Silverman et al. demonstrate that complement activation features prominently in retinitis pigmentosa in close association with activated microglia. This response mediates adaptive neuroprotection for photoreceptors by facilitating a C3-CR3–dependent clearance of apoptotic photoreceptors by microglial phagocytosis.
Abstract
Complement activation has been implicated as contributing to neurodegeneration in retinal and brain pathologies, but its role in retinitis pigmentosa (RP), an inherited and largely incurable photoreceptor degenerative disease, is unclear. We found that multiple complement components were markedly up-regulated in retinas with human RP and the rd10 mouse model, coinciding spatiotemporally with photoreceptor degeneration, with increased C3 expression and activation localizing to activated retinal microglia. Genetic ablation of C3 accelerated structural and functional photoreceptor degeneration and altered retinal inflammatory gene expression. These phenotypes were recapitulated by genetic deletion of CR3, a microglia-expressed receptor for the C3 activation product iC3b, implicating C3-CR3 signaling as a regulator of microglia–photoreceptor interactions. Deficiency of C3 or CR3 decreased microglial phagocytosis of apoptotic photoreceptors and increased microglial neurotoxicity to photoreceptors, demonstrating a novel adaptive role for complement-mediated microglial clearance of apoptotic photoreceptors in RP. These homeostatic neuroinflammatory mechanisms are relevant to the design and interpretation of immunomodulatory therapeutic approaches to retinal degenerative disease.
Introduction
Retinitis pigmentosa (RP), an important blinding disease worldwide, consists of a genetically heterogeneous group of inherited photoreceptor degenerations involving monogenic mutations in genes expressed predominantly in photoreceptors and retinal pigment epithelial (RPE) cells (Daiger et al., 2013). Although gene therapy for one specific causative mutation (RPE65) has recently gained approval (Russell et al., 2017), broadly applicable, efficacious treatment is still unavailable for most affected patients, who typically progress to legal blindness by 40 yr of age and severe loss of peripheral and central vision by 60 yr of age (Hartong et al., 2006). This unmet medical need underscores the need to understand general mechanisms that underlie the progressive loss of photoreceptors to discover therapies that can slow degeneration and preserve visual function. Clinical and laboratory studies of RP have revealed that in addition to the physiological changes occurring within mutation-bearing photoreceptors, the disease is characterized by non–cell-autonomous neuroinflammatory changes in the retina; these changes include the presence of activated microglia (Roque et al., 1996; Gupta et al., 2003; Zhao et al., 2015) and the up-regulation in inflammatory cytokine expression (Yoshida et al., 2013; Sudharsan et al., 2017), which can influence the progression of photoreceptor degeneration (Mustafi et al., 2012; Peng et al., 2014; Zabel et al., 2016). However, the consequences of innate immune system activation in the retina in the context of photoreceptor degeneration, and the molecular and cellular mechanisms underlying its activation and effects, are not well understood.
Complement expression and activation constitute one important aspect of innate immune system activation in central nervous system (CNS) function and pathology, in both the retina and the brain. Although complement function is necessary for normal development of neural circuits through synaptic elimination (Stevens et al., 2007; Schafer et al., 2012), it has been implicated as an etiologic factor in neurodegeneration (Tenner et al., 2018). In the retina, polymorphisms in multiple complement-related genes have been associated with genetic risk for age-related macular degeneration (AMD; Fritsche et al., 2013), a disease featuring photoreceptor apoptosis and atrophy (Dunaief et al., 2002), as well as complement-laden deposits and increased complement activation (Mullins et al., 2000; Crabb et al., 2002). Common to both RP and AMD is the presence of activated microglia that translocate into the photoreceptor layer from the inner retina to come into close proximity to degenerating photoreceptors on histopathological analyses (Gupta et al., 2003; Sennlaub et al., 2013; Lad et al., 2015); these microglia are also thought to be sources of complement and complement regulatory factors within the retina (Ma et al., 2013; Rutar et al., 2014; Natoli et al., 2017). Although clinical investigations involving the inhibition of complement activation as a therapeutic objective have been initiated for the treatment of AMD (Yaspan et al., 2017), the adaptive functions versus deleterious dysregulation of complement activation in retinal pathologies have not been clearly discerned, nor have the underlying mechanisms of action linking complement and neurodegeneration been fully elucidated.
In the current study, we sought to understand the involvement of complement in photoreceptor degeneration by using the rd10 mouse model of RP, in which a missense mutation in exon 13 of the Pde6b gene results in a time-dependent degeneration of rod photoreceptors beginning at postnatal day (P)18 (Chang et al., 2007). We aimed to investigate spatiotemporal patterns of complement activation occurring in the retina and to correlate these to ongoing photoreceptor degeneration and microglial activation. We investigated the function and consequences of complement activation by examining photoreceptor degeneration in rd10 mice that were deficient for either complement component 3 (C3; the central complement component) and complement receptor 3 (CR3; the microglia-expressed receptor to iC3b, a product of C3 activation). We discovered that complement expression and activation in the retina were prominently activated by rd10-related degeneration; in particular, increased C3 expression was found primarily in microglia translocated to the outer nuclear layer (ONL), with a concurrent opsonization of degenerating photoreceptors by iC3b. The net effect of C3 activation appears to be adaptive, as genetic deletion of C3 resulted in accelerated photoreceptor degeneration, an effect that was phenocopied with the genetic ablation of CR3. We found that this adaptive complement function related to the phagocytic clearance of apoptotic photoreceptors by microglia, which helped to promote homeostasis in the degenerating retina and limit collateral effects that induce further non–cell-autonomous photoreceptor degeneration. While dysregulated inflammatory responses involving excessive complement and microglial activation are currently thought to predominantly exacerbate retinal degeneration, our findings here conversely highlight the adaptive functions of complement and microglia that serve to promote homeostasis and limit photoreceptor loss. These factors are likely consequential in the overall understanding of the adaptive versus deleterious effects of neuroimmune activation in the CNS and require consideration in the design and interpretation of complement-targeted therapies for retinal diseases such as RP and AMD.
Results
Complement expression and activation coincide spatiotemporally with photoreceptor degeneration in RP
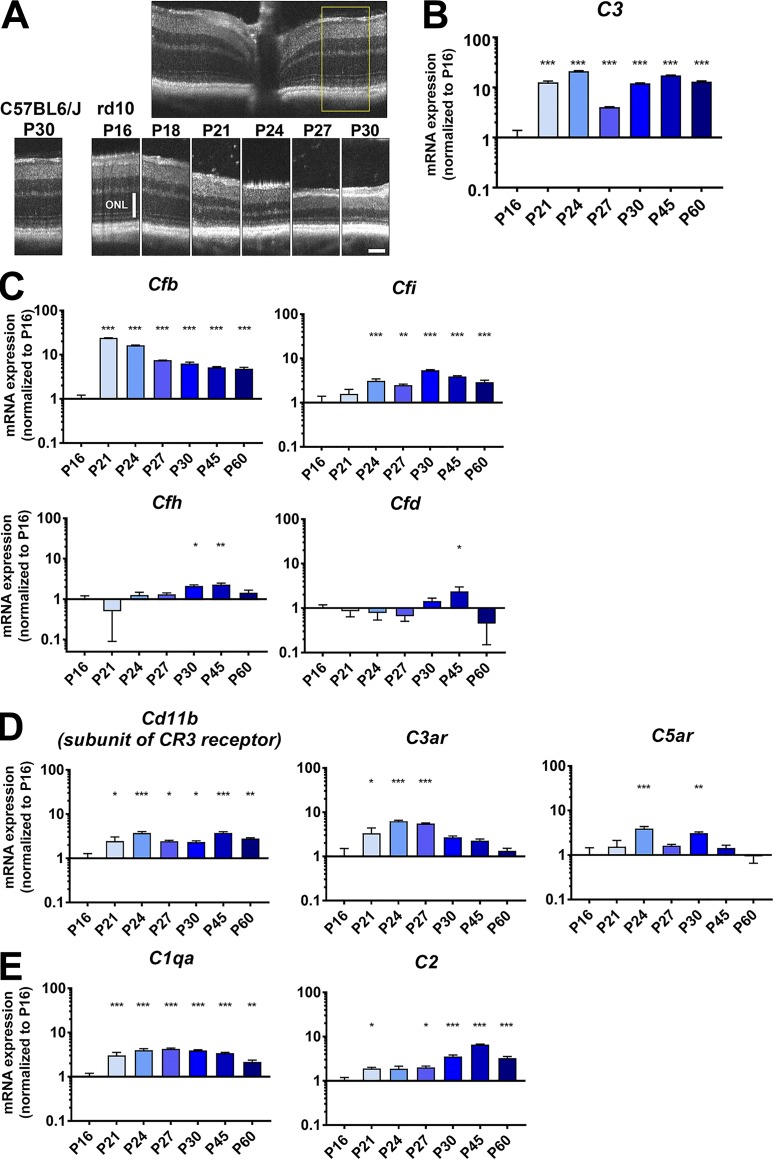
To address the potential contribution of local complement-mediated mechanisms to photoreceptor degeneration, we first investigated how complement expression varied spatiotemporally within the retina across the period of degeneration. We performed quantitative PCR to measure retinal mRNA levels of complement components, regulatory factors, and receptors during the highly stereotypical progression of photoreceptor degeneration in the rd10 mouse. We observed that relative to P16, a time point preceding the onset of rod degeneration, retinal levels of C3 mRNA were prominently up-regulated beginning at P21, coinciding with the onset of rod degeneration, and were maintained at elevated levels (>10-fold) up to P60 (Fig. 1, A and B). Increases in mRNA levels for regulatory proteins in the alternative pathway, Cfb and Cfi, were similarly elevated in magnitude and timing, while those for Cfh and Cfd were smaller and occurred later, peaking at P45 (Fig. 1 C). Expression levels of complement receptors, Cd11b, a component of CR3, as well as C3ar and C5ar, were elevated during rod degeneration (Fig. 1 D), as were components of the classic pathway, C1qa and C2 (Fig. 1 E), underscoring the possibility that complement-mediated signaling to retinal microglia, which express these complement receptors, is occurring in this scenario. Similar mRNA expression changes were not detected for these transcripts over the same developmental time points in WT (C57BL6/J) retina during the first postnatal month (Fig. S1). These findings reveal a prominent up-regulation of multiple complement components that is induced locally in the retina upon the onset and progression of rd10-related degeneration.
Figure 1.
mRNA expression of complement components, regulatory factors, and receptors demonstrate prominent up-regulation during photoreceptor degeneration in the rd10 mouse retina. (A) In vivo OCT of the central retina of rd10 mice demonstrated progressive atrophy of the ONL from P16 to P30. Inset (yellow box) shows the retinal locus of longitudinal comparison. Scale bar, 100 µm. (B) mRNA expression levels of C3 in the rd10 retina from P16 to P60 were analyzed. (C) Analysis of mRNA expression of complement regulatory factors showed early and marked up-regulation for Cfb and Cfi, but not for Cfh or Cfd. (D and E) mRNA levels for receptors of complement components, including Cd11b, C3ar, and C5ar (D), as well as for components of the classic complement pathway, C1qa and C2 (E), were also significantly increased during photoreceptor degeneration. mRNA expression levels at different time points were normalized to levels at P16. P values for comparisons relative to levels at P16: *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA with Dunnett’s multiple comparison test; n = 4 animals per time point; data collected from two independent experiments for each gene. All data shown as mean ± SEM.
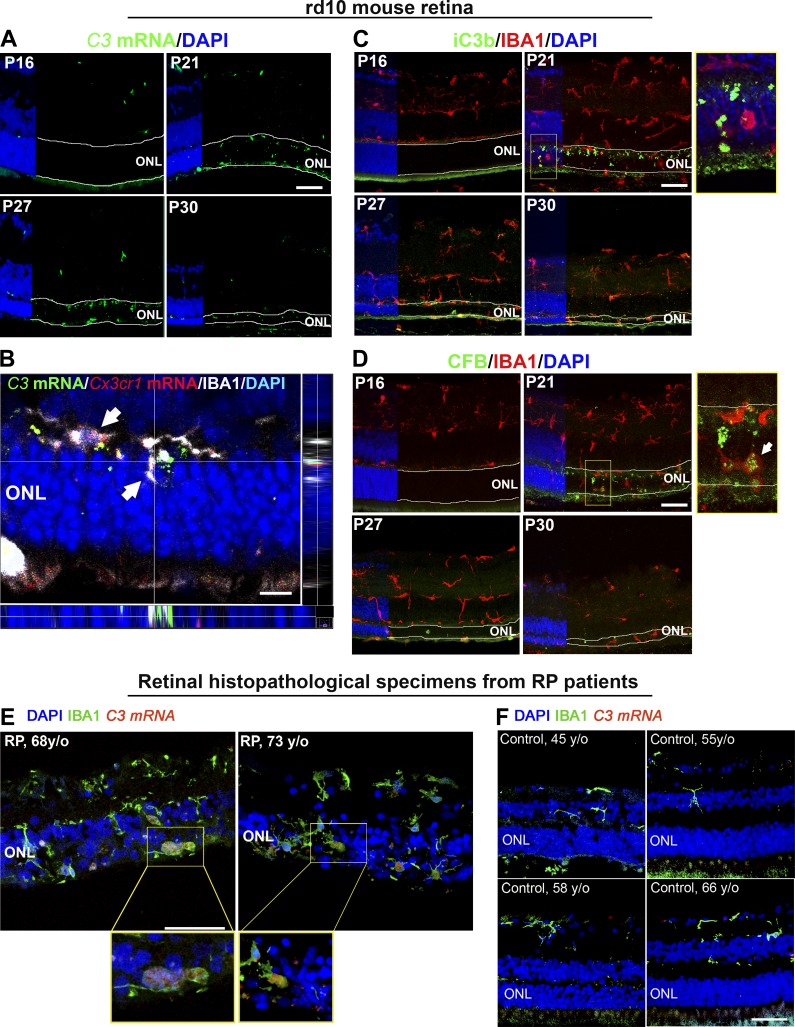
To discover the retinal locus of complement molecule expression, we performed fluorescent in situ hybridization of C3 mRNA in rd10 retinal sections at time points across rod degeneration (P16–P30). While intraretinal C3 expression was not significantly detected at P16, it emerged prominently at P21 within the degenerating ONL of the photoreceptor and was sustained up to P30 (Fig. 2 A). C3 mRNA labeling colocalized predominantly with IBA1+ cells, which also expressed Cx3cr1 mRNA, indicating microglia translocating into the ONL during rd10 degeneration (Zhao et al., 2015) likely constituted the primary source of C3 up-regulation (Fig. 2 B). Immunopositivity for iC3b was detected in the ONL beginning at P21, localizing to photoreceptor nuclei (Fig. 2 C), and the positive regulator of C3 activation complement factor b (CFB) also colocalized with IBA1+ cells in the ONL beginning at P21 (Fig. 2 D). These findings indicate that microglia translocating into the ONL at the start of rod degeneration likely contribute to C3 expression, with resulting C3 activation occurring in close proximity to degenerating photoreceptors. Up-regulated C3 expression within ONL microglia was similarly detected in histopathological specimens from human patients diagnosed with RP, but absent in healthy, non-RP retinas (Fig. 2, E and F). This indicates C3 up-regulation as an inflammatory response in the context of inherited photoreceptor degenerations that is conserved across species.
Figure 2.
Increased complement expression and activation during photoreceptor loss are spatially localized to the degenerating ONL. (A) Analysis of C3 mRNA expression by in situ hybridization in retinal sections from rd10 animals aged P16–P30 demonstrated increased labeling located specifically within the ONL (highlighted by boundary lines) in sections from P21 to P30. (B) High-magnification analysis at P21 demonstrated that labeling for C3 (green) colocalized spatially with regions of Cx3cr1 (red) detected with in situ hybridization, and IBA1 immunopositivity (white), indicating prominent C3 up-regulation within microglia translocating into the degenerating ONL, as indicated by arrows. Orthogonal views demonstrate localization of C3 and Cx3cr1 mRNA within the IBA1+ microglial cytoplasm. Scale bar, 10 µm. (C) Immunohistochemical analysis for iC3b (green) demonstrated minimal immunopositivity at P16, but localized within the ONL beginning at P21. High-magnification analysis (inset) showed iC3b deposition on somata of ONL cells near IBA1+ (red) microglia. (D) Immunohistochemical analysis for CFB (green), a positive regulator of C3 activation, similarly demonstrates increased labeling beginning at P21, localizing in and around IBA1+ (red) microglia in the ONL (inset). n = 3 mice per time point; data collected from two independent experiments. (E) Localization of C3 mRNA expression by in situ hybridization (red) in histopathologic retinal sections from patients with RP (n = 2) demonstrates labeling in IBA1+ microglia (green) that have translocated into the ONL; insets show high-magnification views of ONL microglia with labeling for C3 mRNA within DAPI-labeled nuclei. (F) Similar evaluations of retinal sections from healthy middle-aged control patients without a diagnosis of retinal disease (n = 4) demonstrated an absence of microglia in the ONL or detectable C3 expression. Scale bars, 50 µm. Data collected from two independent experiments each for mouse and human samples.
Photoreceptor degeneration is accelerated with genetic deficiency for C3 in rd10 mice
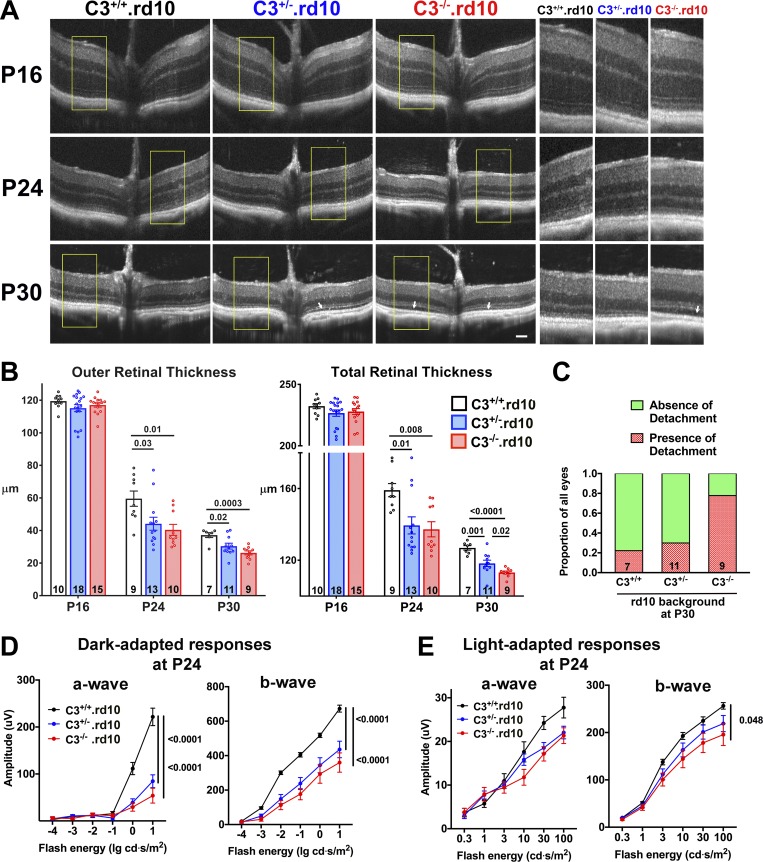
To investigate whether C3 up-regulation and activation constitute an adaptive mechanism or conversely a deleterious contribution to rd10-related photoreceptor degeneration, we crossed mice that were genetically ablated for the C3 gene into the rd10 background, creating within individual litters three genotypes containing both, one, or neither copies of the intact C3 gene: C3+/+.rd10, C3+/−.rd10, and C3−/−.rd10. Quantitative comparison of C3 mRNA levels between these genotypes demonstrated gene dose-dependent reductions in C3+/−.rd10 and C3−/−.rd10 retinas relative to C3+/+.rd10 retinas (0.69 ± 0.17, P = 0.31, and 0.24 ± 0.05, P = 0.01, respectively). Optical coherence tomography (OCT) imaging of central retina at P16 before degeneration onset revealed no significant differences across all three genotypes in the general laminated structure of the retina or in the thicknesses of the overall retina or of the outer retinal layers (Fig. 3, A and B). As rod degeneration progressed at P24 and P30, retinal thicknesses were significantly decreased in animals deleted for one or both copies of C3; mean thicknesses were least in C3−/−.rd10 animals and intermediate in C3+/−.rd10 animals. As rod outer segments degenerate at these time points, shallow areas of detachment of the outer retina from the underlying RPE layer emerged as previously described (Pennesi et al., 2012); these detachments were most prevalent in C3−/−.rd10 animals, followed by C3+/−.rd10 animals (Fig. 3, A and C). Electroretinography (ERG) assessment of dark-adapted responses (which are contributed to by both rod and cone photoreceptors) at P24 demonstrated significantly diminished a- and b-wave amplitudes in C3+/−.rd10 and C3−/−.rd10 animals, relative to C3+/+.rd10 animals (Fig. 3 D), with the magnitude of the mean amplitudes between the genotypes ordered as C3+/+.rd10 > C3+/−.rd10 > C3−/−.rd10, matching those for retinal thicknesses. At P24, cone-dominated light-adapted responses demonstrated no significant differences in a-wave amplitudes and a slight difference in b-wave amplitudes between genotypes (Fig. 3 E). Later, at P35, when rod degeneration is fairly complete and cone degeneration underway (Chang et al., 2007), C3−/−.rd10 animals demonstrated significantly decreased cone responses in both the a- and b-wave amplitudes (Fig. S2), indicating that secondary cone degeneration following rod degeneration was also accelerated with C3 deficiency in the rd10 retina.
Figure 3.
In vivo structural and functional evidence of accelerated photoreceptor degeneration in C3-deficient rd10 mice. (A) OCT imaging was performed in rd10 mice that had been genetically ablated for C3 before (P16) and during (P24, P30) rod degeneration; littermates of the three genotypes were analyzed and compared. Insets (yellow boxes) show magnified and juxtaposed images comparing corresponding retinal areas. (B) Quantification of outer (from OPL to RPE) and total (from vitreal surface to the RPE) retinal thicknesses demonstrated statistical similarity between the three genotypes at P16 but accelerated thinning in C3+/−.rd10 and C3−/−.rd10 animals (number of eyes analyzed per group provided at the bottom of each column). (C) Analysis of retinal detachments observed (white arrows in A); the relative prevalence of such detachments at P30 was highest in C3−/−.rd10 and lowest in C3+/+.rd10 animals. (D) ERG evaluation at P24 demonstrated decreased dark-adapted, rod-dominant a- and b-wave amplitudes in C3+/−.rd10 and C3−/−.rd10 relative to C3+/+.rd10 animals. (E) Light-adapted, cone-mediated responses were similar in a-wave amplitudes, but slightly decreased in b-wave amplitudes for C3−/−.rd10 relative to C3+/+.rd10 animals (number of eyes analyzed: C3+/+.rd10 = 12; C3+/−.rd10 = 11, and C3−/−.rd10 = 9). P values derived from a two-way ANOVA with Tukey’s multiple comparisons test; data collected from five and three independent experiments for OCT and ERG, respectively. All data shown as mean ± SEM.
Corroborating histological assessments in retinal sections revealed that ONL thickness, delineated by immunohistochemical labeling for rod-expressed rhodopsin and nuclear staining with DAPI, demonstrated significantly lower ONL thickness in C3+/−.rd10 and C3−/−.rd10 animals relative to C3+/+.rd10 animals, reflecting accelerated rod degeneration at both P24 and P30 (Fig. S3, A and B). The densities of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)+ photoreceptors in the ONL at P30 were significantly higher in C3+/−.rd10 and C3−/−.rd10 animals, reflecting increased accumulation of apoptotic photoreceptors in C3 deficiency (Fig. S3, C and D).
To ascertain that C3 deficiency accelerated photoreceptor degeneration in a context related specifically to the rd10 mutation, we evaluated the structure and function of retinas from control C3+/+, C3+/−, and C3−/− animals, which do not possess the rd10 mutation across the same P16–P30 time period. While C3 deficiency has been previously associated with late-onset retinal degeneration in aged mice (Yu et al., 2012; Mukai et al., 2018), we did not detect C3-related retinal thinning in the first postnatal month across the same time points evaluated in rd10 mice in OCT evaluations; outer and total retinal thicknesses were similar across all three genotypes at P16 and P30 (Fig. S4 A). ERG assessments at P24 revealed no differences in photoreceptor-dependent a-wave amplitudes, and conversely, slightly increased b-wave amplitudes in C3+/− and C3−/− animals relative to C3+/+ animals (Fig. S4 B), likely reflecting decreased pruning of first-order photoreceptor synapses during retinal development (Stevens et al., 2007). Taken together, these findings show that decreased C3 levels in the specific context of rd10-related changes, rather than with C3 deficiency alone, resulted in an accelerated degeneration of photoreceptors.
Genetic deficiency for CR3, a receptor binding iC3b, accelerates rd10-related photoreceptor degeneration
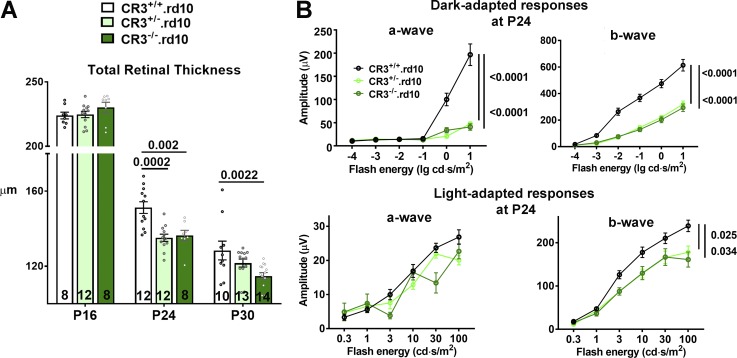
We hypothesized that the mechanism underlying the adaptive function of C3 up-regulation and activation in the rd10 retina involved physiological changes in activated retinal microglia in the photoreceptor layer. Our immunohistochemical studies had detected increased deposition of C3b/iC3b within the ONL in close proximity to activated translocated microglia, in which Cd11b, a component of the microglia receptor CR3, was found to be expressed. As iC3b-CR3 binding constitutes a significant mode of complement–microglia interaction enabling the clearance of opsonized debris by microglial phagocytosis (Hong et al., 2016; Xu et al., 2017), we investigated the adaptive role of CR3-mediated microglia function by crossing mice genetically ablated for Cd11b (hence CR3) into the rd10 background, generating litters containing the three genotypes: CR3+/+.rd10, CR3+/−.rd10, and CR3−/−.rd10. Quantitative comparison of Cd11b mRNA levels between these genotypes demonstrated gene dose-dependent reductions in CR3+/−.rd10 and CR3−/−.rd10 retinas relative to CR3+/+.rd10 retinas (0.26 ± 0.02, P < 0.004, and 0.001 ± 0.0002, P < 0.001, respectively). OCT imaging demonstrated a course of photoreceptor degeneration in CR3-deficient animals that phenocopied that observed in C3-deficient animals; at peak (P24) and late (P30) rod degeneration, CR3-deficient retinas demonstrated significantly greater rates of retinal thinning, with retinal thicknesses decreasing in the following order of genotypes: CR3+/+.rd10 > CR3+/−.rd10 > CR3−/−.rd10 (Fig. 4 A). ERG assessments at P24 similarly demonstrated marked decreases in dark-adapted a- and b-wave amplitudes in CR3-deficient mice relative to their CR3-sufficient littermates, while corresponding cone-mediated light-adapted responses were affected to a lesser degree (Fig. 4 B). This congruence between C3 and CR3 deficiency phenotypes supports the notion that C3b/iC3b signaling to retinal microglia through the CR3 receptor contributes to the complement-mediated adaptive mechanism in rd10-related photoreceptor degeneration. As found for C3 deficiency, examination of CR3−/− control animals lacking the rd10 mutation from P16 to P30 did not reveal evidence for structural and functional retinal degeneration (Fig. S4, C and D) but conversely showed small increases in retinal thickness and b-wave amplitudes. Unlike in the mature retina, where microglia help maintain synaptic structure and function (Wang et al., 2016), postnatal microglia over the time points examined are likely engaged in C3-CR3–mediated developmental synaptic pruning (Stevens et al., 2007), which is expectedly decreased here with CR3 deficiency. These findings indicate C3-CR3 signaling as a mode of complement–microglia interaction constituting an adaptive mechanism that limits rd10-related structural and functional photoreceptor degeneration.
Figure 4.
CR3 deficiency in the rd10 mouse demonstrates an accelerated phenotypic degeneration similar to that observed in C3 deficiency. (A) OCT imaging was performed in mice on the rd10 background (CR3+/+.rd10) and compared with animals in which one (CR3+/−.rd10) or both (CR3−/−.rd10) copies of the Cd11b (CR3) gene was genetically ablated. At P16, total retinal thickness was similar between all three genotypes. At P24 and P30, accelerated degeneration was found in CR3+/−.rd10 and CR3−/−.rd10 animals, relative to CR3+/+.rd10 animals. Number of eyes analyzed in each group is provided at the bottom of each column. (B) ERG evaluation of retinal function at P24 demonstrated significantly and markedly lower dark-adapted, rod-dominant a- and b-wave amplitudes in CR3+/−.rd10 and CR3−/−.rd10 relative to CR3+/+.rd10 animals. Comparison of light-adapted, cone-mediated responses did not show significant differences in a-wave amplitudes, but b-wave amplitudes were decreased in CR3+/−.rd10 and CR3−/−.rd10 animals. Number of eyes analyzed: 14, 16, and 10, for CR3+/+.rd10, CR3+/−.rd10, and CR3−/−.rd10 animals, respectively. P values were derived from a two-way ANOVA with Tukey’s multiple comparisons test; data collected from four independent OCT and ERG experiments. All data shown as mean ± SEM.
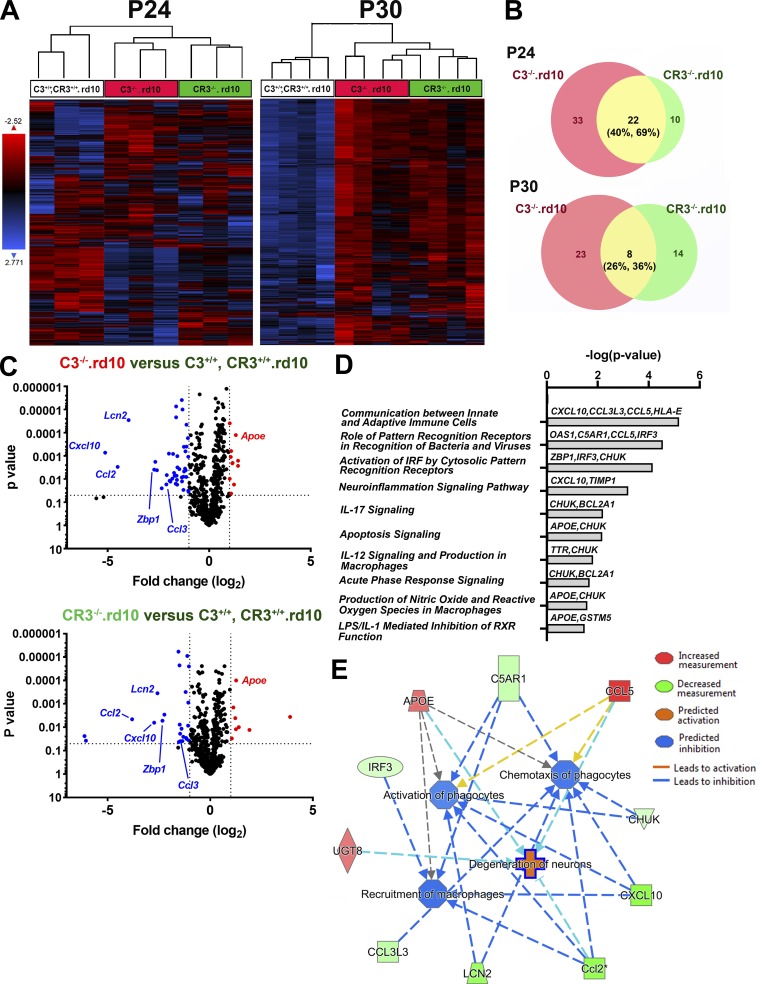
C3- and CR3-deficient rd10 retinas demonstrate similarities in mRNA expression of inflammation-related genes
To further investigate the inflammatory mechanisms involving C3-CR3–mediated adaptation to photoreceptor degeneration, we compared the mRNA expression of 758 inflammatory genes in complement-sufficient rd10 retinas (C3+/+, CR3+/+.rd10) versus either C3- (C3−/−.rd10) or CR3-deficient (CR3−/−.rd10) retinas using the Nanostring profiling platform. At both peak (P24) and late (P30) phases of rod degeneration, unsupervised clustering of all expressed transcripts revealed greater similarity between C3−/−.rd10 and CR3−/−.rd10 retinas compared with C3+/+, CR3+/+.rd10 retinas (Fig. 5 A). Identification of genes differentially expressed (DE; fold-change >2.0; P < 0.05) between (1) C3+/+, CR3+/+.rd10 versus C3−/−.rd10 and (2) C3+/+, CR3+/+.rd10 versus CR3−/−.rd10 retinas at P24 and P30 time points revealed that genes DE as a result of C3 deficiency overlapped considerably with those arising from CR3 deficiency, particularly at P24 (40–44% overlap; Fig. 5 B). A number of these shared DE genes were linked to microglial function and demonstrated patterns of differential expression that were similar in direction and magnitude (Fig. 5 C) between C3 and CR3 deficiency. These included Apoe, which has been broadly implicated in microglial responses to aging and pathological neurodegeneration (Krasemann et al., 2017; Kang et al., 2018); Lcn2, which regulates inflammation, in part through the up-regulation of microglial phagocytosis (Xing et al., 2014; Parmar et al., 2018); and Cxcl10, which influences microglial recruitment and debris clearance (Skripuletz et al., 2013). Gene ontogeny analyses using Ingenuity Pathway Analysis found that shared DE genes at P24 featured in canonical pathways involving interferon regulatory factor signaling, neuroinflammation signaling, and pattern recognition receptor signaling (Fig. 5 D) and were associated with functions of increased neuronal degeneration and decreased phagocyte activation and chemotaxis (Fig. 5 E). These findings indicate a common molecular pattern of inflammatory changes between C3 and CR3 deficiency that appear influential in regulating the activation and phagocytic clearance.
Figure 5.
mRNA profiling of inflammatory-related genes using Nanostring reveals common differentially regulated genes between C3- and CR3-deficient rd10 retinas. Nanostring multiplex analysis was used to profile mRNA expression levels of 757 neuroinflammatory-related genes in retinas from the three genotypic groups at P24 and P30: (1) C3+/+, CR3+/+.rd10, (2) C3−/−.rd10, and (3) CR3−/−.rd10 retinas (n = 3–4 animals in each group at each age). (A) Unsupervised clustering of all genes expressed above background levels demonstrated similar patterns of gene expression between C3−/−.rd10, and CR3−/−.rd10 retinas at both P24 and P30. (B) DE genes (fold-change ≥2.0; P < 0.05) between C3+/+, CR3+/+.rd10 and C3−/−.rd10 retinas (red circles) and between C3+/+, CR3+/+.rd10 and CR3−/−.rd10 retinas (green circles) were identified at P24 and P30, and the number of DE genes in common between the two groups (yellow segments) was counted and expressed as fractional compositions in each group. Data from P24 and P30 collected in separate independent experiments. (C) Volcano plots highlighting the distributions of DE genes at P24 for both cross-group comparisons; annotations highlight DE genes common to both comparisons. (D) Listing of canonical pathways represented by common DE genes at P24 (n = 3). (E) Network analysis of functions attributed to common DE genes at P24.
Microglial phagocytosis of apoptotic photoreceptors is regulated by a C3-CR3–dependent mechanism
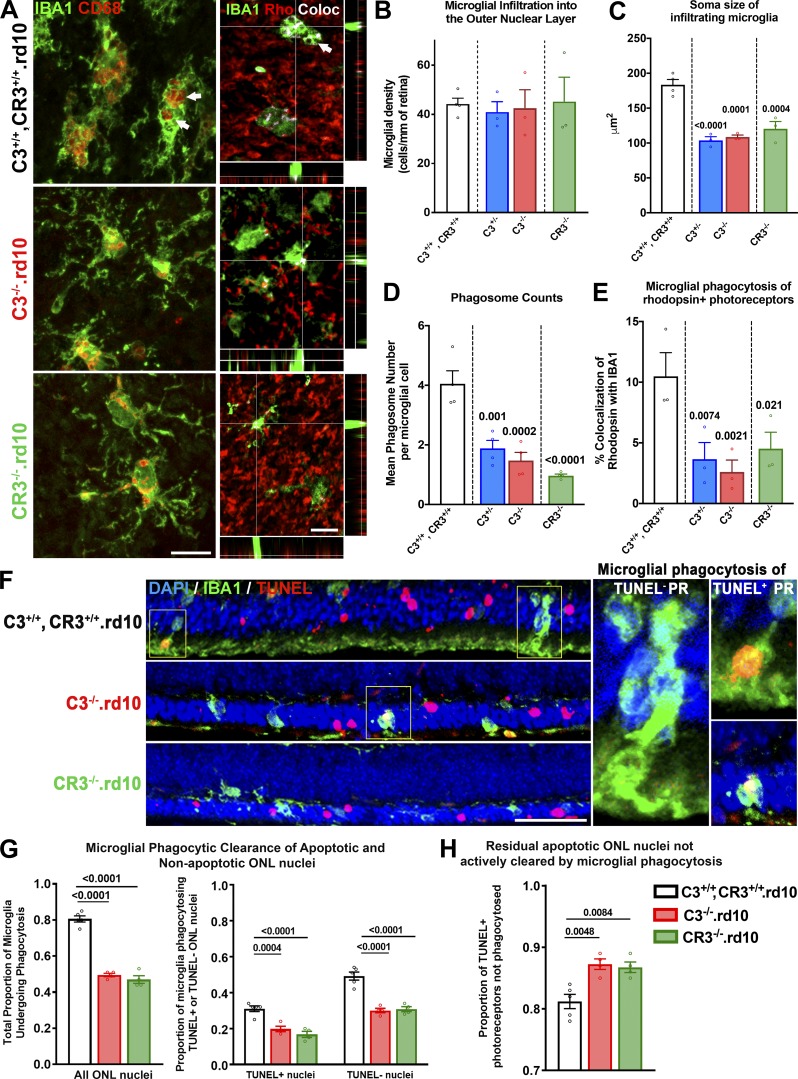
As microglial responses in pathological situations can modulate the rate of neurodegeneration, we investigated how C3 and CR3 deficiency influences microglial morphology and physiology in the rd10 retina. We examined microglia in the ONL of flat-mounted retinas from C3- and CR3-sufficient (C3+/+, CR3+/+.rd10), C3-haploinsufficient (C3+/−.rd10), C3-deficient (C3−/−.rd10), and CR3-deficient (CR3−/−.rd10) animals at P24 (Fig. 6 A). While the density of ONL microglia was similar across all four genotypes (Fig. 6 B), the mean soma size of C3+/+, CR3+/+.rd10 microglia was significantly larger than those of the other three genotypes (Fig. 6 C), corresponding to a higher mean number of phagosomes located in the soma (Fig. 6 D). These differences corroborated with a greater degree of colocalization between rhodopsin immunopositivity and IBA1+ ONL microglia of C3+/+, CR3+/+.rd10 animals, indicating that decreased expression of either C3 or CR3 resulted in lower ability of microglia to internalize degenerating photoreceptors (Fig. 6 E).
Figure 6.
Microglial phagocytosis of apoptotic photoreceptors is decreased with C3 and CR3 deficiency in the rd10 retina. (A–E) Immunohistochemical analysis of microglia translocating into the ONL was performed in flat-mounted retina isolated at P24 from: C3+/+, CR3+/+.rd10, C3−/−.rd10, and CR3−/−.rd10 mice. (A) IBA1+ microglia (green) in the ONL demonstrating a deramified and amoeboid morphology were found in close contact with rhodopsin-expressing rods (red). Many microglia also demonstrated one or more CD68-immunopositive intracellular phagosomes (arrows) that also colocalized with rhodopsin immunopositivity (depicted by white pixels). Scale bars, 10 µm. Densities of ONL microglia were similar between genotypes (B), but microglia from animals with genetic ablation of C3 or CR3 showed significantly smaller mean soma sizes (C) that corresponded to lower mean numbers of intracellular phagosomes per microglia (D) and decreased extents of rhodopsin colocalization within microglia (E), which reflected decreased microglial phagocytosis of rod photoreceptors. (F–H) Comparative analysis of phagocytosis of viable (TUNEL−) and apoptotic (TUNEL+) photoreceptors by ONL microglia was performed for the three genotypes at P24. (F) Phagosomes within IBA1+ microglia were observed to contain both TUNEL+ and TUNEL− nuclei (insets). Scale bar, 50 µm. (G) Decreased phagosomes in C3−/−.rd10 and CR3−/−.rd10 microglia relative to C3+/+, CR3+/+.rd10 microglia was reflected in decreased total phagocytic capacity; these differences were found for the subsets of microglia phagocytosing TUNEL+ photoreceptor nuclei and phagocytosing TUNEL− photoreceptor nuclei. (H) The proportions of TUNEL+ nuclei remaining in the ONL were increased in C3 and in CR3 deficiency, indicating a greater accumulation of uncleared apoptotic photoreceptors in these genotypes. P values indicate comparisons to the C3+/+, CR3+/+.rd10 genotype using a one-way ANOVA with Dunnett’s multiple comparison test; n = 3–4 animals per genotypic group; all data collected from two independent experiments each. All data shown as mean ± SEM.
As microglia in rd10-related degeneration can phagocytose both apoptotic and nonapoptotic photoreceptors, enabling clearance of dead cells and phagoptosis of living cells, respectively (Zhao et al., 2015), we examined the relative phagocytosis of these targets by microglia in the contexts of C3 and CR3 deficiency (Fig. 6 F). We found that the proportion of ONL microglia engaged in photoreceptor phagocytosis (containing one or more photoreceptor-containing phagosomes) was significantly decreased in C3−/−.rd10 and CR3−/−.rd10 animals compared with C3+/+, CR3+/+.rd10 animals; this measure of phagocytotic activity was decreased for both the phagocytosis of apoptotic TUNEL+ photoreceptors and that of TUNEL− photoreceptors (Fig. 6 G). This decreased ability of C3- and CR3-deficient microglia to phagocytose apoptotic photoreceptors also resulted in an increased proportion of TUNEL+ photoreceptors that were left uninternalized in these genotypes (Fig. 6 H).
To confirm that C3-CR3 expression and interaction involving microglia directly regulate microglial phagocytic capacity, we isolated retinal microglia from C3+/+, CR3+/+.rd10, C3−/−.rd10, and CR3−/−.rd10 animals and evaluated their ability to phagocytose fluorescently labeled bovine photoreceptor outer segments (POSs) in vitro. In a medium supplemented with C3-depleted serum, microglia lacking either C3 or CR3 expression demonstrated a lower rate of POS internalization than microglia capable of expressing both C3 and CR3 (Fig. S5, A and B). However, when exogenous C3 was added to the medium (in the form of normal serum), the proportion of microglia phagocytosing POSs increased in the C3−/−.rd10 group to that found in the C3+/+, CR3+/+.rd10 group, while that in the CR3−/−.rd10 group remained unchanged. Together, these findings indicate that microglial expression and secretion of C3 can result in the opsonization of targets such as photoreceptors to promote their phagocytosis via a CR3-dependent mechanism.
C3 and CR3 deficiency in the rd10 retina are associated with increased microglial neurotoxicity
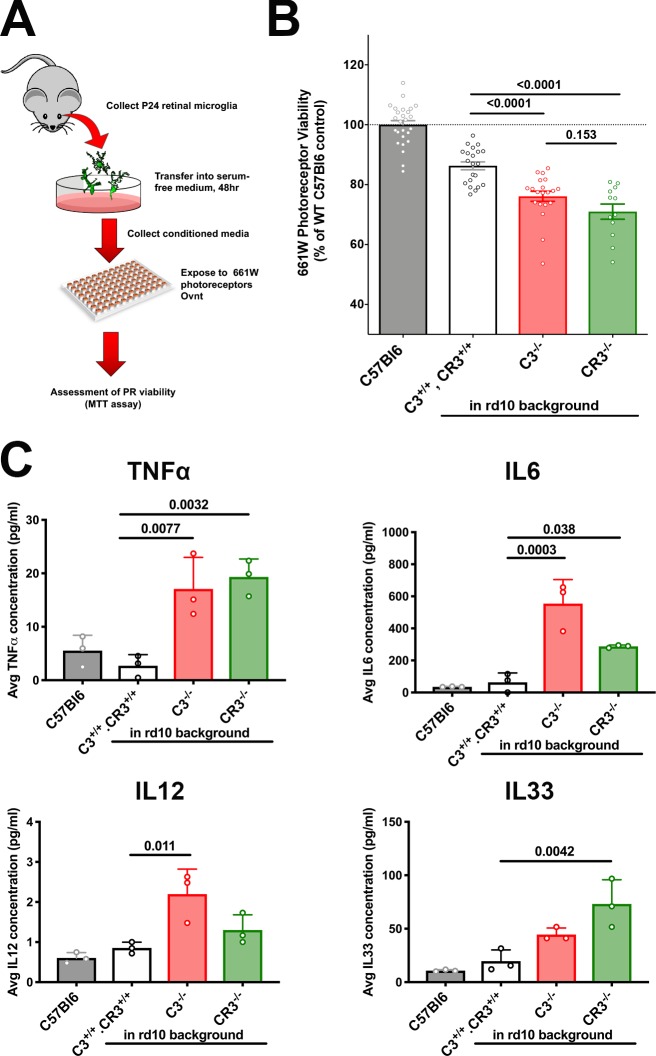
To further explore the relationship between decreased microglial phagocytic clearance and increased photoreceptor degeneration with C3 and CR3 deficiency in the rd10 retina, we examined the ability of retinal microglia cultured from different genotypes of mice to exert neurotoxic effects on photoreceptors. Retinal microglia were isolated from P24 animals from WT (C57BL6), C3+/+, CR3+/+.rd10, C3−/−.rd10, and CR3−/−.rd10 animals and cultured in serum-free media for 48 h. The conditioned media were then exposed to cultured 661W photoreceptors, whose resulting cell viability was assessed using the MTT assay (Fig. 7 A). While conditioned media from C3+/+, CR3+/+.rd10 microglia decreased photoreceptor viability more than microglia from WT nondegenerating retina, conditioned media from C3- or CR3-deficient rd10 retinas exerted the largest negative impact on photoreceptor viability (Fig. 7 B). Consistent with previous studies in which phagocytosis of apoptotic cells was linked with the down-regulation of microglial activation and proinflammatory cytokine expression (Magnus et al., 2001), we found that protein levels of the cytokines TNFα, IL6, IL12, and IL33 were elevated in C3- and/or CR3-deficient rd10 retinas (Fig. 7 C). These findings indicate that while increased nonclearance and accumulation of apoptotic photoreceptors owing to decreased microglial phagocytosis may directly compromise the survival of nearby photoreceptors (Pérez-Garijo et al., 2013), phagocytosis-deficient microglia can exert more neurotoxic effects on nearby photoreceptors, possibly via increased proinflammatory cytokine secretion.
Figure 7.
Decreased microglial phagocytic capacity from C3-CR3 deficiency is correlated with increased microglial neurotoxicity to 661W photoreceptors. (A) Schematic of in vitro neurotoxicity assay. (B) Microglia from degenerating rd10 retinas of all three genotypes was associated with a greater neurotoxic effect than microglia from C57BL6 retinas. Phagocytosis-deficient microglia from C3−/−.rd10 and CR3−/−.rd10 retinas in addition were associated with significantly greater neurotoxicity than C3+/+, CR3+/+.rd10 retinas. (C) Increased neurotoxicity in C3−/−.rd10, and CR3−/−.rd10 microglia was associated with increased secretion of proinflammatory cytokines TNFα, IL6, IL12, and IL33 in their conditioned media, compared with C57Bl6 WT and C3+/+, CR3+/+.rd10 microglia. P values were derived from a one-way ANOVA with Tukey’s multiple comparisons test; n = 14 and 3 replicates per condition for the neurotoxicity assay (A) and cytokine level assay, respectively. Data collected from three independent experiments in B and two independent experiments in C. All data shown as mean ± SEM.
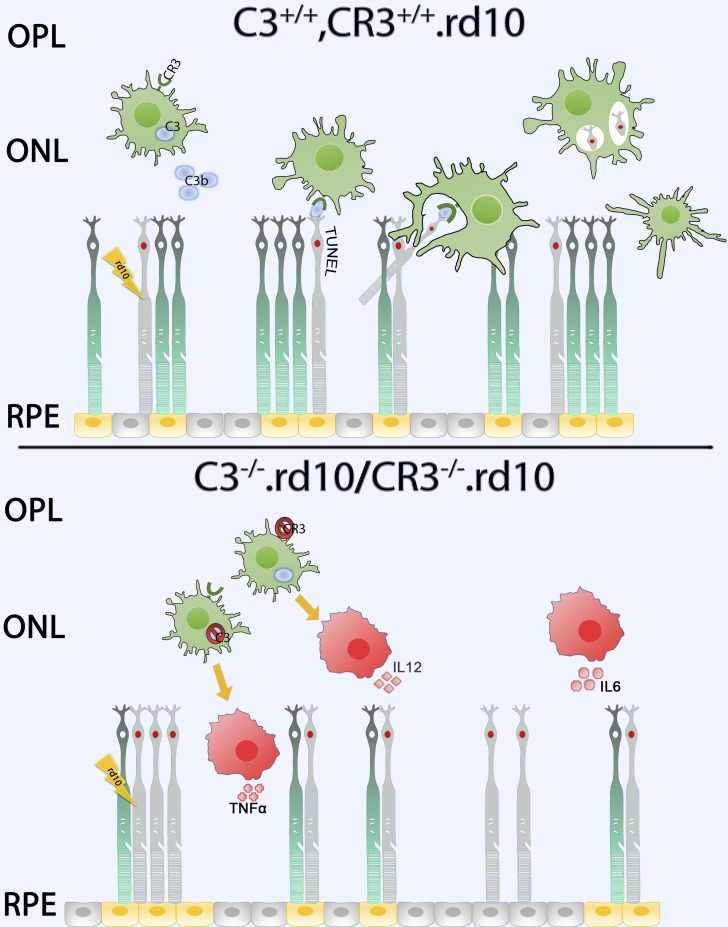
Taken together, the results from our study examining the role of complement in photoreceptor degeneration demonstrate an adaptive, complement-dependent mechanism that is induced upon the onset of degenerative changes that facilitates the clearance of apoptotic photoreceptors by activated retinal microglia expressing C3 and CR3 (Fig. 8). The inhibition of this mechanism can reduce adaptive homeostasis to retinal pathology, inducing further deleterious alterations in the immune environment of the retina, which results in an acceleration of structural and functional degeneration.
Figure 8.
Schematic depicting a C3-CR3–dependent mechanism of microglial phagocytic clearance of apoptotic photoreceptors in the degenerating rd10 retina. Upper panel: In the complement-sufficient rd10 (C3+/+, CR3+/+.rd10) mouse retina, microglia, sensing the degeneration of mutation-bearing rod photoreceptors, translocate into the ONL, up-regulating their expression and secretion of C3. Extracellular activation of C3b results in the opsonization of TUNEL+ apoptotic photoreceptors. CR3-expressing microglia recognize the opsonized targets via iC3b-CR3 binding and clear them via phagocytosis, after which ONL microglia are decreased in their activation status. Lower panel: In rd10 retinas for which either C3 (C3−/−.rd10) or CR3 (CR3−/−.rd10) is genetically deficient, the opsonization of apoptotic photoreceptors or the recognition of opsonized targets is impaired, respectively. Failure of microglial phagocytic clearance results in the accumulation of apoptotic TUNEL+ rods that induce a more rapid, non–cell-autonomous degeneration of nearby rod photoreceptors. Activated microglia potentiate degeneration through increased secretion of proinflammatory cytokines (TNFα, IL6, IL12), augmenting neurotoxicity to photoreceptors, and accelerating the course of structural and functional degeneration in the rd10 retina.
Discussion
We discover here that RP-related photoreceptor degeneration, which arises from rod photoreceptor gene mutation, induces a marked, non–cell-autonomous, neuroinflammatory response that involves microglial activation and local up-regulation of complement expression and activation in the photoreceptor layer. This response is spatiotemporally coincident with photoreceptor degeneration and constitutes an adaptive response aimed at restoring homeostasis by microglial phagocytic clearance of apoptotic photoreceptors, limiting the deleterious sequelae of cell death in the retina. The mechanism underlying this response, comprising microglial secretion of C3 and microglial responses via the receptor CR3, provides insight into how the innate immune system copes with the disruptive influence of intraparenchymal neuronal death and presents opportunities for immunomodulatory therapy to optimize preservation of photoreceptor structure and function in RP and other neurodegenerative retinal diseases.
Studies examining multiple CNS regions have documented complement up-regulation and expression upon neurodegeneration onset in the brain (Hong et al., 2016) and the retina (Rutar et al., 2011), indicating that these constitute a conserved response by the innate immune system to neuronal injury. However, corresponding to the diversity of complement-related effector functions, the net impact of complement activation on neural outcomes has been quite varied, with complement inhibition by genetic or pharmacological means producing both positive and negative outcomes depending on the context. In the majority of studies, genetic and/or pharmacological inhibition of complement components, such as C1q (Silverman et al., 2016; Jiao et al., 2018), C3 (Jha et al., 2011; Natoli et al., 2017; Bosco et al., 2018; Katschke et al., 2018), CFB, and complement factor d (CFD; Sweigard et al., 2015), have decreased retinal neurodegeneration, and genetic ablation of complement components in models of Alzheimer’s disease (Hong et al., 2016; Shi et al., 2017) and frontotemporal dementia (Lui et al., 2016) has decreased neural loss in the brain. These findings indicate that complement-mediated responses triggered in these injury contexts are likely excessive and/or inappropriately regulated and culminate in worsened overall synaptic degeneration and neuronal death.
There is, however, emerging evidence that in other contexts complement activation may conversely serve adaptive immune functions. In the retina, absent any induced injury, long-term genetic ablation of complement molecules has resulted in a slow deterioration of structure and function that gradually becomes evident in aged animals (Coffey et al., 2007; Yu et al., 2012; Hoh Kam et al., 2013; Mukai et al., 2018), indicating that constitutive complement function may be required for long-term tissue homeostasis. In the DBA/2J mouse model of glaucoma, genetic and pharmacological inhibition of C3 increased retinal ganglion cell degeneration (Harder et al., 2017), while in the amyloid precursor protein transgenic Alzheimer’s disease mouse model, C3 ablation and inhibition worsened total Aβ and fibrillar amyloid plaque burden in the brain, altered microglial activation, and increased hippocampal neuronal degeneration (Wyss-Coray et al., 2002; Maier et al., 2008). These examples illustrate that complement can exert positive adaptive effects in CNS pathologies, although the underlying cellular and molecular mechanisms are currently incompletely elucidated.
We discover here that part of the adaptive complement response to photoreceptor degeneration resulting from the rod-specific Pde6b mutation in the rd10 mouse model involves (1) the increased expression of C3, primarily by microglia translocating into the photoreceptor layer at the outset of rod degeneration; (2) the opsonization of photoreceptors by iC3b; and (3) the phagocytic clearance of photoreceptors by microglia via the receptor CR3 (Fig. 8). In the retina, while the expression of C3 has been attributed to astrocytes (Harder et al., 2017) and RPE cells (Luo et al., 2013), we find in the rd10 retina, as well as in human histopathological specimens of RP, that activated microglia in the outer retina comprise the main source of C3 expression, similar to that found in a model of light-induced photoreceptor injury (Rutar et al., 2011). The increased iC3b deposition in the ONL upon the onset of photoreceptor degeneration, elevated retinal expression of Cd11b, a component of the CR3, and the ONL translocation of CR3-expressing microglia, point to increased microglia–photoreceptor interactions via iC3b-CR3 binding (Bajic et al., 2013; Xu et al., 2017) as part of an adaptive complement response. C3–CR3 interaction has been previously demonstrated as a molecular mechanism employed by microglia to eliminate supernumerary synapses during normal brain development (Stevens et al., 2007) and to clear synaptic debris following injury-induced synaptic degeneration (Norris et al., 2018), but which may drive maladaptive phagocytosis of synapses in certain CNS pathologies (Berg et al., 2012; Hong et al., 2016). Additionally, iC3b-CR3 function in microglia has also been shown to mediate beneficial clearance of apoptotic neurons (Elward and Gasque, 2003) and extracellular debris (Fu et al., 2012; Hadas et al., 2012) following neural injury via phagocytosis. In the context of photoreceptor degeneration, the positive versus negative contributions of phagocytosis in microglia have also been mixed. Microglia have been shown to accelerate degeneration by phagocytosing stressed but still living photoreceptors in a process termed phagoptosis (Brown and Neher, 2014); inhibition of this form of phagocytosis in the rd10 model by the blockade of the vitronectin receptor resulted in slowed degeneration (Zhao et al., 2015), while its augmentation in CX3CR1 deficiency accelerated loss of photoreceptor structure and function (Zabel et al., 2016). Conversely, in a model of retinal detachment, retinal microglia appear to support photoreceptor survival; when autofluorescent microglia, which presumably had phagocytosed photoreceptors, were pharmacologically depleted, degeneration was increased (Okunuki et al., 2018). We discover here in the rd10 retina that both C3 and CR3 participate in facilitating microglial phagocytosis of apoptotic photoreceptors; when either of these factors are deficient, decreased microglial phagocytosis was accompanied by increased accumulation of apoptotic cells, increased proinflammatory cytokine expression, and accelerated degeneration (Fig. 8). These current and previous findings indicate that microglia employ a variety of molecular mechanisms to contribute to a balance between the adaptive clearance of apoptotic cells and the maladaptive phagoptosis of stressed but still living photoreceptors.
We found that apoptotic cell accumulation, occurring here from a deficiency in C3-CR3–mediated microglial clearance, was associated with compromised survival of the remaining photoreceptors. The mechanism for this deleterious influence may arise from apoptotic photoreceptors releasing TNFα to induce non–cell-autonomous apoptosis in adjacent cells (Pérez-Garijo et al., 2013; Fuchs and Steller, 2015) and/or increased proinflammatory effects of greater microglial activation, which may be triggered by signals released from the lysis of uncleared cells (Hochreiter-Hufford and Ravichandran, 2013) and decreased down-regulatory effects on microglial activation that typically occur after the successful phagocytosis of apoptotic cells (Magnus et al., 2001). We found support for a microglia-mediated deleterious influence, as phagocytosis-deficient microglia cultured from C3- or CR3-ablated retinas demonstrated greater in vitro neurotoxicity to photoreceptors, an effect that was also associated with increased retinal levels of proinflammatory cytokines, including TNFα and IL6. This altered inflammatory milieu in the rd10 ONL may carry over beyond the period of rod death to secondarily impact cone death as was observed here, underscoring a broad contribution to complement-mediated apoptotic cell clearance to promoting long-term homeostasis in the degenerating retina.
Beyond the involvement of microglia, it is possible that C3 or CR3 deficiency may induce physiological abnormalities in other retinal cells, which may additionally contribute to accelerated photoreceptor degeneration in the rd10 retina. In aged C3−/− animals, increased photoreceptor degeneration and accumulation of subretinal material had been detected and linked to deficient RPE phagocytosis of POSs (Hoh Kam et al., 2013). However, these phenotypes were inapparent in young C3−/− and in C3−/−.rd10 animals examined here. Investigating the potential contribution of complement-related RPE abnormalities to degeneration in this context will require further study in closer detail.
One of the limitations of our study is that relatively early onset of degeneration in the rd10 retina had largely precluded the use of “cell-fate” mapping to differentiate endogenous microglia from potentially infiltrating monocytes, as we had previously done (Ma et al., 2017). Despite this, we have used the term “microglia” to refer to the IBA1+ myeloid cells migrating to the ONL and engaging in C3 up-regulation and phagocytosis of photoreceptors. While studies of other genetic mouse models of photoreceptor degeneration have described the infiltration of CCR2+ monocytes into the subretinal space (Sennlaub et al., 2013; Kohno et al., 2015), we had previously found that in the rd10 retina, these infiltrating macrophages appeared to be largely confined to the subretinal space and did not engage photoreceptors via dynamic phagocytosis as observed for CCR2-negative microglia within the ONL (Zhao et al., 2015). In other models of photoreceptor degeneration involving more acute injury, endogenous microglia, as tracked by cell-fate mapping, were found to translocate from the inner to the outer retina, while monocyte-derived macrophages enter into the inner retina via the retinal circulation; however, these cells were not found substantially in the ONL to contribute to photoreceptor phagocytosis (O’Koren et al., 2016; Ma et al., 2017). While we are unable to completely rule out possible contributions from monocyte-derived macrophages to phagocytic clearance, the overall available evidence indicates endogenous microglia to be the predominant cell type involved.
As a result of studies associating increased complement activation with retinal degeneration and implicating complement-related genes as conferring genetic risks of AMD (Fritsche et al., 2013; Mullins et al., 2017), there is much current interest in strategies to inhibit complement activation as a treatment for degenerative retinal diseases (Xu and Chen, 2016). Accordingly, interventions targeting C3 and positive complement regulatory factors to dampen complement activation in the retina (Boyer et al., 2017; Yaspan et al., 2017) have entered clinical investigation. Our findings in this study may be useful here, as they help define the adaptive contributions of complement in the diseased retina and highlight the possibility that therapeutic down-regulation of complement activation may entail decreased apoptotic cell clearance and potential deleterious effects. While complement activation (Mullins et al., 2014), microglial presence in disease lesions (Gupta et al., 2003), and apoptotic photoreceptors (Dunaief et al., 2002) feature in the histopathology of AMD, as in the rd10 retina, further studies are needed to determine the similarities between these contexts. Together, our findings highlight complexities in the multiple adaptive versus deleterious roles of complement in retinal diseases that may require interventions to exert calibrated modulation, rather than broad and complete inhibition, for optimal therapeutic effects.
Materials and methods
Experimental animals
Experiments were conducted according to protocols approved by a local Institutional Animal Care and Use Committee (National Eye Institute Animal Care and Use Committee) and adhered to the Association for Research in Vision and Ophthalmology Statement on animal use in ophthalmical and vision research. The following strains of mice were obtained from the Jackson Laboratory: mice homozygous for the Pde6brd10 (rd10) mutation (#004297), mice homozygous for C3 mutation (#029661, C3−/−), and mice homozygous for the Itgamtm1Myd targeted mutation (#003991, CR3−/−). Animals were housed in a National Institutes of Health (NIH) animal facility under a 12-h light/dark cycle with food ad libitum. To assess complement deficiency in rd10 animals, mice were crossed with C3−/− or CR3−/− animals to generate animals heterozygous for these mutations in the rd10 background, which were interbred to yield animals of all three genotypes (i.e., C3+/+.rd10, C3+/−.rd10, and C3−/−.rd10) in the same litter. Similarly, C3−/− and CR3−/− were bred with WT C57BL6/J animals to generate all three genotypes in the same litter for age-matched comparisons. All experimental animals were genotyped by gene sequencing to confirm the absence of the rd8 mutation (Mattapallil et al., 2012). CR3−/− mice are identified to be on a mixed C57Bl6/J; C57Bl6/N genetic background, and thus sequencing of Crb1 revealed a heterozygous rd8 mutation present. The rd8 mutation was selectively bred out when CR3−/− animals were crossed with either rd10 or C57Bl6/J mice. Both male and female mice in the age range of P16 to P35 were used as specified in individual experiments.
Human eye tissue
Adult human eyes with clinically diagnosed RP were obtained from the donor programs of the Foundation Fighting Blindness (Columbia, MD). Retinal tissue was obtained from the following archived specimens: FFB-316 and FFB-340. Healthy control eyes without diagnoses of retinal disease were collected by Lions Eye Bank (Omaha, NE), Georgia Eye Bank (Atlanta, GA), Southern Eye Bank (Metairie, LA), and Old Dominion Eye Foundation (North Chesterfield, VA) and distributed through the National Disease Research Interchange (Philadelphia, PA). Eyes were fixed with 2–4% paraformaldehyde (PFA), and the retinas were dissected out and sectioned into 12-µm-thick cryostat sections.
mRNA analysis by quantitative PCR
Mice were euthanized by CO2 inhalation,eyes were enucleated, and their retinas were harvested by dissection. Cells in retina tissue were lysed by trituration and homogenized using QIAshredder spin columns (Qiagen). Total RNA was isolated using the RNeasy Mini kit (Qiagen) according to the manufacturer’s specifications. First-strand synthesis was performed with PrimeScript cDNA synthesis kit (Takara) following the manufacturer’s instructions. Quantitative RT-PCR was performed using a SYBR green RT-PCR kit (Affymetrix) in a CFX96 Real-Time PCR system (Bio-Rad) under the following conditions: denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, and then 60°C for 45 s. Threshold cycle values were calculated and expressed as fold-change determined using the comparative threshold cycle (2ΔΔCT) method. Housekeeping genes Gapdh and Actb were used as internal controls. Oligonucleotide primer pairs used are listed in Table S1.
Detection and localization of mRNA in retinal sections using multiplex in situ hybridization
For in situ hybridization of mRNA probes, frozen retinal tissues from murine and human eyes were sectioned at 12 µm. The resulting sections were heated at 65°C for 1 h and immersed in 4% PFA for 15 min and then in RNAscope Protease III for 30 min at 40°C. In situ detection of mouse C3 (Mm-C3, #417841), mouse Cx3cr1 (Mm-Cx3cr1, #314221), and human C3 (Hs-C3, #430701) was performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics) according to the manufacturer’s specifications. Fluorescent signal amplification was performed with TSA Plus fluorescein or Cy5 (1:1,500, NEL760001KT; PerkinElmer).
Immunohistochemistry and TUNEL labeling of retinal tissue
Enucleated eyes from euthanized mice were dissected to form posterior segment eye cups and fixed in 4% PFA in PBS for 1–2 h at 4°C. Eye cups were either cut into 30-µm-thick sections using a cryostat (Leica CM3050S) or dissected to form retinal flat-mounts. Flat-mounted retinas were blocked for 1 h in 2% Roche blocking reagent (11096176001; Millipore Sigma) in PBS with Tween 20 at room temperature. Sections previously subjected to multiplex in situ hybridization were also analyzed. Primary antibodies, which included IBA1 (1:500, Wako; #019-19741), C3b/iC3b (1:100, Hycult Bio; #HM1078), CFB (1:200, Santa Cruz Biotechnology; #sc-67141), rhodopsin (1:100, EMD Millipore; #MAB5356), and CD68 (1:100, Bio-Rad; #MCA1957 [clone FA-11]), were diluted in blocking buffer and incubated overnight for sections and 3 d for flat mounts at 4°C with gentle shaking. After washing in 1× PBS with Tween 20, retinal samples were incubated for 1 h at room temperature for sections or overnight for flat mounts with secondary antibodies (Alexa Fluor 488/568–conjugated anti-rabbit or mouse IgG, 1:250; Thermo Fisher Scientific) and mounted on slides with Prolong Gold with DAPI (Invitrogen) to label cell nuclei. Apoptosis of retinal cells was assayed using a TUNEL assay (in situ cell death detection kit, TMR red; Roche) according to the manufacturer’s specifications. Stained retinal samples were imaged with confocal microscopy (Zeiss LSM 700 or Zeiss LSM 880). For analysis at high magnification, multiplane z-series were collected using 20× or 40× objectives; each z-series spanned from the outer plexiform layer (OPL) to the subretinal space for retinal flat mounts and over a depth of 30 µm for retinal sections, with each section spaced 1 µm apart. Confocal image stacks were viewed and analyzed with Zen software (Zeiss) and/or ImageJ (NIH).
Histological image analysis
Quantitative histological assessments of labeled retinal sections were used to assess the extent of photoreceptor degeneration in experimental mice. Mean ONL thickness was quantified by calculating the area of the ONL in an imaging field as revealed by nuclear labeling by DAPI and/or rhodopsin labeling of rods and dividing by the length of the field. The density of apoptotic photoreceptors was calculated by manually counting TUNEL+ cells in the ONL within the imaging field and dividing by the area of the ONL.
In vivo OCT imaging of mouse retina
Mice were anesthetized with intraperitoneal ketamine (90 mg/kg) and xylazine (8 mg/kg), and their pupils were dilated. Retinal structure was assessed using an OCT imaging system (Bioptigen; InVivoVue Software). Volume scans were captured consisting of 100 horizontal sequential B-scans (consisting of 1,000 A-scans each), spanning an en face retinal area of 1.4 by 1.4 mm centered on the optic nerve. Retinal thickness measurements were performed in each retinal quadrant at a radial distance of 0.7 mm from the optic nerve using the manufacturer’s software (Bioptigen; Diver) and averaged for each eye. Total retinal thickness, defined as the distance from the nerve fiber layer to the RPE, and outer retinal thickness, defined as the distance from the OPL to the inner surface of the RPE, were measured from OCT images after manual retinal segmentation. The presence of shallow retinal detachments was manually evaluated in the entire scan field and scored as present or absent.
ERG analysis
ERGs were recorded using an Espion E2 system (Diagnosys). Mice were anesthetized as described above after dark adaptation overnight. Pupils were dilated, and a drop of proparacaine hydrochloride (0.5%; Alcon) was applied on the cornea for topical anesthesia. Flash ERG recordings were obtained simultaneously from both eyes with gold wire loop electrodes, with the reference electrode placed in the mouth and the ground subdermal electrode at the tail. ERG responses were obtained at increasing light intensities over the ranges of 1 × 10−4 to 10 cd⋅s/m2 under dark-adapted conditions and 0.3 to 100 cd⋅s/m2 under a rod-saturating background light. The stimulus interval between flashes varied from 5 s at the lowest stimulus to 60 s at the highest ones. 2 to 10 responses were averaged depending on flash intensity. ERG signals were recorded with 0.3-Hz low-frequency and 300-Hz high-frequency cutoffs sampled at 1 kHz. Analysis of a-wave and b-wave amplitudes was performed using Espion ERG Data Analyzer software (version 6.0.54). The a-wave amplitude was measured from the baseline to the negative peak, and the b-wave was measured from the a-wave trough to the maximum positive peak. Statistical comparisons between ERG amplitudes between animals of different genotypes were analyzed using a two-way ANOVA.
Image analysis of microglial features
Microglia characteristics were assessed in immunolabeled retinal flat-mount preparations and frozen sections. For each flat mount of the entire retina, 20× and 40× images were acquired in each of the four quadrants; for each quadrant, one central (located midway between the optic nerve and the equator of the globe) and one peripheral (located midway between the equator of the globe and the peripheral retinal edge) imaging field was obtained. Quantitative image analysis for ONL microglia number, mean microglial soma size, microglial phagosome number, and microglia number containing TUNEL+ and TUNEL− ONL nuclei, were performed manually using computer-assisted software (analyze particles function; ImageJ). Internalization of rhodopsin by IBA1+ microglia was quantified in Imaris (Bitplane) by setting a conserved threshold for both rhodopsin and IBA1 labeling channels and computing the number of colocalized voxels.
mRNA profiling in retinal tissue using Nanostring
mRNA expression in retinal tissue was profiled and analyzed using the Nanostring platform nCounter Mouse Neuroinflammation panel containing 757 neuroinflammatory-related mouse genes and 13 internal reference controls (Nanostring; #115000237). Briefly, the total RNA from a single retina was extracted using the RNeasy kit (Qiagen). A total of 100 ng RNA in a volume of 5 µl was then hybridized to the capture and reporter probe sets at 65°C for 16 h according to the manufacturer’s instructions. The individual hybridization reactions were washed and eluted per protocol at a NIH Core Facility (Center for Cancer Research Genomics Core, National Cancer Institute), and the data were collected using the nCounter Digital Analyzer (Nanostring). Generated data were evaluated using internal QC process, and the resulting data were normalized with the geometric mean of the housekeeping genes using the nSolver 4.0 and Advanced Analysis 2.0 software (Nanostring). Retinas from C3+/+; CR3+/+, C3−/−, and CR3−/− genotypes of animals in the rd10 background at time points P24 and P30, each comprising three to four biological repeats, were analyzed. DE genes were defined as those demonstrating a difference in expression level of fold-change of ≥1.5, with a P value of <0.05 (adjusted P value by t test). Unsupervised hierarchical clustering and heat map analysis were performed using nSolver software. Canonical pathway analyses were performed using Ingenuity Pathway Analysis (Qiagen).
Isolation and culture of primary microglia
Retinal microglia were isolated from P24 C57Bl6/J, C3+/+, C3+/+.rd10, C3−/−.rd10, and CR3−/−.rd10 mice. Enucleated globes were immersed in ice-cold HBSS, and retinas were isolated by dissection before transfer into 0.2% papain solution including glucose (1 mg/ml), DNase1 (100 U/ml; Worthington), superoxide dismutase (5 mg/ml; Worthington), gentamycin (1 µl/ml; Sigma-Aldrich), and catalase (5 mg/ml; Sigma-Aldrich) in HBSS, and incubated at 8°C for 45 min and then at 28°C for 7 min. The digested tissue was dissociated by trituration and centrifuged at 150 g for 5 min at 4°C. The resulting cell pellet was resuspended with neutralization buffer containing glucose (2 mg/ml), DNase1 (100 U/ml), superoxide dismutase (5 mg/ml), catalase (5 mg/ml), antipain (50 mg/ml; Roche), d-α-tocopheryl acetate (10 mg/ml; Sigma-Aldrich), albumin (40 mg/ml), and gentamycin (1 ml/ml; Sigma-Aldrich) and again centrifuged at 150 g for 5 min at 4°C. The resulting pellet was resuspended in DMEM/Nutrient Mixture F-12 (1:1) medium with 10% FBS (Gibco) and 1× minimum essential medium nonessential amino acids solution (Sigma-Aldrich) before transfer into 75-cm2 flasks. The resulting mixed cell cultures that adhered to the bottom of the flasks were allowed to grow to confluence. Microglial cells were detached from the adherent cell layer by shaking, collected, and transferred to a new 12-well plate containing sterile glass coverslips. The culture medium with floating microglia cells was collected every 3 d and replaced with fresh culture medium. Experimentation was conducted when cumulative microglia attained 40–50% confluence.
In vitro assay for microglial phagocytosis of POSs
Microglial phagocytosis of bovine rod POSs was evaluated and compared between retinal microglia isolated from mice of different genotypes. Bovine POS preparations (InVision Bioresources) were diluted in serum-free DMEM/F12 (1:1; Gibco) to a concentration of 106 segments/ml and fluorescently labeled with the lipophilic dye DiI (Vybrant Cell-Labeling Solutions; Invitrogen) according to the manufacturer’s instructions. Labeled POSs were incubated in either 20% complete mouse serum or C3-depleted mouse serum for 1 h at 37°C and then transferred back into serum-free medium. The resulting POS suspension (105 segments in 100 µl) was then added to cultured retinal microglia (50% confluence, grown on coverslips in a 12-well plate in 1 ml of DMEM/F12) and incubated at 37°C for 2 h to allow for phagocytosis. Nonphagocytosed POSs were removed by washing, and microglia on the coverslips were fixed in 4% PFA, stained with DAPI to label nuclei, and then mounted and imaged with a Zeiss LSM 880 confocal microscope. The total number of microglia per field and the proportion of microglia incorporating fluorescently labeled POSs were measured.
In vitro assay for microglial neurotoxicity to photoreceptors
661W photoreceptor cells (gift from Dr. Muayyad Al-Ubaidi; University of Oklahoma, Oklahoma City, OK) were maintained in 96-well plates. Cultured retinal microglia from animals of different genotypes were allowed to condition in serum-free medium (DMEM/F12) for 48 h. Conditioned medium (100 µl) was then added to 661W cells for 16 h, and the resulting cell viability was assessed using an MTT cell assay kit (ATCC) following the manufacturer’s specifications. Protein analysis of conditioned medium was performed using a customized ProcartaPlex Multiplex Immunoassay panel (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, magnetic beads were seeded in triplicate onto a 96-well plate, after which conditioned media (50 µl) from retinal microglia cultured from C57BL6/J, C3+/+; C3+/+.rd10, C3−/−.rd10, and CR3−/−.rd10 animals were added to each well and incubated overnight at 4°C. The beads were washed before the addition of the detecting antibodies followed by Streptavidin-PE addition and quantification on a Luminex 200 instrument (Luminex).
Statistics
Statistical analyses were performed using Prism 7.0d (GraphPad). For comparisons involving two data columns, t tests (paired or unpaired) or nonparametric tests (Mann–Whitney U) were used, depending on whether the data followed a Gaussian distribution as determined by normality tests. A normality test (D’Agostino and Pearson) was used to analyze the distribution of all datasets. For comparisons involving three or more data columns, a one-way ANOVA (with Dunnett’s multiple-comparisons test) was used if the data followed a Gaussian distribution, and a nonparametric Kruskal–Wallis test (with Dunn’s multiple-comparisons test) was used if it did not. Datasets from different genotypes were compared using a two-way ANOVA.
Study approval
Experiments were conducted according to protocols approved by a local Institutional Animal Care and Use Committee and adhered to the Association for Research in Vision and Ophthalmology Statement animal use in ophthalmical and vision research. Eye tissue was collected under applicable regulations and guidelines with proper consent, protection of human subjects, and donor confidentiality. Eye tissue was collected under applicable regulations and guidelines with proper consent, protection of human subjects, and donor confidentiality, with approval from the Cleveland Clinic Institutional Review Board (protocol #14-057).
Online supplemental material
Fig. S1 shows analysis of complement expression in age-matched non-rd10 controls. Fig. S2 demonstrates accelerated functional cone degeneration in C3-deficient rd10 mice. Fig. S3 shows histological confirmation of accelerated photoreceptor degeneration. Fig. S4 confirms that deficiency of C3 and CR3 does not contribute to retinal deficits. Fig. S5 demonstrates in vitro evidence that C3-mediated opsonization is necessary for phagocytosis. Table S1 shows information on oligonucleotide sequences used for these studies.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Drs. Joe Hollyfield and Vera Bonilha (Cleveland Clinic Cole Eye Institute Department of Ophthalmical Research) for providing access to the Foundation Fighting Blindness human RP eye collection, and Dr. Robert Fariss (National Eye Institute, Bethesda, MD) for the gift of human control retinal tissues. The 661W cell line was obtained through a material transfer agreement with the Board of Regents of the University of Oklahoma.
This study is supported by funds from the National Eye Institute Intramural Research Program (to W.T. Wong).
The authors declare no competing financial interests.
Author contributions: S.M. Silverman and W.T. Wong designed the study. S.M. Silverman, W. Ma, L. Zhao, and X. Wang developed the methodologies and acquired in vivo data. S.M. Silverman and X. Wang performed cell culture and in vitro assays. S.M. Silverman, X. Wang, and W.T. Wong analyzed and interpreted the data. S.M. Silverman and W.T. Wong wrote and edited the manuscript. W.T. Wong supervised the study.
References
- Bajic G., Yatime L., Sim R.B., Vorup-Jensen T., and Andersen G.R.. 2013. Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. Proc. Natl. Acad. Sci. USA. 110:16426–16431. 10.1073/pnas.1311261110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg A., Zelano J., Stephan A., Thams S., Barres B.A., Pekny M., Pekna M., and Cullheim S.. 2012. Reduced removal of synaptic terminals from axotomized spinal motoneurons in the absence of complement C3. Exp. Neurol. 237:8–17. 10.1016/j.expneurol.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Bosco A., Anderson S.R., Breen K.T., Romero C.O., Steele M.R., Chiodo V.A., Boye S.L., Hauswirth W.W., Tomlinson S., and Vetter M.L.. 2018. Complement C3-Targeted Gene Therapy Restricts Onset and Progression of Neurodegeneration in Chronic Mouse Glaucoma. Mol. Ther. 26:2379–2396. 10.1016/j.ymthe.2018.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer D.S., Schmidt-Erfurth U., van Lookeren Campagne M., Henry E.C., and Brittain C.. 2017. The Pathophysiology of Geographic Atrophy Secondary to Age-Related Macular Degeneration and the Complement Pathway as a Therapeutic Target. Retina. 37:819–835. 10.1097/IAE.0000000000001392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.C., and Neher J.J.. 2014. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci. 15:209–216. 10.1038/nrn3710 [DOI] [PubMed] [Google Scholar]
- Chang B., Hawes N.L., Pardue M.T., German A.M., Hurd R.E., Davisson M.T., Nusinowitz S., Rengarajan K., Boyd A.P., Sidney S.S., et al. . 2007. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 47:624–633. 10.1016/j.visres.2006.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey P.J., Gias C., McDermott C.J., Lundh P., Pickering M.C., Sethi C., Bird A., Fitzke F.W., Maass A., Chen L.L., et al. . 2007. Complement factor H deficiency in aged mice causes retinal abnormalities and visual dysfunction. Proc. Natl. Acad. Sci. USA. 104:16651–16656. 10.1073/pnas.0705079104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb J.W., Miyagi M., Gu X., Shadrach K., West K.A., Sakaguchi H., Kamei M., Hasan A., Yan L., Rayborn M.E., et al. . 2002. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 99:14682–14687. 10.1073/pnas.222551899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger S.P., Sullivan L.S., and Bowne S.J.. 2013. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 84:132–141. 10.1111/cge.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief J.L., Dentchev T., Ying G.S., and Milam A.H.. 2002. The role of apoptosis in age-related macular degeneration. Arch. Ophthalmol. 120:1435–1442. 10.1001/archopht.120.11.1435 [DOI] [PubMed] [Google Scholar]
- Elward K., and Gasque P.. 2003. “Eat me” and “don’t eat me” signals govern the innate immune response and tissue repair in the CNS: emphasis on the critical role of the complement system. Mol. Immunol. 40:85–94. 10.1016/S0161-5890(03)00109-3 [DOI] [PubMed] [Google Scholar]
- Fritsche L.G., Chen W., Schu M., Yaspan B.L., Yu Y., Thorleifsson G., Zack D.J., Arakawa S., Cipriani V., Ripke S., et al. AMD Gene Consortium . 2013. Seven new loci associated with age-related macular degeneration. Nat. Genet. 45:433–439: e1–e2. 10.1038/ng.2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Liu B., Frost J.L., Hong S., Jin M., Ostaszewski B., Shankar G.M., Costantino I.M., Carroll M.C., Mayadas T.N., and Lemere C.A.. 2012. Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Aβ by microglia. Glia. 60:993–1003. 10.1002/glia.22331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y., and Steller H.. 2015. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 16:329–344. 10.1038/nrm3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Brown K.E., and Milam A.H.. 2003. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp. Eye Res. 76:463–471. 10.1016/S0014-4835(02)00332-9 [DOI] [PubMed] [Google Scholar]
- Hadas S., Spira M., Hanisch U.K., Reichert F., and Rotshenker S.. 2012. Complement receptor-3 negatively regulates the phagocytosis of degenerated myelin through tyrosine kinase Syk and cofilin. J. Neuroinflammation. 9:166 10.1186/1742-2094-9-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J.M., Braine C.E., Williams P.A., Zhu X., MacNicoll K.H., Sousa G.L., Buchanan R.A., Smith R.S., Libby R.T., Howell G.R., and John S.W.M.. 2017. Early immune responses are independent of RGC dysfunction in glaucoma with complement component C3 being protective. Proc. Natl. Acad. Sci. USA. 114:E3839–E3848. 10.1073/pnas.1608769114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong D.T., Berson E.L., and Dryja T.P.. 2006. Retinitis pigmentosa. Lancet. 368:1795–1809. 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford A., and Ravichandran K.S.. 2013. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 5:a008748 10.1101/cshperspect.a008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh Kam J., Lenassi E., Malik T.H., Pickering M.C., and Jeffery G.. 2013. Complement component C3 plays a critical role in protecting the aging retina in a murine model of age-related macular degeneration. Am. J. Pathol. 183:480–492. 10.1016/j.ajpath.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. . 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352:712–716. 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P., Banda H., Tytarenko R., Bora P.S., and Bora N.S.. 2011. Complement mediated apoptosis leads to the loss of retinal ganglion cells in animal model of glaucoma. Mol. Immunol. 48:2151–2158. 10.1016/j.molimm.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H., Rutar M., Fernando N., Yednock T., Sankaranarayanan S., Aggio-Bruce R., Provis J., and Natoli R.. 2018. Subretinal macrophages produce classical complement activator C1q leading to the progression of focal retinal degeneration. Mol. Neurodegener. 13:45 10.1186/s13024-018-0278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.S., Ebbert M.T.W., Baker K.E., Cook C., Wang X., Sens J.P., Kocher J.P., Petrucelli L., and Fryer J.D.. 2018. Microglial translational profiling reveals a convergent APOE pathway from aging, amyloid, and tau. J. Exp. Med. 215:2235–2245. 10.1084/jem.20180653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katschke K.J. Jr., Xi H., Cox C., Truong T., Malato Y., Lee W.P., McKenzie B., Arceo R., Tao J., Rangell L., et al. . 2018. Classical and alternative complement activation on photoreceptor outer segments drives monocyte-dependent retinal atrophy. Sci. Rep. 8:7348 10.1038/s41598-018-25557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H., Koso H., Okano K., Sundermeier T.R., Saito S., Watanabe S., Tsuneoka H., and Sakai T.. 2015. Expression pattern of Ccr2 and Cx3cr1 in inherited retinal degeneration. J. Neuroinflammation. 12:188 10.1186/s12974-015-0408-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. . 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad E.M., Cousins S.W., Van Arnam J.S., and Proia A.D.. 2015. Abundance of infiltrating CD163+ cells in the retina of postmortem eyes with dry and neovascular age-related macular degeneration. Graefes Arch. Clin. Exp. Ophthalmol. 253:1941–1945. 10.1007/s00417-015-3094-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.Y., Shang Y., Oldham M.C., Martens L.H., Gao F., et al. . 2016. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 165:921–935. 10.1016/j.cell.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Zhao J., Madden A., Chen M., and Xu H.. 2013. Complement expression in retinal pigment epithelial cells is modulated by activated macrophages. Exp. Eye Res. 112:93–101. 10.1016/j.exer.2013.04.016 [DOI] [PubMed] [Google Scholar]
- Ma W., Cojocaru R., Gotoh N., Gieser L., Villasmil R., Cogliati T., Swaroop A., and Wong W.T.. 2013. Gene expression changes in aging retinal microglia: relationship to microglial support functions and regulation of activation. Neurobiol. Aging. 34:2310–2321. 10.1016/j.neurobiolaging.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Zhang Y., Gao C., Fariss R.N., Tam J., and Wong W.T.. 2017. Monocyte infiltration and proliferation reestablish myeloid cell homeostasis in the mouse retina following retinal pigment epithelial cell injury. Sci. Rep. 7:8433 10.1038/s41598-017-08702-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus T., Chan A., Grauer O., Toyka K.V., and Gold R.. 2001. Microglial phagocytosis of apoptotic inflammatory T cells leads to down-regulation of microglial immune activation. J. Immunol. 167:5004–5010. 10.4049/jimmunol.167.9.5004 [DOI] [PubMed] [Google Scholar]
- Maier M., Peng Y., Jiang L., Seabrook T.J., Carroll M.C., and Lemere C.A.. 2008. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J. Neurosci. 28:6333–6341. 10.1523/JNEUROSCI.0829-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil M.J., Wawrousek E.F., Chan C.C., Zhao H., Roychoudhury J., Ferguson T.A., and Caspi R.R.. 2012. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest. Ophthalmol. Vis. Sci. 53:2921–2927. 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai R., Okunuki Y., Husain D., Kim C.B., Lambris J.D., and Connor K.M.. 2018. The Complement System Is Critical in Maintaining Retinal Integrity during Aging. Front. Aging Neurosci. 10:15 10.3389/fnagi.2018.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.F., Russell S.R., Anderson D.H., and Hageman G.S.. 2000. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 14:835–846. 10.1096/fasebj.14.7.835 [DOI] [PubMed] [Google Scholar]
- Mullins R.F., Schoo D.P., Sohn E.H., Flamme-Wiese M.J., Workamelahu G., Johnston R.M., Wang K., Tucker B.A., and Stone E.M.. 2014. The membrane attack complex in aging human choriocapillaris: relationship to macular degeneration and choroidal thinning. Am. J. Pathol. 184:3142–3153. 10.1016/j.ajpath.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.F., Warwick A.N., Sohn E.H., and Lotery A.J.. 2017. From compliment to insult: genetics of the complement system in physiology and disease in the human retina. Hum. Mol. Genet. 26(R1):R51–R57. 10.1093/hmg/ddx181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D., Maeda T., Kohno H., Nadeau J.H., and Palczewski K.. 2012. Inflammatory priming predisposes mice to age-related retinal degeneration. J. Clin. Invest. 122:2989–3001. 10.1172/JCI64427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli R., Fernando N., Jiao H., Racic T., Madigan M., Barnett N.L., Chu-Tan J.A., Valter K., Provis J., and Rutar M.. 2017. Retinal Macrophages Synthesize C3 and Activate Complement in AMD and in Models of Focal Retinal Degeneration. Invest. Ophthalmol. Vis. Sci. 58:2977–2990. 10.1167/iovs.17-21672 [DOI] [PubMed] [Google Scholar]
- Norris G.T., Smirnov I., Filiano A.J., Shadowen H.M., Cody K.R., Thompson J.A., Harris T.H., Gaultier A., Overall C.C., and Kipnis J.. 2018. Neuronal integrity and complement control synaptic material clearance by microglia after CNS injury. J. Exp. Med. 215:1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Koren E.G., Mathew R., and Saban D.R.. 2016. Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci. Rep. 6:20636 10.1038/srep20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okunuki Y., Mukai R., Pearsall E.A., Klokman G., Husain D., Park D.H., Korobkina E., Weiner H.L., Butovsky O., Ksander B.R., et al. . 2018. Microglia inhibit photoreceptor cell death and regulate immune cell infiltration in response to retinal detachment. Proc. Natl. Acad. Sci. USA. 115:E6264–E6273. 10.1073/pnas.1719601115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar T., Parmar V.M., Perusek L., Georges A., Takahashi M., Crabb J.W., and Maeda A.. 2018. Lipocalin 2 Plays an Important Role in Regulating Inflammation in Retinal Degeneration. J. Immunol. 200:3128–3141. 10.4049/jimmunol.1701573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B., Xiao J., Wang K., So K.F., Tipoe G.L., and Lin B.. 2014. Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J. Neurosci. 34:8139–8150. 10.1523/JNEUROSCI.5200-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi M.E., Michaels K.V., Magee S.S., Maricle A., Davin S.P., Garg A.K., Gale M.J., Tu D.C., Wen Y., Erker L.R., and Francis P.J.. 2012. Long-term characterization of retinal degeneration in rd1 and rd10 mice using spectral domain optical coherence tomography. Invest. Ophthalmol. Vis. Sci. 53:4644–4656. 10.1167/iovs.12-9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A., Fuchs Y., and Steller H.. 2013. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. eLife. 2:e01004 10.7554/eLife.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque R.S., Imperial C.J., and Caldwell R.B.. 1996. Microglial cells invade the outer retina as photoreceptors degenerate in Royal College of Surgeons rats. Invest. Ophthalmol. Vis. Sci. 37:196–203. [PubMed] [Google Scholar]
- Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S., et al. . 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 390:849–860. 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutar M., Natoli R., Kozulin P., Valter K., Gatenby P., and Provis J.M.. 2011. Analysis of complement expression in light-induced retinal degeneration: synthesis and deposition of C3 by microglia/macrophages is associated with focal photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 52:5347–5358. 10.1167/iovs.10-7119 [DOI] [PubMed] [Google Scholar]
- Rutar M., Valter K., Natoli R., and Provis J.M.. 2014. Synthesis and propagation of complement C3 by microglia/monocytes in the aging retina. PLoS One. 9:e93343 10.1371/journal.pone.0093343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., and Stevens B.. 2012. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 74:691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennlaub F., Auvynet C., Calippe B., Lavalette S., Poupel L., Hu S.J., Dominguez E., Camelo S., Levy O., Guyon E., et al. . 2013. CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1 deficient mice. EMBO Mol. Med. 5:1775–1793. 10.1002/emmm.201302692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Chowdhury S., Ma R., Le K.X., Hong S., Caldarone B.J., Stevens B., and Lemere C.A.. 2017. Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci. Transl. Med. 9:eaaf6295 10.1126/scitranslmed.aaf6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S.M., Kim B.J., Howell G.R., Miller J., John S.W., Wordinger R.J., and Clark A.F.. 2016. C1q propagates microglial activation and neurodegeneration in the visual axis following retinal ischemia/reperfusion injury. Mol. Neurodegener. 11:24 10.1186/s13024-016-0089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripuletz T., Hackstette D., Bauer K., Gudi V., Pul R., Voss E., Berger K., Kipp M., Baumgärtner W., and Stangel M.. 2013. Astrocytes regulate myelin clearance through recruitment of microglia during cuprizone-induced demyelination. Brain. 136:147–167. 10.1093/brain/aws262 [DOI] [PubMed] [Google Scholar]
- Stevens B., Allen N.J., Vazquez L.E., Howell G.R., Christopherson K.S., Nouri N., Micheva K.D., Mehalow A.K., Huberman A.D., Stafford B., et al. . 2007. The classical complement cascade mediates CNS synapse elimination. Cell. 131:1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Sudharsan R., Beiting D.P., Aguirre G.D., and Beltran W.A.. 2017. Involvement of Innate Immune System in Late Stages of Inherited Photoreceptor Degeneration. Sci. Rep. 7:17897 10.1038/s41598-017-18236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard J.H., Matsumoto H., Smith K.E., Kim L.A., Paschalis E.I., Okonuki Y., Castillejos A., Kataoka K., Hasegawa E., Yanai R., et al. . 2015. Inhibition of the alternative complement pathway preserves photoreceptors after retinal injury. Sci. Transl. Med. 7:297ra116 10.1126/scitranslmed.aab1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenner A.J., Stevens B., and Woodruff T.M.. 2018. New tricks for an ancient system: Physiological and pathological roles of complement in the CNS. Mol. Immunol. 102:3–13. 10.1016/j.molimm.2018.06.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao L., Zhang J., Fariss R.N., Ma W., Kretschmer F., Wang M., Qian H.H., Badea T.C., Diamond J.S., et al. . 2016. Requirement for Microglia for the Maintenance of Synaptic Function and Integrity in the Mature Retina. J. Neurosci. 36:2827–2842. 10.1523/JNEUROSCI.3575-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T., Yan F., Lin A.H., Lambris J.D., Alexander J.J., Quigg R.J., and Masliah E.. 2002. Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer’s mice. Proc. Natl. Acad. Sci. USA. 99:10837–10842. 10.1073/pnas.162350199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C., Wang X., Cheng C., Montaner J., Mandeville E., Leung W., van Leyen K., Lok J., Wang X., and Lo E.H.. 2014. Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke. 45:2085–2092. 10.1161/STROKEAHA.114.005733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., and Chen M.. 2016. Targeting the complement system for the management of retinal inflammatory and degenerative diseases. Eur. J. Pharmacol. 787:94–104. 10.1016/j.ejphar.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang J., Wang J.H., and Springer T.A.. 2017. Distinct recognition of complement iC3b by integrins αXβ2 and αMβ2. Proc. Natl. Acad. Sci. USA. 114:3403–3408. 10.1073/pnas.1620881114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaspan B.L., Williams D.F., Holz F.G., Regillo C.D., Li Z., Dressen A., van Lookeren Campagne M., Le K.N., Graham R.R., Beres T., et al. MAHALO Study Investigators . 2017. Targeting factor D of the alternative complement pathway reduces geographic atrophy progression secondary to age-related macular degeneration. Sci. Transl. Med. 9:eaaf1443 10.1126/scitranslmed.aaf1443 [DOI] [PubMed] [Google Scholar]
- Yoshida N., Ikeda Y., Notomi S., Ishikawa K., Murakami Y., Hisatomi T., Enaida H., and Ishibashi T.. 2013. Clinical evidence of sustained chronic inflammatory reaction in retinitis pigmentosa. Ophthalmology. 120:100–105. 10.1016/j.ophtha.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Yu M., Zou W., Peachey N.S., McIntyre T.M., and Liu J.. 2012. A novel role of complement in retinal degeneration. Invest. Ophthalmol. Vis. Sci. 53:7684–7692. 10.1167/iovs.12-10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel M.K., Zhao L., Zhang Y., Gonzalez S.R., Ma W., Wang X., Fariss R.N., and Wong W.T.. 2016. Microglial phagocytosis and activation underlying photoreceptor degeneration is regulated by CX3CL1-CX3CR1 signaling in a mouse model of retinitis pigmentosa. Glia. 64:1479–1491. 10.1002/glia.23016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zabel M.K., Wang X., Ma W., Shah P., Fariss R.N., Qian H., Parkhurst C.N., Gan W.B., and Wong W.T.. 2015. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol. Med. 7:1179–1197. 10.15252/emmm.201505298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.