Abstract
Differences in HLA-C expression are inversely correlated with HIV viral load set-point and slower progression to AIDS, linked to enhanced cytotoxic T cell immunity. Yet, beyond T cells, HLA-C serves as a dominant ligand for natural killer (NK) cell killer immunoglobulin-like receptors (KIR). Thus, we speculated that HLA-C expression levels may also impact NK activity, thereby modulating HIV antiviral control. Phenotypic and functional profiling was performed on freshly isolated PBMCs. HLA-C expression was linked to changes in NK subset distribution and licensing, particularly in HLA-C1/C1, KIR2DL3+2DL2-individuals. Moreover, high levels of HLA-C, were associated with reduced frequencies of anergic CD56neg NKs and lower frequencies of KIR2DL1/2/3+ NK cells, pointing to an HLA-C induced influence on the NK cell development in the absence of disease. In HIV infection, several spontaneous controllers, that expressed higher levels of HLA-C demonstrated robust NK-IFN-γ secretion in response to target cells, highlighting a second disease induced licensing phenotype. Thus this population study points to a potential role for HLA-C levels both in NK cell education and development.
Keywords: HLA-C, KIR, Natural killer cells, HIV-1
1. Introduction
Several host genetic determinants have been associated with control of HIV-1 infection, identified either through classic candidate-gene approaches or, more recently, through genomewide association studies (GWAS) [1], able to identify novel genetic variants in an unbiased manner. The first reported GWAS identified a number of variants associated with viral control including a previously defined specific human leukocyte antigen-B (HLA-B), as well as a single nucleotide polymorphism (SNP) 35 kilobases (Kb) upstream of the HLA-C locus (rs9264942) that showed the most significant association with viral load (VL) control [2]. Moreover, in subsequent GWAS analyses, the impact of the SNP upstream of HLA-C was confirmed in cohorts of subjects that spontaneously control HIV, in the absence of HAART [3]. Using data from 1698 patients of European ancestry, this SNP was shown to associate with differences in HLA-C mRNA levels, which was later confirmed at the protein level on primary T cells from European Americans [4]. Moreover, HLA-C expression was directly linked to the magnitude of the cytotoxic CD8+ T cell response, suggesting a critical role for HLA-C level in driving enhanced viral control through the induction of superior cytotoxic T cell activity.
However, beyond its role in presenting peptides to T cells, HLA-C is also the primary ligand for several natural killer (NK) cell receptors, called the 2-domain killer cell immunoglobulin-like receptors (KIR2Ds) [5,6]. Specifically, the low-level, but persistent expression of HLA-C, which is upregulated in the setting of inflammation, has been directly implicated in both: a) NK cell licensing during NK cell development and b) tonic inhibition of NK cells through self-recognition on mature NK cells via inhibitory KIRs [7]. Yet, how the baseline expression level of HLA-C impacts NK cells has been incompletely defined.
Here we performed an in-depth analysis of NK cell phenotype and function in a large cohort (154) of healthy HIV-uninfected subjects and 28 HIV-infected individuals to determine whether variation in HLA-C expression directly impacted NK cell profiles. While changes in HLA-C expression showed some evidence of altered NK cell licensing in healthy individuals, elevated HLA-C levels were also associated with reduced levels of KIR2D+ NK cells as well as anergic CD56neg NK cells in healthy individuals but increased numbers of cytolytic CD56dim cells and enhanced IFN-γ licensing signature among HIV+ subjects. These data point to a plausible role for HLA-C expression levels on NK cell development and function that may contribute to the pre-infection presence of more functional and less inhibited NK cells that are then able to respond more aggressively upon infection thereby contributing to enhanced antiviral control and the evolution of more effective T cell immunity.
2. Methods
2.1. Subjects and genotyping
Healthy HIV uninfected subjects were drawn from a living biobank of >1600 healthy adult blood donors between the ages of 18 and 50 who were recruited from the Boston area as part of the Brigham and Women’s Hospital Pheno Genetic Project. Demographic details of the cohort were previously described [8]. The Institutional Review Board approved the study and all subjects signed informed consent. HIV+ subjects were recruited from the Ragon Institute cohort and patients were classified into one of three disease categories: elite controller (EC, individuals that spontaneously control HIV to undetectable levels); viremic controllers (VC, individuals that spontaneously suppress viremia to below 2000 copies/ml of blood), chronic progressors (CRO, non-controllers with viral loads above 2000 copies/ml). None of the subjects received anti-retroviral treatment at the time of the study. HLA class I typing and KIR genotyping were performed as previously described [9]. HLA-C expression levels were determined using a predictive model based on HLA-C genotypes, as previously described [10]. The model predicts HLA-C expression levels based on HLA-C alleles for which HLA-C expression levels were determined in an independent cohort of 150 African Americans. The model was subsequently validated in 100 subjects of European-American and African-American descent and thus, only subjects of these races were included in the study. Importantly, because of the variation in HLA-C expression among donors encoding the same HLA-C alleles, likely related to heterozygosity, the model derived a single HLA-C expression value for each allele from this large HLA-C expression study, as a substitute for directly measuring HLA-C expression.
2.2. Flow cytometry
Phenotypic and functional profiling was performed on freshly isolated PBMCs using multiparametric flow cytometry. PBMCs were isolated from fresh whole blood using Ficoll-Hypaque density gradient centrifugation, and PBMCs were stained for CD3 (UCHT1), CD16 (3G8), CD56 (NCAM16.2), CD161 (DX12), NKG2D (1D11), CD94 (HP-3D9), NKp46 (9E2), NKp44 (p44–8), 2-domain KIR by antibody cocktail of CD158a recognizing KIR2DL1, 2DS1 (HP-3E4) and CD158b recognizing KIR2DL2/DL3 (CH-L), 3-domain KIR by NKB1 antibody recognizing KIR3DL1 (DX9), 2B4 (2–69), CD69 (FN50), HLA-DR (G46–6), CD58 (1C3), perforin (γG9), granzyme (GB11) (BD Biosciences), NKG2A (Z199, Beckman Coulter) in four panels (106 cells per panel) to dissect the phenotypic signatures associated with variation in HLA-C levels. In parallel, NK cell functional activity was assessed following stimulation of PBMCs with the following NK target cell lines, each at a 10:1 effector:target ratio: i) p815 cells (a mouse leukemic cell line, ATCC) pre- incubated for 1 h with 1 μg/ml p815-specific Ab (Abcam, Cambridge, MA) as a measure of antibody-dependent cell-mediated cytotoxicity; ii) K562 cells (ATCC), a MHC-devoid target cell line; and iii) 721.221 cells (ATCC), a MHC class I-deficient B cell line. As a positive control, the PBMCs were treated with PMA (2.5 μg/ml) and ionomycin (0.5 μg/ml) (Sigma-Aldrich), and a condition with media alone or p815 cells lacking the coating Ab as a negative control. All cells were stained for CD107a (H4A3) prior to and throughout the 4 h of stimulation in the presence of Brefeldin A (10 μg/ml; Sigma-Aldrich) and monensin (6 μg/ml; GolgiStop; BD Biosciences). The cells were then stained with 4 different immunophenotypic panels (using the markers described above), fixed/permeabilized, and stained for intracellular interferon-γ (IFN-γ, 25723.11). Data were acquired on a BD LSRII flow cytometer and analyzed using FlowJo software v9.3.
2.3. Statistical analysis
Correlation analyses using the Pearson product-moment correlations were utilized to examine associations between HLA-C expression levels and NK phenotype and functions in uninfected controls. Adjustment for multiple testing was made by Bonferroni correction. Kruskal-Wallis with Dunn’s multiple comparison test was used to examine differences among HIV-infected patient groups. Reported p-values are two sided and values of p ⩽ 0.05 were considered significant.
3. Results
3.1. HLA-C and KIR gene profiles
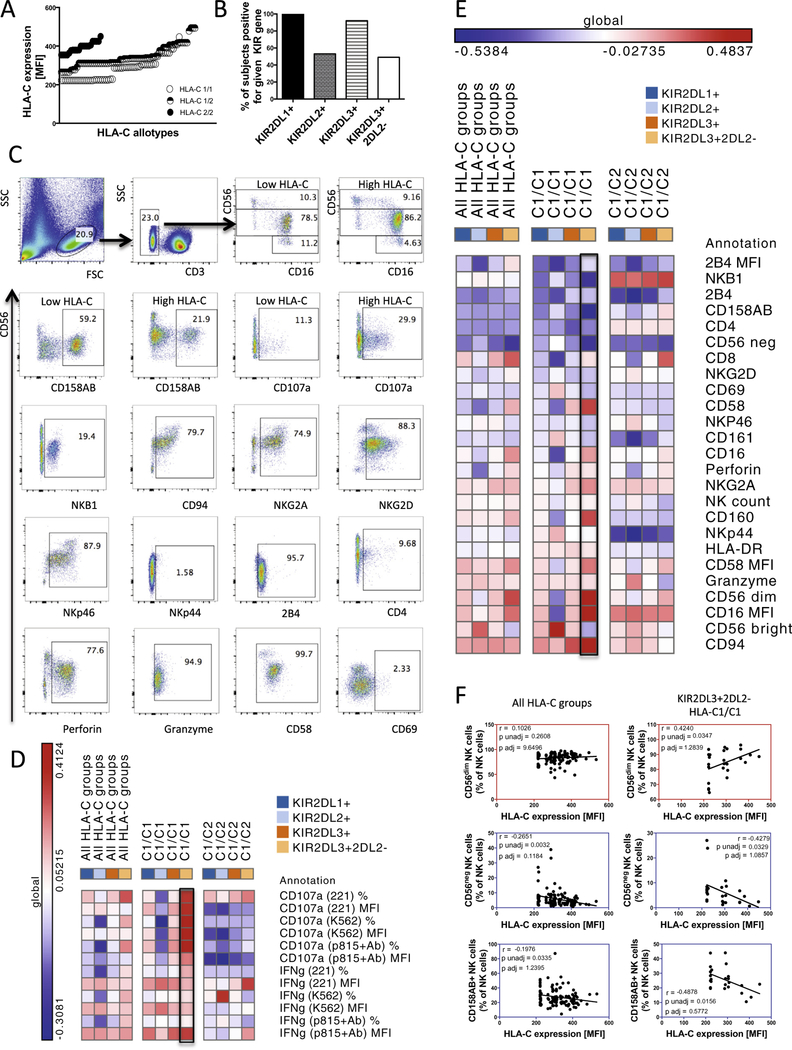
HLA-C expression levels, linked to HLA-C genotype, were previously defined in a cohort of 1698 individuals of European ancestry to generate a predictive model of HLA-C expression [10]. Using this model, HLA-C expression levels were predicted for our cohort, demonstrating a continuous distribution across the healthy study subjects. Predicted HLA-C expression was elevated in subjects homozygous for HLA-C2 groups in contrast to subjects homozygous for HLA-C1 or HLA-C1/C2 heterozygotes, nevertheless continuous distribution was observed in all HLA-C groups (Fig. 1A). Because HLA-C interacts with three inhibitory KIR receptors: KIR2DL1, KIR2DL2 and KIR2DL3, that act as the dominant ligands for HLA-C, we next examined the distribution of all 3 KIRs across our patient groups by KIR genotyping as previously described [11]. As expected, all healthy subjects (154) encoded KIR2DL1. KIR2DL3 was encoded by 92% of healthy subjects, among which, 49% lacked KIR2DL2. Conversely, 53% healthy patients encoded KIR2DL2 (Fig. 1B).
Fig. 1.
HLA-C expression modulates NK cell function and subset distribution in healthy subjects in a HLA-C1/C1/KIR2DL3+2DL2− haplotype dependent manner. (A) Predicted HLA-C expression levels are depicted based on individual HLA-C allotypes, showing a continuous distribution within HLA-C1/C1, HLA-C1/C2 and HLA-C2/C2 groups. The KIR gene profiles of the cohort are illustrated in (B). The gating strategy as well as examples of flow cytometric stains are illustrated in (C). The heat-map illustrates positive (red) or negative (blue) associations (r values) with HLA-C expression levels and various NK cell functions (D) and phenotypes (E) (listed per row) in all HLA-C group subjects, HLA-C1/C1 subjects or HLA-C1/C2 subjects encoding different KIR receptors (listed as columns). Correlation plots in (F) reveal associations between HLA-C expression levels and the frequency of CD56dim (top panels, red framed plots), CD56neg (middle panels, blue framed plots) or KIR2D+ (bottom panels, blue framed plots) NK cells in all HLA-C group subjects (left panels) or HLA-C1/C1/KIR3DL3+2DL2− subjects (right panels). In the heatmap, red depicts positive correlations and blue depicts negative correlations, with color intensity indicating the intensity of that relationship.
3.2. HLA-C expression levels alter NK cell licensing in subjects restricted by specific HLA-C/KIR haplotypes
During development, only NK cells expressing inhibitory receptors for self-HLA [7], which ensures self-inhibition, are functionally “licensed” to respond to MHC-I low/deficient target cells [7,12,13]. Moreover, the strength of licensing is selectively regulated by the potency and level of HLA expression as demonstrated in the Ly49-mouse model system [14]. Although the higher dose of HLA-C associated with the rs9264942 SNP has not been shown to influence NK cell education in terms of degranulation and cytokine production in response to HLA-deficient target cells [15], we speculated that a continuous distribution model of HLA-C expression may more accurately reveal links to NK cell licensing, or that enhanced HLA-C expression may drive the accumulation of more functional, optimally licensed NK cell subsets, able to control HIV viremia more effectively. Moreover, given that NK cells may both kill via direct recognition of infected/HLA-low cells by licensed NK cells or indirectly kill via unlicensed (KIR-negative, NKG2A-negative) NK cells through antibody mediated cellular cytotoxicity (ADCC) [16], here we sought to investigate the impact of HLA-C on both direct and indirect NK cell activity.
To address this hypothesis, PBMCs from healthy donors were stimulated with 3 different target cell lines that trigger NK cell activation through disparate pathways, ultimately allowing for an unbiased view of the NK cell functionality. Degranulation and cytokine secretion were both measured. We observed that HLA-C expression levels did not impact global NK cell functional competence upon PMA/ionomycin stimulation (data not shown) but associated with NK cell degranulation or IFN-γ secretion following stimulation with all three target cell lines. Increased NK-function in high HLA-C expressors was observed only in subjects encoding KIR2DL3 and lacking KIR2DL2, homozygous for HLA-C1 group alleles (Fig. 1D, framed heatmap column). Overall, these data point to an effect of HLA-C expression levels on NK cell function, restricted to particular HLA-C/KIR haplotype backgrounds.
3.3. HLA-C expression levels modulate NK cell subsets
NK cells can be divided into three subsets based on their expression of CD56 and CD16 (Fig. 1C). In peripheral blood, the cytolytic mature CD56dimCD16+ NK cells dominate, whereas more immature cytokine producing CD56brightCD16− NK cells make up a smaller population [17]. Anergic CD56neg NK cells are rare in healthy individuals and represent at most a few percent of total NK cells in the blood but expand in chronic viral infections [18,19]. While total NK cell numbers were unaffected by HLA-C expression levels, correlation analyses revealed changes in several NK cell subsets in the setting of differential HLA-C expression levels. Elevated HLA-C expression levels were inversely associated with the overall abundance of anergic CD56neg NK cells (Fig. 1E and F). Moreover, even fewer anergic CD56neg NK cells were observed in high HLA-C expressors within HLA-C1/C1 background that encoded KIR2DL3 and lacked KIR2DL2 (r = −0.4279, Fig. 1E and F). In contrast, positive relationships were observed between CD56dim NK cells and HLA-C MFI (Fig. 1E and F), while no effects were observed in CD56bright populations. Furthermore, a number of additional associations between HLA-C MFI and NK cell phenotype were observed in HLA-C1/C1/KIR2DL3+KIR2DL2− subjects, such as a decrease in the frequency of 2-domain KIR+ NK cells (Fig. 1E and F), 3-domain KIR+ NK cells (Fig. 1E), and an increase in FcγRIIIa expression on NK cells (Fig. 1E) suggesting that enhanced HLA-C expression may have a broader impact on NK cell development in specific HLA-C/KIR backgrounds. Thus these data point to a novel role of KIR/HLA-C interactions in NK cell development, and specifically in generation of functional NK cells.
3.4. High HLA-C expression levels, in HIV-1 infection, are linked to licensing
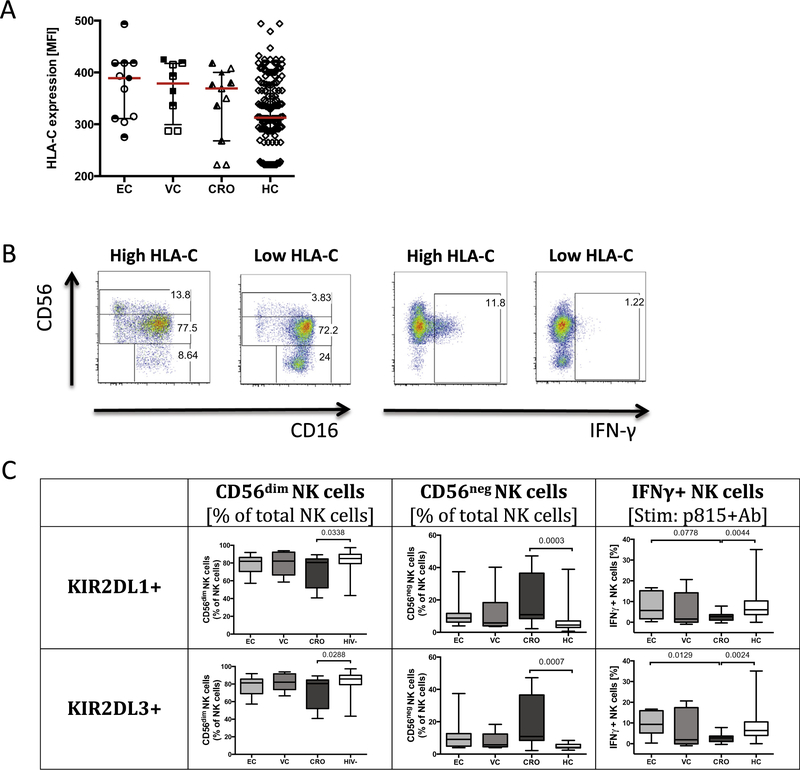
While HLA-C expression appeared to affect NK cell subset distribution and function in healthy individuals, enhanced NK cell function has also been linked to heightened antiviral activity in HIV infection [9]. Thus we next sought to define whether HLA-C levels affected NK cell biology post infection. Using the predictive model of HLA-C expression [10], HLA-C levels were imputed for a cohort of HIV-1 infected individuals including a spectrum of disease progression profiles: spontaneous controllers and normal progressors. As expected, a range of HLA-C expression profiles were observed across the patient groups, with the lowest levels of HLA-C among chronically infected progressors (CRO) (Fig. 2A). In addition, in contrast to spontaneous controllers, chronically infected subjects with high viral load are prone to further HLA-C downregulation by HIV Vpu [20]. Interestingly, low levels of HLA-C expression were also associated with an expansion of CD56neg NK cells in chronic progressors as compared to controllers (Fig. 2B), suggesting that pre-infection CD56neg expansions are amplified following infection. Moreover, similar to the healthy cohort, the preservation of CD56dim NK cells in HIV-infected subjects was dependent on the presence of KIR2DL3 (Fig. 2C), confirming the importance of HLA-C/KIR interactions in shaping the evolution of NK cell subset development. We next examined the relationship of HLA-C expression and NK cell function within HIV infected subjects. Unique to the HIV-infected cohort, high HLA-C expression, particularly in the presence of KIR2DL3, was associated with robust IFN-γ secretion, not observed in chronic progressors (Fig. 2C) following stimulation with antibody-opsonized targets. These data point to a role for HLA-C/KIR interactions following infection in shaping NK cell activity, either related to: a) the rapid unleashing of more functional NK cells following infection, or b) the generation of new NK cells after infection with enhanced antiviral activity that may contribute to antiviral control or the generation of more functional T cell immunity.
Fig. 2.
Impact of HLA-C expression on NK cell subset distribution and function among HIV-1 infected subjects. Predicted HLA-C expression levels are depicted based on individual HLA-C allotypes (A) and show differential levels among clinical subgroups including elite controllers (EC), n = 11; viremic controllers (VC), n = 8; chronic progressors (CRO), n = 10, healthy controls (HC), n = 154. Open shapes indicate subjects in the HLA-C1/C1 group, half-open shapes represent subjects in the HLA-C1/C2 group; and closed shapes represent subjects in the HLA-C2/C2. The flow plots illustrate the differences in NK cell subset distribution and IFN-γ secretion in subjects with low or high HLA-C expression (B). The differences in NK subset distribution and IFN-γ secretion are illustrated in subjects expressing KIR2DL1 and KIR2DL3 (C).
4. Discussion
Elevated HLA-C expression has been linked to enhanced viral control and slower progression to AIDS linked to increased HIV-specific CTL responses [10]. However, because HLA-C is the dominant ligand for KIR receptors, we aimed to define whether elevated HLA-C expression may also impact HIV control via alterations in NK cell antiviral profiles. Interestingly, HLA-C expression significantly impacted NK cell functional activity among healthy subjects encoding HLA-C1/C1 and KIR2DL3 in the absence of KIR2DL2 (Fig. 1). Moreover, spontaneous HIV control was associated with robust IFN-γ secretion among HIV-infected subjects especially in the presence of KIR2DL3 (Fig. 2), arguing for a disease-induced HLA-C-dependent induction of highly functional NK cells. Yet, irrespective of health or disease, HLA-C expression levels, in combination with select KIRs on a HLA-C1/C1 haplotype background, had a profound impact on NK cell subset distribution and specifically on the accumulation of CD56dim and CD56neg NK cells (Fig.1). Moreover, HLA-C expression had an effect on distribution of KIR2DL+ and KIR3DL+ NK cells, suggesting the involvement of HLA-C-KIR interaction in limiting the selection and expression of KIRs during NK cell development Thus these data highlight the unexpected influence of HLA-C expression levels not only on shaping T cell immunity [10], but also in influencing NK cell development towards more functional mature NK cells that may then be able to respond more aggressively upon infection.
While, specific combinations of KIR/HLA haplotypes have been shown to influence the outcome of HIV-1 infection [21] with well-established protective effects of certain HLA-Bw4/KIR3DL1/S1 haplotypes [22,23], less attention has been given to HLA/KIR interactions involving KIR2DL ligands. The identification of KIR2DL2-associated HIV-1 sequence polymorphisms provided the first evidence for KIR-associated NK-cell-mediated immune pressure on HIV [24] and recent studies have identified increased frequencies and function of KIR2DL1–3+ NK cells in primary HIV-1 infection that are dependent on HLA-C haplotypes [25]. Our data further extend the critical role of KIR2D/HLA-C interactions that may not only be shaped by the presence of the receptor-ligand in modeling KIR+ NK cell repertoires [26], but also by the level of ligand, HLA-C expression. Interestingly, the data reported here argue that prior to infection, HLA-C levels may influence the development of NK cells, in such a way that HLA-C signals through KIR2DL3 may maintain NK cells in a more functionally mature state, limit the numbers of inhibitory KIR expressed on NK cells, and augment the level of FcγRIIIa. These KIR− NK cells can be activated more rapidly via activating NK cell receptors including FcγRIIIa due to the absence of an inhibitory HLA/KIR signal [27,28]. Importantly, uneducated NK cell responses have been shown to play a critical beneficial role in tumor control in patients undergoing monoclonal antibody therapeutic treatment via the induction of ADCC [29] as well as in response to murine CMV infection [30]. Therefore, HLA-C driven accumulation of more mature, functional, inhibitory KIR-negative, FcγRIIIa+ NK cells may result in the generation of a pool of NK cells that may respond more rapidly and effectively to antibodies following HIV infection. Furthermore, upon HIV infection, we observed that these mature functional NK cells also retained the ability to respond more effectively to antibody opsonized material, an immunological function that has been recently linked to protection from HIV acquisition in the first moderately successful HIV vaccine trial [31] as well as in the context of natural control of HIV infection [32].
KIR2DL1 binds C2 HLA-C alleles with high affinity [33], whereas KIR2DL2 and KIR2DL3 preferentially bind to group C1 HLA-C alleles, although KIR2DL2 can also bind to C2 alleles [34]. Importantly, KIR2DL2 binds to HLA-C1 with stronger affinity than does KIR2DL3 [33]. Thus, the stronger KIR2DL2 binding to HLA-C in subjects encoding this KIR may diminish the impact of variation in HLA-C expression on NK cell profile dynamics during NK cell education caused by KIR2DL3/HLA-C1 interaction. By contrast, subjects encoding KIR2DL3, lacking KIR2DL2, may be more susceptible to variation in HLA-C expression, where elevated levels of the ligand may alter NK cell potency via the programming of more effective NK cells, that are not anergic CD56neg cells that typically accumulate in chronic viral infections [18,19].
While predictive HLA-C expression modeling pointed to NK cell phenotypic and functional associations related to HLA-C/KIR backgrounds, it is plausible that the direct assessment of HLA-C expression levels could provide enhanced resolution on NK cell clonal dynamics associated with HLA-C expression in future studies. Moreover, beyond HLA-C expression, other factors influence NK cell development, including cytomegalovirus (CMV) infection. Specifically, CMV infection is associated with the accumulation of a fraction of KIR+NKG2C+ NK cells that significantly impacts the overall NK cell clonal diversity within infected individuals [35–38]. Moreover, CMV remains the clearest example of infection-driven expansion of mature CD57+NKG2C+CD158b+ NK cells that produce significant IFN-γ after stimulation with MHC Class I-deficient target cells [39,40]. Thus it is likely that combinations of KIR/HLA genotypes, KIR gene copy number variation [41,42], and co-infections may all contribute to NK cell clonal diversity, function, and bioactivity warranting larger population based studies in the future to probe the influence of all these parameters on NK cell selection, development, education, and activity following malignant transformation and/or infection.
Whether elevated HLA-C expression alters NK cell immunophenotypic properties in the bone marrow during development, or in the periphery, through tonic interactions with inhibitory self-sensing KIR, is unknown. However, these data suggest that changes in HLA-C expression have a significant impact on qualitative changes in NK cell phenotype and function, linked to more effective NK cell profiles that may aide in the development of a highly effective, rapid innate immune responses upon viral exposure. Thus future larger studies aimed at defining the functional consequences of different HLA-C expression levels, in the context of co-infection, may identify critical divergences in the NK cell response soon after infection, particularly during the most acute window of disease, both in the ability to contain virally infected cells, but also in tuning the evolving adaptive immune response.
Acknowledgements
We thank the subjects of the PhenoGenetic Project at Brigham & Women’s Hospital for the samples that they have provided (RC2 GM093080). This work was supported by the Ragon Institute, the National Institute of Health (R01 AI080289) and NIH Harvard Center for AIDS Research (P30 AI060354–02). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
G.A., P.L.D.J., M.C., Y.S. M.A. designed and supervised the study. M.S., Q.L., M. G. did the acquisition and/or did analysis of data. M. S. and G.A. have written the manuscript. M.S., M.D., C.T.B., T.J.S, P.B., G.A. have discussed and interpreted the data and revised the manuscript critically for its important intellectual content. M.S., Q.L., M.D., M.G., C.T.B., T.J.S., Y.S., M.C., M.A., B.D.W., P.B., P.L.D.J., G.A. provided the final approval of the final version of the manuscript.
Abbreviations:
- NK cell
natural killer cell
- KIR
killer cell immunoglobulin-like receptors
- HLA
human leukocyte antigen
- IFN-γ
interferon-γ
Footnotes
Disclosure
The authors declare no conflict of interest.
References
- [1].van Manen D, van’t Wout AB, Schuitemaker H, Genome-wide association studies on HIV susceptibility, pathogenesis and pharmacogenomics, Retrovirology 9 (2012) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. , A whole-genome association study of major determinants for host control of HIV-1, Science 317 (5840) (2007) 944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. , The major genetic determinants of HIV-1 control affect HLA class I peptide presentation, Science 330 (6010) (2010) 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Thomas R, Apps R, Qi Y, Gao X, Male V, O’HUigin C, et al. , HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C, Nat. Genet 41 (12) (2009) 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Blais ME, Dong T, Rowland-Jones S, HLA-C as a mediator of natural killer and T-cell activation: spectator or key player?, Immunology 133 (1) (2011) 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL, HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells, Proc. Natl. Acad. Sci. U.S.A 90 (24) (1993) 12000–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. , Licensing of natural killer cells by host major histocompatibility complex class I molecules, Nature 436 (7051) (2005) 709–713. [DOI] [PubMed] [Google Scholar]
- [8].Xia Z, Liu Q, Berger CT, Keenan BT, Kaliszewska A, Cheney PC, et al. , A 17q12 allele is associated with altered NK cell subsets and function, J. Immunol 188 (7) (2012) 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. , Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes, J. Exp. Med 204 (12) (2007) 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, et al. , Influence of HLA-C expression level on HIV control, Science 340 (6128) (2013) 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. , Human diversity in killer cell inhibitory receptor genes, Immunity 7 (6) (1997) 753–763. [DOI] [PubMed] [Google Scholar]
- [12].Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. , Human NK cell education by inhibitory receptors for MHC class I, Immunity 25 (2) (2006) 331–342. [DOI] [PubMed] [Google Scholar]
- [13].Hoglund P, Brodin P, Current perspectives of natural killer cell education by MHC class I molecules, Nat. Rev. Immunol 10 (10) (2010) 724–734. [DOI] [PubMed] [Google Scholar]
- [14].Brodin P, Lakshmikanth T, Johansson S, Karre K, Hoglund P, The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells, Blood 113 (11) (2009) 2434–2441. [DOI] [PubMed] [Google Scholar]
- [15].Charoudeh HN, Schmied L, Gonzalez A, Terszowski G, Czaja K, Schmitter K, et al. , Quantity of HLA-C surface expression and licensing of KIR2DL+ natural killer cells, Immunogenetics 64 (10) (2012) 739–745. [DOI] [PubMed] [Google Scholar]
- [16].Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH, A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules, Blood 105 (11) (2005) 4416–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. , Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset, Blood 97 (10) (2001) 3146–3151. [DOI] [PubMed] [Google Scholar]
- [18].Hu PF, Hultin LE, Hultin P, Hausner MA, Hirji K, Jewett A, et al. , Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity, J. Acquir. Immune Defic. Syndr. Hum. Retrovirol 10 (3) (1995) 331–340. [PubMed] [Google Scholar]
- [19].Gonzalez VD, Falconer K, Bjorkstrom NK, Blom KG, Weiland O, Ljunggren HG, et al. , Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment, J. Immunol 183 (10) (2009) 6612–6618. [DOI] [PubMed] [Google Scholar]
- [20].Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA, et al. , HIV-1 Vpu mediates HLA-C downregulation, Cell Host Microbe. 19 (5) (2016) 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bashirova AA, Thomas R, Carrington M, HLA/KIR restraint of HIV: surviving the fittest, Annu. Rev. Immunol 29 (2011) 295–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Song R, Lisovsky I, Lebouche B, Routy JP, Bruneau J, Bernard NF, HIV protective KIR3DL1/S1-HLA-B genotypes influence NK cell-mediated inhibition of HIV replication in autologous CD4 targets, PLoS Pathog.10 (1) (2014) e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. , Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1, Nat. Genet 39 (6) (2007) 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, et al. , HIV-1 adaptation to NK-cell-mediated immune pressure, Nature 476 (7358) (2011) 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Korner C, Granoff ME, Amero MA, Sirignano MN, Vaidya SA, Jost S, et al. , Increased frequency and function of KIR2DL1–3+ NK cells in primary HIV-1 infection are determined by HLA-C group haplotypes, Eur. J. Immunol 44 (10) (2014) 2938–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M, Analyses of HLA-C-specific KIR repertoires in donors with group A and B haplotypes suggest a ligand-instructed model of NK cell receptor acquisition, Blood 117(1) (2011) 98–107. [DOI] [PubMed] [Google Scholar]
- [27].Raulet DH, Vance RE, Self-tolerance of natural killer cells, Nat. Rev. Immunol 6 (7) (2006) 520–531. [DOI] [PubMed] [Google Scholar]
- [28].Jaeger BN, Vivier E, When NK cells overcome their lack of education, J. Clin. Investig 122 (9) (2012) 3053–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. , Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment, J. Clin. Investig 122 (9) (2012) 3260–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Orr MT, Murphy WJ, Lanier LL, ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection, Nat. Immunol 11 (4) (2010) 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, et al. , Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines, Sci. Trans. Med 6 (228) (2014) 228ra38. [DOI] [PubMed] [Google Scholar]
- [32].Lewis GK, Role of Fc-mediated antibody function in protective immunity against HIV-1, Immunology 142 (1) (2014) 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P, Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3, J. Immunol 180(6) (2008) 3969–3979. [DOI] [PubMed] [Google Scholar]
- [34].Uhrberg M, Parham P, Wernet P, Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes, Immunogenetics 54 (4) (2002) 221–229. [DOI] [PubMed] [Google Scholar]
- [35].Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. , Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function, Immunity 42 (3) (2015) 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. , NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs, Blood 121 (14) (2013) 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. , Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection, Proc. Natl. Acad. Sci. U.S.A. 108 (36) (2011) 14725–14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M, Imprint of human cytomegalovirus infection on the NK cell receptor repertoire, Blood 104 (12) (2004) 3664–3671. [DOI] [PubMed] [Google Scholar]
- [39].Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. , Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function, Blood 119 (11) (2012) 2665–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu Z, Sinzger C, Frascaroli G, Reichel J, Bayer C, Wang L, et al. , Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity, J. Virol 87 (13) (2013) 7717–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, et al. , Influence of KIR gene copy number on natural killer cell education, Blood 121(23) (2013) 4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, et al. , Copy number variation of KIR genes influences HIV-1 control, PLoS Biol. 9 (11) (2011) e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]