Abstract
Alternative splicing is a co-transcriptional mechanism that generates protein diversity by including or excluding exons in different combinations, thereby expanding the diversity of protein isoforms of a single gene. Abnormalities in this process can result in deleterious effects to human health, and several xenobiotics are known to interfere with splicing regulation through multiple mechanisms. These changes could lead to human diseases such as cancer, neurological disorders, autoimmune diseases, and developmental disorders. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is an environmental contaminant generated as a byproduct of various industrial activities. Exposure to this dioxin has been linked to a wide range of pathologies through the alteration of multiple cellular processes. However, the effects of TCDD exposure on alternative splicing have not yet been studied. Here, we investigated whether a single po. dose of 5 μg/kg or 500 μg/kg TCDD influence hepatic alternative splicing in adult male C57BL/6Kou mouse. We identified several genes whose alternative splicing of precursor messenger RNAs was modified following TCDD exposure. In particular, we demonstrated that alternative splicing of Cyp1a1, Ahrr, and Actn1 was significantly altered after TCDD treatment. These findings show that the exposure to TCDD has an impact on alternative-splicing, and suggest a new avenue for understanding TCDD-mediated toxicity and pathogenesis.
Introduction
Dioxins are a group of environmentally persistent contaminants, which include polychlorinated dibenzo-p-dioxins (PCDDs), polychlorinated dibenzofurans (PCDFs) and 11 dioxin-like polychlorinated biphenyls (PCBs). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is the most toxic dioxin and is a product of anthropogenic activity generated from waste incineration, chlorine paper bleaching and pesticide manufacture [1]. Human exposure to TCDD has been linked to a variety of malignancies, such as chloracne and hepatotoxicity [2]. In animal models, TCDD exposure produces an exceedingly broad range of toxic effects including wasting syndrome, thymic atrophy, endocrine disruption, immune suppression, behavioral alterations, teratogenicity, cancer and death [3, 4]. The precise mechanisms through which TCDD induces toxicity are not fully understood; however, almost all of its effects are mediated through the aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor that is a member of the bHLH-PAS (basic-helix-loop-helix-Per-ARNT-Sim) superfamily. Upon binding to TCDD, AhR activates two pathways. The genomic pathway as a transcription factor, binding to xenobiotic response elements (XREs) located in the promoter regions of its target genes, resulting in an up- or down-regulation of the abundance of a battery of genes encoding xenobiotic-metabolizing enzymes [5] among many others [6–8]. And the nongenomic pathway, exerted through a variety of ways, including crosstalk with other transcription factor (such as estrogen receptor alpha, androgen receptor, NF-κB, and ß-catenin) [9–12], altering histone marks or DNA methylation patterns [13, 14], modifying the activation of Src tyrosine kinase [15], or increasing the intracellular calcium concentration [16].
Benzo[a]pyrene, an AhR agonist, disrupt nephrogenesis by modulating the splicing of Wilms’s tumor suppressor [17]. Moreover, CD44 splicing variants were found after Benzo[a]pyrene treatment [18]. On the other hand, the effects of TCDD exposure on alternative splicing mechanisms have not been investigated, nor more broadly has the influence of AhR activation. However, there are some observations that suggest it. Human ALDH3 gene present three alternative splice acceptor sites at the 3’-end of intron 1. Interestingly, the splice variants derived from these sites were observed in controls but not in HepG2-TCDD treated cells, suggesting that this dioxin might modify alternative splicing [19]. Alternative splicing occurs co-transcriptionally and increases protein diversity by including or excluding exons in different combinations, resulting in the production of a diverse array of proteins from a single gene. In humans, ~92–94% of genes are alternatively spliced, allowing for the generation of more than 100,000 protein isoforms [20, 21]. This process is carried out by the splicing code, which consists of three elements: the cis-acting elements of the transcript, the spliceosome (which assembles step-wise on the precursor mRNA (pre-mRNA) substrates), and auxiliary factors. Each of these components can be targeted by external agents, causing modifications to the alternative splicing of pre-mRNAs. In particular, xenobiotics can interfere with splicing through multiple mechanisms including inhibition of spliceosome assembly, interference with the recognition of alternative exons, or modification of the abundances of splicing factors [22]. Alterations in the splicing process can result in significant deleterious effects on human health. In particular, alternative splicing abnormalities have been linked to cancer, neurological disorders, autoimmune diseases, and developmental disorders, among others [23, 24]. Therefore, identifying agents with the potential to alter alternative splicing is crucial for understanding many human health problems.
To address this, we have investigated the effects of TCDD exposure on alternative splicing using a mouse model known to be sensitive to TCDD toxicities. In particular, we have evaluated the transcriptome of liver from adult male mice exposed to a low or high dose of TCDD (5 or 500 μg/kg) or vehicle control, focusing on differential abundance of alternatively spliced transcripts. We identified several genes whose splicing processes were modified by TCDD exposure and performed validation of key findings using real-time quantitative PCR (qPCR).
Materials and methods
Animal handling
Male C57BL/6Kou mice at the age of 13–15 weeks were obtained from the National Public Health Institute, Division of Environmental Health Institute, Kuopio, Finland. The animals were housed in a pathogen-free facility at 21°C with 50 ± 10% relative humidity and artificial illumination on a 12/12 hour light/dark cycle and tap water ad libitum. This study was approved by the Finnish National Animal Experiment Board (Eläinkoelautakunta, ELLA; permit code: ESLH-2008-07223/Ym-23).
Experimental design
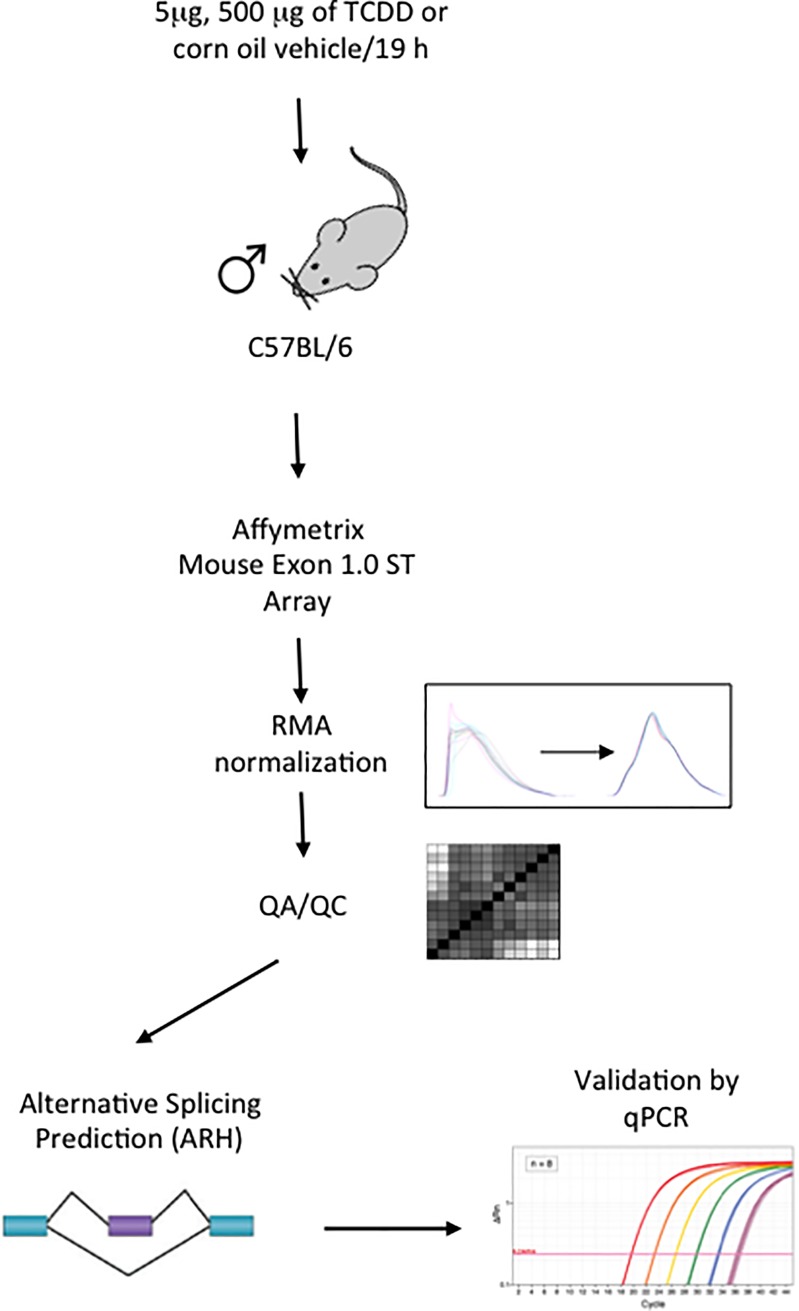
Animals were divided into 3 groups of 4 mice each and treated by oral gavage with a 5 μg/kg or 500 μg/kg dose of TCDD dissolved in corn oil or corn oil alone (10 mL/kg). Mice were observed 2 times after exposure for their general appearance and behavior. Animals were euthanized at 19 hours post-treatment with carbon dioxide followed immediately by cardiac exsanguination. Livers were immediately excised and frozen in liquid nitrogen. The overall experimental design is outlined in Fig 1.
Fig 1. Experimental design.
Twelve total male C57BL/6Kou mice at the age of 13–15 weeks were divided into 3 groups of 4 mice each, and treated by oral gavage with a 5 or 500 μg/kg dose of TCDD dissolved in corn oil or corn oil vehicle alone (10 mL/kg). Liver tissue was collected 19 h post-treatment. Affymetrix Mouse Exon 1.0 ST Arrays were used to assess RNA abundance. Data were normalized using RMA and homogeneity assessed; no outliers were detected. Alternative splicing events were predicted using an entropy base measure (ARH), and transcripts with significantly differential abundance were validated using RT-qPCR.
RNA isolation and exon array hybridization
Liver samples were ground to a fine powder in liquid nitrogen using a mortar and pestle, followed by homogenization in a lysis buffer using a Polytron. RNA was isolated using an RNeasy Mini Kit (Qiagen, Mississauga, Canada) following the manufacturer’s instructions. RNA was quantified using a NanoDrop UV spectrophotometer (Thermo Scientific, Mississauga, Canada) and its integrity verified using RNA 6000 Nano chips on an Agilent 2100 Bioanalyzer (Agilent Technologies, Mississauga, Canada). All samples with an RNA integrity number (RNI) ≥ 8.5 were used for subsequent analysis. RNA was assayed on an Affymetrix Mouse Exon 1.0 ST Array at the Center for the Applied Genomics at the Hospital for Sick Children (Toronto, Canada) according to manufacturer’s protocols.
Exon array preprocessing
Raw array data (CEL files) were loaded into the R statistical environment (v3.1.2) using the oligo package (v3.6) from the Bioconductor library [25]. Gene and ProbeSet annotation was performed using moex10sttranscriptcluster.db (v3.6) and moex10stprobeset.db (v3.6) packages. Data were normalized using the RMA algorithm [26]. Data were assessed for distributional homogeneity and visualized using the BPG package [27] with the lattice (v0.20–33) and latticeExtra (v0.60–26) packages in R. Raw data is available in GEO at GSE126328, and in the TCDD transcriptomics package (v2.2.5) for R (download from: https://labs.oicr.on.ca/boutros-lab/tcdd-transcriptomics)[28].
Alternative splicing and statistical analysis
Only ProbeSets with annotation core databases were used for the analysis. Differential abundance analysis was performed using the limma package (v3.2) for R. Linear modeling was performed to contrast each TCDD dose to control. Standard errors of coefficients were adjusted using an empirical Bayes model. Model-based t-tests were applied to test for significance, followed by false discovery rate adjustments for multiple testing. To detect alternative splicing events, an entropy base measure for splicing prediction was performed using the ARH package as well as the exon variations within a gene [29].
Real-time quantitative PCR (qPCR) analysis
Complementary DNA (cDNA) for the qPCR assay was prepared from 3 μg of total liver RNA using random primers and SuperScript First-Strand Synthesis (Invitrogen, Carlsbad, CA, USA). The real-time polymerase reaction (PCR) was performed in a StepOne Real-Time PCR System with TaqMan Universal PCR Master Mix (Applied Biosystems, Branchburg, NJ, USA) according to the manufacturer’s protocol. Relative transcript abundance was quantified using the comparative threshold cycle (Ct) method. The probes used for each exon were obtained from Integrated DNA Technologies (IDT, Skokie, IL, USA). The gene encoding the 18S ribosomal RNA (rRNA, endogenous) with identification number ID Mm00507222_s1 (IDT) was used to normalize the results.
Statistical analysis
Real Time quantitative PCR results are presented as the mean values ± standard deviation (S.D.) of three replicates. The statistical significance of the data was evaluated using the Student’s t-test. In all cases, the differences between groups were considered to be statistically significant when the p value was less than 0.05.
Results
Alterations in the splicing process can result in significant deleterious effects on human health. Therefore, identifying agents with the potential to alter alternative splicing is necessary. TCDD are known to impact the transcriptome, however the effects on alternative splicing are unclear. To address this, exon arrays were used to evaluate the alternative splicing of livers from adult male mice exposed to TCDD. The experimental outline is presented in Fig 1.
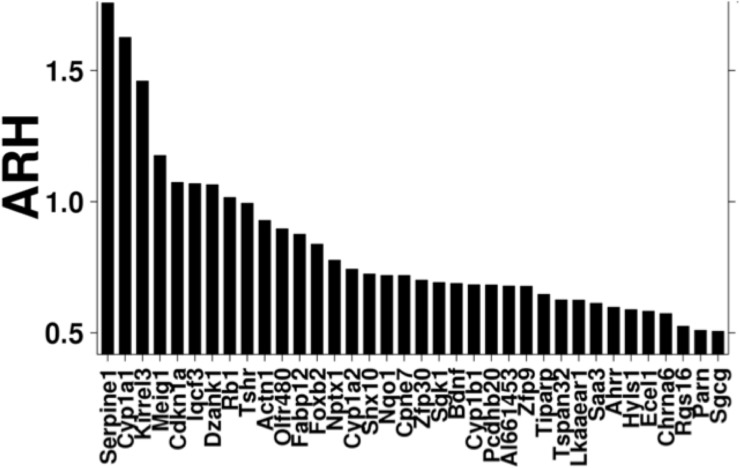
After pre-processing the data, splicing assessment was performed via ARH analysis. Using a threshold of ≥ 0.5, we identified 36 genes with differential splicing (Fig 2).
Fig 2. Alternative splicing observed following TCDD exposure.
ARH analysis was performed to identify differentially spliced transcripts between 500 μg/Kg TCDD-treated mice and controls. Thirty six genes with ARH values of 0.5 or higher were identified.
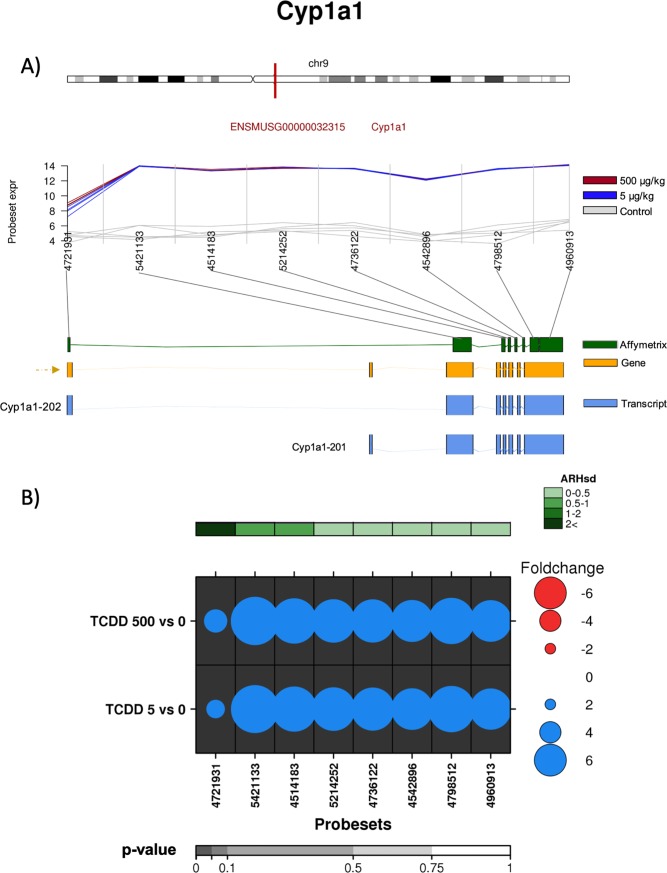
From this list, several genes whose abundances were known to be regulated by AhR were identified, including cytochrome P450s 1a1 (Cyp1a1), 1a2 (Cyp1a2), 1b1 (Cyp1b1), NAD(P)H quinone dehydrogenase (Nqo1), TCDD inducible poly(ADP-ribose) polymerase (Tiparp), aryl hydrocarbon receptor repressor (Ahrr) and serum amyloid A (Saa) [30, 31]. From these gene sets we determined whether TCDD modified the alternative splicing of the pre-mRNAs of Cyp1a1 and Ahrr. The Cyp1a1 gene is known to be highly induced following exposure to TCDD. It is composed of 8 exons, 6 of which are coding exons (NCBI) and has been reported to have two different transcripts, Cyp1a1-201 with 7 exons (6 coding exons) and NM_009992.4 (also named Cyp1a1-202) also with 7 exons (6 coding exons) (Fig 3A).
Fig 3. Analysis of the effect of TCDD on exonic abundances of Cyp1a1.
A) Localization and normalized abundances of ProbeSets targeting distinct exons within the Cyp1a1 gene. Upper panel: chromosome 9 structure and Cyp1a1 localization (red line). Middle panel: Log2 of the intensities for each of the probes targeting regions within this gene, control (gray lines), low dose TCDD 5 μg/kg (fuchsia lines) and high dose TCDD 500 μg/kg (brown lines). Bottom panel: Comparative structure of known Cyp1a1 isoforms. Gene reference, including direction of the transcription (yellow), transcripts as reported by Ensembl (blue) and Affymetrix microarray ProbeSets (green). B) RNA abundance of each ProbeSet was contrasted between TCDD treated mouse liver (5 or 500 μg/kg separately) and controls. Dotmap shows the differential abundances for each ProbeSet for each group; such that red dots show reduced abundance following TCDD exposure, while blue shows increased abundance. Dot size represents the magnitude of change. Background shading indicates significant of change, as quantified by the bottom gray bar (FDR adjusted p-value). Top green covariate bar represents the ARH value (with darker shading indicating higher intensity).
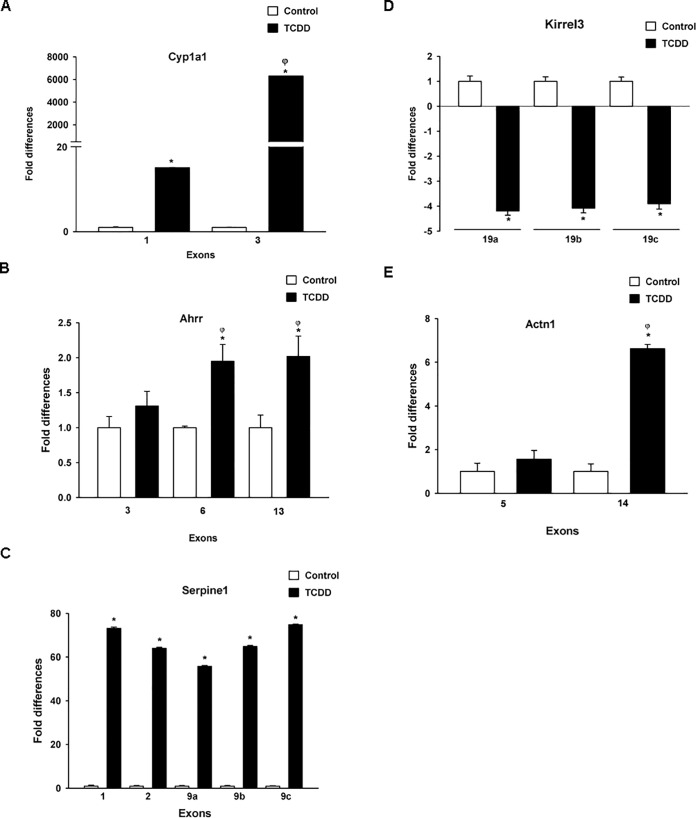
According to our analysis, at both high and low dose of TCDD (5 and 500 μg/kg), we detected consistently increased abundance of all Cyp1a1 exons. Interestingly exon 1 showed considerably lower abundance than other exons (Fig 3B), indicating that TCDD treatment changed the relative abundance of the splice variant predominantly to variant 2 (NM_001136059.2). This variant excluded exon 1, which alters the 5’ UTR. These results may also suggest an alternative transcriptional start site located between exon 1 and exon 2; however, in silico analysis failed to identify XREs in this region (ALGGEN PROMO database [http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3], TH = 85). Validation of the exon abundance was done via qRT-PCR using probes for exons 1 and 3. As Fig 4A shows, 500 μg/kg TCDD treatment resulted in ~6,000-fold induction of exon 3. In contrast, less than 20-fold induction was observed for exon 1, confirming the exon array results.
Fig 4. Validation of TCDD effects on exon abundances.
Total mRNA was extracted from the liver from mice treated with corn oil or TCDD (500 μg/kg). A) Cyp1a1 exon 1 (ProbeSet 4721931) and exon 3 (ProbeSet 5421133). B) Ahrr exon 3 (ProbeSet 4469533), exon 6 (ProbeSet 5258969) and exon 13 (ProbeSet 4848271). C) Serpine1 exon 1 (ProbeSet 4403932), exon 2 (ProbeSet 5024287), exon 9a (ProbeSet 4354131), exon 9b (ProbeSet 5079297) and exon 9c (ProbeSet 5082204). D) Kirrel3 exon 19a (ProbeSet 5043467), exon 19b (ProbeSet 5047088), and exon 19c (ProbeSet 4806289). E) Actn1 exon 5 (ProbeSet 5008356) and exon 14 (ProbeSet 5597164). mRNA levels were determined by RT-qPCR and normalized to 18S ribosomal RNA. The results are expressed as the mean ± S.E. of samples from three different mice. *p < 0.05, treatment vs. control. ȹp < 0.05, between treatments.
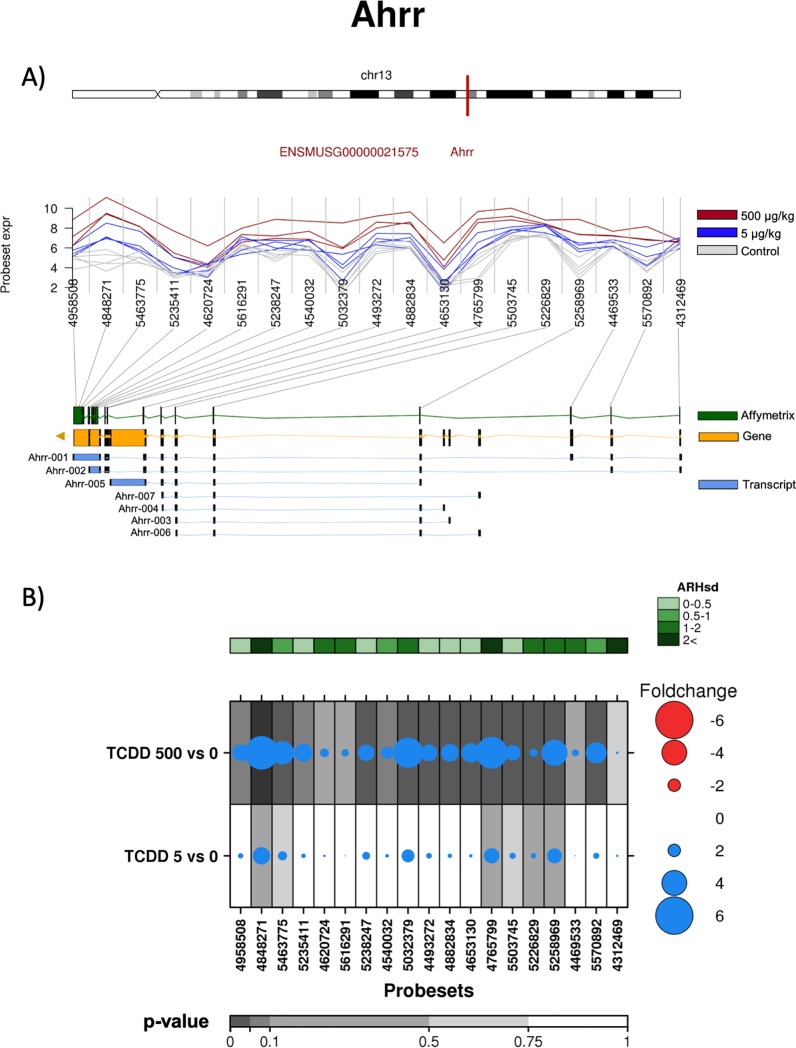
Another interesting gene identified as differentially abundant by ARH was Ahrr. The Ahrr gene is encoded by 13 exons (Fig 5A), and Ensembl, identified 7 different splice variants for this gene. Based on differential abundance of ProbeSets, we observed preferential expression following TCDD treatment of a splice variant that excluded exons 1 and 3, but included exon 13 (Fig 5B).
Fig 5. Analysis of the effect of TCDD on exonic abundances of Ahrr.
A) Localization and normalized abundances of ProbeSets targeting distinct exons within the Ahrr gene. Upper panel: chromosome 13 structure and Ahrr localization (red line). Middle panel: Log2 of the intensities for each of the probes targeting regions within this gene, control (gray lines), low dose TCDD 5 μg/kg (fuchsia lines) and high dose TCDD 500 μg/kg (brown lines). Bottom panel: Comparative structure of known Ahrr isoforms. Gene reference, including direction of the transcription (yellow), transcripts as reported by Ensembl (blue) and Affymetrix microarray ProbeSets (green). B) RNA abundance of each ProbeSet was contrasted between TCDD treated mouse liver (5 or 500 μg/kg separately) and controls. Dotmap shows the differential abundances for each ProbeSet for each group; such that red dots show reduced abundance following TCDD exposure, while blue shows increased abundance. Dot size represents the magnitude of change. Background shading indicates significant of change, as quantified by the bottom gray bar (FDR adjusted p-value). Top green covariate bar represents the ARH value (with darker shading indicating higher intensity).
This transcript does not correspond to any reported splice variant, so may represent a novel isoform, or a complex combination of isoforms. The RT-qPCR verifies that after TCDD treatment the abundance of exon 3 is indeed lower than that of exons 6 and 13 (Fig 4B).
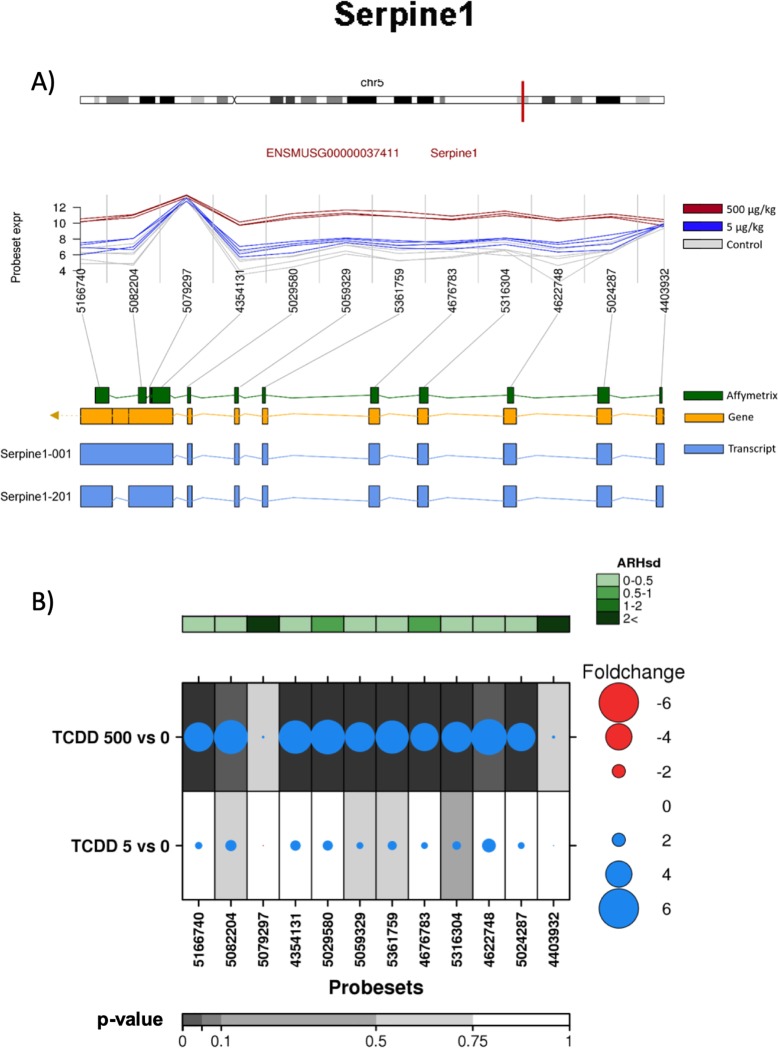
In addition to known AHR-core genes, ARH detected significant differential splicing of 29 other genes. Of these, Serpine1 and Kirrel3 demonstrated the highest ARH index. The Serpine1 gene encodes for plasminogen activator inhibitor 1. It contains 9 exons, and two transcripts for this gene have been reported, Serpine1-001 and Serpine1-201 (Fig 6A).
Fig 6. Analysis of the effect of TCDD on exonic abundances of Serpine 1.
A) Localization and normalized abundances of ProbeSets targeting distinct exons within the Serpine 1 gene. Upper panel: chromosome 5 structure and Serpine 1 localization (red line). Middle panel: Log2 of the intensities for each of the probes targeting regions within this gene, control (gray lines), low dose TCDD 5 μg/kg (fuchsia lines) and high dose TCDD 500 μg/kg (brown lines). Bottom panel: Comparative structure of known Serpine 1 isoforms. Gene reference, including direction of the transcription (yellow), transcripts as reported by Ensembl (blue) and Affymetrix microarray ProbeSets (green). B) RNA abundance of each ProbeSet was contrasted between TCDD treated mouse liver (5 or 500 μg/kg separately) and controls. Dotmap shows the differential abundances for each ProbeSet for each group; such that red dots show reduced abundance following TCDD exposure, while blue shows increased abundance. Dot size represents the magnitude of change. Background shading indicates significant of change, as quantified by the bottom gray bar (FDR adjusted p-value). Top green covariate bar represents the ARH value (with darker shading indicating higher intensity).
The splice variant corresponds to a cassette-exon 9 skipping. Exon array analysis indicates that TCDD treatment increased abundance of all ProbeSets, excluding the 4403932 ProbeSet located at exon 1 and the 5079297 ProbeSet at exon 9 (Fig 6B). Results from qRT-PCR validation demonstrated that TCDD increased the abundance of all exons tested, including exons 1, 2 and 9. The latter was determined by the use of three different ProbeSets. With respect to the magnitude of their induction, no significant differences were observed between the exons analyzed (Fig 4C).
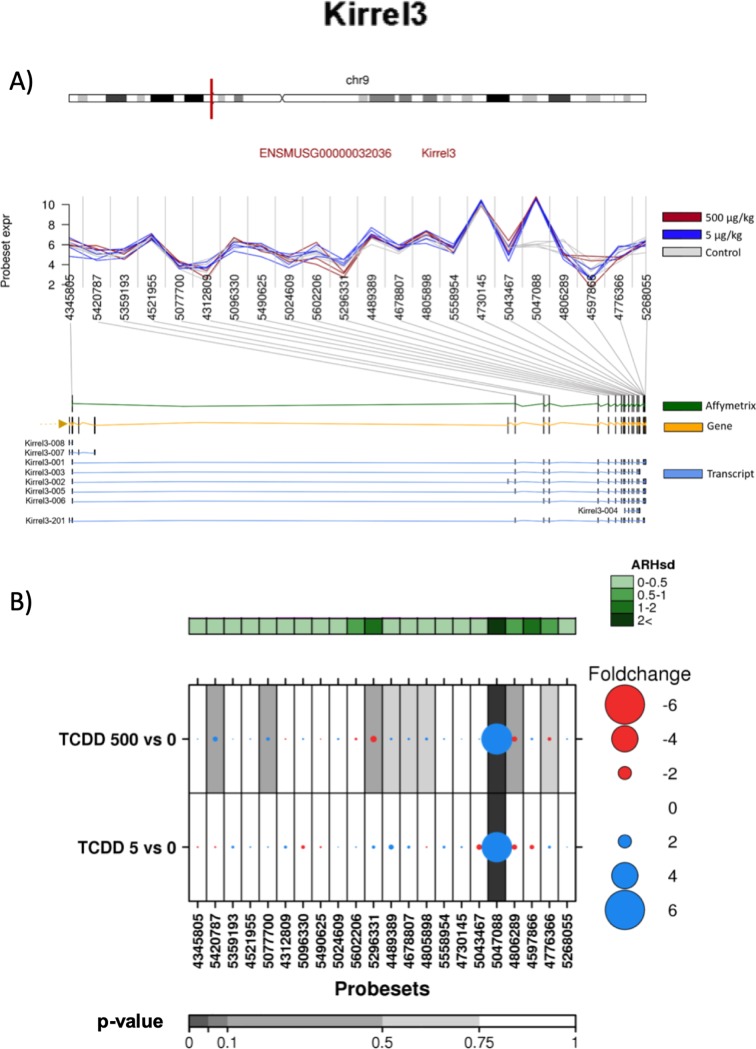
Kirrel3 represents the most complex gene on which we focused. The protein encoded by the Kirrel3 gene is Kirrel-like nephrin family adhesion molecule 3. It contains 19 exons, and 9 different splice variants have been identified (Fig 7A).
Fig 7. Analysis of the effect of TCDD on exonic abundances of Kirrel3.
A) Localization and normalized abundances of ProbeSets targeting distinct exons within the Kirrel3 gene. Upper panel: chromosome 9 structure and Kirrel3 localization (red line). Middle panel: Log2 of the intensities for each of the probes targeting regions within this gene, control (gray lines), low dose TCDD 5 μg/kg (fuchsia lines) and high dose TCDD 500 μg/kg (brown lines). Bottom panel: Comparative structure of known Kirrel3 isoforms. Gene reference, including direction of the transcription (yellow), transcripts as reported by Ensembl (blue) and Affymetrix microarray ProbeSets (green). B) RNA abundance of each ProbeSet was contrasted between TCDD treated mouse liver (5 or 500 μg/kg separately) and controls. Dotmap shows the differential abundances for each ProbeSet for each group; such that red dots show reduced abundance following TCDD exposure, while blue shows increased abundance. Dot size represents the magnitude of change. Background shading indicates significant of change, as quantified by the bottom gray bar (FDR adjusted p-value). Top green covariate bar represents the ARH value (with darker shading indicating higher intensity).
Exon array data indicated that TCDD treatment only induced the abundance of the 5047088 ProbeSet located at exon 19. In contrast, ProbeSets 4806289 and 5043467, located in the same exon adjacent to 5047088 ProbeSet, did not exhibit any change after TCDD treatment (Fig 7B), suggesting that dioxin treatment induced the alternative splicing of exon 19. However, the RT-qPCR analysis indicated that TCDD treatment resulted in a decrease in the abundance of all three regions located on exon 19, without differences between them (Fig 4D).
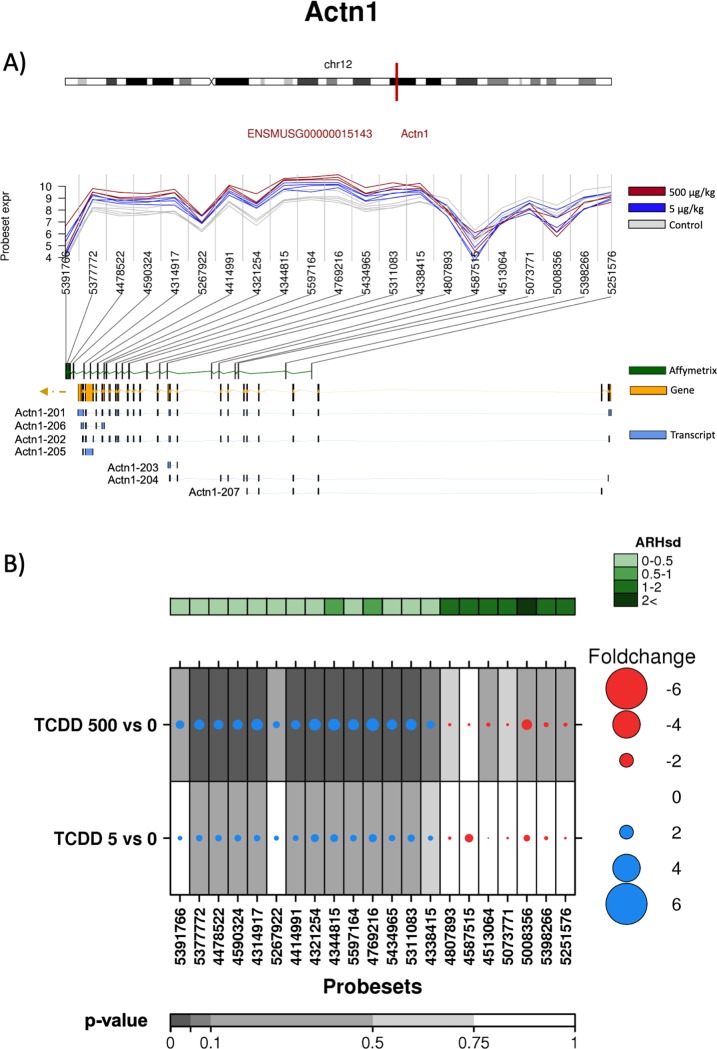
Finally, the effect of TCDD on α-Actn1 alternative splicing was evaluated. α-Actn1 gene encodes for alpha actinin 1, and contains 22 exons and has 7 identified splicing variants (Fig 8A).
Fig 8. Analysis of the effect of TCDD on exonic abundances of Actn1.
A) Localization and normalized abundances of ProbeSets targeting distinct exons within the Actn1 gene. Upper panel: chromosome 12 structure and Actn1 localization (red line). Middle panel: Log2 of the intensities for each of the probes targeting regions within this gene, control (gray lines), low dose TCDD 5 μg/kg (fuchsia lines) and high dose TCDD 500 μg/kg (brown lines). Bottom panel: Comparative structure of known Actn1 isoforms. Gene reference, including direction of the transcription (yellow), transcripts as reported by Ensembl (blue) and Affymetrix microarray ProbeSets (green). B) RNA abundance of each ProbeSet was contrasted between TCDD treated mouse liver (5 or 500 μg/kg separately) and controls. Dotmap shows the differential abundances for each ProbeSet for each group; such that red dots show reduced abundance following TCDD exposure, while blue shows increased abundance. Dot size represents the magnitude of change. Background shading indicates significant of change, as quantified by the bottom gray bar (FDR adjusted p-value). Top green covariate bar represents the ARH value (with darker shading indicating higher intensity).
The exon array data suggested that TCDD induced the abundance of a α-Actn1 variant constituted predominantly by the region encompassing exons 11 to 20 (Fig 8B). These results were supported by RT-qPCR, which indicated a 6-fold induction of exon 14 upon TCDD exposure, while no effect was observed on exon 5 (Fig 4E).
Discussion
Exposure to TCDD results in a wide range of deleterious effects on human health by altering cell processes, such as the cell cycle and differentiation [32], among many others. To date, it has not been shown whether toxicants like TCDD can alter splice site selection and lead to changes in transcript variant abundances. In the current study, the effect of TCDD on alternative splicing was evaluated through the use of exon arrays. ARH analysis of the exon arrays indicated that the alternative splicing of several liver mouse transcripts was modified by dioxin exposure (Fig 2). Several genes that are transcriptionally regulated by the AhR in response to TCDD exposure were determined to have altered splicing. Two of these (Cyp1a1 and Ahrr), were selected for validation using RT-qPCR. In humans, several CYP1A1 splice variants have been identified, some with functional effects. A variant with deletion of exon 6 has been identified in the human brain, but not in the liver, and it is unable to bioactivate PAHs [33]. In contrast, a CYP1A1 splice variant formed via excision of an 84-bp intron within exon 2 is enzymatically active and is localized exclusively in the nucleus and mitochondria [34]. In mice, a variant that affects the functionality and/or localization of the protein has not yet been reported. The present data indicates that TCDD promotes the abundance of transcript Cyp1a1-201, but it also induces the abundance of the 5’ UTR variant (Cyp1a1-202). Both transcript variants generate the same protein; therefore CYP1A1 enzyme function should remain the same.
Regarding Ahrr, it has been reported a human splice variant resulting in modifications of AHRR functions. This variant, not yet included in NCBI database, has been identified as the active form of this protein and lacks exon 8. It is present several tissues, including human liver, lung, heart, spleen, kidney and brain [35]. No report has indicated that any of mouse splice variants result in modifications of Ahrr functions. Based on the exon array analysis and the RT-qPCR data indicates that after TCDD treated samples have lower abundance of exon 3 than that of exons 6 and 13, suggesting a novel Ahrr splice variant. The functional consequences of this variant require future investigation.
According to the exon array analysis, TCDD treatment also modifies α-Actn1 alternative splicing, favoring production of a variant lacking the first 8 exons. This observation was partially confirmed by RT-qPCR, which showed that TCDD induces exon 14 abundance 6-fold compared to untreated samples, while no effect was observed for exon 5.
α-Actinin1 is coded by α-Actn1 gene. Through its ability to interact with actin filaments, α-Actinin1 plays an important role in the regulation of intracellular infrastructure, and therefore in cell adhesion, cytokinesis and cell migration [36]. Calcium binding to the EF-hand domain reduces its affinity for actin filaments. Splicing at the EF-hand domain results in 2 splice variants. Inclusion of exon 19a, but not 19b leads to expression of the non-muscle α-Actinin1 with an active EF-hand domain. In contrast, when exon 19b, but not 19a is included, the calcium-insensitive smooth muscle isoform is expressed [37], which has been associated with cancer cells [38] [39]. As indicated above, TCDD promotes the expression of an α-Actinin1 variant without exon 5, which is located at the N-terminal actin binding-domain region [40]. Recently, it has been reported that mutations in the actin-binding domain of α-Actinin1 increase its association with actin [41]. Therefore, the TCDD-induced splice variant might have altered α-Actinin1-actin interactions.
ARH analysis also predicted that TCDD treatment increased the abundance of exon 19 splice variants of Kirrel3. However, validation was not consistent with this result. Moreover, RT-qPCR assays indicated that TCDD treatment resulted in a decrease in exon 19 abundance relative to control samples. Although no splicing event was detected, clearly TCDD treatment altered Kirrel3 abundance. Kirrel3, also known as Neph2, is localized in the pre- and post-synapse in the hippocampus. In particular, Kirrel3 regulates the output specificity of hippocampal DG axons [42], and disruption of its locus has been associated with neurodevelopmental disorders [43]. Kirrel3 null mice present a reduction in mossy fibers filipodia formation in the hippocampus, resulting in the hyperexcitability of CA3 neurons [44]. Furthermore, a previous study showed that TCDD decreased the side of the pyramidal mossy fiber field of the hippocampus [45]. Therefore, our current results suggest that the effect of TCDD on the mossy fiber field of the hippocampus is through decreasing Kirrel3 abundance.
Validation of Serpine1 by qRT-PCR did not show a splicing event as the ARH analysis predicted. However, TCDD treatment induced the expression of exons 1, 2 and 9 more than 50-fold when compared to nontreated samples. The Serpine1 gene encodes the plasminogen activator inhibitor type I (PAI-1), which inhibits the activation of plasminogen into plasmin, and therefore the proteolysis of proteins in connective tissue, basement membranes and blood clots. The present data suggest that TCDD, through the induction of Serpine1, might alter mechanisms involved in tissue remodeling, contributing to the development of several pathologic processes, such as metastasis and intravascular thrombosis [46] [47].
Finally, although the ARH method correctly predicted the alternative splicing of some pre-mRNAs, the analysis also resulted in false positive data as with Kirrel3 and Serpine1, pointing out the need for the improvement of the prediction methods used to evaluate alternative splicing when exon arrays are used. RNA sequencing is also used for the identification of alternative splicing. Both methods have advantages and disadvantages. Nazrev and collaborators in a comparative analysis conclude that combining the use of both platforms for the identification of alternative splicing will be more reliable [48].
In conclusion, this study demonstrated that TCDD exposure leads to modify the alternative splicing of several transcripts. The mechanism of these effects is still unknown and will require future investigation focused on the effects of TCDD on spliceosome assembly, on the recognition of alternative exons, and/or on the levels of splicing factors. Additionally, more studies will be necessary to elucidate whether the AhR plays a role in these splicing changes.
Acknowledgments
Dr. María Cabañas-Cortés for its technical support in the RT-qPCR.
Data Availability
Raw data is available in GEO at GSE126328, and in the TCDD transcriptomics package (v2.2.5) for R (download from https://github.com/BoutrosLaboratory/TCDD.Transcriptomics or https://labs.oicr.on.ca/boutros-lab/tcdd-transcriptomics).
Funding Statement
This study was supported by Terry Fox Research Institute and CIHR New Investigator Awards, to PCB; and Consejo Nacional de Ciencia y Tecnología grant 153377 and 280897, GE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aylward LL, Hays SM. Temporal trends in human TCDD body burden: decreases over three decades and implications for exposure levels. Journal of exposure analysis and environmental epidemiology. 2002;12(5):319–28. Epub 2002/08/29. 10.1038/sj.jea.7500233 . [DOI] [PubMed] [Google Scholar]
- 2.Manz A, Berger J, Dwyer JH, Flesch-Janys D, Nagel S, Waltsgott H. Cancer mortality among workers in chemical plant contaminated with dioxin. Lancet. 1991;338(8773):959–64. Epub 1991/10/19. 10.1016/0140-6736(91)91835-i . [DOI] [PubMed] [Google Scholar]
- 3.Pohjanvirta R, Tuomisto J. Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: effects, mechanisms, and animal models. Pharmacological reviews. 1994;46(4):483–549. Epub 1994/12/01. . [PubMed] [Google Scholar]
- 4.White SS, Birnbaum LS. An overview of the effects of dioxins and dioxin-like compounds on vertebrates, as documented in human and ecological epidemiology. Journal of environmental science and health Part C, Environmental carcinogenesis & ecotoxicology reviews. 2009;27(4):197–211. Epub 2009/12/03. 10.1080/10590500903310047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug metabolism and disposition: the biological fate of chemicals. 1998;26(12):1194–8. Epub 1998/12/23. . [PubMed] [Google Scholar]
- 6.Boutros PC, Yao CQ, Watson JD, Wu AH, Moffat ID, Prokopec SD, et al. Hepatic transcriptomic responses to TCDD in dioxin-sensitive and dioxin-resistant rats during the onset of toxicity. Toxicol Appl Pharmacol. 2011;251(2):119–29. 10.1016/j.taap.2010.12.010 . [DOI] [PubMed] [Google Scholar]
- 7.Moffat ID, Boutros PC, Chen H, Okey AB, Pohjanvirta R. Aryl hydrocarbon receptor (AHR)-regulated transcriptomic changes in rats sensitive or resistant to major dioxin toxicities. BMC Genomics. 2010;11:263 10.1186/1471-2164-11-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69(1):140–53. 10.1124/mol.105.018705 . [DOI] [PubMed] [Google Scholar]
- 9.Kumar MB, Tarpey RW, Perdew GH. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. The Journal of biological chemistry. 1999;274(32):22155–64. Epub 1999/07/31. 10.1074/jbc.274.32.22155 . [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Baumgarten SC, Zhou P, Stocco C. Testosterone-dependent interaction between androgen receptor and aryl hydrocarbon receptor induces liver receptor homolog 1 expression in rat granulosa cells. Molecular and cellular biology. 2013;33(15):2817–28. Epub 2013/05/22. 10.1128/MCB.00011-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. The Journal of biological chemistry. 1999;274(1):510–5. Epub 1998/12/29. 10.1074/jbc.274.1.510 . [DOI] [PubMed] [Google Scholar]
- 12.Prochazkova J, Kabatkova M, Bryja V, Umannova L, Bernatik O, Kozubik A, et al. The interplay of the aryl hydrocarbon receptor and beta-catenin alters both AhR-dependent transcription and Wnt/beta-catenin signaling in liver progenitors. Toxicological sciences: an official journal of the Society of Toxicology. 2011;122(2):349–60. Epub 2011/05/24. 10.1093/toxsci/kfr129 . [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Yuan XG, Liu CP, Zhai SN, Zhang DW, Fu YX. Preliminary research on DNA methylation changes during murine palatogenesis induced by TCDD. J Craniomaxillofac Surg. 2017;45(5):678–84. 10.1016/j.jcms.2017.02.004 . [DOI] [PubMed] [Google Scholar]
- 14.Yuan X, Qiu L, Pu Y, Liu C, Zhang X, Wang C, et al. Histone acetylation is involved in TCDDinduced cleft palate formation in fetal mice. Mol Med Rep. 2016;14(2):1139–45. 10.3892/mmr.2016.5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enan E, Matsumura F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochemical pharmacology. 1996;52(10):1599–612. Epub 1996/11/22. 10.1016/s0006-2952(96)00566-7 . [DOI] [PubMed] [Google Scholar]
- 16.Puga A, Hoffer A, Zhou S, Bohm JM, Leikauf GD, Shertzer HG. Sustained increase in intracellular free calcium and activation of cyclooxygenase-2 expression in mouse hepatoma cells treated with dioxin. Biochemical pharmacology. 1997;54(12):1287–96. Epub 1997/12/11. 10.1016/s0006-2952(97)00417-6 . [DOI] [PubMed] [Google Scholar]
- 17.Falahatpisheh MH, Ramos KS. Ligand-activated Ahr signaling leads to disruption of nephrogenesis and altered Wilms' tumor suppressor mRNA splicing. Oncogene. 2003;22(14):2160–71. 10.1038/sj.onc.1206238 . [DOI] [PubMed] [Google Scholar]
- 18.Yan C, Wu W, Li H, Zhang G, Duerksen-Huehes P, Zhu X. Benzo(a)pyrene treatment leads to changes in nuclear protein expression and alternative splicing. Mutation Research. 2010; 686 (1–2): 47–56. 10.1016/j.mrfmmm.2010.01.015 . [DOI] [PubMed] [Google Scholar]
- 19.Hsu LC, Chang WC, Chang C, Tsukamoto N, Yoshida A. The human aldehyde dehydrogenase 3 gene (ALDH3): identification of a new exon and diverse mRNA isoforms, and functional analysis of the promoter. Gene Expression. 1996; 6(2): 87–99. . [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–63. Epub 2010/01/30. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6. Epub 2008/11/04. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaharieva E, Chipman JK, Soller M. Alternative splicing interference by xenobiotics. Toxicology. 2012;296(1–3):1–12. Epub 2012/02/11. 10.1016/j.tox.2012.01.014 . [DOI] [PubMed] [Google Scholar]
- 23.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature reviews Genetics. 2007;8(10):749–61. Epub 2007/08/30. 10.1038/nrg2164 . [DOI] [PubMed] [Google Scholar]
- 24.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nature reviews Genetics. 2016;17(1):19–32. Epub 2015/11/26. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–7. Epub 2010/08/07. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. Epub 2003/08/20. 10.1093/biostatistics/4.2.249 . [DOI] [PubMed] [Google Scholar]
- 27.P'ng C, Green J, Chong LC, Waggott D, Prokopec SD, Shamsi M, et al. BPG: Seamless, automated and interactive visualization of scientific data. BMC Bioinformatics. 2019;20(1):42 10.1186/s12859-019-2610-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prokopec SD, Houlahan KE, Sun RX, Watson JD, Yao CQ, Lee J, et al. Compendium of TCDD-mediated transcriptomic response datasets in mammalian model systems. BMC Genomics. 2017;18(1):78 10.1186/s12864-016-3446-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasche A, Herwig R. ARH: predicting splice variants from genome-wide data with modified entropy. Bioinformatics. 2010;26(1):84–90. Epub 2009/11/06. 10.1093/bioinformatics/btp626 . [DOI] [PubMed] [Google Scholar]
- 30.Rowlands JC, Gustafsson JA. Aryl hydrocarbon receptor-mediated signal transduction. Crit Rev Toxicol. 1997;27(2):109–34. 10.3109/10408449709021615 . [DOI] [PubMed] [Google Scholar]
- 31.Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab Invest. 2009;89(6):695–707. 10.1038/labinvest.2009.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chopra M, Schrenk D. Dioxin toxicity, aryl hydrocarbon receptor signaling, and apoptosis-persistent pollutants affect programmed cell death. Crit Rev Toxicol. 2011;41(4):292–320. 10.3109/10408444.2010.524635 . [DOI] [PubMed] [Google Scholar]
- 33.Kommaddi RP, Turman CM, Moorthy B, Wang L, Strobel HW, Ravindranath V. An alternatively spliced cytochrome P4501A1 in human brain fails to bioactivate polycyclic aromatic hydrocarbons to DNA-reactive metabolites. J Neurochem. 2007;102(3):867–77. 10.1111/j.1471-4159.2007.04599.x . [DOI] [PubMed] [Google Scholar]
- 34.Leung YK, Lau KM, Mobley J, Jiang Z, Ho SM. Overexpression of cytochrome P450 1A1 and its novel spliced variant in ovarian cancer cells: alternative subcellular enzyme compartmentation may contribute to carcinogenesis. Cancer Res. 2005;65(9):3726–34. 10.1158/0008-5472.CAN-04-3771 . [DOI] [PubMed] [Google Scholar]
- 35.Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, et al. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Molecular and cellular biology. 2009;29(13):3465–77. 10.1128/MCB.00206-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58(2):104–11. 10.1002/cm.20007 . [DOI] [PubMed] [Google Scholar]
- 37.Southby J, Gooding C, Smith CW. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutally exclusive exons. Molecular and cellular biology. 1999;19(4):2699–711. 10.1128/mcb.19.4.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325 10.1186/1471-2164-7-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7(7):1214–24. 10.1074/mcp.M700590-MCP200 . [DOI] [PubMed] [Google Scholar]
- 40.Waites GT, Graham IR, Jackson P, Millake DB, Patel B, Blanchard AD, et al. Mutually exclusive splicing of calcium-binding domain exons in chick alpha-actinin. The Journal of biological chemistry. 1992;267(9):6263–71. . [PubMed] [Google Scholar]
- 41.Murphy AC, Lindsay AJ, McCaffrey MW, Djinovic-Carugo K, Young PW. Congenital macrothrombocytopenia-linked mutations in the actin-binding domain of alpha-actinin-1 enhance F-actin association. FEBS Lett. 2016;590(6):685–95. 10.1002/1873-3468.12101 . [DOI] [PubMed] [Google Scholar]
- 42.Rawson RL, Martin EA, Williams ME. Mechanisms of input and output synaptic specificity: finding partners, building synapses, and fine-tuning communication. Curr Opin Neurobiol. 2017;45:39–44. 10.1016/j.conb.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–37. 10.1016/j.cell.2012.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin EA, Muralidhar S, Wang Z, Cervantes DC, Basu R, Taylor MR, et al. The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. Elife. 2015;4:e09395 10.7554/eLife.09395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers BE, Lin TM, Vanka A, Peterson RE, Juraska JM, Schantz SL. Tetrachlorodibenzo-p-dioxin exposure alters radial arm maze performance and hippocampal morphology in female AhR mice. Genes Brain Behav. 2005;4(1):51–9. 10.1111/j.1601-183X.2004.00098.x . [DOI] [PubMed] [Google Scholar]
- 46.Placencio VR, DeClerck YA. Plasminogen Activator Inhibitor-1 in Cancer: Rationale and Insight for Future Therapeutic Testing. Cancer Research. 2015;75(15):2969–74. 10.1158/0008-5472.CAN-15-0876 PMCID: PMC4613764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen R, Yan J, Liu P, Wang Z, Wang C. Plasminogen activator inhibitor links obesity and thrombotic cerebrovascular diseases: The roles of PAI-1 and obesity on stroke. Metab Brain Dis. 2017;32(3):667–73. 10.1007/s11011-017-0007-3 [DOI] [PubMed] [Google Scholar]
- 48.Nazarov PV, Muller A, Kaoma T, Nicot N, Maximo C. RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples. BMC Genomics. 2017;18(1):1–18. 10.1186/s12864-016-3406-7 PMCID: PMC5461714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data is available in GEO at GSE126328, and in the TCDD transcriptomics package (v2.2.5) for R (download from https://github.com/BoutrosLaboratory/TCDD.Transcriptomics or https://labs.oicr.on.ca/boutros-lab/tcdd-transcriptomics).