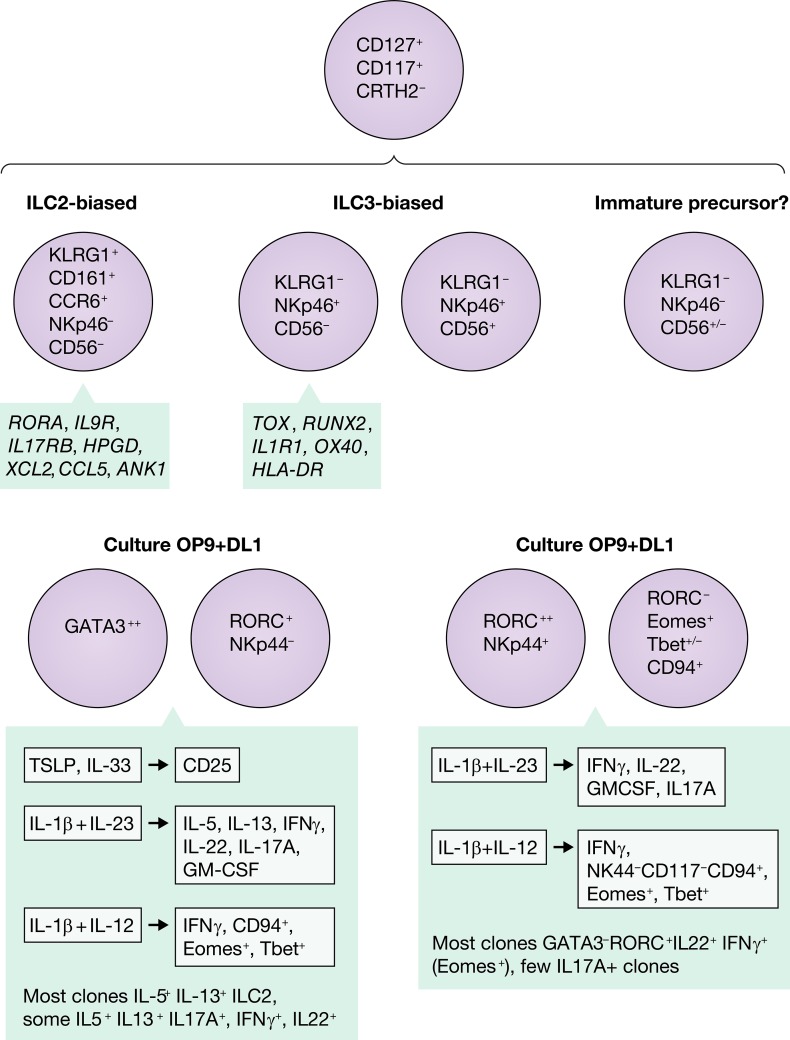
Human ILC precursors contain populations of KLRG1+ ILCs biased towards the ILC2 lineage and NKp46+ ILCs biased towards the ILC3 lineage.
Abstract
In this issue of JEM, Nagasawa et al. (https://doi.org/10.1084/jem.20190490) have undertaken a detailed study of the cell surface phenotype, transcriptional profile, and cytokine secretion of ILC progenitor populations in human peripheral blood and tissues and describe markers that highlight the heterogeneity of these chameleons.
The family of innate lymphoid cells (ILCs) is remarkably plastic in its adaptation to environmental clues, making definitive assignment to distinct “subsets” difficult. While genetically introduced fate-tracking reporter and adoptive cell transfer experiments have revealed conversion between the ILC types in mouse models (Vivier et al., 2018), precisely characterizing human ILC types is challenging.

Insights from Lewis L. Lanier.
ILCs have been classified into five subsets—natural killer (NK) cells, ILC1, ILC2, ILC3, and lymphoid tissue inducer cells—based on transcription factors driving their maturation and their secretion of cytokines, paralleling the T helper cell subsets. In simpler times, human NK cells were identified as CD3−CD56+ or CD3−NKp46+ lymphocytes, yet NKp46 and CD56 are now known to be expressed on subsets of ILCs. ILCs are discriminated from NK cells by expression of CD127 on ILCs, but not mature NK cells; however, immature NK cells may express CD127. ILC1 cells are the most difficult to distinguish from NK cells, as they are very similar (Spits et al., 2016). CD200R1 is expressed at high levels on mouse ILC1 and not NK cells (Weizman et al., 2017), but Nagasawa et al. detected that CD200R1 on all human ILC and human CD56+CD200R1+ cells may give rise to NK cells, ILC1, and ILC3 (Chen et al., 2018). Similarly, while expression of Eomes is useful to discriminate mouse NK cells from mouse ILC1, Eomes expression appears to be more variable in humans. CRTH2 and CD161 are prototypic markers of ILC2, but CD161 is also expressed by NK cell subsets. A further complication arises due to phenotypic differences in ILC populations based on their tissue residency.
In this issue of JEM, Nagasawa, et al. (2019) analyze the subset of human CD127+ lymphocytes lacking mature “lineage” markers isolated from blood, tonsil, and nasal polyps. Starting with a population expressing CD117 that has previously been suggested to contain ILC progenitor populations, unsupervised clustering based on multidimensional flow cytometry analysis revealed three subsets: one characterized by expression of NKp46 and CD56, another by expression of KLRG1, and a third lacking NKp46 and KLRG1 and CD56+/−. The KLRG1−CD56+/−NKp46− population is proposed to be composed of immature precursor cells, in that single cell clones derived from the subset were capable of making all combinations of cytokines (IL-5, IL-13, IL17A, IL-22, and IFNγ). The KLG1+ population was clearly skewed toward the ILC2 differentiation pathway and never acquired expression of CD56 or NKp46. By contrast, the KLRG1-negative cells expressing NKp46 and/or CD56 were biased toward ILC3 commitment and were unable to be pushed into an ILC2 phenotype in response to different culture conditions. This study was a tour de force in combining high-dimensional flow cytometry, transcriptional microarray assays, assay for transposase-accessible chromatin using sequencing (ATAC-Seq), single cell cloning, and functional analysis of ILC isolated from peripheral blood, tonsil, and nasal polyps. The functional studies involved culturing these ILC precursors with OP9 feeder cells in the presence or absence of delta-like 1 and stimulating the different populations or single cell clones with IL-1b, IL-12, IL-23, IL-33, and thymic stromal lymphopoietin. Upon exposure to IL-12 and IL-1b, some of both the ILC2-biased or ILC3-biased populations up-regulated CD94, Eomes, and Tbet and secreted IFNγ, similar to an ILC1 or NK cell phenotype. No transcription factor gene could definitively assign a cell population to a particular ILC1, ILC2, or ILC3 lineage in which in certain conditions, all of these ILCs were capable of expressing RORC, GATA3, Tbet, or EOMES. A caveat to these in vitro experiments is whether, in vivo, these progenitor cells would ever encounter such a simple environment with only one or two cytokines at relevant concentrations to induce these changes. These in vitro experiments clearly demonstrate the potential for these outcomes, but it will be necessary to document how these cell types further mature in vivo to validate their physiological relevance. Fortunately, with recent advances in the ability to perform RNA sequencing and ATAC-Seq on single cells, it will be possible to explore whether these cells types are expanded during infection, inflammation, or cancer with even small biopsy samples available.
Plasticity of ILC precursors and differentiation into ILC2 and ILC3. Nagasawa, et al. (2019) identify four subsets of human ILC precursor or progenitor cells based on expression of KLRG1 and NKp46, highlight discriminating transcription factors in the populations, and demonstrate their plasticity when stimulated with DL1 and polarizing cytokine environments.
Collectively, the message from this elegant work by Nagasawa, et al. (2019) is that while ILCs can be pigeon-holed into “ILC2” or “ILC3” bins based on a few markers, in reality there are many transitional states blurring these distinctions, and these ILCs have remarkable plasticity that allows them to adapt to their environment to mediate appropriate biological responses.
Acknowledgments
With acknowledgments to Jefferson Airplane.
References
- Chen L., et al. . 2018. Immunity. 49:464–476.e4. 10.1016/j.immuni.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, M., et al. J. Exp. Med. 2019 doi: 10.1084/jem.20190490. [DOI] [Google Scholar]
- Spits H., et al. . 2016. Nat. Immunol. 17:758–764. 10.1038/ni.3482 [DOI] [PubMed] [Google Scholar]
- Vivier E., et al. . 2018. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Weizman, et al. . 2017. Cell. 171:795–808.e12. 10.1016/j.cell.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]