Abstract
Fibrin hydrogels are used as a model system for studying cell-ECM biophysical interactions. Bulk mechanical stiffness of these hydrogels has been correlated to mechanotransduction and downstream signaling. However, stiffness values proximal to cells can vary by orders of magnitude at the length scale of microns. Patterning of matrix stiffness at this spatial scale can be useful in studying such interactions. Here we present and evaluate a technique to selectively stiffen defined regions within a fibrin hydrogel. Laser scanning illumination activates ruthenium-catalyzed crosslinking of fibrin tyrosine residues, resulting in tunable stiffness changes spanning distances as small as a few microns and a localized compaction of the material. As probed by active microrheology, stiffness increases by as much as 25X, similar to previously observed stiffness changes around single cells in 3D culture. In summary, our method allows for selective modification of fibrin stiffness at the micron scale with the potential to create complex patterns, which could be valuable for the investigation of mechanotransduction in a biologically meaningful way.
Keywords: Microrheology, Stiffness, Tissue Mechanics, Extracellular Matrix, Hydrogel, Fibrin
1. Introduction
Many cell types sense and respond to the stiffness of their surrounding extracellular matrix (ECM). In 2D, this has been well demonstrated with initial substrate stiffness affecting critical cellular processes ranging from migration [1] to differentiation [2]. Interactions between cells and their surrounding ECM can be studied using 3D hydrogels as a model system for the ECM. While these hydrogel systems are far removed from in vivo tissue, they can be tuned and/or functionalized in ways appropriate for testing specific hypotheses. Hydrogels can be composed of natural, synthetic, or composite materials with tunable material stiffness [3], porosity [4], density [5], susceptibility to enzymes [6], and adhesion ligand concentrations [7]. For the case of naturally derived ECMs, it may be challenging to independently modulate subsets of these features. Our group has previously demonstrated concentration independent modulation of fibrin stiffness and establishment of stiffness gradients using strain fields instead of chemical modifications [8]. We have also used optical tweezers active microrheology (AMR) to map these stiffness gradients in both cell-free and cell-containing systems. In the case of the latter, we have shown pericellular stiffness distributions are dependent on cell mediated matrix remodeling and strain hardening [9]. A new method is needed to exogenously reproduce these distributions in situ.
While engineered synthetic materials allow for control over matrix stiffness with minimal influence on other ECM properties [10,11], they typically are not fibrous and thus lack a fundamental property of biological tissues. Further, mesh sizes are orders of magnitude smaller than natural tissues and essentially present themselves as a continuous barrier with respect to cell confinement and biotransport. Synthetic hydrogel systems incorporating light-mediated crosslinking sites permitting stiffening by both wide field and patterned illumination have been described [12–14]. Such hydrogels have been utilized in the study of endothelial cells [15] and stem cells [16,17], showing that cells respond differently to hydrogels of different stiffness.
In this paper we present a technique for patterning the stiffness within a naturally derived fibrous matrix: fibrin. Fibrin is a naturally occurring fibrous protein that is an essential component of blood clots, making up a significant portion of the provisional ECM during wound healing [18]. In vitro fibrin hydrogels have been used as a model ECM for studying angiogenesis [19], stem cells [20,21], macrophages [22,23], and cancer cell lines [24]. As a hydrogel, fibrin forms a fibrous, viscoelastic, randomly interconnected matrix with pores on the length scale of one micron. Compared to cell-free systems, fibrin stiffness proximal to cells [8] and sprouting endothelial vessels [27] is elevated and more heterogenous, spanning orders-of-magnitude at the subcellular spatial scale. This phenomenon is possible because fibrin is permissive to cell mediated remodeling by enzymatic breakdown, deposition of new matrix, and other modalities of ECM modification [25,26].
Methods for increasing the stiffness of fibrin hydrogels have been described in literature, including chemical crosslinking via incubation in glutaraldehyde [28] or exposure to UV [29]. Of note, Bjork et al. demonstrated crosslinking of fibrin via ruthenium-catalyzed photocrosslinking (RCP), and showed RCP did not reduce cell viability [30]. For RCP, a light activated ruthenium compound and sodium persulfate (SPS) oxidizes tyrosine residues in the fibrin forming tyrosine radicals, which can react with other tyrosine residues to form dityrosine in the presence of persulfate [31].
Here we present and evaluate an extension of this technique, where we selectively pattern crosslinking within the volume of 3D fibrin hydrogels using a laser scanning confocal microscope and measure induced stiffness changes via AMR. With our method, we have achieved selective stiffening within fibrin hydrogels at scales as small as a few microns, which may be beneficial for future studies on cell-ECM interactions.
2. Materials and Methods
2.1. Hydrogel Preparation
Fibrin hydrogels were prepared with 2.0 μm carboxylated silica microbeads (Bangs Laboratories) dispersed within the hydrogels prior to gelation. Bovine fibrinogen stock (Sigma) was dissolved in phosphate buffered saline (PBS, Gibco) and sterile filtered (0.22 μm, Olympus) before use. Gelation was conducted by mixing fibrinogen/microbead/PBS solution with bovine thrombin (Sigma), to achieve a 1 mL hydrogel with final concentrations of 2.5 mg/mL fibrin, 0.8% (w/v) microbeads, and 4 U/mL thrombin (Sigma). Hydrogels were cast into 35 mm glass bottom dishes (MatTek), resulting in a hydrogel approximately 500 μm thick at the center, where experiments were conducted. Hydrogels were kept in an incubator (37° C, 5% CO2) for 25 minutes after which they were hydrated with culture medium: 2 mL of DMEM (Gibco) + 10% FBS (Gibco) + 1% Penicillin-Streptomycin (Gibco). Fibrin hydrogels were then stored for 24 hours in the incubator before use.
2.2. Selective and Bulk Photocrosslinking
For photocrosslinking, 200 μL of culture medium was removed and substituted with 200 μL of a concentrated crosslinker solution, producing a solution of 0.15 mg/mL tris(2,2′-bipyridyl) dichlororuthenium (II) hexahydrate (Sigma) and 0.24 mg/mL sodium persulfate (Sigma). A Fluoview 1200 laser scanning confocal microscope (Olympus) was used for selective-crosslinking. With the built-in functionality of the Fluoview system, the user can draw arbitrary ROIs to expose specific regions to controlled amounts of 488 nm laser illumination. In all cases presented here, the 488 nm source was used at 0.94 mW power. Scans were conducted at 2 μs/pixel with a total scan resolution of 1600 pixel by 1600 pixel over a 211.2 μm by 211.2 μm area. A 60X 1.45 NA oil immersion microscope objective (Olympus) was used, at a focus plane approximately 35 μm from the cover glass. For full construct crosslinking, dishes were held 1 cm above an array of 4 blue LEDs (460 nm, SparkFun). Each LED was powered at an operating current of 20 mA. Following crosslinking, hydrogels were washed 3 times with culture medium to prevent any further light-activated crosslinking.
2.3. Microstructural Assessment
Confocal reflection microscopy (CRM) was performed to assess structural differences within the fibrin mesh. Imaging was performed using the 559 nm laser line of the FV1200 operating at 0.72 mW. An optical flow algorithm based on the Farneback method [32] was utilized to analyze successive frames of these images and capture deformation dynamics across the entire crosslinking process by summing deformations throughout the time series. Summed optical flow results were down sampled via bilinear interpolation by a factor of 80, resulting in nodes being spaced 10.6 μm apart. Additionally, fiber density was analyzed in the CRM images. Images were binarized using the Matlab built-in adaptive binarize function and objects smaller than 10 pixels were removed. Bright pixels were then counted in annular areas emanating from the center of the crosslinked region.
2.4. Active Microrheology
AMR was conducted within the volume of the fibrin hydrogels. Our AMR instrument has been described previously [8,9]. Briefly, probe beads within the hydrogel are oscillated via a 1064 nm laser trap, steered by oscillating galvanometer mirrors. The bead acts as a lens to deflect a second, stationary laser beam (785 nm), which is focused onto a quadrant photodiode. The complex-valued material response (α*) is calculated from the phase-amplitude relationship between trapping forces and probe bead displacement [33,34]. Here we report 1/α’, which is the inverse of the real component of α* and can be considered a metric of stiffness. Beads were probed at 50 Hz and were located approximately 35 μm from the bottom cover glass. Potential effects of cover glass proximity on measurements were controlled by performing all measurements for different treatments at the same approximate height.
2.5. Live-Dead Assay
Normal human dermal fibroblasts (NHDF, Lonza) were cultured in culture medium (described above) in an incubator (37° C, 5% CO2). 2.5 mg/mL fibrinogen with 150K/mL NHDF cells in PBS and 4 U/mL thrombin were cast in 35 mm glass bottom dishes and hydrated with culture medium. After 24 hours incubation, crosslinking solution was added and crosslinked via 1000 scans of the 488 nm laser in a 100μm diameter circular region of interest, conducted using settings in Section 2.2. The crosslinking solution was washed out with 3 washes of culture medium and then fibrin hydrogels were placed back into the incubator for 4 hours. “No crosslinker controls” received the same treatment (including the washout) as the crosslinked samples except 200 μL PBS was added rather than crosslinker solution. Live (positive) controls were not treated. Finally, dead (negative) controls were treated with 70% ethanol to verify ethidium homodimer stain was detectable in dead cells. Medium was aspirated and replaced with 2 μM Calcein-AM (Invitrogen) and 4 μM ethidium homodimers (Invitrogen) in PBS. Cells were placed back into the incubator for 10 minutes before being imaged in Fluoview with a 10X 0.3 NA air objective. Calcein-AM was excited with 488 nm laser and 505–540 nm emission was collected. Ethidium homodimer was excited with 559 nm laser and 655–755 nm emission was collected. 5 by 5 tiled images were taken for all assays with Fluoview multiarea controller and tiled with ImageJ.
2.6. Statistical Analysis
Statistical analysis was conducted in Origin Pro. Sample sets were tested using non-parametric statistical methods. Mann-Whitney U-tests (α = 0.05) with a Bonferroni correction was conducted for all statistical comparisons presented in this paper. Data in the manuscript are presented as mean ± standard deviation.
3. Results
3.1. Selective Crosslinking Results in Structural Changes to Fibrin Mesh
A schematic of RCP is shown in Figure 1A, along with a cartoon of two crosslinking modalities used in this paper; bulk (Figure 1B) and selective (Figure 1C). Figure 1C highlights the utility of our technique, which is to selectively crosslink fibrin near cells. The crosslinking solution is comprised of a Ru compound and SPS and is activated by blue light (Figure 1A), forming Ru (III) and a sulfate radical [31]. The Ru compound oxidizes tyrosine residues already present within fibrin to form a dityrosine crosslink mediated by sulfate [35].
FIGURE 1.
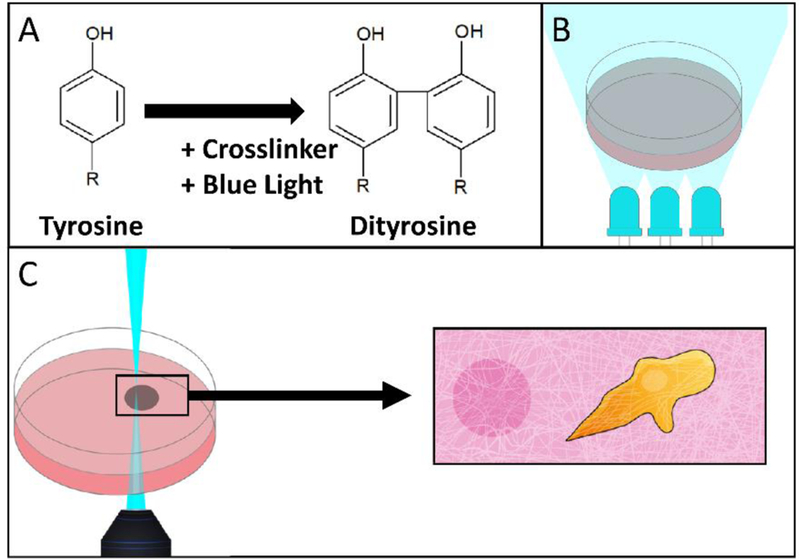
Schematic of two crosslinking modalities: laser scanning confocal microscopy and bulk LED illumination. (A) Tyrosine residues are crosslinked to form dityrosine by blue light activated crosslinker. (B) An entire hydrogel can be crosslinked by illumination with 460 nm LEDs. (C) Illustration depicting the intended usage of selective photocrosslinking, where regions proximal to cells could be selectively stiffened by a laser scanning confocal microscope.
To demonstrate the selectivity in crosslinking, fibrin constructs were imaged before and after 488 nm laser treatment, which is absorbed by the Ru compound to initiate the crosslinking reaction. Circular treatment regions having diameter of either 10 μm or 100 μm are referred to as T10 and T100, respectively. Larger fields of view containing a treated region were imaged by CRM using the 559 nm laser line. Representative fields of view are shown in Figure 2A and 2B in both transmitted light (i, ii, iii) and CRM imaging modalities (iv, v, vi).
FIGURE 2.
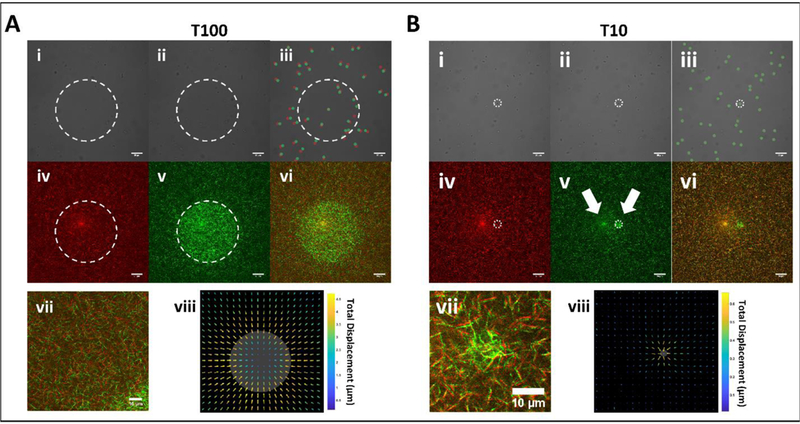
Transmitted light and CRM imagery of 100 and 10 μm (referred to as T100 and T10, respectively) diameter treatment spot sizes within fibrin hydrogels before and after photoactivation by 488 nm illumination. (A) Transmitted light images of T100 treatment spot (i) before, (ii) after, and (iii) composite. Probe bead locations in the composite are marked red and green for before and after, respectively. CRM images (iv) before, (v) after, and (vi) composite. (vii) Zoomed inset of the composite image suggests contraction toward treated area, confirmed by (viii) optical flow analysis. Gray area denotes the treated region. Note: Arrow length was scaled for visualization, and color indicates displacement magnitude. (B) Corresponding images and analysis for T10 treatments. Note that the leftmost arrow in (v) points to an imaging artifact common to CRM while the rightmost points to the treated spot. Boundaries of treatment spots are marked with white dashed lines. Scale bars = 20 μm.
Following T100 treatment, embedded microbeads within and beyond the region of treatment were displaced in the transverse plane by several microns (Figure 2A, i–iii). CRM images show elevated pixel intensities in the treated region as compared to non-treated (Figure 2A, iv, v). Pseudo-colored composite images of the mesh before and following treatment show that fibers were displaced, notably outside of the treated region (Figure 2A, vi, vii). To capture full frame videos of mesh dynamics during the crosslinking process, we implemented a CRM imaging strategy during which full field-of-view imaging at 559 nm was recorded in between every 100 sequential circular scans at 488 nm. Videos show a symmetric contraction of the mesh toward the treated area (Supplementary Videos 1 and 2). Optical flow analysis confirms displacements were predominantly directed radially toward the center of the treated area in T100 treated samples (Figure 2A, viii). Peak displacement was located just outside the treatment region and decreased radially in either direction. Fiber density increased inside the crosslinked region, with maximum change at the boundary (Figure S1).
We next investigated such displacements for a T10 treated region (10-micron diameter treatment). As compared to T100 treatment, microbeads within and beyond the treatment region were displaced by no more than 500 nm in the transverse plane (Figure 2B, i, ii, iii). CRM imagery confirms the effect on the matrix is highly localized to the region of treatment (Figure 2B, iv, v, vi, vii). Outside the treated area, displacement was confined to a region immediately proximal boundary as detected by optical flow analysis (Figure 2B, viii).
3.2. Stiffness changes in Fibrin hydrogels
Next, we quantified the increase in stiffness within treated areas. To assess the dependence of stiffening on duration of light exposure, AMR was conducted on the same 8 beads in a region subjected to sequential crosslinking treatments (Figure 3A and 3B; 100 scans per treatment with 488 nm, see Methods). Stiffness increased with each sequential treatment and after 1000 scans reached values comparable to LED crosslinking (Figure 3A; Figure 3C, p=0.54, Mann-Whitney Test). LED crosslinked samples appeared to have a denser mesh (Figure S2) similar to that observed within selectively crosslinked regions (Figure 2). To assess changes to the stiffness and structure that may be caused by 559 nm imaging and exposure to the crosslinking solution, a set of crosslinking experiments were conducted without 488 nm light treatment. Only minute changes in stiffness (Figure S3A) and displacement (Figure S3B) were observed following 10 scans of 559 nm imaging.
Figure 3.
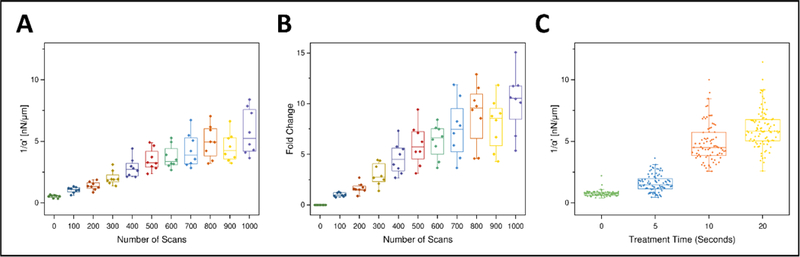
Stiffness increases with treatment time within a fibrin hydrogel after either selective or bulk crosslinking. (A) Stiffness (1/ α’, nN/μm) probed by AMR using the same probe particles (nbeads=8) after successive laser scans of a T100 treatment. Probe particles are all located within the treatment region. (B) Data from (A) represented as fold change over the pretreatment stiffness for each bead. (C) Bulk crosslinking via LEDs shows increase in stiffness with light exposure (nsamples=2, per treatment time).
Significant stiffening occurred outside T100, but not T10 treated regions (Figure 4), a result consistent with the displacement fields (Figure 2A). Representative stiffness maps are shown in Figure 4. Prior to crosslinking, stiffness is low and heterogeneous throughout the field of view (Figure 4A & 4D). Following treatment, stiffness is notably increased within the treated areas and, for the case of T100 only, outside the treatment area as well (Figure 4B & 4E). The complete set of measurements is shown in Figure 4C and 4F. On average, increases in stiffness within the treated region were 11.6 ± 7.0 nN/μm and 6.5 ± 3.6 nN/μm for T100 and T10, respectively. Outside of the treated regions, stiffness changes were substantially lower: 1.3 ± 1.3 nN/μm and 0.2 ± 0.4 nN/μm, for T100 and T10, respectively. For all treatments, stiffness increases were statistically significant (p < 0.05, Mann-Whitney test, with Bonferroni Correction), except for T10 Sample 1.
Figure 4.
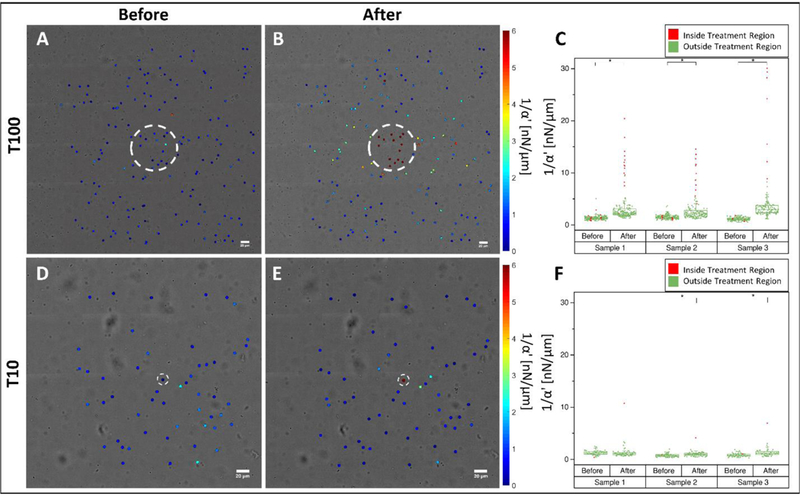
Distribution of stiffness within a fibrin hydrogel probed by AMR before and after T100 and T10 treatments. Stiffness maps (A) before and (B) after T100 treatment. (C) Scatter plot of all T100 samples before and after treatment. Stiffness maps (D) before and (E) after T10 treatment. (F) Scatter plot of all T10 samples before and after treatment. (*) denotes significance at 0.05 level via a Mann-Whitney U-Test. Color maps in (A, B, D, E) saturate at 6 nN/μm. Scale bars = 20 μm.
Changes in stiffness as a function of radial distance to the center point of the treated region are plotted in Figure 5. Within both T100 and T10 treatments, stiffness change was most prominent within the boundaries of the treated region (Figure 5A and 5B), with fold changes as high as 25X (Figure 5C and 5D). In T100 treatments, stiffness change extends beyond the treated region, decreasing with distance from the boundary for at least 50 μm. This stiffening lacks radial symmetry and is greater to the left and right of the crosslinked region (Figure 4B), suggesting radial-circumferential stiffness anisotropy. To investigate potential anisotropy, the position of each bead was cast into a coordinate system with the origin at the center of the treated region, and a horizontal x-axis passing through the centroid of that region. The first coordinate is the distance to the origin and the second is the absolute value of angular deviation from the x-axis, defined here as ϴ (Figure 6A). This angular deviation is important because our AMR system exclusively oscillates beads in the horizontal axis (x-axis). Thus, anisotropy would present as a dependency of stiffness on ϴ, as was observed (Figure 6B and Figure 6C). This trend is highlighted when data is binned by ϴ and distance, as shown for T100 (Figure 6D) and T10 (Figure 6E).
Figure 5.
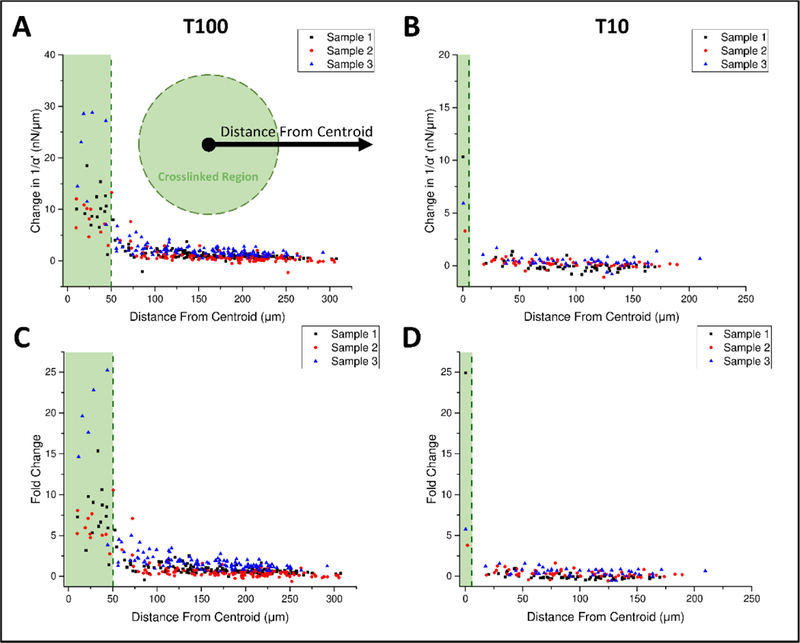
Stiffening decreases with radial distance from treated region center. Stiffening for (A) T100 and (B) T10 samples as a function of distance from the treatment area center. Treatment region shaded in green. Fold change presented in (C) and (D) for T100 and T10, respectively. Dotted lines mark the outer radius of the treatment area. (nsamples= 3, per treatment type).
Figure 6.
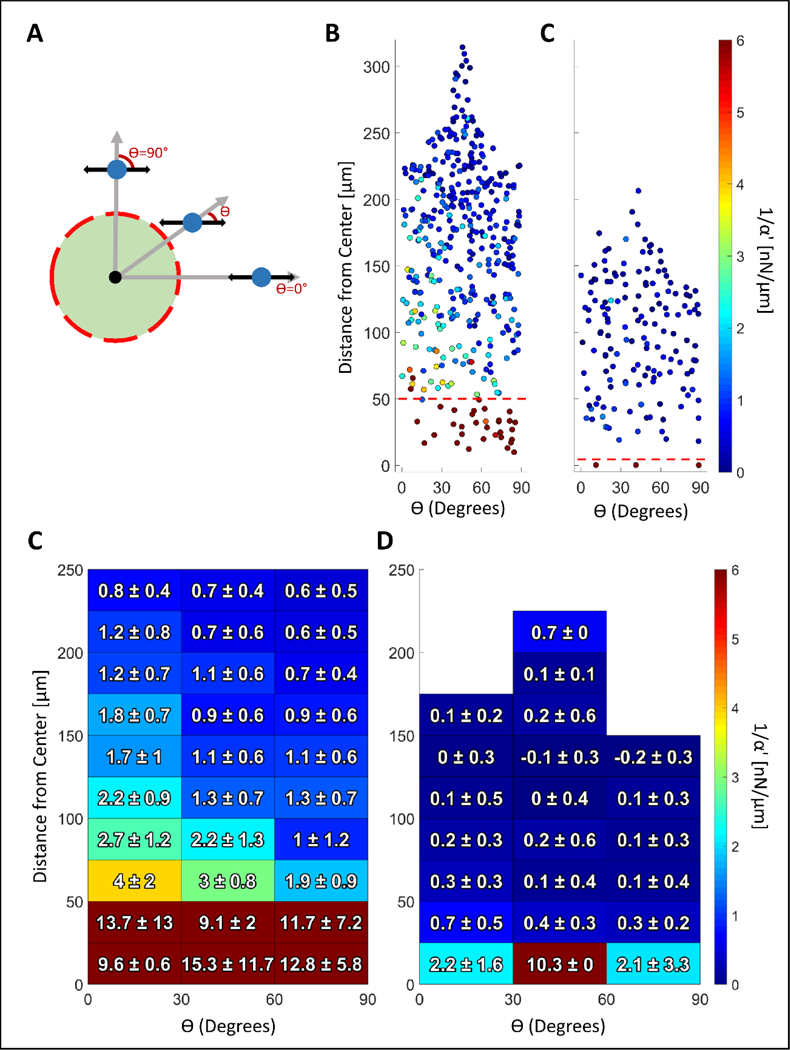
Analysis of stiffness anisotropy. (A) The angle (ϴ) between the axis of bead oscillation and a radial line emanating from the treatment centroid is a function of bead position. AMR was conducted exclusively along the horizontal axis shown by the black arrows. Beads located at ϴ = 90 degrees are oscillated circumferentially, whereas beads located at ϴ= 0 degrees radially. Stiffness data from (B) T100 samples (nsamples = 3) and (C) T10 samples (nsamples = 3) plotted as a function of absolute value of ϴ. Red dashed line marks the radial boundary of the treatment zone. Data from (A) and (B) was binned and presented in (D) and (E), respectively. Figure S6A and S6B display bead count per bin.
3.3. Cell Viability after selective crosslinking
Selective crosslinking (T100) was performed near NHDFs embedded within fibrin hydrogels. Cells immediately proximal to the treated region appear viable 4 hours after crosslinking (Figure 7A). Zoomed in views of replicate experiments (n = 6) are shown in Figure 7B. Figure 7C shows results for areas surrounding (1) a T100 crosslinked region, (2) a control T100 without crosslinker, and for (3) live and (4) dead controls.
Figure 7.
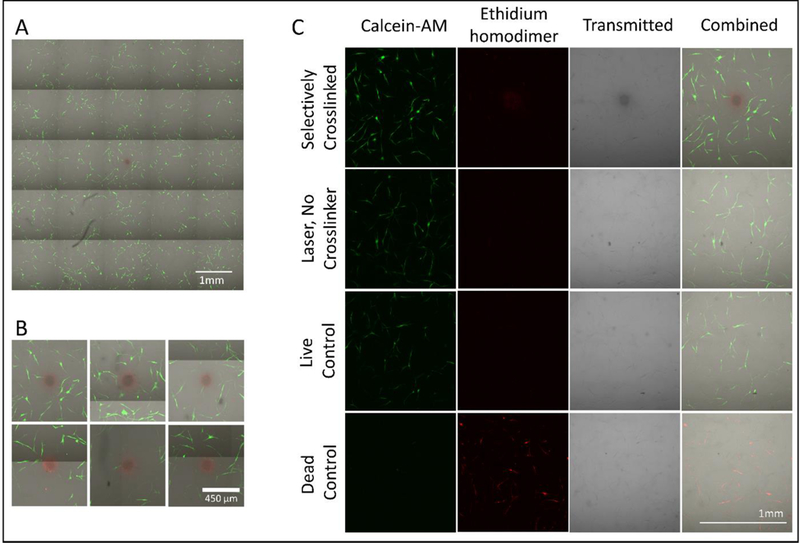
Live-dead assay of fibrin hydrogel containing NHDF cells 4 hours after T100 treatment. Calcein-AM (green) stains live cells and ethidium homodimer (red) stains dead cells. (A) Representative 5 by 5 field-of-view montage. (B) Zoomed in view around the crosslinked region for 6 replicates. (C) Areas surrounding (1) a T100 crosslinked region, (2) a control T100 without crosslinker, and (3) live and (4) dead controls. Note: The crosslinked region shows signal in the red channel even in cell-free systems with no ethidium homodimer.
4. Discussion and Conclusion
Here we present a method for tuning ECM stiffness within a fibrin hydrogel in select regions. Important to the study of mechanobiology, our method can stiffen local regions at the resolution and dynamic range observed around single cells and cell colonies [9,27]. In many cell-ECM biophysical studies, the initial stiffness of an ECM has been linked to phenotypical changes including stem cell fate [2,36]. However, within a permissive environment (such as fibrin), stiffness, architecture, and composition can be modified by cells over time [37–39], such that the system no longer reflects initial conditions. This holds true for synthetic hydrogels, engineered to be degradable or otherwise, which are also subject to deposition of new matrix by cells [40,41]. These studies motivated the need for the tool presented in this manuscript, which can make stiffness modifications to fibrin systems on the same order of magnitude that cells can.
In this study we used a photocrosslinking method coupled with a laser scanning confocal microscope to generate patterns of crosslinked areas within a fibrin hydrogel (additional patterns shown in Figure S5). Crosslinking was indicated by increased pixel brightness and effects on local stiffness were investigated via AMR. Degree of stiffening in the treated region was observed to be as much as 25-fold, which may be significant in the study of roles for stiffness in cell-ECM physical interactions. In fact, we have previously observed cells stiffening their pericellular matrix by as much as 35-fold. For example, in one system, the stiffness proximal to a cell ranged from 16.9 ± 8.0 nN/μm as compared to its cell-free control 0.8 ± 0.5 nN/μm [9]. Our laser-mediated crosslinking method can produce a similar range of stiffness at the sub-cellular spatial scale, allowing for exogenous control of local stiffness in a biologically meaningful way.
Interestingly, stiffening was also observed beyond the treatment region where photo-activated crosslinking was not conducted. We examined changes to fibrin mesh architecture outside the T100 treated region (Figure 5A) to understand the nature of this stiffness change. One possibility is that activated crosslinker diffuses beyond the treated region to generate a gradient of crosslinking. However, crosslinked fibrin is brighter in CRM and regions of increased brightness exclusively overlap the region of 488 nm illumination (Figure 2) indicating that the crosslinking stays confined. Our group has previously shown that fibrin stiffness, as assessed by AMR, increases with fibrin density, which could explain the stiffening [8]. Fibrin density decreased, or remained unchanged, in the stiffened area beyond the treated region. By contrast within the treated region, both fibrin density and stiffness increased near the treatment boundary (Figure S1). These results suggest other mechanisms are at play, notably stretch-mediated hardening which is supported by the radial inward bead displacement shown in Figure 2Aiii. Such a displacement could stiffen the surrounding material through strain hardening, an important characteristic of fibrin and other natural ECMs [42]. Both cell-mediated and exogenous strain have been shown to stiffen these matrices as measured by ourselves [8,9] and others [43,44]. In support of strain hardening, the stiffened region (beyond the treatment area) is co-located with the largest displacements of the fibrin mesh (Figure 2A, viii; Figure 5A). Also, because displacements are overwhelmingly radial, it would be expected that stretch-mediated stiffening should be anisotropic with lesser stiffening in the circumferential direction. While for each bead AMR was exclusively performed along the x-axis, the population of probed beads indicates anisotropy. Specifically, probe beads located near the x-axis are oscillated radially with respect to the treated region, whereas beads located at ± 90 degrees are oscillated circumferentially. Consistent with anisotropy, the radial oscillations reported greater stiffness (Figure S6) with an apparent decrease in stiffness with an angular deviation from the x-axis (Figure 6A). Taken together, these results imply that crosslinking a large area can induce stretch mediated hardening in regions not directly affected by the photo crosslinking chemistry, a phenomenon that may be useful in the study of cellular mechanobiology. We also observed matrix remodeling surrounding the treated region for T10 samples, as assessed by confocal imaging (Figure 2B, vii) and optical flow analysis (Figure 2B, viii). However, we did not detect stretch-mediated stiffening where fiber displacement was detected, in part due to limitations of the AMR method. Specifically, beads were spaced too far from one another to provide sufficient spatial resolution to allow characterization of local changes in stiffness relative to the treatment spot size. In our experience, the bead density required to achieve bead proximity on the order of several microns alters the structural characteristics of a fibrin gel thus precluding stretch-hardening analysis.
Lastly, because our method is intended for use in the study of mechanobiology, we sought to verify the conditions used for crosslinking were not cytotoxic. We verified the exact conditions used in the AMR study did not appear to affect viability of NHDFs, even those adjacent to the treatment zone. It is important to note that methods here will likely require fine tuning to be extended to other cell-ECM experimental systems. There are many factors to consider including: hydrogel composition and density, cell type, cell-ECM ligand interactions and accessibility, culture media, range of stiffness, and required stiffness resolution. To account for these one will likely need to optimize the shape of the crosslinked region, crosslinker concentration, and light dose.
In summary, we report a method to selectively stiffen fibrin matrices at small spatial scales. Presently, we are working on extending these results to other natural matrices such as collagen and beginning to analyze cellular response to changes in their local pericellular stiffness by our method.
Supplementary Material
6. Acknowledgments
We thank Linda McCarthy for her assistance in cell culture. We thank the funding sources that made this research possible National Institute of Health (NIH) T32 pre-doctoral Training Grant HL116270, and the Laser Microbeam and Medical Program, a National Biomedical Technology Resource (NIH P41-EB015890). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Disclosures
The authors declare no competing financial interests
7. References
- 1.Isenberg BC, DiMilla PA, Walker M, Kim S, and Wong JY, “Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength,” Biophys. J. 97(5), 1313–1322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, and Discher DE, “Matrix Elasticity Directs Stem Cell Lineage Specification,” Cell 126(4), 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Wells RG, “The role of matrix stiffness in regulating cell behavior,” Hepatology 47(4), 1394–1400 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lien SM, Ko LY, and Huang TJ, “Effect of pore size on ECM secretion and cell growth in gelatin scaffold for articular cartilage tissue engineering,” Acta Biomater. 5(2), 670–679 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Kniazeva E and Putnam AJ, “Endothelial cell traction and ECM density influence both capillary morphogenesis and maintenance in 3-D,” AJP Cell Physiol. 297(1), C179–C187 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Trappmann B, Baker BM, Polacheck WJ, Choi CK, Burdick JA, and Chen CS, “Matrix degradability controls multicellularity of 3D cell migration,” Nat. Commun. 8(1), 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GOBIN AS and WEST JL, “Cell migration through defined, synthetic ECM analogs,” FASEB J. 16(7), 751–753 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Kotlarchyk MA, Shreim SG, Alvarez-Elizondo MB, Estrada LC, Singh R, Valdevit L, Kniazeva E, Gratton E, Putnam AJ, and Botvinick EL, “Concentration independent modulation of local micromechanics in a fibrin gel,” PLoS One 6(5), (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keating M, Kurup A, Alvarez-Elizondo M, Levine AJ, and Botvinick E, “Spatial distributions of pericellular stiffness in natural extracellular matrices are dependent on cell-mediated proteolysis and contractility,” Acta Biomater. 57, 304–312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonker AM, Bode SA, Kusters AH, Van Hest JCM, and Löwik DWPM, “Soft PEG-Hydrogels with Independently Tunable Stiffness and RGDS-Content for Cell Adhesion Studies,” Macromol. Biosci. 15(10), 1338–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Rape AD, Zibinsky M, Murthy N, and Kumar S, “A synthetic hydrogel for the high-throughput study of cell-ECM interactions,” Nat. Commun. 6(May), 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemir S, Hayenga HN, and West JL, “PEGDA hydrogels with patterned elasticity: Novel tools for the study of cell response to substrate rigidity,” Biotechnol. Bioeng. 105(3), 636–644 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Kloxin AM, Tibbitt MW, and Anseth KS, “Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms,” Nat. Protoc. 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh Y-C, Corbin EA, Caliari SR, Ouyang L, Vega SL, Truitt R, Han L, Margulies KB, and Burdick JA, “Mechanically dynamic PDMS substrates to investigate changing cell environments,” Biomaterials 145, 23–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lampi MC, Guvendiren M, Burdick JA, and Reinhart-King CA, “Photopatterned Hydrogels to Investigate the Endothelial Cell Response to Matrix Stiffness Heterogeneity,” ACS Biomater. Sci. Eng. 3(11), 3007–3016 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Marklein RA and Burdick JA, “Spatially controlled hydrogel mechanics to modulate stem cell interactions,” Soft Matter 6(1), 136–143 (2009). [Google Scholar]
- 17.Yang C, DelRio FW, Ma H, Killaars AR, Basta LP, Kyburz KA, and Anseth KS, “Spatially patterned matrix elasticity directs stem cell fate,” Proc. Natl. Acad. Sci. 113(31), E4439–E4445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark RAF, “Fibrin and wound healing.,” Ann. N. Y. Acad. Sci. 936, 355–367 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Nakatsu MN, Davis J, and Hughes CCW, “Optimized fibrin gel bead assay for the study of angiogenesis.,” J. Vis. Exp. (3), 186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolehmainen K and Willerth SM, “Preparation of 3D fibrin scaffolds for stem cell culture applications.,” J. Vis. Exp. (61), e3641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arulmoli J, Pathak MM, McDonnell LP, Nourse JL, Tombola F, Earthman JC, and Flanagan L. a., “Static stretch affects neural stem cell differentiation in an extracellular matrix-dependent manner,” Sci. Rep. 5, 8499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colvin RB and Dvorak HF, “Fibrinogen/fibrin on the surface of macrophages: detection, distribution, binding requirements, and possible role in macrophage adherence phenomena,” J. Exp. Med. 142(6), 1377–1390 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh JY, Smith TD, Meli VS, Tran TN, Botvinick EL, and Liu WF, “Differential regulation of macrophage inflammatory activation by fibrin and fibrinogen,” Acta Biomater. 47, 14–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Tan Y, Zhang H, Zhang Y, Xu P, Chen J, Poh Y-C, Tang K, Wang N, and Huang B, “Soft fibrin gels promote selection and growth of tumorigenic cells.,” Nat. Mater. 11(8), 734–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu P, Takai K, Weaver VM, and Werb Z, “Extracellular matrix degradation and remodeling in development and disease.,” Cold Spring Harb Perspect Biol 3(12), 1–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey JD, Dufresne ER, and Schwartz MA, “Mechanotransduction and extracellular matrix homeostasis,” Nat. Rev. Mol. Cell Biol. 15(12), 802–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliar BA, Keating MT, Kong YP, Botvinick EL, and Putnam AJ, “Sprouting angiogenesis induces significant mechanical heterogeneities and ECM stiffening across length scales in fibrin hydrogels,” Biomaterials 162, 99–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charulatha V and Rajaram A, “Influence of different crosslinking treatments on the physical properties of collagen membranes.,” Biomaterials 24(5), 759–67 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Cornwell KG and Pins GD, “Discrete crosslinked fibrin microthread scaffolds for tissue regeneration,” J. Biomed. Mater. Res. Part A 82A(1), 104–112 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Bjork JW, Johnson SL, and Tranquillo RT, “Ruthenium-catalyzed photo cross-linking of fibrin-based engineered tissue,” Biomaterials 32(10), 2479–2488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fancy DA and Kodadek T, “Chemistry for the analysis of protein–protein interactions: Rapid and efficient cross-linking triggered by long wavelength light,” Chem. Biochem. 96(3), 6020–6024 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farnebäck G, “Two-frame motion estimation based on polynomial expansion,” Image Anal. 2003(1), 363–370 (2003). [Google Scholar]
- 33.Brau RR, Ferrer JM, Lee H, Castro CE, Tam BK, Tarsa PB, Matsudaira P, Boyce MC, Kamm RD, and Lang MJ, “Passive and active microrheology with optical tweezers,” J. Opt. A Pure Appl. Opt. 9(8), S103–S112 (2007). [Google Scholar]
- 34.Mizuno D, Tardin C, Schmidt CF, and Mackintosh FC, “Nonequilibrium mechanics of active cytoskeletal networks.,” Science 315(5810), 370–3 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Syedain ZH, Bjork J, Sando L, and Tranquillo RT, “Controlled compaction with ruthenium-catalyzed photochemical cross-linking of fibrin-based engineered connective tissue,” Biomaterials 30(35), 6695–6701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, and Mooney DJ, “Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate.,” Nat. Mater. 9(6), 518–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birkedal-Hansen H, “Proteolytic remodeling of extracellular matrix,” Curr. Opin. Cell Biol. 7(5), 728–735 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Matrisian LM, “Metalloproteinases and their inhibitors in matrix remodeling,” Trends Genet. 6(4), 121–125 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Larsen M, Artym VV, Green JA, and Yamada KM, “The matrix reorganized: extracellular matrix remodeling and integrin signaling,” Curr. Opin. Cell Biol. 18(5), 463–471 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Caliari SR, Vega SL, Kwon M, Soulas EM, and Burdick JA, “Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments,” Biomaterials 103, 314–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicodemus GD, Skaalure SC, and Bryant SJ, “Gel structure has an impact on pericellular and extracellular matrix deposition, which subsequently alters metabolic activities in chondrocyte-laden PEG hydrogels,” Acta Biomater. 7(2), 492–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah JV and Janmey PA, “Strain hardening of fibrin gels and plasma clots,” Rheol. Acta 36(3), 262–268 (1997). [Google Scholar]
- 43.Jansen KA, Bacabac RG, Piechocka IK, and Koenderink GH, “Cells actively stiffen fibrin networks by generating contractile stress,” Biophys. J. 105(10), 2240–2251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winer JP, Oake S, and Janmey PA, “Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation,” PLoS One 4(7), (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


