aberrant expression of other chemokine receptors (Figs. 4 E and S4 E). Indeed, the CD69Tg eliminated appearance of TCR−CD8+ cells in the periphery of E2ΔISP mice (Fig. 4, E and F), indicating that escape of TCR-unsignaled E-deficient thymocytes into the periphery required S1P1 expression.
Together, these results identify absent CXCR4 expression and aberrant S1P1 expression as both contributing to the escape of TCR-unsignaled E-deficient thymocytes into the periphery, indicating that E-proteins are required to properly coordinate chemokine receptor expression during thymocyte development. In addition these results identify a contribution of CXCR4 to retention of TCR-unsignaled cells in the thymus.
CXCR4–CXCL12 adheres preselection thymocytes to cortical thymic epithelium
We were intrigued to have found that CXCR4 contributes to thymic retention of TCR-unsignaled cells and further examined this finding in E-protein–sufficient wild-type mice. Specifically, we examined if CXCR4-mediated thymic retention was the result of its interaction with its chemokine ligand CXCL12 (also known as SDF-1) that is secreted and bound to the surface of cortical TECs (Bleul et al., 1996; Zlotnik and Yoshie, 2000; Lucas et al., 2017). To experimentally assess the consequences of interrupting CXCR4–CXCL12 interactions specifically in the thymus, we used Foxn1-Cre (Rossi et al., 2007) to conditionally delete Cxcl12 in TECs. We refer to such mice as SDFΔTEC and observed that intrathymic deletion of CXCL12 increased thymocyte surface expression of its receptor, CXCR4, as a result of absent receptor–ligand interactions (Fig. 5 A). However, releasing TCR-unsignaled thymocytes from cortical epithelium was not itself sufficient for thymocytes to leave the thymic cortex, as TCR-unsignaled DP (RORγt+CD4+CD8+) thymocytes were still absent from the thymic medulla of SDFΔTEC mice (Fig. S4 F), as was previously observed in CXCR4-deficient mice (Lucas et al., 2017). Consequently, to determine if CXCL12 deletion actually released TCR-unsignaled thymocytes from cortical epithelium, we introduced an S1P1 Tg (S1P1Tg; Chi and Flavell, 2005; Fig. S4 G) that would permit released thymocytes to escape and appear in the periphery.
Figure 5.
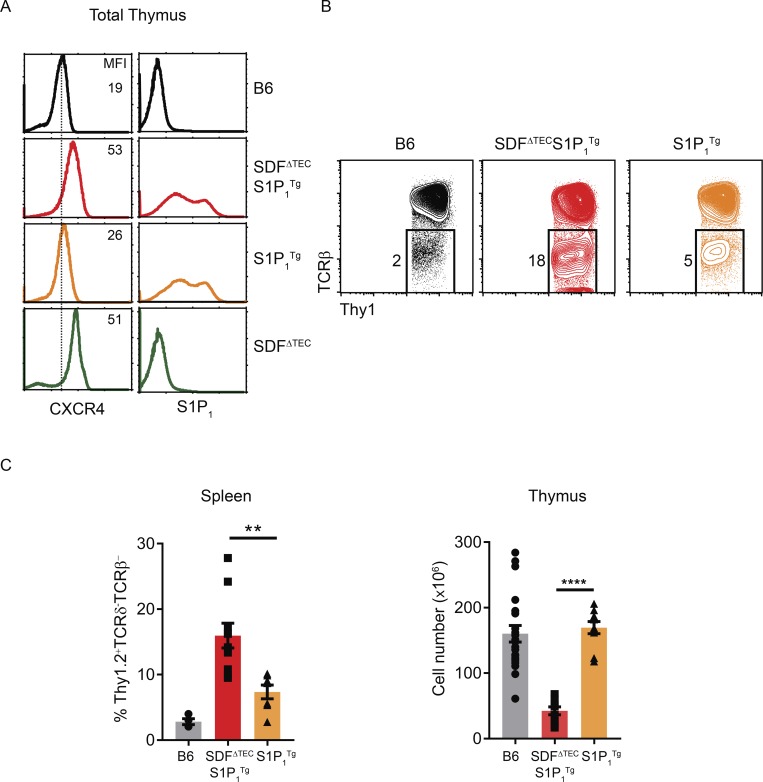
CXCR4 interaction with CXCL12 (SDF-1) prevents preselection thymocytes from leaving the thymus. (A) Thymocyte profiles of CXCR4 and S1P1 expression from the indicated mouse strains. Numbers within the profiles indicate the mean fluorescence intensity (MFI) of CXCR4 staining. (B) Flow cytometry profiles of TCRβ versus Thy1.2 expression on Thy1.2+TCRγδ− spleen cells from B6, SDF1ΔTECS1P1Tg, and S1P1Tg mice. (C) Bar graphs displaying frequencies of Thy1.2+TCRγδ−TCRβ− spleen cells and total thymus cellularity in the indicated mouse strains. Mean ± SEM are shown. **, P < 0.01; ****, P < 0.0001 determined by Student’s t test. Data are from 2–4 experiments with 4–10 mice per genotype.
The S1P1Tg was sufficient by itself to cause some TCR-unsignaled TCR−Thy1.2+ T cells to appear in the periphery (Fig. 5 B). Importantly, however, compared with S1P1Tg mice with intact CXCL12 in the thymus, SDFΔTECS1P1Tg mice with deletion of CXCL12 in the thymus had significantly more TCR-unsignaled TCR−Thy1.2+ cells in the periphery despite significantly fewer cells remaining in the thymus (Fig. 5, B and C). In these mice, peripheral TCR−Thy1.2+ cells consisted of both TCR−CD8+ and TCR−DP cells (Fig. S4 H). We conclude that CXCR4–CXCL12 interactions adhere TCR-unsignaled thymocytes to the thymic cortical epithelium.
TCR positive selection signals extinguish CXCR4 expression
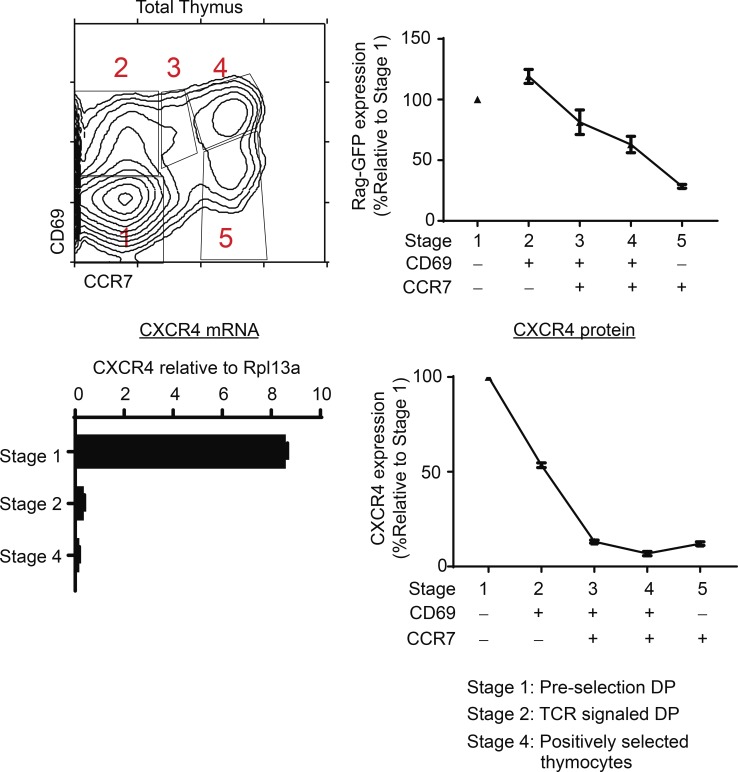
If CXCR4–CXCL12 interactions promote adherence of thymocytes to cortical TECs, persistent CXCR4 expression would inhibit migration of thymocytes out of the thymic cortex. Consequently, we wished to determine in normal wild-type mice if TCR-mediated positive selection signals extinguish CXCR4 gene expression. We did this by tracking the differentiation of preselection DP and TCR-signaled thymocytes through five sequential developmental stages of positive selection by differential expression of CD69 and CCR7 (Kimura et al., 2016; Fig. 6, top left). As previously reported, TCR-unsignaled preselection DP thymocytes are stage 1 (CD69−CCR7−) cells, whereas just-signaled DP thymocytes are stage 2 (CD69+CCR7−) cells, which then sequentially progress through stages 3–5, as revealed by linearly decreasing levels of Rag-GFP (Kimura et al., 2016; Fig. 6, top right). Assessment of mRNA expression in purified sorted cells at developmental stages 1, 2, and 4 reveals that CXCR4 mRNA expression is high in preselection stage 1 DP cells but is then abruptly extinguished in stage 2 TCR-signaled DP thymocytes (Fig. 6, bottom left). Unlike the abrupt extinction of CXCR4 mRNA expression, CXCR4 surface protein expression declines more gradually on positively selected thymocytes, to disappear from the cell surface at stage 3 when thymocytes detectably become CCR7+ (Fig. 6, bottom right). These findings reveal that TCR-mediated positive selection signals do extinguish CXCR4 gene expression to cause loss of CXCR4 surface protein expression during positive selection.
Figure 6.
TCR-mediated positive selection signals extinguish CXCR4 expression in normal thymocytes. Positive selection of wild-type B6 thymocytes can be divided into five distinct developmental stages, referred to as stages 1–5, by differential CD69 and CCR7 expression (top left). Rag-GFP expression (top right) and CXCR4 surface expression (bottom right) on thymocytes at stages 1–5 of positive selection are displayed as normalized values relative to stage 1 (stage 1 Rag-GFP MFI = 1,301; stage 1 CXCR4 MFI = 89) from three and five individual mice, respectively. CXCR4 mRNA expression in electronically sorted stage 1 (CD69−CCR7−), stage 2 (CD69+CCR7−), and stage 4 (CD69+CCR7+) thymocytes as determined by quantitative RT-PCR is shown (bottom left). Mean ± SEM from three to five mice in two to four experiments.
Migration to the medulla requires extinction of CXCR4 expression
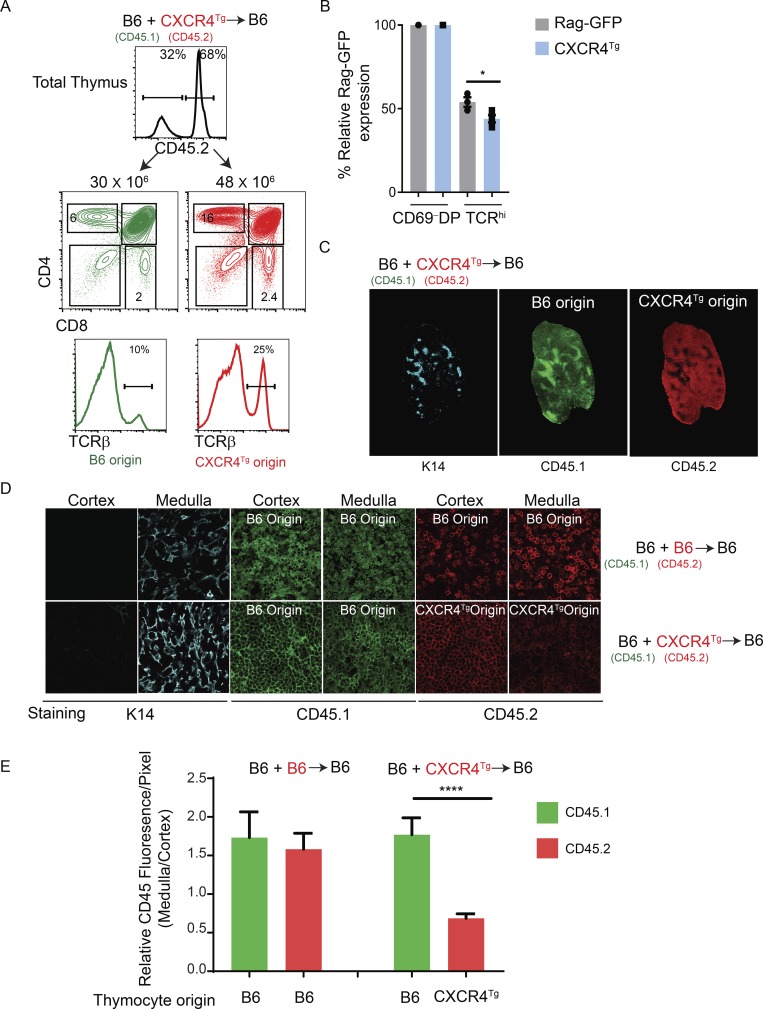
To experimentally test if migration of TCR-signaled thymocytes into the medulla would be inhibited by persistent CXCR4 expression, we used CXCR4Tg to induce transgenic CXCR4 expression on all thymocytes, including TCR-signaled thymocytes undergoing positive selection that express CCR7 (Fig. S3 C). We constructed B6+CXCR4Tg->B6 mixed-donor bone marrow radiation chimeras that simultaneously contained both wild-type B6 (CD45.1) and CXCR4Tg (CD45.2) thymocytes in the same individual thymi. In the thymi of B6+CXCR4Tg->B6 mixed-donor chimeras, B6 origin (CD45.1) and CXCR4Tg origin (CD45.2) thymocytes both differentiated into all thymocyte subsets identified by CD4/CD8 expression, including TCRβhi cells (Fig. 7 A). Notably, mature SP and TCRβhi thymocyte subsets were more prevalent among CXCR4Tg-origin thymocytes than B6-origin thymocytes (Fig. 7 A, bottom). As an explanation, we found that mature CXCR4Tg-origin thymocytes displayed significantly lower Rag-GFP expression than mature B6-origin thymocytes (McCaughtry et al., 2007; Fig. 7 B), indicating more prolonged thymic residency time and greater accumulation of mature CXCR4Tg thymocytes in the thymus despite a higher frequency of S1P1+ cells (Fig. S5 A). Because CXCR4Tg cells were selectively retained in the chimeric thymus, relatively fewer CXCR4Tg-origin T cells were present in the chimeric periphery than B6-origin T cells (Fig. S5 B). Interestingly, low-magnification CD45 staining of thymic sections revealed that CXCR4Tg-origin cells were much more densely packed in the thymic cortex than thymic medulla, whereas the opposite was true of wild-type B6-origin thymocytes, which were more densely packed in the thymic medulla than thymic cortex (Fig. 7 C). Notably, thymocyte CD45 expression increased during positive selection of both CXCR4Tg- and B6-origin thymocytes (Fig. S5 C), so both CD45.1 and CD45.2 staining would be expected to be more intense in the medulla, contrary to what we observed with CXCR4Tg-origin thymocytes (Fig. 7 C). Higher magnification of histological sections confirmed that the density of CXCR4Tg-origin (CD45.2) thymocytes was visibly lower in the thymic medulla than in the thymic cortex, but this was not the case for B6-origin (CD45.1 or CD45.2) thymocytes (Fig. 7 D). To quantify this finding, we determined the ratio of CD45 fluorescence in the medulla versus cortex for each donor thymocyte population in multiple chimeric mice and confirmed that the medulla/cortex ratio was significantly lower for CXCR4Tg-origin than B6-origin thymocytes in the same thymus (Fig. 7 E). These results document that persistent CXCR4 expression inhibits release of TCR-signaled thymocytes from the thymic cortex, increasing thymic residency time and impairing thymocyte migration into the thymic medulla. We conclude that migration of TCR-signaled thymocytes from the cortex into the thymic medulla requires extinction of CXCR4 expression.
Figure 7.
Transgenic CXCR4 expression inhibits migration of TCR-signaled thymocytes from the cortex into the medulla. (A) Mixed donor radiation bone marrow chimers were constructed by injecting B6 (CD45.1) and CXCR4Tg (CD45.2) bone marrow into lethally irradiated (950 rad) B6 (CD45.1) host mice. Top profile shows CD45.2 staining of thymocytes 8–10 wk after bone marrow reconstitution and reveals the frequency of thymocytes derived from each donor bone marrow. Middle panel shows CD4/CD8 profiles of B6 and CXCR4Tg donor origin thymocytes. Numbers on top of histograms indicate number of donor origin thymocytes. Numbers within the boxes indicate frequency of thymocytes within that box. Bottom panel shows TCRβ profiles on total thymus of the indicated origin from radiation mixed chimeric mice, with the frequency of TCRβhi cells indicated. (B) Bar graph displaying relative Rag-GFP expression on postselection TCRhi thymocytes normalized to preselection CD69− DP thymocytes from each strain. (C) Whole-tissue mosaics of thymus sections (10×) immunolabeled for K14 (cyan), CD45.1 (green), and CD45.2 (red) from B6+CXCR4Tg radiation mixed bone marrow chimera. (D) Higher-magnification (63×) fluorescence images of thymus sections immunolabeled for K14 (cyan), CD45.1 (green), and CD45.2 (red) from the indicated radiation mixed chimeric mice. (E) MFI of CD45 staining in the medulla was normalized to that in the cortex of individual mixed-donor bone marrow chimeric thymi. Each bar represents 7–13 individual measurements from 2–3 different thymi. The normalized signal intensities for both CD45.2 and CD45.1 are displayed as bar graphs. *, P < 0.05; ****, P < 0.0001 determined by Student’s t test. Data are from two to five experiments with five mice per genotype (A–C).
Discussion
The present study documents that CXCR4 expression on preselection thymocytes adheres them to the thymic cortex and inhibits their leaving. As a result, CXCR4 expression must be extinguished by TCR-mediated positive selection signaling to allow positively selected thymocytes to leave the thymic cortex and migrate into the thymic medulla. CXCR4-mediated adherence of preselection thymocytes to the thymic cortex results from CXCR4 interaction with its ligand CXCL12 on the surface of cortical TECs (Bleul et al., 1996). It is because E2A/HEB-deficient preselection thymocytes fail to express CXCR4 that they can leave the thymic cortex, which contributes to their escape from the thymus into the periphery. Thus, the E-protein transcription factors E2A and HEB regulate the ordered expression pattern of chemokine receptors during differentiation in the thymus, and it is their regulation of the chemokine receptor CXCR4 that is responsible for the retention of TCR-unsignaled preselection thymocytes in the thymic cortex.
The present study provides compelling evidence that E-protein deficiency results in disordered chemokine receptor expression on preselection thymocytes and that aberrant chemokine receptor expression is responsible for their escape into the periphery of E-protein–deficient mice (schematically illustrated in Fig. S5 D). In wild-type mice, TCR− preselection thymocytes express CXCR4, which binds to CXCL12 (SDF-1) on cortical TECs and potentially adheres preselection thymocytes to the thymic cortex (Bleul et al., 1996; Lucas et al., 2017), whereas TCR− preselection thymocytes in E2A/HEB-deficient mice do not express CXCR4 but instead aberrantly express the chemokine receptor S1P1 (and CCR7). Because they do not express CXCR4, TCR− preselection E-protein–deficient thymocytes do not adhere to the thymic cortex, and because they aberrantly express S1P1, their nonadherent preselection thymocytes are able to migrate out of the thymus and into the periphery. That this explains the appearance of preselection TCR−CD8+ T cells into the periphery of E-protein–deficient mice is supported by the fact that similar chemokine receptor alterations cause preselection TCR−CD8+ T cells in wild-type (E-protein–sufficient) mice to also appear in the periphery. We induced TCR−CD8+ preselection CXCR4+ thymocytes in wild-type mice to express transgenic S1P1 and prevented their adherence to the thymic cortex by deleting CXCL12 (SDF-1) expression in TECs. In wild-type mice, S1P1Tg expression was sufficient to cause appearance of peripheral TCR−CD8+ T cells, but additional disruption of CXCR4–CXCL12 (SDF-1) interactions in the thymic cortex significantly enhanced the appearance of TCR−CD8+ T cells in the periphery. Thus, it was not necessary for E-protein deficiency to do more than induce aberrant chemokine receptor expression on preselection thymocytes to cause the appearance of peripheral TCR−CD8+ T cells.
The present study also demonstrates in wild-type B6 mice that migration of TCR-signaled thymocytes from cortex to the medulla requires extinction of CXCR4 expression, and that CXCR4 expression is extinguished by TCR-mediated positive selection signals. Using CXCR4Tg mice, we demonstrate that persistent expression of CXCR4 on TCR-signaled cells impairs their migration toward the medulla, despite their expression of CCR7 that normally attracts TCR-signaled thymocytes into the medulla (Ueno et al., 2004). Thus, positive-selecting TCR signals must extinguish CXCR4 expression to allow positively selected thymocytes to leave the thymic cortex and migrate into the thymic medulla. While CXCR4 adheres preselection thymocytes to the thymic cortex and its extinction is necessary for emigration out of the thymic cortex, our results do not exclude the possibility that migration of TCR-signaled thymocytes out of the thymic cortex additionally requires disruption of other chemokine receptor signaling pathways. For example, in vitro migration studies have suggested that TCR-signaled DP thymocytes must express plexinD1 and be signaled by semaphorin-3E to prevent CCR9-CCL25 signaling from inhibiting emigration of positively selected thymocytes out of the thymic cortex (Choi et al., 2008). Notably, while apparently important for migration of TCR-signaled thymocytes out of the cortex, CCR9-CCL25 signaling is not known to occur in preselection thymocytes or to contribute to their adherence to the thymic cortex (Choi et al., 2008). In any event, CXCR4–CXCL12 interactions must be the predominant signaling pathway for cortical retention of preselection thymocytes, because we found that disruption of in vivo CXCR4–CXCL12 interactions is sufficient to release preselection thymocytes from the thymic cortex.
As a chemokine receptor, CXCR4 is primarily thought to be important for its role in directing migration of DN thymocytes across the thymic cortex to the subcapsular region and back (Plotkin et al., 2003; Trampont et al., 2010). However, other functions for CXCR4 have also been proposed. It has been suggested that CXCR4 functions as a costimulatory molecule to induce optimal β-selection signaling in developing DN thymocytes (Trampont et al., 2010); it has been suggested that CXCR4 signals the chemorepulsion of CD4SP thymocytes out of the fetal thymus (Poznansky et al., 2000; Vianello et al., 2005); and it has been suggested that CXCR4 possesses adherence function from in vitro thymic slice experiments in which CXCR4 was found to promote localization of human DP thymocytes to the thymic cortex (Halkias et al., 2013). The present study now directly demonstrates in vivo that CXCR4 adheres preselection thymocytes to the thymic cortex.
In conclusion, the primary function of the thymus is to generate T cells with a functional TCR on their surface. This primary function of the thymus is critically dependent on the E-protein transcription factors E2A and HEB proteins, because they regulate the ordered expression of chemokine receptors during thymocyte development so that preselection thymocytes express CXCR4 and are retained in the thymic cortex until they are TCR-signaled to extinguish CXCR4, which allows them to leave the thymic cortex and migrate to the thymic medulla.
Materials and methods
Animals
Mice with Tcf3fl and Tcf12fl (Jones and Zhuang, 2007) alleles were provided by Y. Zhuang and were bred to CD4-Cre and E8III-Cre mice to obtain E2ΔISP and E2ΔDP mice, respectively. SOCS1Tg mice (Yoshimura et al., 2007) were provided by M. Kubo (Tokyo University of Science, Tokyo, Japan); S1P1 transgenic mice (Chi and Flavell, 2005) by H. Chi (St. Jude Children's Research Hospital, Memphis, TN); CXCL12fl/fl mice (Ding and Morrison, 2013) by S. Morrison (University of Texas Southwestern Medical Center, Dallas, TX); CD69 transgenic mice (Feng et al., 2002) by P. Love (National Institutes of Health, Bethesda, MD); Rag-GFP mice by M. Nussenzweig (Rockefeller University, New York, NY); and Foxn1-cre mice (Rossi et al., 2007) by G. Hollander (University of Oxford, Oxford, UK). cDNA encoding CXCR4 was cloned into human CD2-based vector and was used to generate CXCR4-transgenic mice. ZAPKO mice were bred in our own colony. C57BL/6 (B6) mice were obtained from the Frederick National Laboratory for Cancer Research (Frederick, MD). Animal experiments were approved by the National Cancer Institute Animal Care and Use Committee, and mice were cared for in accordance with National Institutes of Health guidelines.
Flow cytometry
Monoclonal antibodies with the following specificities were used: CD184 (CXCR4, clone 2B11), CD197 (CCR7, clone 4B12), CD4 (RM4-5), and TCRβ (H57-597) from eBioscience; Thy1 (HIS51), CD45.2 (104), and CD69 (H1.2F3) from BioLegend; RORγt (clone Q31-378) from BD Biosciences; CD8 (5H10) from Invitrogen; and S1P1 (713412) from R&D Systems.
Single-cell suspensions of thymus and LNs were obtained by tweezing the organs with forceps. Cells were stained at 4°C in HBSS supplemented with 0.5% BSA and 0.5% NaN3 and first incubated with anti-FcR (2.4G2) followed by staining with fluorochrome-conjugated antibodies. Cells were acquired using an LSRII or LSRFortessa (Becton Dickinson). Doublets and dead cells were excluded from analysis by forward light scatter and propidium iodide gating. Data were analyzed using EIB-Flow software developed at the National Institutes of Health and FlowJo software (TreeStar).
Electronic cell sorting
Cells were electronically sorted on a FACSAria II. To obtain DP thymocytes, whole thymocytes were stained for CD4 and CD8 and electronically sorted to obtain purified CD4+CD8+ cells. For E2ΔDP mice, whole thymocytes were stained with CD4, CD8, and CXCR4 and electronically sorted to obtain CXCR4−CD4+CD8+ thymocytes. To obtain preselection, TCR-signaled, and postselection thymocytes, whole thymocytes were stained for CD4, CD8, Qa2, CD69, and CCR7. Only CD4+ thymocytes were electronically sorted to purify preselection CD69−CCR7− (stage 1), TCR-signaled CD69+CCR7− (stage 2), and postselection CD69+CCR7+ (stage 4) cells. To eliminate the contamination of peripheral recirculating mature CD4+ T cells, Qa2hi cells were excluded from the sorting gates of stages 1 and 2.
Quantitative real-time PCR
Total RNA was isolated using RNeasy Mini or RNeasy Micro kits (Qiagen), and cDNA was synthesized using SuperScript III with oligo(dT) primers (Invitrogen). SYBR Green detection system (Qiagen) or Taqman probes (Thermo Fisher Scientific) were used for real-time PCR, and samples were analyzed on an ABI PRISM 7900HT Sequence Detection System or QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). All mRNA values were determined by quantitative PCR and are expressed relative to Rpl13a or Actb.
ChIP analysis
30 × 106 DPs were sorted and pooled from multiple 3–5-wk-old mice and then fixed with 1% formaldehyde and 1.5 mM ethylene glycol-bis (succinic acid N-hydroxysuccinimide ester). Cross-linked cells were lysed, and nuclei were extracted, sonicated using Bioruptor Plus (Diagenode), and immunoprecipitated with E2A antibody (V-18, Lot G0814; Santa Cruz Biotechnology). DNA was purified using ChIP DNA Clean and Concentrator Kit (Zymoresearch). Libraries were prepared with the NEBNext primer set as per manufacturer’s recommendation. Sequencing was done on a HiSeq4000 platform (Illumina).
Bone marrow chimeras
Irradiation mixed-donor bone marrow chimeras were constructed by reconstituting lethally irradiated (950 rad) host mice with a mixture of B6+B6 (1:1) or B6+CXCR4Tg (1:5) T-depleted bone marrow cells (10–15 × 106 bone marrow cells). Chimeric mice were analyzed 8–10 wk after reconstitution.
Immunofluorescence microscopy
8–10 wk after irradiation, thymic lobes from mixed bone marrow chimeras were mounted in optimal cutting temperature compound before snap freezing. 6-µm cryosections were prepared, air dried for 15 min, and then incubated for 2–3 h with optimal dilution of primary rabbit polyclonal anti-keratin 14 (Covance Research Products) and anti-CD45.1 and anti-CD45.2 antibodies (Fig. 7) or with anti-CD4, anti-CD8, and anti-Rorγt antibodies (Fig. S4 F). Tissues were washed and, after an amplification step (anti-rabbit Alexa Fluor 546), mounted on microscopic slides and imaged. Fluorescence images were acquired using an AxioObserver Z1 wide-field epifluorescence microscope (Carl Zeiss Microscopy) equipped with a 10× plan-apochromat (NA 0.45) objective lens, pE-4000 LED light source (CoolLED), and ORCA Flash 4.0 v2 sCMOS camera (Hamamatsu Photonics). Extended field of view tile images were collected with 10% image overlap and stitched together using Zeiss Zen Blue v2.3 image acquisition and processing software. The resultant stitched images were analyzed using the Measure module of Zen blue software. For higher-magnification images (63×), thymic medulla and cortex images were similarly adjusted for display purposes, with identical settings used for a given channel across the samples. For intensity measurements (performed using Fiji software), corresponding channels of original (raw) images were only background-subtracted. Then, the average intensity/pixel of the region containing medulla was normalized against that of cortex.
Statistical analysis
Student's t test with two-tailed distributions was performed using GraphPad Prism 7 software. P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows thymocyte profiles and cell numbers from B6, E2ΔISP, and E2ΔDP mice. Fig. S2 quantifies IL-7R and SOCS1 gene expression in DP thymocytes from wild-type and E-deficient mice and assesses the impact of SOCS1Tg expression on their frequency of TCR+CD8+ thymocytes. Fig. S3 shows the effect of positive selection signaling on CXCR4 surface expression in B6 and CXCR4Tg mice, ChIP of the Cxcr4 gene locus in DP thymocytes, and altered CXCR4 and CCR7 expression on ZAPKO E-sufficient and E-deficient thymocytes. Fig. S4 displays profiles of TCR− thymocytes from CXCR4Tg mice, reveals the effect of CXCR4Tg expression and CCR7 deficiency on in vivo distribution of TCR−CD8+ T cells in E-deficient mice, shows the failure of CD69Tg expression to alter chemokine receptor expression on wild-type and E-deficient thymocytes, shows histological thymic sections from B6 and SDFΔTEC mice, and displays thymocyte profiles and peripheral T cells in S1P1Tg mice. Fig. S5 shows S1P1 expression on thymocytes from B6 and CXCR4Tg mice, shows the in vivo distribution of TCRβhi T cells in thymus and periphery of B6+CXCR4Tg->B6 irradiation mixed bone marrow chimeras, and shows a schematic model depicting the impact of chemokine receptor expression on thymocyte migration in the thymi of the various experimental mice used in this study.
Supplementary Material
Acknowledgments
We thank Yousuke Takahama for helpful suggestions and critical review of the manuscript and Susan Sharrow, Larry Granger, Tony Adams, and Assiatu Crossman for flow cytometry.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
The authors declare no competing financial interests.
Author contributions: T. Kadakia designed the study, performed experiments, analyzed data, and contributed to the writing of the manuscript; X. Tai, M. Kruhlak, J. Wisniewski, I-Y. Hwang, S. Roy, T.I. Guinter, A. Alag, J.H. Kehrl, and Y. Zhuang performed experiments, analyzed data, and provided helpful discussions; and A. Singer designed and supervised the study, analyzed data, and wrote the manuscript.
References
- Bankovich A.J., Shiow L.R., and Cyster J.G.. 2010. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J. Biol. Chem. 285:22328–22337. 10.1074/jbc.M110.123299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle I., and Zhuang Y.. 2014. E proteins in lymphocyte development and lymphoid diseases. Curr. Top. Dev. Biol. 110:153–187. 10.1016/B978-0-12-405943-6.00004-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul C.C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., and Springer T.A.. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 382:829–833. 10.1038/382829a0 [DOI] [PubMed] [Google Scholar]
- Bunting M.D., Comerford I., and McColl S.R.. 2011. Finding their niche: chemokines directing cell migration in the thymus. Immunol. Cell Biol. 89:185–196. 10.1038/icb.2010.142 [DOI] [PubMed] [Google Scholar]
- Campbell J.J., Pan J., and Butcher E.C.. 1999. Cutting edge: developmental switches in chemokine responses during T cell maturation. J. Immunol. 163:2353–2357. [PubMed] [Google Scholar]
- Chi H., and Flavell R.A.. 2005. Cutting edge: regulation of T cell trafficking and primary immune responses by sphingosine 1-phosphate receptor 1. J. Immunol. 174:2485–2488. 10.4049/jimmunol.174.5.2485 [DOI] [PubMed] [Google Scholar]
- Choi Y.I., Duke-Cohan J.S., Ahmed W.B., Handley M.A., Mann F., Epstein J.A., Clayton L.K., and Reinherz E.L.. 2008. PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity. 29:888–898. 10.1016/j.immuni.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pooter R.F., and Kee B.L.. 2010. E proteins and the regulation of early lymphocyte development. Immunol. Rev. 238:93–109. 10.1111/j.1600-065X.2010.00957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Tillman R.E., Naoe Y., Taniuchi I., and Littman D.R.. 2007. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204:1945–1957. 10.1084/jem.20070133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., and Murre C.. 2001. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 1:193–199. 10.1038/35105060 [DOI] [PubMed] [Google Scholar]
- Etzensperger R., Kadakia T., Tai X., Alag A., Guinter T.I., Egawa T., Erman B., and Singer A.. 2017. Identification of lineage-specifying cytokines that signal all CD8+-cytotoxic-lineage-fate ‘decisions’ in the thymus. Nat. Immunol. 18:1218–1227. 10.1038/ni.3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Woodside K.J., Vance B.A., El-Khoury D., Canelles M., Lee J., Gress R., Fowlkes B.J., Shores E.W., and Love P.E.. 2002. A potential role for CD69 in thymocyte emigration. Int. Immunol. 14:535–544. 10.1093/intimm/dxf020 [DOI] [PubMed] [Google Scholar]
- Gegonne A., Chen Q.R., Dey A., Etzensperger R., Tai X., Singer A., Meerzaman D., Ozato K., and Singer D.S.. 2018. Immature CD8 Single-Positive Thymocytes Are a Molecularly Distinct Subpopulation, Selectively Dependent on BRD4 for Their Differentiation. Cell Reports. 24:117–129. 10.1016/j.celrep.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkias J., Melichar H.J., Taylor K.T., Ross J.O., Yen B., Cooper S.B., Winoto A., and Robey E.A.. 2013. Opposing chemokine gradients control human thymocyte migration in situ. J. Clin. Invest. 123:2131–2142. 10.1172/JCI67175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Yoshida T., Kinjyo I., Minoguchi S., Yasukawa H., Kato S., Mimata H., Nomura Y., Seki Y., Kubo M., and Yoshimura A.. 2001. A mutant form of JAB/SOCS1 augments the cytokine-induced JAK/STAT pathway by accelerating degradation of wild-type JAB/CIS family proteins through the SOCS-box. J. Biol. Chem. 276:40746–40754. 10.1074/jbc.M106139200 [DOI] [PubMed] [Google Scholar]
- Jones M.E., and Zhuang Y.. 2007. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 27:860–870. 10.1016/j.immuni.2007.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlecek T.A., van Oers N.S., Lefrancois L., Olson S., Finlay D., Chu D.H., Connolly K., Killeen N., and Weiss A.. 1998. Differential requirements for ZAP-70 in TCR signaling and T cell development. J. Immunol. 161:4688–4694. [PubMed] [Google Scholar]
- Kimura M.Y., Thomas J., Tai X., Guinter T.I., Shinzawa M., Etzensperger R., Li Z., Love P., Nakayama T., and Singer A.. 2016. Timing and duration of MHC I positive selection signals are adjusted in the thymus to prevent lineage errors. Nat. Immunol. 17:1415–1423. 10.1038/ni.3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N., Arakaki R., Hayashi Y., Kitagawa T., Lipp M., et al. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 24:165–177. 10.1016/j.immuni.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Lancaster J.N., Li Y., and Ehrlich L.I.R.. 2018. Chemokine-Mediated Choreography of Thymocyte Development and Selection. Trends Immunol. 39:86–98. 10.1016/j.it.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Love P.E., and Bhandoola A.. 2011. Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 11:469–477. 10.1038/nri2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas B., White A.J., Parnell S.M., Henley P.M., Jenkinson W.E., and Anderson G.. 2017. Progressive Changes in CXCR4 Expression That Define Thymocyte Positive Selection Are Dispensable For Both Innate and Conventional αβT-cell Development. Sci. Rep. 7:5068 10.1038/s41598-017-05182-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M., Lo C.G., Cinamon G., Lesneski M.J., Xu Y., Brinkmann V., Allende M.L., Proia R.L., and Cyster J.G.. 2004. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 427:355–360. 10.1038/nature02284 [DOI] [PubMed] [Google Scholar]
- McCaughtry T.M., Wilken M.S., and Hogquist K.A.. 2007. Thymic emigration revisited. J. Exp. Med. 204:2513–2520. 10.1084/jem.20070601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry T.M., Etzensperger R., Alag A., Tai X., Kurtulus S., Park J.H., Grinberg A., Love P., Feigenbaum L., Erman B., and Singer A.. 2012. Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus. J. Exp. Med. 209:2263–2276. 10.1084/jem.20121505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo V.N., Tang H.L., and Cyster J.G.. 1998. Epstein-Barr virus-induced molecule 1 ligand chemokine is expressed by dendritic cells in lymphoid tissues and strongly attracts naive T cells and activated B cells. J. Exp. Med. 188:181–191. 10.1084/jem.188.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Adoro S., Guinter T., Erman B., Alag A.S., Catalfamo M., Kimura M.Y., Cui Y., Lucas P.J., Gress R.E., et al. 2010. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 11:257–264. 10.1038/ni.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie H.T., and Zúñiga-Pflücker J.C.. 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25:649–679. 10.1146/annurev.immunol.23.021704.115715 [DOI] [PubMed] [Google Scholar]
- Plotkin J., Prockop S.E., Lepique A., and Petrie H.T.. 2003. Critical role for CXCR4 signaling in progenitor localization and T cell differentiation in the postnatal thymus. J. Immunol. 171:4521–4527. 10.4049/jimmunol.171.9.4521 [DOI] [PubMed] [Google Scholar]
- Poznansky M.C., Olszak I.T., Foxall R., Evans R.H., Luster A.D., and Scadden D.T.. 2000. Active movement of T cells away from a chemokine. Nat. Med. 6:543–548. 10.1038/75022 [DOI] [PubMed] [Google Scholar]
- Rossi S.W., Jeker L.T., Ueno T., Kuse S., Keller M.P., Zuklys S., Gudkov A.V., Takahama Y., Krenger W., Blazar B.R., and Holländer G.A.. 2007. Keratinocyte growth factor (KGF) enhances postnatal T-cell development via enhancements in proliferation and function of thymic epithelial cells. Blood. 109:3803–3811. 10.1182/blood-2006-10-049767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Nakata Y., Dan Y., Uzawa A., Nakagawa K., Saito T., Mita K., and Shirasawa T.. 1998. Loss of SDF-1 receptor expression during positive selection in the thymus. Int. Immunol. 10:1049–1056. 10.1093/intimm/10.8.1049 [DOI] [PubMed] [Google Scholar]
- Suzuki G., Sawa H., Kobayashi Y., Nakata Y., Nakagawa Ki., Uzawa A., Sakiyama H., Kakinuma S., Iwabuchi K., and Nagashima K.. 1999. Pertussis toxin-sensitive signal controls the trafficking of thymocytes across the corticomedullary junction in the thymus. J. Immunol. 162:5981–5985. [PubMed] [Google Scholar]
- Takahama Y. 2006. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6:127–135. 10.1038/nri1781 [DOI] [PubMed] [Google Scholar]
- Trampont P.C., Tosello-Trampont A.C., Shen Y., Duley A.K., Sutherland A.E., Bender T.P., Littman D.R., and Ravichandran K.S.. 2010. CXCR4 acts as a costimulator during thymic beta-selection. Nat. Immunol. 11:162–170. 10.1038/ni.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Saito F., Gray D.H., Kuse S., Hieshima K., Nakano H., Kakiuchi T., Lipp M., Boyd R.L., and Takahama Y.. 2004. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200:493–505. 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianello F., Kraft P., Mok Y.T., Hart W.K., White N., and Poznansky M.C.. 2005. A CXCR4-dependent chemorepellent signal contributes to the emigration of mature single-positive CD4 cells from the fetal thymus. J. Immunol. 175:5115–5125. 10.4049/jimmunol.175.8.5115 [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Naka T., and Kubo M.. 2007. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 7:454–465. 10.1038/nri2093 [DOI] [PubMed] [Google Scholar]
- Zlotnik A., and Yoshie O.. 2000. Chemokines: a new classification system and their role in immunity. Immunity. 12:121–127. 10.1016/S1074-7613(00)80165-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.