This study reveals a novel signaling function of CX3CL1: its intracellular domain controls adult neurogenesis and reverses pathological development of amyloid pathologies in Alzheimer’s mouse brains. Ectopic expression of this domain is likely a novel therapeutic strategy in humans.
Abstract
The membrane-anchored CX3CL1 is best known to exert its signaling function through binding its receptor CX3CR1. This study demonstrates a novel function that CX3CL1 exerts. CX3CL1 is sequentially cleaved by α-, β-, and γ-secretase, and the released CX3CL1 intracellular domain (CX3CL1-ICD) would translocate into the cell nucleus to alter gene expression due to this back-signaling function. Amyloid deposition and neuronal loss were significantly reduced when membrane-anchored CX3CL1 C-terminal fragment (CX3CL1-ct) was overexpressed in Alzheimer’s 5xFAD mouse model. The reversal of neuronal loss in 5xFAD can be attributed to increased neurogenesis by CX3CL1-ICD, as revealed by morphological and unbiased RNA-sequencing analyses. Mechanistically, this CX3CL1 back-signal likely enhances developmental and adult neurogenesis through the TGFβ2/3-Smad2/3 pathway and other genes important for neurogenesis. Induction of CX3CL1 back-signaling may not only be a promising novel mechanism to replenish neuronal loss but also for reducing amyloid deposition for Alzheimer’s treatment.
Introduction
CX3CL1, also known as fractalkine, is a type I transmembrane chemokine with the C-XXX-C fingerprint (Bazan et al., 1997; Pan et al., 1997). It functions as a cellular adhesion molecule when anchored on a membrane. The soluble form, generated through ectodomain shedding by members of the ADAM (a disintegrin and metalloproteinase) protein family such as ADAM-10 (Hundhausen et al., 2003; Schulte et al., 2007; Hurst et al., 2012) is a chemotactic signaling molecule. This secreted N-terminal fragment binds to its sole CX3CR1 receptor via the containing C-XXX-C domain (Imai et al., 1997). The high-affinity binding to CX3CR1 also requires the positively charged residues such as Lys-7 and Arg-47 in the chemokine domain of CX3CL1 (Harrison et al., 2001). For >20 yr, knowledge of CX3CL1 has been centered on the CX3CL1–CX3CR1 interaction to transduce signals between cells for regulating inflammatory responses, leukocyte capture and infiltration, and other neuroinflammatory functions (D’Haese et al., 2012; Sheridan and Murphy, 2013; Gyoneva and Ransohoff, 2015; Liu et al., 2016).
Here we report a novel physiological function of CX3CL1 that is independent from the C-XXX-C domain. We show that CX3CL1 is cleaved not only by metalloproteases, but also by BACE1, and the resulting membrane-anchored CX3CL1 C-terminal fragment (CX3CL1-ct) is subsequently further cleaved by γ-secretase to release the intracellular domain of CX3CL1 (CX3CL1-ICD), which can be translocated into the cell nucleus, thereby inducing transcriptional regulation of a set of genes important for cell growth and differentiation.
To understand the physiological role of CX3CL1-ct in vivo, we generated transgenic mice overexpressing this fragment (Tg-CX3CL1-ct). While Tg-CX3CL1-ct mice exhibit no overt growth, an Alzheimer’s disease (AD) mouse model (5xFAD) overexpressing CX3CL1-ct exhibited significantly reduced amyloid deposition and neuronal loss. The reversal of neuronal loss in 5xFAD mice was likely due to enhanced neurogenesis by overexpressed CX3CL1-ct. We showed that overexpressed CX3CL1-ct facilitates neurogenesis not only in the subgranular zone (SGZ), but also in the subventricular zone (SVZ). Our mechanistic study revealed that CX3CL1-ct induces transcriptional activation of many genes important for neurogenesis. The prominently validated pathway is the TGFβ3 and Smad (proteins homologous to the Sma and Mad proteins from Caenorhabditis elegans and Drosophila melanogaster) signaling pathway. Collectively, our study reveals for the first time that CX3CL1-ICD has a robust back-signaling function that can reverse neuronal loss and reduce amyloid deposition in AD mice.
Results
CX3CL1 is cleaved by α-, β-, and γ-secretases
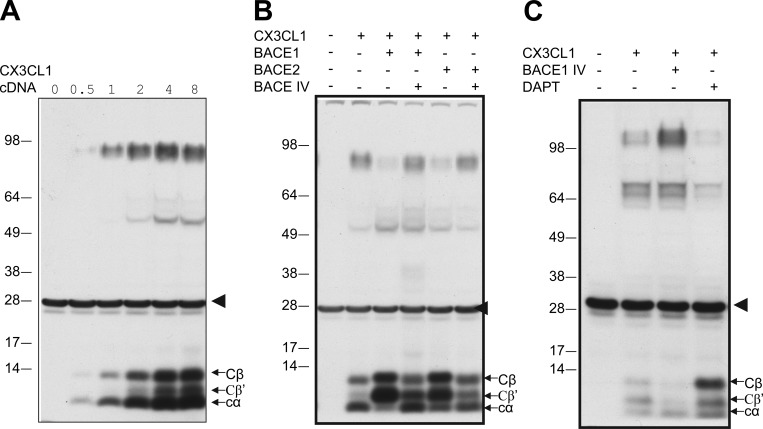
CX3CL1 is a type I transmembrane signaling molecule and is a potential substrate of BACE1, as previously suggested (Kuhn et al., 2012). To test this, we transiently transfected CX3CL1 in BACE1-overexpressing HEK-293 cells and noted two C-terminal fragments at low expression levels and three fragments at higher levels (Fig. 1 A), resembling the sequential cleavage of amyloid precursor protein (APP) by α- and β-secretase (BACE1). Moreover, transient transfection of either BACE1 or BACE2 could similarly produce two larger C-terminal fragments (Fig. 1 B). We designated these two as Cβ (migrating near 12 kD on the Western blot) and Cβ′ (migrating near 10 kD), because treatment with a potent BACE inhibitor (BACE1 inhibitor IV) diminished the levels of these two fragments (Fig. 1 B). A slight increase in the slower-migrating band after BACE inhibition was also noted, which is consistent with prior reports that CX3CL1 was shed by α-secretase (Hundhausen et al., 2003; Hurst et al., 2012).
Figure 1.
Cleavage of CX3CL1 by β- and γ-secretases. (A and B) Myc-tagged CX3CL1 was expressed in BACE1-overexpressing HM cells in a dose-dependent manner. Two clonal stable cell lines (c1 and c2) were also selected from mouse neuroblastoma Neuro-2A cells transfected with myc-tagged CX3CL1. The myc antibody detected three C-terminal fragments: β and β′ fragments for BACE1 cleavages and the α-secretase–cleaved fragment. The arrowhead indicates nonspecific reacted bands with the myc antibody. (B) Both BACE1 and BACE2 similarly cleave CX3CL1, as demonstrated by coexpression of indicated plasmid DNA in HEK-293 cells. Inhibition of BACE activity by compound BACE IV diminished the levels of β and β′ fragments. (C) CX3CL1 was cleaved by γ-secretase, shown by inhibition of γ-secretase with the specific inhibitor DAPT to increase levels of fragments from BACE1 and α-secretase cleavages.
Many type I transmembrane proteins are natural γ-secretase substrates (Haapasalo and Kovacs, 2011). To determine the potential cleavage of CX3CL1 by γ-secretase, we treated transfected cells with N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), which will inhibit γ-secretase cleavages of membrane bound C-terminal fragments. Consequently, DAPT inhibition significantly increased all three C-terminal fragments (Fig. 1 C). Hence, we demonstrate that CX3CL1 is cleavable by α-, β-, and γ-secretases.
Nuclear translocation of the γ-secretase–cleaved CX3CL1-ct
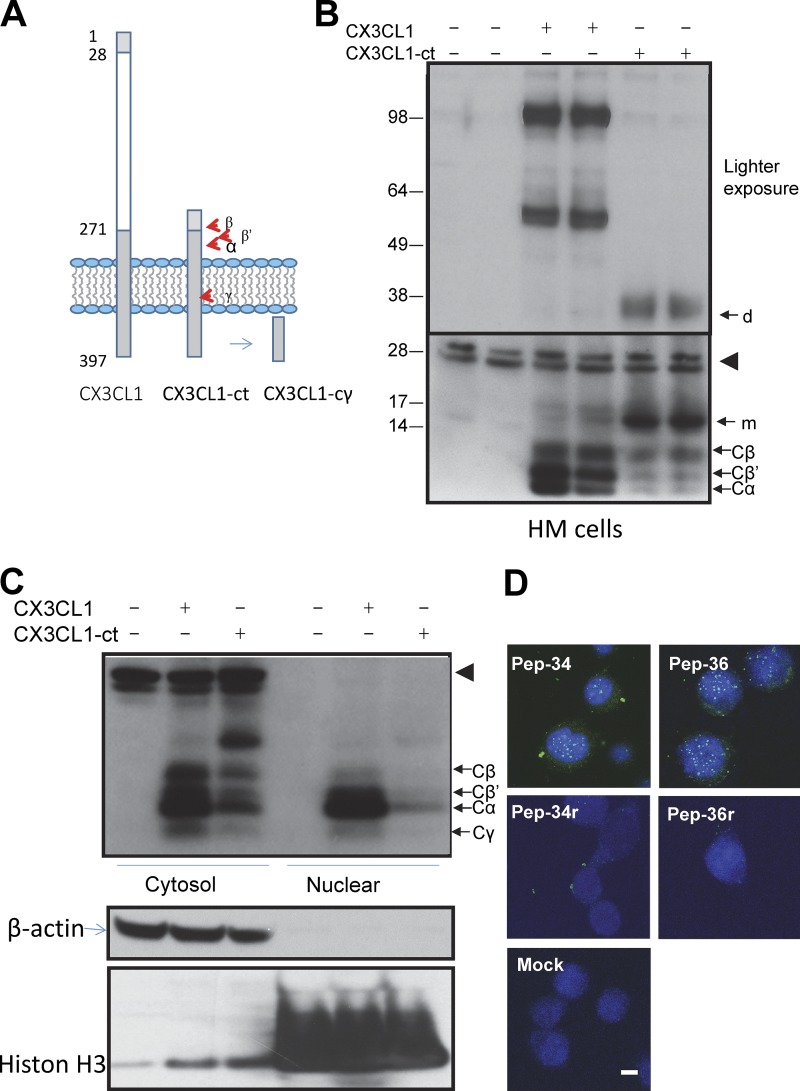
To explore a potential function of the γ-secretase–cleaved CX3CL1-ct, we generated an expression construct named CX3CL1-ct, in which the N-terminal residues from 28–271 were deleted (Fig. 2 A). This CX3CL1-ct construct lacks the C-XXX-C domain but retains the signal peptide sequence and the potential secretase cleavage sites. Expression of CX3CL1-ct in BACE1-overexpressing HEK-293 cells produced all three of the same C-terminal fragments (Fig. 2 B), indicating that all three secretases can faithfully cleave CX3CL1-ct.
Figure 2.
Expression of CX3CL1-ct in cells. (A) Illustration of CX3CL1-ct construction and a potentially cleaved fragment by γ-secretase. (B) CX3CL1 or CX3CL1-ct constructs were expressed in BACE1-overexpressing cells, and all C-terminal fragments were detected in cytosolic fractions. The arrowhead indicates nonspecific reacted bands with the myc-antibody. m indicates the monomer of CX3CL1; d indicates a band that is presumably a dimer of CX3CL1-ct based on the molecular weight. (C) Cytosolic and nuclear fractions were separated from cells expressing either CX3CL1 or CX3CL1-ct. The γ-cleaved CX3CL1 fragment CX3CL1-cγ was detectable. β-Actin was detected mainly from the cytosol fractions, while histone H3 was enriched in the nuclear fractions. Nonspecific bands are indicated by arrowheads. (D) After transfection for 24 h, CX3CL1-ICD peptides Pep-34 and Pep-36, detected by CX3CL1 C-terminal antibody in green, were localized in mouse neuro-2a cell nucleus, marked by Topro in blue. Scale bar, 5 µm.
We then asked whether the γ-cleaved CX3CL1 will release CX3CL1 intracellular domain (CX3CL1-Cγ) to regulate gene expression resembling the Notch intracellular domain (Kopan and Ilagan, 2009). Lysates prepared from the cell cytosol, perinuclear region, and nucleus were examined by Western blot analyses. Noticeably, a 6-kD CX3CL1-Cγ was detectable in the nucleus of CX3CL1-overexpressing HEK-293 cells (Fig. 2 C). The level of CX3CL1-Cγ was relatively lower in cells expressing CX3CL1-ct, likely due to weak transfection of this construct, but it was detectable after a significantly longer exposure (data not shown).
To determine whether CX3CL1-Cγ will mediate nuclear translocation via the intracellular domain (CX3CL1-ICD), we examined cell nuclei from HEK-293 cells transfected with CX3CL1-ct by confocal staining of a nucleus-enriched preparation. We showed that the antibody specific to the CX3CL1 C-terminus readily detected CX3CL1-ICD in the nucleus (Fig. S1). CX3CL1-ICD in the cell nucleus was confirmed by partial colocalization with Topro nuclear staining, while the same antibody showed no signals in control nuclei transfected by the empty vector.
Alternatively, we delivered a synthetic CX3CL1-ICD, spanning the entire intracellular domain of CX3CL1, into mouse Neuro-2A cells by using Chariot Transfection Reagent. After introduction for ∼24 h, we clearly detected CX3CL1-ICD peptide (Pep-34) in the nucleus, while reversely ordered peptide Pep-34r had nonspecific puncta in the cytosol (Fig. 2 D). A modified version of CX3CL1-ICD peptide was also synthesized by fusing SV40 nuclear signal peptide to Pep-34 (named Pep-36). We showed similar nuclear translocation but not its reverse version (Pep-36r; Fig. 2 D). Hence, by using multiple methods, we showed that CX3CL1-ICD is capable of translocating into the cell nucleus.
Overexpression of CX3CL1-ct in 5xFAD mice
To understand the potential physiological function of CX3CL1-ct in vivo, we generated a transgenic mouse model, named Tg-CX3CL1-ct, in which expression of CX3CL1-ct transgene was under the control of tetracycline (Tet-Off) responsive element (Fig. S2). To drive the expression of CX3CL1-ct in neurons, we bred Tg-CX3CL1-ct mice with Tg-CX3CL1-ct/tTA mice, which express tetracycline-controlled transactivator protein (tTA) by the CaMKIIα promoter (Mayford et al., 1996). Tg-CaMKIIα/tTA mice produce low levels of tTA beginning at embryonic day 18 and higher levels after early postnatal days in forebrain neurons. In postnatal day 6 (P6) brains, we noted strong expression of CX3CL1-ct in broad regions of the hippocampus and neurons in cortical layers II–V in the bigenic Tg-CX3CL1-ct/tTA mice (data not shown). Mice overexpressing Tg-CX3CL1-ct in neurons were fertile and healthy, with no visible differences in growth or behaviors compared with control littermates.
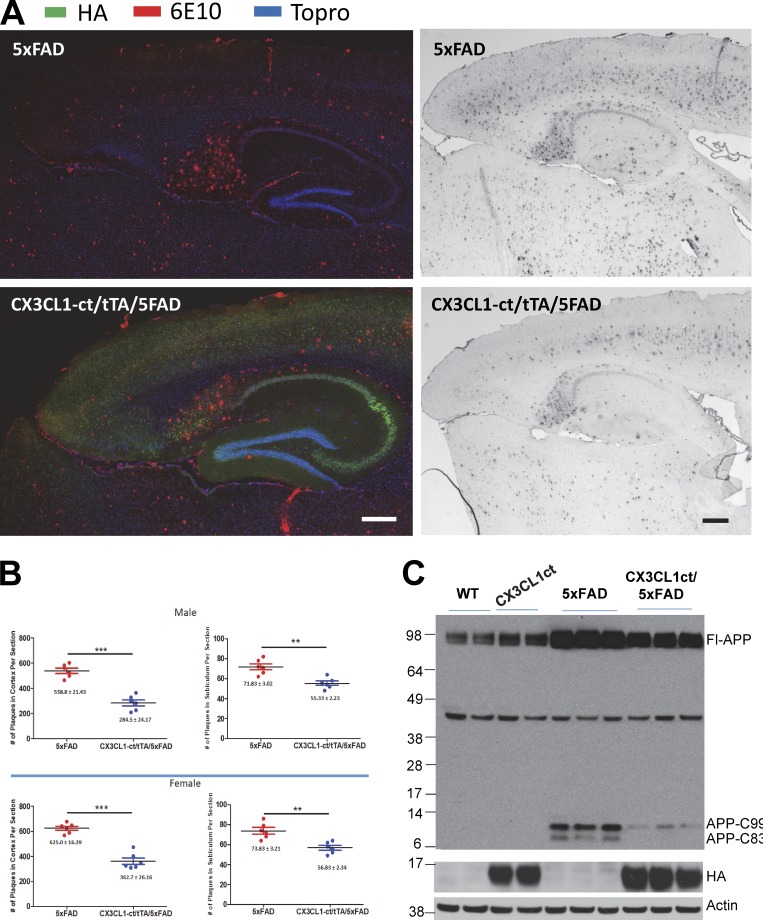
CX3CL1 was previously shown to alter amyloid deposition (Lee et al., 2014). To explore whether this C-terminal fragment has any effect on amyloid deposition, we then bred Tg-CX3CL1-ct/tTA mice with 5xFAD mice, which develop amyloid plaques after P60 (Oakley et al., 2006) found that enhanced expression of CX3CL1-ct was sufficient to reduce amyloid deposition in broad brain regions (Fig. 3 A). Further quantification of amyloid deposits in cortical and subiculum confirmed significant reduction of amyloid plaques in both 9-mo-old male and female Tg-CX3CL1-ct/tTA/5xFAD mice compared with 5xFAD littermates (Fig. 3 B; reduction by ∼23% in subiculum and 45% in cortical region, n = 6). Biochemical analyses of protein lysates showed that ectopic expression of CX3CL1-ct decreased total levels of APP-C99 and C83, products of full-length APP cleaved by BACE1 and α-secretase, respectively (Fig. 3 C). Reduction of both APP C-terminal fragments would correlate with the decrease in amyloid deposition.
Figure 3.
Overexpression of CX3CL1-ct in 5xFAD transgenic mice reduces amyloid deposition. (A) Two different pairs of Tg-5xFAD and Tg-CX3CL1-ct/tTA/5xFAD mice were used for comparing the density of amyloid plaques by confocal or DAB staining. Antibody 6E10 recognizes human Aβ peptides and amyloid plaques, while HA-tagged CX3CL1-ct transgene was detected with HA antibody. Cell nucleus was marked by Topro. Scale bars, 200 µm. (B) Densities of amyloid plaques from subiculum or cortex were quantified from six mice in each pair. Both male and female mice were used for comparisons. Error bars are ± SEM. (C) APP and its cleavage product C99 and C83 were examined by antibody 8717 (n = 3 experiments). **, P < 0.01; ***, P < 0.001.
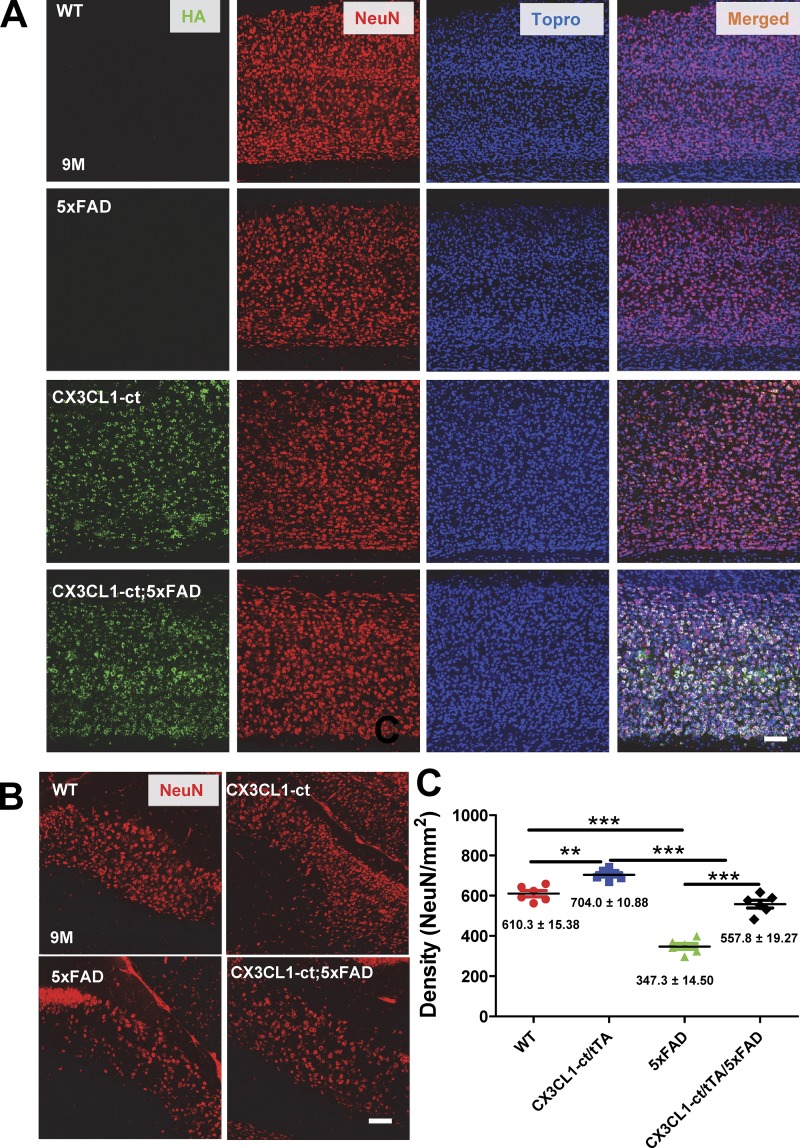
Neuronal loss in 9-mo-old of 5xFAD mice was noted by Eimer and Vassar (2013), and this was confirmed in our histochemical examinations. We showed that overexpressed CX3CL1-ct reversed such neuronal loss in both subiculum (Fig. 4 A) and cortical (Fig. 4 B) layers. This reversal of neuronal loss was confirmed by quantification of neuronal nuclear antigen (NeuN)–positive neurons (Fig. 4 C; 557.8 ± 19.27 of Tg-CX3CL1-ct/tTA/5xFAD vs. 347.3 ± 14.50 of 5xFAD subiculum neurons/mm2; 704.0 ± 10.88 of Tg-CX3CL1-ct/tTA vs. 610.3 ± 15.8 of WT subiculum neurons/mm2; n = 6 mice; P < 0.01 and P < 0.001, ANOVA).
Figure 4.
Ectopic expression of CX3CL1-ct in 5xFAD mice reverses neuronal losses. (A and B) Neuronal loss was observed in the 5xFAD subiculum (A) and cortical layers (B) compared with WT controls. Higher neuronal density, marked by NeuN antibody, was noted in mice overexpressing CX3CL1-ct, suggesting a potential increase in neurogenesis. Ectopic expression of CX3CL1-ct in 5xFAD brains showed more neurons compared with 5xFAD comparable regions. Scale bar, 30 µm. (C) NeuN-marked neurons in the subiculum region were counted for comparison, one in every 10 sections per mouse. Each dot represents one animal (n = 6; **, P < 0.01; ***, P < 0.001, Student’s t test); error bars are ± SEM.
Overexpressed CX3CL1-ct enhances neurogenesis in mouse SGZ and SVZ
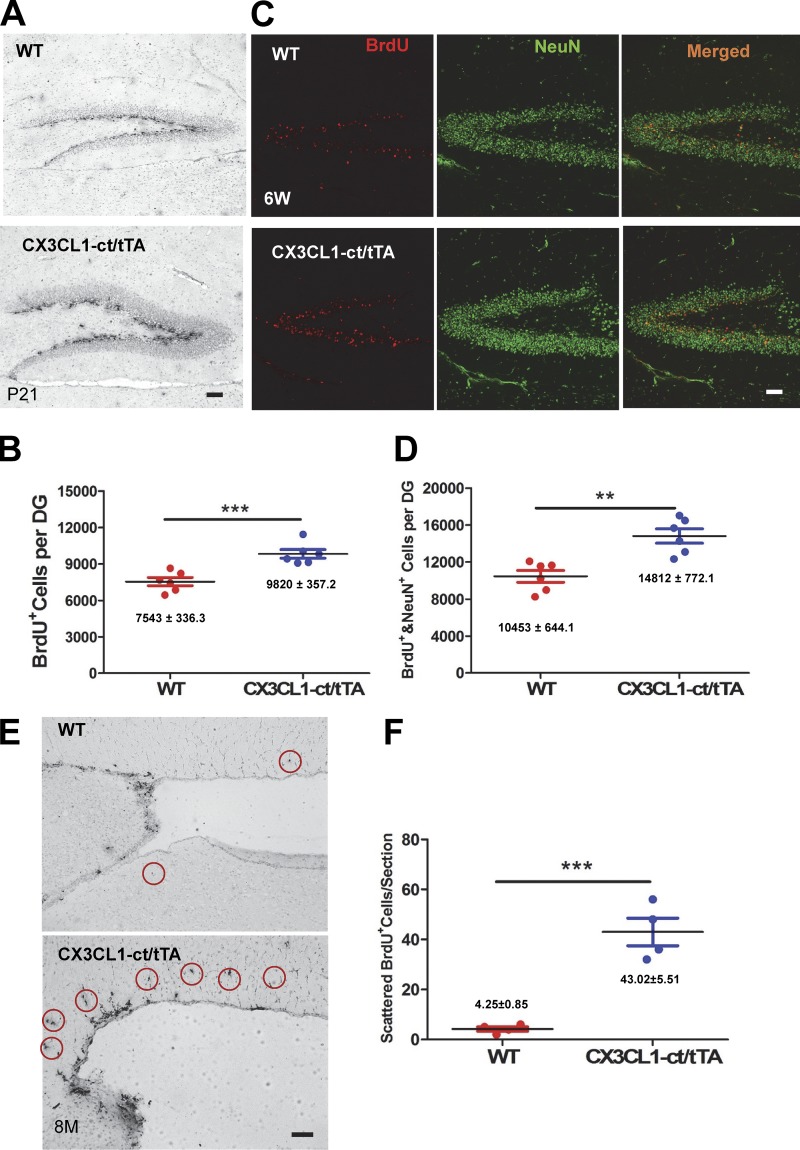
In our phenotypical analyses of Tg-CX3CL1-ct/tTA mice, we noted a ∼15% increase in NeuN-positive neurons as compared with WT littermates, suggesting a potential increase in neurogenesis by the overexpression of CX3CL1-ct. To confirm enhanced neurogenesis, we conducted BrdU pulse labeling experiments in P21 mice using our previously published procedures (Hu et al., 2013). We showed that the number of BrdU-positive cells in the bigenic Tg-CX3CL1-ct/tTA mouse SGZ was indeed greater than that in nontransgene-expressing littermates (Fig. 5 A). Further stereological quantification confirmed the increase in BrdU-positive cells in bigenic Tg-CX3CL1-ct/tTA mouse SGZ by ∼30% (Fig. 5 B; 9,820 ± 357.2 of Tg-CX3CL1-ct/tTA vs. 7,543 ± 336.3 of WT SGZ BrdU+ cells; n = 6; P < 0.001). In 6-wk-old mice examined for fate determination by measuring BrdU- and NeuN double-positive cells, more mature neurons derived from Tg-CX3CL1-ct/tTA mouse SGZ multipotent radial glia appeared (Fig. 5 C). We confirmed this by quantification (Fig. 5 D; 10,453 ± 644.1 BrdU+/NeuN+-cells in Tg-CX3CL1-ct/tTA vs. 14,812 ± 772.1 BrdU+/NeuN+-cells per dentate WT; n = 6; P < 0.001).
Figure 5.
Enhanced neurogenesis by CX3CL1-ct in transgenic mice. (A) Immunostaining of brain sections from BrdU pulse-labeled mice at P21 was performed using BrdU antibody. Scale bar, 30 µm. (B) Stereological quantification confirmed enhanced neurogenesis in the neurogenic niche of the SGZ of Tg-CX3cl1-ct/tTA mice. (C) Mice were BrdU-labeled starting at P11 for 5 d and then examined at 6 wk of age. Brain sections were fixed and costained with BrdU (red) and NeuN (green) antibodies. Scale bar, 30 μm. (D) Quantification of both BrdU-positive cells and mature neurons in granular cell layers in Tg-CX3cl1-ct/tTA mice and WT control littermates was conducted, and the results were plotted (n = 6 pairs; **, P < 0.01, Student’s t test). (E and F) Newly dividing cells in the SVZ were labeled for BrdU, marked with circles (E), and quantitatively compared from one in every six fixed sections (F; n = 4; ***, P < 0.001, Student’s t test). Error bars are ± SEM.
This increased neurogenesis was not restricted to the SGZ, as enhanced neurogenesis in the SVZ was also evident. BrdU-positive cells in the same P21 Tg-CX3CL1-ct/tTA mouse SVZ were visibly greater (data not shown). However, BrdU-positive cells became visibly more numerous in 8-mo-old adult SVZ (Fig. 5 E). We showed that neurogenesis is much less active and sparsely labeled in nontransgenic adult SVZ while significantly more active in adult Tg-CX3CL1-ct/tTA SVZ. For quantification of BrdU-positive cells in SVZ, we chose one of every six sections and quantified those BrdU-positive cells scattered along the rostral migratory stream in reflecting cells migrating out of the SVZ neurogenic niche. Our data showed that BrdU-positive cells were significantly more numerous in Tg-CX3CL1-ct/tTA SVZ compared with controls (Fig. 5, E and F; 43.02 ± 5.51 vs. 4.25 ± 0.85 BrdU+ cells per average section; n = 4 animals; P < 0.001).
For further confirmation, we also examined cells expressing minichromosome maintenance protein 2 (MCM2), which is a proliferation marker in neurogenesis (von Bohlen und Halbach, 2011), and found significantly more MCM2-positive cells in the P6 Tg-CX-CL1-ct/tTA SGZ (Fig. S3, A and B) and SVZ (Fig. S3, C and D) compared with nontransgene-expressing littermates. MCM2-expressing cells in adult brains (6 wk old) are normally fewer compared with early developmental stages, and CX3CL1-ct markedly increased cells positive for MCM2 in both the SGZ (Fig. S4, A and B) and the SVZ (Fig. S4 C), consistent with the enhanced neurogenesis induced by this molecule in the adult mouse. Altogether, we demonstrate the enhanced neurogenesis by overexpressed CX3CL1-ct in both the SVZ and SGZ.
Expression of CX3CL1-ct in the adult is sufficient to induce mouse adult neurogenesis
To determine whether CXCL1-ct induces neurogenesis in adult mice, we suppressed expression of CX3CL1-ct transgene in Tg-CX3CL1-ct/tTA mice during early development with doxycycline (Dox) treatment. Mice were treated with Dox beginning during the mating period, and this treatment lasted until newborn Tg-CX3CL1-ct/tTA offspring reached the age of 2 mo. Removal of Dox treatment induced expression of transgene, and mice were left untreated for an additional 4 mo so that transgene was expressed only during the adult stage. Biochemical and phenotypic analyses were conducted on mice at the age of 6 mo. Compared with nontransgene-expressing littermates, Tg-CX3CL1-ct/tTA mice expressed high levels of CX3CL1-ct, as confirmed by the hemagglutinin (HA) tag antibody (data not shown).
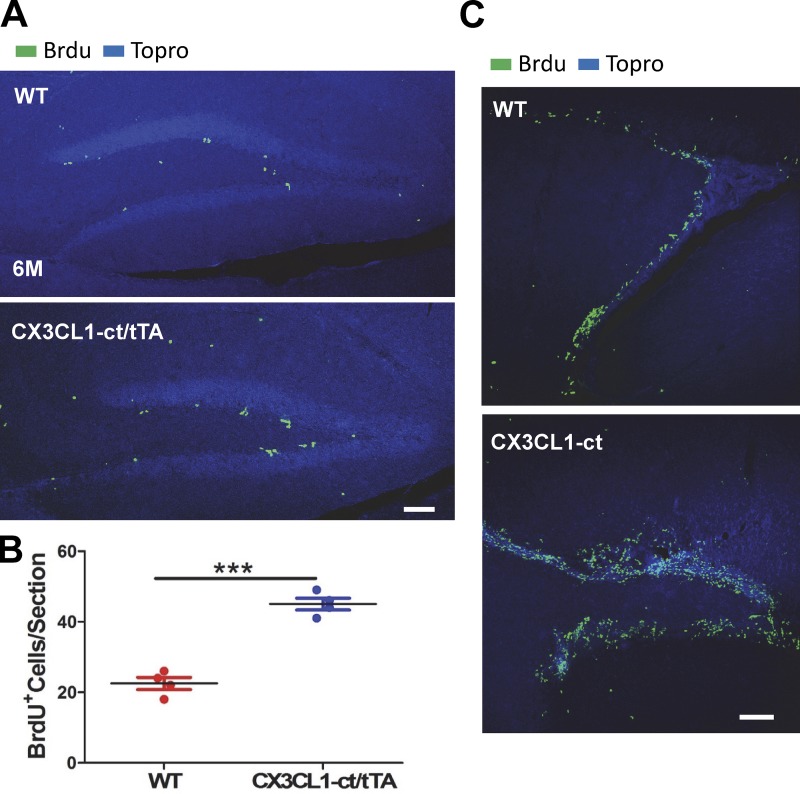
We then examined adult neurogenesis by conducting BrdU pulse labeling experiments in mice at the age of 6 mo after transgene was turned on for 4 mo. BrdU-positive cells were clearly increased in Tg-CX3CL1-ct/tTA SGZ (Fig. 6 A), as confirmed by quantification (Fig. 6 B). In the SVZ, BrdU-positive cells were densely distributed in the WT SVZ; however, they were more diffuse and greater in number in the Tg-CX3CL1-ct/tTA SVZ (Fig. 6 C), consistent with more active neurogenesis in Tg-CX3CL1-ct/tTA brains.
Figure 6.
CX3CL1-ct enhances neurogenesis in adult transgenic mice. (A) Tg-CX3CL1-ct/tTA mice that were treated with Dox for 2 mo (2M) and left untreated for another 4 mo (4M) were pulse labeled for examination of adult neurogenesis. Fixed brain sections were reacted with antibody for BrdU and Topro for the nucleus. (B) Semiquantification was conducted using one in every six sections to compare BrdU-positive cells in the Tg-CX3CL1-ct/tTA SGZ with that in WT (n = 4; ***, P < 0.001, Student’s t test). Each animal is shown in one dot. Error bars are ± SEM. (C) BrdU+ cells in Tg-CX3CL1-ct/tTA SVZ were significantly increased compared with WT SVZ. Scale bars, 60 µm.
CX3CL1-ct controls expression of genes important for neurogenesis
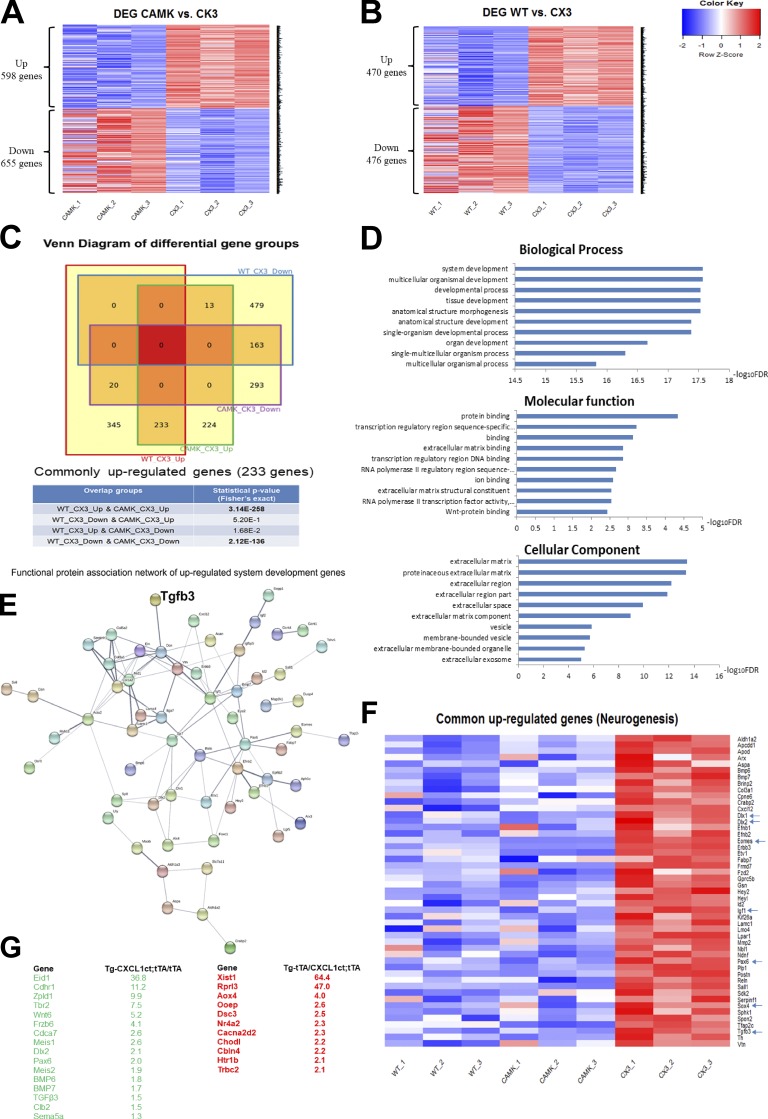
To explore the molecular mechanisms underlying the effect of CX3CL1-ct on neurogenesis, we conducted unbiased genome-wide RNA sequencing (RNA-seq) experiments using P5 mouse brains in which neurogenesis is active. We compared gene expression filings from three pairs of WT and Tg-CX3CL1-ct/tTA mice and observed 1,253 genes that were significantly altered (Fig. 7 A; adjusted P < 0.05; n = 3 pairs). When comparing Tg-CamKII-tTA with Tg-CX3CL1-ct/tTA mice, we found 946 genes significantly altered (Fig. 7 B; adjusted P < 0.05; n = 3 pairs). Remarkably, 233 genes were commonly up-regulated (P = 3.14 × 10−258) and 163 down-regulated (P = 2.12 × 10−136) in Tg-CX3CL1-ct/tTA compared with WT or Tg-CamKII-tTA samples (Fig. 7 C).
Figure 7.
Molecular signatures of newborn mice in response to CX3CL1-ct expression. (A and B) Hippocampi from newborn pups were used for RNA-seq analyses; heatmap shows significant changes (P < 0.05) in genes between WT and Tg-CX3C1-ct/tTA (CX3) samples (A), and between Tg-Camk2a-tTA (CAMK) and Tg-CX3C1-ct/tTA samples (B). (C) Overlap statistical analysis uncovered similar transcriptional alterations shown in Fig. 8, A and B and revealed 233 up-regulated and 163 down-regulated genes in Tg-CX3C1-ct/tTA samples compared with both WT and Tg Camk2a tTA. (D) Among 233 commonly up-regulated genes, the top biological pathways indicates the control of system and multicellular organismal development. (E) TGFβ3 appears to be one of the top genes in the protein functional network involving system development. (F) Among 233 commonly up-regulated genes, ∼50 genes are important for the control of neurogenesis, including those well-documented genes highlighted by arrows. (G) Selected genes with significant changes (P < 0.001) between Tg-tTA and Tg-CX3C1-ct/tTA are listed, and some of them are known to control neurogenesis.
Among these commonly up-regulated genes, many of them are known to control system or multicellular organismal development, cell differentiation, and regulation of extracellular matrix cell signaling, analyzed based on the pathway cloud using the Reactome, Kyoto Encyclopedia of Genes and Genomes, and Gene Ontology databases, respectively (see gene ontology terms in Fig. 7 D). Specifically, TGFβ3 was found to be one of the top up-regulated genes in system development gene network by CX3CL1-ct overexpression (Fig. 7 E), consistent with the above cellular and biochemical analyses.
In related to the observed neurogenesis induced by CX3CL1-ct, we found that >50 of 233 commonly up-regulated genes have documented important roles in neurogenesis (see heatmap in Fig. 7 F). Among this list, bone morphogenetic proteins (BMPs) including BMP6 and BMP7, Tbr2 (also known as Eomes), homeobox transcription factor Dlx 1 and 2 (neuroblasts express distal-less 1 and 2), Pax 6, and Sox4 are known to regulate neurogenesis. Other noted elevated genes include Eid-1, which was elevated by ∼30-fold in Tg-CX3CL1-ct/tTA compared with WT samples. This protein is recognized as a nodal point that couples cell cycle exit to the transcriptional activation of genes required for neuronal differentiation (Miyake et al., 2000). Genes controlling the frizzle-winless signaling pathways, such as WNT5a, WNT6, WNT7, frizzle-related protein, and cadherin-related family protein, were also significantly elevated in Tg-CX3CL1-ct/tTA mouse hippocampi. Hence, our data suggest that CX3CL1-ICD is capable of regulating large sets of signaling genes that control neurogenesis in mice.
Overexpression of CX3CL1-ct in mice enhances TGFβ2/β3-Smad2 signaling
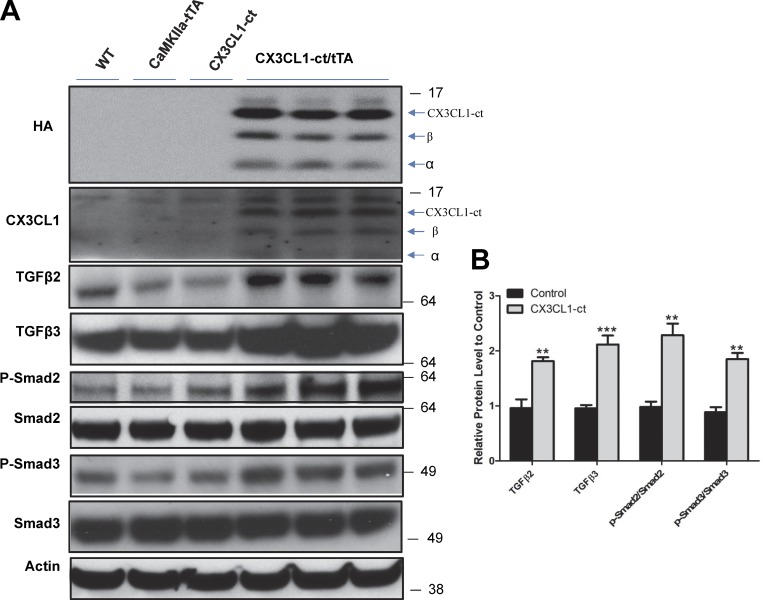
To confirm changes of inducible genes by CX3CL1-ct, we first overexpressed either full-length CX3CL1 or CX3CL1-ct in HEK-293 cells and found significant elevation of both TGFβ2 and TGFβ3 but not TGFβ1 (Fig. S5). Moreover, elevated levels of TGFβ2 and TGFβ3 significantly enhanced phosphorylation of Smad2 and Smad3 in HEK293 cells, while the levels of total Smad2 and Smad3 were marginally increased. We further validated elevated expression of both TGFβ2 and TGFβ3 in 2-mo-old bigenic mouse hippocampi compared with all other control groups (WT or either single transgenic line), which were expressed at similar levels (Fig. 8, A and B). In the hippocampus, the downstream molecules Smad2 and Smad3 were expressed but not significantly altered among different groups (Fig. 8, A and B). However, phosphorylated Smad2 and Smad3 were evidently elevated, consistent with the activation of the TGFβ2/3-Smad2/3 pathway. Altogether, our data suggest that CX3Cl1 likely controls adult neurogenesis by activating the TGFβ2/3-Smad2/3 pathway and contributes to the reversed neuronal loss in AD mouse models.
Figure 8.
Increased TGFβ signaling and neurogenesis by expressing CX3CL1-ct in adult mice. (A) Protein levels were compared between transgenic mice (Tg-CX3cl1-ct/tTA) and their control littermates (WT, Tg-Cx3CL1-ct and Tg-CamKIIa-tTA) using the indicated antibodies. Expression of transgene CX3CL1-ct was validated by HA and CX3CL1 antibody in compound mice. BACE1-cleaved C-terminal fragment was marked by β, and ADAM-cleaved fragment, by α. (B) Bar graphs confirm that neuronal expression of CX3CL1-ct significantly increases levels of TGFβ2, TGFβ3, p-Smad2, and p-Smad3 compared with controls (n = 3 experiments; **, P < 0.01; ***, P < 0.001, Student’s t test; error bars are ± SEM).
Discussion
While previous studies have centered on the role of CX3CL1 via its interaction with CX3CR1, we demonstrated here that the intracellular C-terminal domain of CX3CL1, CX3CL1-ICD, derived from γ-secretase cleavage of CX3CL1-ct, could translocate into the cell nucleus and regulate expression of large numbers of genes. It should be noted that prior study has focused on the cleavage of CX3CL1 by ADAM10 and ADAM17, and CX3CL1 is not obviously cleaved by BACE1 in cultures (Schulte et al., 2007). We showed that CX3CL1 can readily be cleaved by overexpressed BACE1 (Fig. 1). The discrepancy is likely related to the expression level of BACE1 in cells; BACE1 activity is high in neurons, suggesting a natural processing of CX3CL1 by both α- and β-secretase in neurons. We examined the intrinsic physiological function of CX3CL1-ICD by using Tg-CX3CL1-ct mice under the tetracycline-controlled promoter. Expression of CX3CL1-ct in the mouse forebrains by the CamKII driver appears not to cause discernible abnormality based on visual inspection and body weight monitoring. Strikingly, this expression in Alzheimer’s 5xFAD mouse model not only reduces amyloid deposition by down-regulating expression of APP but also significantly reverses neuronal loss, indicating a potential therapeutic application of this fragment in patients.
In this study, we also reveal for the first time that CX3CL1 controls TGFβ/BMP/Smad signaling pathways through its C-terminal domain, which is not able to bind CX3CR1 due to the lack of the epidermal growth factor–like domain and topological restraints. We have confirmed the induced expression of TGFβ2 and TGFβ3 in both cell lines and transgenic mice through this fragment. Their increases, in turn, enhance phosphorylation of Smad2 and Smad3; phosphorylated Smad2 and Smad3 translocate to the cell nucleus and form a heterodimer as active transcription factors (Shi and Massagué, 2003). Our RNA-seq results also revealed up-regulation of multiple members of the BMP family by CX3CL1-ICD, and additional biochemical characterization will be needed to validate these gene expressions.
Tg-CX3CL1-ct/tTA mice exhibit enhanced neurogenesis, not only during early development, but also during adulthood. Enhanced neurogenesis is not restricted to the SGZ, but also occurs in the SVZ. How CX3CL1-ICD promotes neurogenesis is an important mechanistic question. While RNA-seq analyses identified many up-regulated genes important for the control of neurogenesis, we saw that the TGFβ3–Smad2 pathway is likely one of the top candidate pathways, as TGFβ3 was consistently shown to be up-regulated in both cellular and mouse models. It has been previously shown that Smad2-mediated transcription is critically required for brain development, function, and/or maintenance of neuronal cells (Lu et al., 2005;Wang et al., 2011); phospho-Smad2 appears to be an effector of this pathway (Massagué and Xi, 2012).
In addition, CX3CL1-ICD may induce expression of many genes directly or indirectly through effects of the TGFβ superfamily. One such molecule is Pax6, which is known to be expressed by radial glial progenitors and is required for the unidirectionality of lineage commitment toward neuronal differentiation (Manuel et al., 2015). In P5 Tg-CX3CL1-ct/tTA mouse brains, Pax6 was elevated by approximately twofold (see our array results). Expression of Pax6 can be up-regulated by transcriptional factors in the super TGFβ family (Grocott et al., 2007). Meis1 and Meis2, cofactors of Pax6, were elevated by 2.6- and 1.98-fold, respectively, in Tg-CX3CL1-ct mouse brains compared with WT controls (Fig. 7 G). It was shown that Meis2 interacts with Pax6 in SVZ-derived neural stem and progenitor cells to drive generic neurogenesis in the adult SVZ system (Brill et al., 2008; Agoston et al., 2014). Dxl2 is also complexed with Pax6 and Meis2 for driving both neurogenesis and proliferation in the postnatal SVZ (Suh et al., 2009), and expression of Dlx2 is also strongly elevated in Tg-CX3CL1-ct/tTA mouse brains. Since Pax6 can spatially and temporarily control differentiation of multiple late-born neuronal cell types (Remez et al., 2017), its precise expression in Tg-CX3CL1-ct will be more fully investigated in the future. Other noted genes such as insulin growth factor-1 (Igf1) and calretinin are known important players in adult neurogenesis (Brandt et al., 2003), and they were both significantly elevated by CX3CL1-ICD for promoting adult neurogenesis in broad brain regions (SGZ and SVZ).
While genes important for neurogenesis are altered in Tg-CX3CL-ct/tTA mice, it remains to be determined how and what altered genes can decrease amyloid deposits in 5xFAD mice in our future studies. Nevertheless, our study provides clear evidence that the effect of CX3CL1 does not have to occur through interactions with its cognate receptor CX3CR1 as recently suggested (Lee et al., 2010). We postulate that enhancing the expression of CX3CL1-ct in the adult, for example, by introducing a molecular mimic of this CX3CL1-ct, will benefit AD patients by reversing neuronal loss and reducing amyloid deposition.
Materials and methods
Generation of Tg-CX3CL1-ct mice
C-terminal fragment–derived CX3CL1, including the signal peptide sequences as illustrated in Fig. 1 C, was subcloned into the BamHI and NotI sites of pTRE2hyg vector (Clontech Laboratories). A linearized NheI fragment containing the transgene was used for transgenic mouse production. Five Tg-CX3CL1-ct founders on the C57BL/6-CBA(J) background were identified by PCR with primers (forward, 5′-TATCTCTGTCGTGGCTGCT-3′; reverse, 5′-GCAGGATGATTACCAGGATGTAG-3′) and further confirmed by Southern blotting. Tg-CX3CL1-ct mice were backcrossed with C57BL/6J mice for six generations before crossing with CaMKIIα-tTA mice (The Jackson Laboratory; 007004).
Mice were housed in designated animal rooms at 23°C on a 12-h light/dark cycle with food and water available ad libitum. For Dox (Sigma-Aldrich) treatment, the drug was added to drinking water at 0.5 mg/ml, supplemented with 2% sucrose. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Lerner Research Institute in compliance with the guidelines established by the Public Health Service Guide for the Care and Use of Laboratory Animals.
Western blotting and antibodies
Mouse tissues were freshly dissected, and total proteins were extracted using modified radioimmunoprecipitation assay buffer. At least two or three mice from each group were used for Western blot analysis. In the study of nuclear proteins, cytoplasmic and nuclear fractions of proteins were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions. Equal amounts of protein (50 µg) were resolved on a NuPAGE Bis-Tris gel (Invitrogen) and transferred onto a nitrocellulose membrane (Invitrogen). After protein transfer, the blot was incubated with the following antibodies: p-Smad2 (#3104, RRID: AB_390732), Smad2/3 (#8685; RRID: AB_10889933), p-Smad1 (9553s; RRID: AB_2107775), Smad1 (#6944; RRID: AB_10860070), TGFβ1 (SC-146; RRID: AB_632486), TGFβ2 (SC-90; RRID: AB_2303237), TGFβ3 (SC-82; RRID: AB_2202303), and BrdU (ab6326; RRID: AB_305426; Abcam); NeuN (MAB377; RRID: AB_2298772; EMD Millipore); HA (#1867423; RRID: AB_10094468; Roche); CX3CL1 (SC7225, RRID: AB_2087136; Santa Cruz); 6E-10 (9320–10, RRID: AB_662804; Signet); MCM2 (ab4461, RRID: AB_304470; Abcam); and APP-C (A8717, RRID: AB_258409; Sigma-Aldrich). HRP-conjugated secondary antibodies were used and visualized using enhanced chemiluminescence (Thermo Fisher Scientific).
Immunohistochemistry
Standard immunohistochemical or confocal experiments were conducted. Briefly, serial sections (12-µm thickness from fixed mouse brains) were fixed with 4% paraformaldehyde in PBS for 20 min. Sections were permeabilized with 0.3% Triton X-100 in PBS containing 0.3% H2O2 for 30 min. After being rinsed in PBS two times to remove detergent, the sections were treated by microwave in 0.05 M citrate-buffered saline (pH 6.0) for 5 min and immunostained with the indicated specific primary antibodies. For 3,3′-diaminobenzidine (DAB) staining, sections were subsequently reacted with the specified primary antibody and corresponding biotinylated secondary antibodies (1:200) and developed according to the protocol from the Vector ABC kit (1:400; Vector Laboratories). For the confocal experiment, fluorescence-labeled second antibody was applied for visualization on a microscope.
Peptide synthesis and transfection
Peptides were custom-synthesized (GenScript USA) and solubilized in 1× PBS to attain a working solution concentration of 2 µg/µl. Multiple aliquots were prepared to avoid repeated freeze/thaw cycles and were stored at −80°C before use. Sequences of the peptides used are given in Sequences of peptides, below. Monolayers of N2A cells were grown to 70% confluence in either 2-well chamber slides (Thermo Fisher Scientific) or 100 × 20-mm tissue culture treated dishes (Corning) before transfection with peptides. Cells were transfected with 1 and 20 µg of peptides for immunostaining and protein analysis, respectively. Peptide transfection was performed using Chariot (Active Motif) according to the manufacturer’s instructions. Briefly, cells were conditioned in serum-free medium for 2 h before transfection. Peptides diluted in 1× PBS were incubated with Chariot for 30 min at room temperature for Chariot–peptide complex formation and then added to the cells in fresh serum-free medium. Cells were then incubated at 37°C, 5% CO2 for different time points before analysis.
Sequences of peptides
Pep-34 has the sequence of RKMAGEMAEGLRYIPRSCGSNSYVLVPV from CX3CL1 C-terminus, while Pep-34r has the reverse order of this sequence. Pep-36 has the sequence of SV40 nucleus translocation sequence at N terminal side of Pep-34 (PKKKRKVEDPYCRKMAGEMAEGLRYIPRSCGSNSYVLVPV). Pep-36r has the order of nucleus translocation sequence Pep-34r (PKKKRKVEDPYCRKMAGEMAEGLRYIPRSCGSNSYVLVPV).
Mouse strains and breeding strategy
The AD mouse model, 5xFAD, is a transgenic mouse model with five mutations (APPSw,FlLon,PSEN1*M146L*L286V) and purchased from the Jackson Laboratory. We bred Tg-CX3CL1-ct/tTA mice with 5xFAD mice to obtain mice heterozygous for the transgenic 5xFAD, Tg-CX3CL1-ct/tTA, and Tg-CX3CL1-ct/tTA/5xFAD. For most experimental comparisons such as amyloid deposition and neuronal losses, we use 5xFAD and Tg-CX3CL1-ct/tTA/5xFAD mice.
RNA-seq data generation and analyses
Hippocampi were dissected from P4 newborn pups and immediately snap-frozen in dry ice. Total RNA was prepared from the frozen tissue and used to generate a sequencing library by the Lerner Research Institute Core Facility using the Illumina MiSeq desktop sequencer (for pilot run) and the Illumina HiSeq2500 (for production run). Raw fastq data (101-bp paired-end) were mapped to mm10 mouse genome assembly using Tophat alignment tool, and Cuffdiff pipeline (Trapnell et al., 2013) was used for differential expression analysis with Ensembl mouse gene annotation. Statistical significance was based on adjusted P values of 0.05. Gene overlap Venn diagram and statistical calculation were generated using NeuVenn web tool (http://bioinfo5pilm46.mit.edu:318/neuvenn). Gene ontology analysis was performed using STRING web tool. Genes enriched in neurogenesis were identified using ToppGene web tool. Expression heatmaps were generated using R script with color gradient representing z-score of fragments per kilobase of transcript per million mapped reads. Functional protein association network was visualized using STRING web tool. The RNA-seq Gene Expression Omnibus accession no. is GSE129722.
Neurogenesis assays
The proliferation rate of neural stem cells in CX3CL1-ct mice (n = 6) was compared with that of control animals (n = 6) and assessed using BrdU immunohistochemistry. For this, mice were intraperitoneally injected with 50 mg/kg of BrdU (B9285; Sigma-Aldrich) at P21 twice at 12-h intervals. The animals were then sacrificed 24 h after the first injection of BrdU. Brain samples were fixed in 4% paraformaldehyde at 4°C overnight and then transferred into 30% sucrose in PBS for 2 d. Sections were pretreated with serial 1 and 2 M HCl, reacted with primary antibody BrdU, and then processed by using VectaStain-ABC Kits (Vector Laboratories) and SigmaFAST DAB with metal enhancer tablets (Sigma-Aldrich) according to the manufacturer’s instructions. After color development, the sections were dehydrated by incubation for 3 min in each of 70, 95, and 100% ethanol, followed by 5 min in two changes of xylenes, and mounted using Permount. Labeled cells were quantified by stereology. Every third section (of a total of 30 sections) was counted under a 100× objective, and the sum was multiplied by 3 to estimate the total number of BrdU-positive cells in the region. Cells were counted if they were in or touching the SGZ, and cells were excluded if they were more than two cell diameters from the granule cell layer. To analyze neural differentiation, 15 mg/kg of BrdU was injected once daily for 5 d beginning at P11. The animals were sacrificed after an additional 21 d, and brain sections were examined with double labeling using the primary antibodies BrdU (1:100) and NeuN (1:1,000).
Statistical analysis
Quantitative data are presented as mean ± SEM. All experiments were independently repeated at least three times. Statistical analyses were conducted using Prism 6 software (GraphPad). Statistical comparisons between groups were analyzed for significance by one-way ANOVA with Tukey’s post hoc test and Student’s t tests. Significant P values are denoted by the use of asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Error bars in each case represent SEM.
Online supplemental material
Fig. S1 shows the translocation of CX3CL1-ICD into the cell nucleus. Fig. S2 shows the generation of transgenic mice inducibly overexpressing CX3CL1-ct. Fig. S3 depicts enhanced neurogenesis in early developmental transgenic mice expressing CX3CL1-ct protein. Fig. S4 depicts enhanced neurogenesis in adult transgenic mice expressing CX3CL1-ct protein. Fig. S5 shows that expression of CX3CL1-ct induces TGFβ signaling activation in cells.
Supplementary Material
Acknowledgments
The authors thank Dr. Chris Nelson for his critical reading and editing of this manuscript.
R. Yan is supported by the National Institutes of Health (grants RF1AG058261, AG025493, NS074256, and AG046929). L.-H. Tsai is supported by the National Institute on Aging (RF1AG054012), the JPB Foundation, the Robert A. and Rene E. Belfer Family Foundation, and the Alana Foundation. Both the Yan and Tsai laboratories are also supported by the Cure Alzheimer’s Fund.
The authors declare no competing financial interests.
Author contributions: Q. Fan performed experiments and contributed most of results in this study; M. Gayen, N. Singh, and W. He performed experiments and contributed various biochemical and morphological data; F. Gao contributed to the bioinformatic data analyses; X. Hu, L-H. Tsai, and R. Yan contributed to data analyses. Q. Fan and R. Yan wrote discussed the study and wrote the manuscript.
References
- Agoston Z., Heine P., Brill M.S., Grebbin B.M., Hau A.C., Kallenborn-Gerhardt W., Schramm J., Götz M., and Schulte D.. 2014. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 141:28–38. 10.1242/dev.097295 [DOI] [PubMed] [Google Scholar]
- Bazan J.F., Bacon K.B., Hardiman G., Wang W., Soo K., Rossi D., Greaves D.R., Zlotnik A., and Schall T.J.. 1997. A new class of membrane-bound chemokine with a CX3C motif. Nature. 385:640–644. 10.1038/385640a0 [DOI] [PubMed] [Google Scholar]
- Brandt M.D., Jessberger S., Steiner B., Kronenberg G., Reuter K., Bick-Sander A., von der Behrens W., and Kempermann G.. 2003. Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Mol. Cell. Neurosci. 24:603–613. 10.1016/S1044-7431(03)00207-0 [DOI] [PubMed] [Google Scholar]
- Brill M.S., Snapyan M., Wohlfrom H., Ninkovic J., Jawerka M., Mastick G.S., Ashery-Padan R., Saghatelyan A., Berninger B., and Götz M.. 2008. A dlx2- and pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J. Neurosci. 28:6439–6452. 10.1523/JNEUROSCI.0700-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haese J.G., Friess H., and Ceyhan G.O.. 2012. Therapeutic potential of the chemokine-receptor duo fractalkine/CX3CR1: an update. Expert Opin. Ther. Targets. 16:613–618. 10.1517/14728222.2012.682574 [DOI] [PubMed] [Google Scholar]
- Eimer W.A., and Vassar R.. 2013. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Aβ42 accumulation and Caspase-3 activation. Mol. Neurodegener. 8:2 10.1186/1750-1326-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocott T., Frost V., Maillard M., Johansen T., Wheeler G.N., Dawes L.J., Wormstone I.M., and Chantry A.. 2007. The MH1 domain of Smad3 interacts with Pax6 and represses autoregulation of the Pax6 P1 promoter. Nucleic Acids Res. 35:890–901. 10.1093/nar/gkl1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoneva S., and Ransohoff R.M.. 2015. Inflammatory reaction after traumatic brain injury: therapeutic potential of targeting cell-cell communication by chemokines. Trends Pharmacol. Sci. 36:471–480. 10.1016/j.tips.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapasalo A., and Kovacs D.M.. 2011. The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 25:3–28. 10.3233/JAD-2011-101065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.K., Fong A.M., Swain P.A., Chen S., Yu Y.R., Salafranca M.N., Greenleaf W.B., Imai T., and Patel D.D.. 2001. Mutational analysis of the fractalkine chemokine domain. Basic amino acid residues differentially contribute to CX3CR1 binding, signaling, and cell adhesion. J. Biol. Chem. 276:21632–21641. 10.1074/jbc.M010261200 [DOI] [PubMed] [Google Scholar]
- Hu X., He W., Luo X., Tsubota K.E., and Yan R.. 2013. BACE1 regulates hippocampal astrogenesis via the Jagged1-Notch pathway. Cell Reports. 4:40–49. 10.1016/j.celrep.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundhausen C., Misztela D., Berkhout T.A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., et al. . 2003. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 102:1186–1195. 10.1182/blood-2002-12-3775 [DOI] [PubMed] [Google Scholar]
- Hurst L.A., Bunning R.A., Sharrack B., and Woodroofe M.N.. 2012. siRNA knockdown of ADAM-10, but not ADAM-17, significantly reduces fractalkine shedding following pro-inflammatory cytokine treatment in a human adult brain endothelial cell line. Neurosci. Lett. 521:52–56. 10.1016/j.neulet.2012.05.057 [DOI] [PubMed] [Google Scholar]
- Imai T., Hieshima K., Haskell C., Baba M., Nagira M., Nishimura M., Kakizaki M., Takagi S., Nomiyama H., Schall T.J., and Yoshie O.. 1997. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell. 91:521–530. 10.1016/S0092-8674(00)80438-9 [DOI] [PubMed] [Google Scholar]
- Kopan R., and Ilagan M.X.. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P.H., Koroniak K., Hogl S., Colombo A., Zeitschel U., Willem M., Volbracht C., Schepers U., Imhof A., Hoffmeister A., et al. . 2012. Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 31:3157–3168. 10.1038/emboj.2012.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Varvel N.H., Konerth M.E., Xu G., Cardona A.E., Ransohoff R.M., and Lamb B.T.. 2010. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am. J. Pathol. 177:2549–2562. 10.2353/ajpath.2010.100265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Xu G., Jay T.R., Bhatta S., Kim K.W., Jung S., Landreth G.E., Ransohoff R.M., and Lamb B.T.. 2014. Opposing effects of membrane-anchored CX3CL1 on amyloid and tau pathologies via the p38 MAPK pathway. J. Neurosci. 34:12538–12546. 10.1523/JNEUROSCI.0853-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Jiang L., Bian C., Liang Y., Xing R., Yishakea M., and Dong J.. 2016. Role of CX3CL1 in Diseases. Arch. Immunol. Ther. Exp. (Warsz.). 64:371–383. 10.1007/s00005-016-0395-9 [DOI] [PubMed] [Google Scholar]
- Lu J., Wu Y., Sousa N., and Almeida O.F.. 2005. SMAD pathway mediation of BDNF and TGF beta 2 regulation of proliferation and differentiation of hippocampal granule neurons. Development. 132:3231–3242. 10.1242/dev.01893 [DOI] [PubMed] [Google Scholar]
- Manuel M.N., Mi D., Mason J.O., and Price D.J.. 2015. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 9:70 10.3389/fncel.2015.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., and Xi Q.. 2012. TGF-β control of stem cell differentiation genes. FEBS Lett. 586:1953–1958. 10.1016/j.febslet.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M., Bach M.E., Huang Y.Y., Wang L., Hawkins R.D., and Kandel E.R.. 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science. 274:1678–1683. 10.1126/science.274.5293.1678 [DOI] [PubMed] [Google Scholar]
- Miyake S., Sellers W.R., Safran M., Li X., Zhao W., Grossman S.R., Gan J., DeCaprio J.A., Adams P.D., and Kaelin W.G. Jr. 2000. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 20:8889–8902. 10.1128/MCB.20.23.8889-8902.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. . 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Lloyd C., Zhou H., Dolich S., Deeds J., Gonzalo J.A., Vath J., Gosselin M., Ma J., Dussault B., et al. . 1997. Neurotactin, a membrane-anchored chemokine upregulated in brain inflammation. Nature. 387:611–617. 10.1038/42491 [DOI] [PubMed] [Google Scholar]
- Remez L.A., Onishi A., Menuchin-Lasowski Y., Biran A., Blackshaw S., Wahlin K.J., Zack D.J., and Ashery-Padan R.. 2017. Pax6 is essential for the generation of late-born retinal neurons and for inhibition of photoreceptor-fate during late stages of retinogenesis. Dev. Biol. 432:140–150. 10.1016/j.ydbio.2017.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte A., Schulz B., Andrzejewski M.G., Hundhausen C., Mletzko S., Achilles J., Reiss K., Paliga K., Weber C., John S.R., and Ludwig A.. 2007. Sequential processing of the transmembrane chemokines CX3CL1 and CXCL16 by alpha- and gamma-secretases. Biochem. Biophys. Res. Commun. 358:233–240. 10.1016/j.bbrc.2007.04.100 [DOI] [PubMed] [Google Scholar]
- Sheridan G.K., and Murphy K.J.. 2013. Neuron-glia crosstalk in health and disease: fractalkine and CX3CR1 take centre stage. Open Biol. 3:130181 10.1098/rsob.130181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., and Massagué J.. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 113:685–700. 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- Suh Y., Obernier K., Hölzl-Wenig G., Mandl C., Herrmann A., Wörner K., Eckstein V., and Ciccolini F.. 2009. Interaction between DLX2 and EGFR regulates proliferation and neurogenesis of SVZ precursors. Mol. Cell. Neurosci. 42:308–314. 10.1016/j.mcn.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., and Pachter L.. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31:46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bohlen und Halbach O. 2011. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res. 345:1–19. 10.1007/s00441-011-1196-4 [DOI] [PubMed] [Google Scholar]
- Wang L., Nomura M., Goto Y., Tanaka K., Sakamoto R., Abe I., Sakamoto S., Shibata A., Enciso P.L., Adachi M., et al. . 2011. Smad2 protein disruption in the central nervous system leads to aberrant cerebellar development and early postnatal ataxia in mice. J. Biol. Chem. 286:18766–18774. 10.1074/jbc.M111.223271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.