Abstract
Purpose
Lung cancer in never smokers is recognized as a distinct molecular, clinicopathologic and epidemiologic entity. The aim of the study was to investigate the molecular profile in Swiss never smokers with lung adenocarcinoma and to correlate the mutation status with clinicopathologic and demographic patient characteristics and outcome.
Methods
One hundred thirty-eight never smokers diagnosed with lung adenocarcinoma at the University Hospital Zurich between 2011–2018 were included in the study. Data from the electronic medical records were reviewed to characterize clinicopathologic and demographic features, molecular profile, treatment and outcome.
Results
The majority of patients were female (58.7%) with a median age at diagnosis of 64.5 years (range, 27.1–94.2 years). The most common mutations were EGFR (58.7%) followed by ALK (12.3%), TP53 (5.8%), MET (5.8%), KRAS (4.3%), ERBB2 (4.3%), PIK3CA (2.9%), BRAF (2.2%), ROS1 (1.4%), RET (1.4%), CTNNB1 (0.7%), PARP1 (0.7%), TET1 (0.7%) and PIK3CG (0.7%). Median overall survival (mOS) was 51.0 months (mo). Early clinical stage (p = 0.002) and treatment with targeted therapy (HR 2.53, 95% CI 1.35–4.74, p = 0.004) were independently associated with longer mOS. Patients with oncogenic driver mutations had significantly longer mOS (52.2 mo) compared to patients without mutations (16.9 mo) (HR 3.38, 95% CI 1.52–7.55, p = 0.003). Besides, patients with EGFR mutated (57.8 mo) or ALK rearranged (59.9 mo) tumors had significantly longer mOS compared to the EGFR wildtype (35.0 mo), ALK wildtype (46.5 mo) and pan-negative (16.9 mo) cohorts (HR 2.35, 95% CI 1.37–4.04, p = 0.002; HR 7.80, 95% CI 3.28–18.55, p < 0.001; HR 3.96, 95% CI 1.21–12.95, p = 0.023 and HR 34.78, 95% CI 3.48–34.65, p = 0.003).
Conclusion
Never smokers with lung adenocarcinoma display distinct clinicopathologic and molecular features and are characterized by a high incidence of targetable mutations. Never smokers with targetable mutations have significantly longer survival compared to patients without mutations.
Introduction
Although the majority of lung cancer cases are associated with tobacco smoking, a considerable proportion (~10–40%) of patients develop lung cancer without a significant personal history of tobacco use [1–5]. Lung cancer in never smokers (LCINS) constitutes an entity with distinct gender, geographic, clinicopathologic and molecular characteristics compared to lung cancer occurring in smokers [2,3]. The majority (~65%-87% [1,6–8]) of never smoker patients with lung cancer are women, and the incidence of LCINS is significantly higher in certain geographic regions, including East Asia, than in the United States and in Europe [1,9,10]. Molecular profiling studies have shown that never smokers with lung adenocarcinoma harbor significantly lower somatic mutation burden than smokers with the same disease [11]. Besides, C > A transversions are more common in smokers, while C > T transitions occur more frequently in never smokers [12]. EGFR mutations and ALK rearrangements appear to be more frequent in never smokers than in smokers, whereas KRAS and BRAF mutations are less common in this subset of patients [6,13,14]. The majority of previous studies that have characterized the genomic alterations of LCINS have come from East Asia due to the high incidence of LCINS in this geographic region. In contrast, data on the molecular characteristics of LCINS in Western populations are scarce [1,6].
The aim of the current study was to analyze the molecular features of Swiss never smoker patients with lung adenocarcinoma and to correlate the mutation status with demographic and clinicopathologic characteristics and outcome. Because lung adenocarcinoma is the most common histologic subtype in never smokers with non-small cell lung cancer (NSCLC) accounting for up to 94% of LCINS [7], we focused on never smokers with adenocarcinoma histology.
Methods
Patients and data collection
We performed a retrospective analysis of demographic, clinical and pathologic data stored in the electronic medical record system at the University Hospital Zurich of all never smokers with a pathologic diagnosis of stage I-IV lung adenocarcinoma, radio-chemotherapy and targeted therapy naïve, diagnosed at our institution between January 2011 and January 2018. Inclusion criteria were: 1) being a self-declared never smoker; 2) a diagnosis of histologically and/or cytologically confirmed lung adenocarcinoma; 3) radio-chemotherapy and targeted therapy naïve; and 4) tissue blocks/cell blocks with adequate tumor cellularity. Exclusion criteria were: 1) non-adenocarcinoma histology; 2) previous chemotherapy, targeted therapy or radiotherapy; and 3) insufficient tumor material. Never smokers were defined as individuals who had smoked less than 100 cigarettes in their lifetime. The following data were retrieved from the electronic patient record system: gender, ethnicity, age at diagnosis, clinical stage at diagnosis, TNM stage at diagnosis (according to the Union for International Cancer Control (UICC) TNM classification of malignant tumors, 8th edition [15]), tumor location, tumor size, type and duration of systemic treatment, and survival history. Overall survival was measured from the date of pathologic diagnosis until the date of death. Patients were censored on May 31, 2018 if they were alive. Patients without a known date of death were censored at the time of last follow-up. The diagnosis of lung adenocarcinoma was made on hematoxylin and eosin stained and immunostained formalin-fixed, paraffin-embedded tissue sections from resection or biopsy specimens and/or on cytologic samples based on the 2015 World Health Organization classification for lung tumors [16]. The study was approved by the Cantonal Ethics Committee of Zurich (StV-No. 2018–01919), and written informed consent was obtained from all patients, including written permission of two patients to publish their PET-CT scans.
Molecular analysis and immunohistochemistry
Tumor tissues from core biopsies (58.7%), resection specimens (30.4%) and cytologic samples (10.9%) were used to perform all molecular analyses according to the National Comprehensive Cancer Network (NCCN) and Swiss Society of Pathology (SSPath) guidelines, as previously described [17]. Specific genotyping methods utilized included Sanger sequencing (SS) of EGFR, KRAS, ERBB2 and BRAF, immunohistochemistry (IHC)/immunocytochemistry (ICC) assays for ALK and ROS1, break-apart fluorescence in situ hybridization (FISH) for ALK, ROS1 and RET and targeted DNA- and RNA-based next-generation sequencing (NGS). Targeted DNA- and RNA-based NGS was performed in 69 patients using different customer panels during the study period, including the Ion AmpliSeq Colon and Lung Cancer panel 2 (CLP2), Ion AmpliSeq Fusion Lung Cancer Research panel (LFP), and Oncomine DNA panel for Solid Tumors and Fusion Transcripts (Thermo Fisher Scientific/Life Technologies, Carlsbad, California, USA). We used the Ion Library Quantitation kit (Thermo Fisher Scientific) for quantification of DNA and RNA libraries, the Ion One Touch 200 Template Kit v2 DL (lately replaced by the Ion Hi-Q Chef Kit and the Ion Chef System) (Thermo Fisher Scientific) for emulsion polymerase chain reaction (PCR) and template preparation, and the Ion Personal Genome Machine 200 Kit v2 (lately replaced by the Ion Personal Genome Machine Hi-Q Sequencing Kit) (Thermo Fisher Scientific) as sequencing platform. For Sanger sequencing, we used the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, Buckinghamshire, UK) for purification of amplified DNA fragments, the Genetic Analyzer 3130x1 (Applied Biosystems, Foster City, CA, USA) for sequencing and the Sequencher 5.1 (Gene Code Corporation, Ann Arbor, MI, USA) for data analysis. ALK and ROS1 immunohistochemistry (IHC)/immunocytochemistry (ICC) was performed on the automated immunostainer DiscoveryUltra (Roche Ventana) using a mouse anti-human ALK monoclonal antibody (clone 5A4, Leica Biosystems) and a rabbit anti-human ROS1 monoclonal antibody (clone D4D6, Cell Signaling Technology). ALK or ROS1 IHC/ICC positive cases were confirmed by FISH using the Vysis LSI ALK Dual Color Break Apart Rearrangement Probe (Abbott Molecular, Baar, Switzerland) and the ZytoLight SPEC ROS1 Dual Color Break Apart Probe (Zytovision GmbH, Bremerhaven, Germany). FISH testing for RET rearrangement was performed using the ZytoLight SPEC RET Dual Break Apart Probe (Zytovision GmbH, Bremerhaven, Germany).
Statistical analysis
Descriptive statistics were applied to describe the patient characteristics of the study cohort. The results are presented as frequencies and percentages for categorical variables and as mean ± standard deviation, median and range for continuous variables. Univariate analysis was performed to assess associations between mutation status and clinicopathologic and demographic characteristics, using chi-square test or Fisher exact test for categorical variables and t test or non-parametric Mann-Whitney test for continuous variables. Median overall survival (mOS) was estimated using the Kaplan-Meier method, and survival curves were compared with the log-rank test. Adjusted hazard ratios were calculated using Cox regression models (with a backward stepwise selection method) that included age, gender, clinical stage and treatment as independent variables. P-values < 0.05 were considered statistically significant. All statistical analyses were performed using SPSS Statistics software (version 24.0, IBM, Ehningen, Germany).
Results
Patients
We identified 138 never smokers who fulfilled the inclusion criteria. Patient and tumor characteristics are summarized in Table 1. Most patients were female (81/138, 58.7%), with a median age at diagnosis of 64.5 years (mean, 63.3 ± 13.1 years; range, 27.1–94.2 years), and presented with clinical stage IV (95/138, 68.8%). 15.2% (21/138) of patients had brain metastases at diagnosis, and 12.3% (17/138) of patients developed brain metastases during follow-up. Of the entire study population, 8 (8/138, 5.8%) patients were lost to follow-up, and 130 (130/138, 94.2%) patients, including 116 patients with oncogenic driver mutations, were followed up for a median time of 28.5 months (mo) (mean, 31.6 ± 19.0 mo, range, 2–84 mo). 56 (56/130, 43.1%) patients died during follow-up, while 74 (74/130, 56.9%) patients were alive at last follow-up, including 45 (45/130, 34.6%) patients with stable disease and 29 (29/130, 22.3%) patients with progressive disease. Median OS for the evaluable patients was 51.0 mo (mean, 52.0 ± 3.2 mo). 46 (46/130, 35.4%) patients received one treatment modality, while 84 (84/130, 64.6%) patients were treated with combined therapies. Treatment consisted of surgery in 54 (54/130, 41.5%) patients, chemotherapy in 88 (88/130, 67.7%) patients, radiotherapy in 40 (40/130, 30.8%) patients and molecular targeted therapy in 61 (61/130, 46.9%) patients. Patients with EGFR mutation and ALK rearrangement were treated with targeted therapy in 59.8% (55/92). The majority of patients with stage IV disease for whom information about treatment was available (86/89, 96.6%) received first-line systemic (targeted or chemotherapy) treatment. Most of the chemotherapy-treated patients (including those who underwent chemo-radiotherapy) received platinum-based regimens (94%). Among individuals for whom information about treatment was available, 92.9% (26/28) of patients with stage I-IIIA, 89.7% (35/39) of patients with stage I-IIIB and 87.8% (36/41) of patients with stage I-IIIC underwent surgical resection.
Table 1. Patient and tumor characteristics.
| Clinical and demographic characteristics | |||
| All patients (n = 138) | All patients (n = 138) | ||
| Age (years) | Distribution | ||
| Median (range) | 64.5 (27.1–94.2) | Central | 32 (23.2) |
| Mean | 63.3 ± 13.1 | Peripheral | 83 (60.1) |
| Gender | Central and peripheral | 23 (16.7) | |
| Male | 57 (41.3) | T stage | |
| Female | 81 (58.7) | T1 | 19 (13.8) |
| Ethnicity | T2 | 39 (28.3) | |
| Caucasian | 130 (94.2) | T3 | 25 (18.1) |
| Asian | 4 (2.9) | T4 | 55 (39.9) |
| Other | 4 (2.9) | Lymph node involvement | 105 (76.1) |
| Clinical stage at diagnosis | N stage | ||
| I | 9 (6.5) | N0 | 33 (23.9) |
| II | 11 (8.0) | N1 | 16 (11.6) |
| III | 23 (16.7) | N2 | 39 (28.3) |
| IV | 95 (68.8) | N3 | 50 (36.2) |
| Localization | Extrathoracic metastasis/-es | 65 (47.1) | |
| Right upper lobe | 31 (22.5) | M stage | |
| Right lower lobe | 14 (10.1) | M0 | 43 (31.2) |
| Middle lobe | 8 (5.8) | M1a | 30 (21.7) |
| Left upper lobe | 27 (19.6) | M1b | 18 (13.0) |
| Left lower lobe | 22 (15.9) | M1c | 47 (34.1) |
| Lingula | 2 (1.4) | Brain metastases at diagnosis | 21 (15.2) |
| Involvement of two lobes | 34 (24.6) | Brain metastases at diagnosis and during follow-up |
38 (27.5) |
| Size (mm) | |||
| Mean | 46.3 ± 24.5 | Malignant pleural effusion | 40 (29.0) |
| Molecular characteristics | |||
| All patients (n = 138) | Patients tested† | ||
| EGFR | 81/138 (58.7) | EGFR | 81/138 (58.7) |
| ALK | 17/138 (12.3) | ALK | 17/108 (15.7) |
| MET | 8/138 (5.8) | MET | 8/53 (15.1)‡ |
| KRAS | 6/138 (4.3) | KRAS | 6/114 (5.3) |
| ERBB2 | 6/138 (4.3) | ERBB2 | 6/74 (8.1) |
| PIK3CA | 4/138 (2.9) | PIK3CA | 4/58 (6.9) |
| BRAF | 3/138 (2.2) | BRAF | 3/72 (4.2) |
| ROS1 | 2/138 (1.4) | ROS1 | 2/78 (2.6) |
| RET | 2/138 (1.4) | RET | 2/52 (3.8) |
| Other | 10/138 (7.2) | Other | 10/56 (17.9) |
| Triple negative | 34/138 (24.6) | Triple negative | 34/104 (32.7) |
| Pan-negative | 13/138 (9.4) | Pan-negative | 13/56 (23.2) |
| Treatment | |||
| Patients with information about treatment (n = 130) | |||
| Surgery | 54 (41.5) | ||
| Chemotherapy | 88 (67.7) | ||
| Radiotherapy | 40 (30.8) | ||
| Targeted therapy | 61 (46.9) | ||
| EGFR | 45/76 (59.2) | ||
| ALK | 10/16 (62.5) | ||
| ERBB2 | 2/6 (33.3) | ||
| BRAF | 1/3 (33.3) | ||
| MET | 2/8 (25.0) | ||
| PIK3CA | 1/1 (100) | ||
Data are mean values ± standard deviations for continuous variables and number of patients with percentages in parentheses for categorical variables.
†Percentages in parentheses refer to the number of tested patients.
‡Exclusively patients tested for MET mutations (including MET exon 14 skipping mutations) with NGS.
Mutation analyses
At least one mutation was present in 125 (90.6%) of 138 analyzed tumors. EGFR (exon 19 deletions– 45/81, 55.6%, exon 21 p.L858R point mutation– 26/81, 32.1%) was the most common mutation (81/138, 58.7%, NGS: 33, SS: 48) (Table 2), followed by ALK (17/138, 12.3%, NGS: 3, FISH: 14), TP53 (8/138, 5.8%, NGS: 8), MET (8/138, 5.8%, NGS: 8), KRAS (6/138, 4.3%, NGS: 3, SS: 3), ERBB2 (6/138, 4.3%, NGS: 3, SS: 3), PIK3CA (4/138, 2.9%, NGS: 4), BRAF (3/138, 2.2%, NGS: 2, SS: 1), ROS1 (2/138, 1.4%, NGS: 2), RET (2/138, 1.4%, NGS: 1, FISH: 1), CTNNB1 (1/138, 0.7%, NGS: 1), PARP1 (1/138, 0.7%, NGS: 1), TET1 (1/138, 0.7%, NGS: 1) and PIK3CG (1/138, 0.7%, NGS: 1) (Table 1). Doublet EGFR mutations were present in 9 (9/138, 6.5%) tumors, including 4 tumors with p.L858R and non-p.L858R missense mutations and 5 tumors with two non-p.L858R missense mutations (S1 Table). Of 21 tumors with multiple mutations, 17 (17/21, 81.0%) had EGFR as a co-mutation (S1 Table). Mutations were not detected in 13 (9.4%) of 138 tumors (pan-negative), and 34 (24.6%) of 138 tumors were triple-negative (EGFR negative/ALK negative/KRAS negative) (Table 1). KRAS mutations were most frequently located in exon 2/codon 12 (5/6, 83.3%), and p.G12V (3/6, 50.0%) was the most common subtype (Table 2). ERBB2 mutations were exclusively exon 20 insertions/duplications, and the most frequent ERBB2 mutation was p.A771_M774dup (2/6, 33.3%) (Table 2). Of 21 patients with brain metastases at diagnosis, 15 (15/21, 71.4%) had EGFR mutation, 2 (2/21, 9.5%) were pan-negative, 2 (2/21, 9.5%) had ALK translocation, 1 (1/21, 4.8%) harbored PIK3CA mutation and 1 (1/21, 4.8%) harbored MET exon 14 skipping mutation.
Table 2. Oncogenic driver mutations in never smokers.
| EGFR mutation (n = 81) | ||||
| cDNA change | Amino acid change | Frequency | Percentage | |
| Exon 21 | c.2573T>G | p.L858R | 26 | 32.1 |
| Exon 21 | c.2497T>G | p.L833V | 1 | 1.2 |
| Exon 21 | c.2560A>G | p.T854A | 1 | 1.2 |
| Exon 21 | c.2579A>T | p.K860I | 1 | 1.2 |
| Exon 20 | c.2303_2311dup | p.S768_D770dup | 2 | 2.5 |
| Exon 20 | c.2303G>T | p.S768I | 2 | 2.5 |
| Exon 20 | c.2320G>A | p.V774M | 1 | 1.2 |
| Exon 20 | c.2389T>A | p.C797S | 1 | 1.2 |
| Exon 20 | c.2320_2321insCACATG | p.H773_V774insAH | 1 | 1.2 |
| Exon 20 | c.2310_2311insGGGC | p.D770_N771insG | 1 | 1.2 |
| Exon 20 | c.2317_2322 | p.H773_V774dup | 1 | 1.2 |
| Exon 19 | c.2235_2249del/c.2236_2250del† | p.E746_A750del | 26 | 32.1 |
| Exon 19 | c.2240_2257del | p.L747_P753delinsS | 5 | 6.2 |
| Exon 19 | c.2237_2255delinsT | p.E746_S752delinsV | 3 | 3.7 |
| Exon 19 | c.2254_2277del | p.S752_I759del | 2 | 2.5 |
| Exon 19 | c.2240_2254del | p.L747_T751del | 2 | 2.5 |
| Exon 19 | c.2239_2256del | p.L747_S752del | 2 | 2.5 |
| Exon 19 | c.2239_2252delinsCA | p.L747_T751delinsQ | 1 | 1.2 |
| Exon 19 | c.2239_2248delinsC | p.L747_A750delinsP | 1 | 1.2 |
| Exon 19 | c.2239_2258delinsCA | p.L747_P753delinsQ | 1 | 1.2 |
| Exon 19 | c.2239_2251delinsC | p.L747_T751delinsP | 1 | 1.2 |
| Exon 19 | c.2251_2277del | p.T751_I759delinsS | 1 | 1.2 |
| Exon 19 | c.2281G>T | p.D761Y | 1 | 1.2 |
| Exon 18 | c.2156G>C | p.G719A | 2 | 2.5 |
| Exon 18 | c.2126A>C | p.E709A | 1 | 1.2 |
| Exon 18 | c.2126A>G | p.E709G | 1 | 1.2 |
| Exon 18 | c.2155G>T | p.G719C | 1 | 1.2 |
| Exon 18 | c.2142G>C | p.K714N | 1 | 1.2 |
| KRAS mutations (n = 6) | ||||
| cDNA change | Amino acid change | Frequency | Percentage | |
| Codon 12/Exon 2 | c.35G>T | p.G12V | 3 | 50.0 |
| Codon 12/Exon 2 | c.34G>T | p.G12C | 1 | 16.7 |
| Codon 12/Exon 2 | c.35G>A | p.G12D | 1 | 16.7 |
| Codon 61/Exon 3 | c.182A>T | p.Q61L | 1 | 16.7 |
| BRAF mutations (n = 3) | ||||
| cDNA change | Amino acid change | Frequency | Percentage | |
| Exon 15 | c.1799T>A | p.V600E | 2 | 66.7 |
| Exon 15 | c.1799T>A | p.V600G | 1 | 33.3 |
| ERBB2 mutations (n = 6) | ||||
| cDNA change | Amino acid change | Frequency | Percentage | |
| Exon 20 | c.2324_2325ins | p.E770_A771ins | 1 | 16.7 |
| Exon 20 | c.2313_2324dup | p.A771_M774dup | 2 | 33.3 |
| Exon 20 | c.2326_2327insTGT | p.G776delinsVC | 1 | 16.7 |
| Exon 20 | c.2310_2311ins12 | p.E770_A771insAYVM | 1 | 16.7 |
| Exon 20 | c.2331_2339dup | p.G778_P780dup | 1 | 16.7 |
| PIK3CA (n = 4) | ||||
| cDNA change | Amino acid change | Frequency | Percentage | |
| Exon 10 | c.1633G>A | p.E545K | 2 | 50.0 |
| Exon 10 | c.1624G>A | p.E542K | 1 | 25.0 |
| Exon 21 | c.3140A>G | p.H1047R | 1 | 25.0 |
†c.2235_2249del: n = 18; c.2236_2250del: n = 8.
Correlation with clinicopathologic characteristics
No statistically significant differences were found between male and female never smokers with respect to clinical stage, TNM stage, presence of brain metastasis at diagnosis and during follow-up, tumor location, mean tumor size, mean patient age at diagnosis and the frequency of oncogenic driver mutations (S2 Table). Comparative analyses of EGFR mutated tumors with EGFR wildtype, pan-negative and ALK positive tumors showed that EGFR mutated tumors more frequently had extrathoracic metastases at diagnosis compared to EGFR wildtype tumors (55.6% vs. 35.1%, p = 0.018) and were more commonly located in the right upper lobe compared to EGFR wildtype, pan-negative and ALK positive tumors (28.4% vs. 14.0%, p = 0.047; 28.4% vs. 0.0%, p = 0.033 and 28.4% vs. 0.0%, p = 0.010) (Table 3). There was a significant difference in mean age between patients with EGFR mutated tumors and those without identifiable mutations (61.9 ± 13.4 vs. 71.9 ± 9.2 years, p = 0.011) (Table 3) as well as between patients with mutations in their tumors and the pan-negative cohort (62.4 ± 13.2 vs. 71.9 ± 9.2 years, p = 0.012) (S3 Table). ALK rearranged lung adenocarcinomas were less frequently located in the right upper lobe (0.0% vs. 25.6%, p = 0.013) compared to ALK wildtype tumors and more commonly showed ipsilateral mediastinal or subcarinal lymph node metastasis (N2) compared to EGFR mutated and ALK wildtype tumors (52.9% vs. 22.2%, p = 0.016 and 52.9% vs. 24.8%, p = 0.022) (Tables 3 and 4) (Fig 1). Comparative analysis of patients with tumors harboring exon 21 p.L858R point mutation and patients with tumors carrying exon 19 deletions showed significant differences in mean age, tumor location (central vs. peripheral) and the frequency of ipsilateral mediastinal or subcarinal lymph node metastasis (N2) and malignant pleural effusion (S4 Table). Patients with brain metastases at diagnosis more frequently had T4 tumors and contralateral mediastinal, hilar or supraclavicular lymph node metastasis (N3) compared to patients without brain metastases at diagnosis (61.9% vs. 35.9%, p = 0.025 and 61.9% vs. 31.6%, p = 0.008) (S5 Table). Likewise, patients with brain metastases at diagnosis and during follow-up more commonly showed lymph node metastases compared to patients without brain metastases (92.1% vs. 70.0%, p = 0.007) (S6 Table). Lung adenocarcinoma diagnosed in patients < 45 years was more commonly associated with lymph node metastasis (100.0% vs. 73.4%, p = 0.022), notably contralateral mediastinal, hilar or supraclavicular lymph node metastasis (N3) (64.3% vs. 33.1%, p = 0.021), and extrathoracic metastases (78.6% vs. 43.5%, p = 0.013) compared to tumors diagnosed in patients > 45 years (S7 Table).
Table 3. Comparison of EGFR mutated tumors with EGFR wildtype, pan-negative and ALK positive tumors.
| Variable |
EGFR mt (n = 81) |
EGFR wt (n = 57) |
p | Pan-negative (n = 13) |
p |
ALK positive (n = 17) |
p |
|---|---|---|---|---|---|---|---|
| Age (years) | 61.9 ± 13.4 | 65.3 ± 12.6 | 0.133 | 71.9 ± 9.2 | 0.011 | 64.2 ± 14.0 | 0.515 |
| Gender | 0.225 | 0.250 | 0.097 | ||||
| Male | 30 (37.0) | 27 (47.4) | 7 (53.8) | 10 (58.8) | |||
| Female | 51 (63.0) | 30 (52.6) | 6 (46.2) | 7 (41.2) | |||
| Clinical stage | |||||||
| I | 6 (7.4) | 3 (5.3) | 0.736 | 1 (7.7) | 0.971 | 0 (0.0) | 0.586 |
| II | 5 (6.2) | 6 (10.5) | 0.361 | 1 (7.7) | 0.839 | 2 (11.8) | 0.601 |
| III | 10 (12.3) | 13 (22.8) | 0.104 | 3 (23.1) | 0.381 | 5 (29.4) | 0.130 |
| IV | 60 (74.1) | 35 (61.4) | 0.114 | 8 (61.5) | 0.339 | 10 (58.8) | 0.242 |
| T stage | |||||||
| T1 | 11 (13.6) | 8 (14.0) | 0.939 | 0 (0.0) | 0.352 | 3 (17.6) | 0.705 |
| T2 | 25 (30.9) | 14 (24.6) | 0.418 | 4 (30.8) | 0.995 | 4 (23.5) | 0.547 |
| T3 | 18 (22.2) | 7 (12.3) | 0.135 | 1 (7.7) | 0.455 | 2 (11.8) | 0.670 |
| T4 | 27 (33.3) | 28 (49.1) | 0.062 | 8 (61.5) | 0.066 | 2 (11.8) | 0.537 |
| LN meta | 59 (72.8) | 46 (80.7) | 0.286 | 10 (76.9) | 0.754 | 15 (88.2) | 0.228 |
| N stage | |||||||
| N0 | 22 (27.2) | 11 (19.3) | 0.286 | 3 (23.1) | 0.754 | 2 (11.8) | 0.228 |
| N1 | 10 (12.3) | 6 (10.5) | 0.742 | 2 (15.4) | 0.670 | 1 (5.9) | 0.683 |
| N2 | 18 (22.2) | 21 (36.8) | 0.060 | 3 (23.1) | 0.945 | 9 (52.9) | 0.016 |
| N3 | 31 (38.3) | 19 (33.3) | 0.552 | 5 (38.5) | 0.990 | 5 (29.4) | 0.491 |
| Extrathoracic meta | 45 (55.6) | 20 (35.1) | 0.018 | 5 (38.5) | 0.252 | 5 (29.4) | 0.050 |
| M stage | |||||||
| M0 | 21 (25.9) | 22 (38.6) | 0.114 | 5 (38.5) | 0.339 | 7 (41.2) | 0.242 |
| M1a | 15 (18.5) | 15 (26.3) | 0.274 | 5 (38.5) | 0.709 | 5 (29.4) | 0.330 |
| M1b | 12 (14.8) | 6 (10.5) | 0.461 | 1 (7.7) | 0.686 | 0 (0.0) | 0.119 |
| M1c | 33 (40.7) | 14 (24.6) | 0.048 | 4 (30.8) | 0.495 | 5 (29.4) | 0.383 |
| Localization | |||||||
| Right upper lobe | 23 (28.4) | 8 (14.0) | 0.047 | 0 (0.0) | 0.033 | 0 (0.0) | 0.010 |
| Right lower lobe | 6 (7.4) | 8 (14.0) | 0.204 | 3 (23.1) | 0.107 | 2 (11.8) | 0.624 |
| Middle lobe | 5 (6.2) | 3 (5.3) | 0.821 | 1 (7.7) | 0.839 | 2 (11.8) | 0.601 |
| Left upper lobe | 15 (18.5) | 12 (21.1) | 0.712 | 3 (23.1) | 0.709 | 4 (23.5) | 0.736 |
| Left lower lobe | 12 (14.8) | 10 (17.5) | 0.666 | 2 (15.4) | 0.957 | 3 (17.6) | 0.721 |
| Lingula | 0 (0.0) | 2 (3.5) | 0.169 | 0 (0.0) | - | 1 (5.9) | 0.173 |
| Mixed | 20 (24.7) | 14 (24.6) | 0.986 | 4 (30.8) | 0.733 | 5 (29.4) | 0.761 |
| Distribution | |||||||
| Central | 18 (22,2) | 14 (24.6) | 0.749 | 3 (23.1) | 0.945 | 6 (35.3) | 0.351 |
| Peripheral | 50 (61.7) | 33 (57.9) | 0.651 | 6 (46.2) | 0.288 | 7 (41.2) | 0.118 |
| Central and peripheral | 13 (16.0) | 10 (17.5) | 0.817 | 4 (30.8) | 0.243 | 4 (23.5) | 0.487 |
| Malignant PE | 28 (34.6 | 12 (21.1) | 0.085 | 4 (30.8) | 0.787 | 4 (23.5) | 0.378 |
| Size (mm) | 45.7 ± 21.9 | 47.1 ± 28.0 | 0.755 | 55.0 ± 28.9 | 0.180 | 50.4 ± 36.9 | 0.486 |
| Brain meta at diagnosis | 15 (18.5) | 6 (10.5) | 0.198 | 2 (15.4) | 0.782 | 2 (11.8) | 0.729 |
| Brain meta at diagnosis and during follow-up |
23 (28.4) | 15 (26.3) | 0.788 | 4 (30.8) | 0.861 | 6 (35.3) | 0.571 |
Data are mean values ± standard deviations for continuous variables and number of patients with percentages in parentheses for categorical variables.
Bold numbers indicate significant p-values (< 0.05).
Table 4. Comparison of ALK positive tumors with ALK negative and pan-negative tumors.
| Variable |
ALK positive (n = 17) |
ALK negative† (n = 121) |
p |
ALK negative‡ (n = 91) |
p | Pan-negative (n = 13) |
p |
|---|---|---|---|---|---|---|---|
| Age (years) | 64.2 ± 14.0 | 63.1 ± 13.1 | 0.753 | 63.9 ± 12.5 | 0.929 | 71.9 ± 9.2 | 0.098 |
| Gender | 0.117 | 0.166 | 7 (53.8) | 0.785 | |||
| Male | 10 (58.8) | 47 (38.8) | 37 (40.7) | 6 (46.2) | |||
| Female | 7 (41.2) | 74 (61.2) | 54 (59.3) | ||||
| Clinical stage | |||||||
| I | 0 (0.0) | 9 (7.4) | 0.601 | 7 (7.7) | 0.594 | 1 (7.7) | 0.433 |
| II | 2 (11.8) | 9 (7.4) | 0.626 | 7 (7.7) | 0.631 | 1 (7.7) | 0.709 |
| III | 5 (29.4) | 18 (14.9) | 0.162 | 16 (17.6) | 0.316 | 3 (23.1) | 0.696 |
| IV | 10 (58.8) | 85 (70.2) | 0.341 | 61 (67.0) | 0.513 | 8 (61.5) | 0.880 |
| T stage | |||||||
| T1 | 3 (17.6) | 16 (13.2) | 0.705 | 13 (14.3) | 0.715 | 0 (0.0) | 0.238 |
| T2 | 4 (23.5) | 35 (28.9) | 0.779 | 23 (25.3) | 0.878 | 4 (30.8) | 0.698 |
| T3 | 2 (11.8) | 22 (18.2) | 0.957 | 15 (16.5) | 0.907 | 1 (7.7) | 0.613 |
| T4 | 2 (11.8) | 48 (39.7) | 0.905 | 40 (44.0) | 0.832 | 8 (61.5) | 0.269 |
| LN meta | 15 (88.2) | 90 (74.4) | 0.361 | 65 (71.4) | 0.228 | 10 (76.9) | 0.628 |
| N stage | |||||||
| N0 | 2 (11.8) | 31 (25.6) | 0.361 | 26 (28.6) | 0.228 | 3 (23.1) | 0.628 |
| N1 | 1 (5.9) | 15 (12.4) | 0.693 | 8 (8.8) | 0.678 | 2 (15.4) | 0.565 |
| N2 | 9 (52.9) | 30 (24.8) | 0.022 | 25 (27.5) | 0.038 | 3 (23.1) | 0.098 |
| N3 | 5 (29.4) | 45 (37.2) | 0.532 | 32 (35.2) | 0.646 | 5 (38.5) | 0.705 |
| Extrathoracic meta | 5 (29.4) | 60 (49.6) | 0.119 | 39 (42.9) | 0.300 | 5 (38.5) | 0.705 |
| M stage | |||||||
| M0 | 7 (41.2) | 36 (29.8) | 0.341 | 30 (33.0) | 0.513 | 5 (38.5) | 0.880 |
| M1a | 5 (29.4) | 25 (20.7) | 0.529 | 22 (24.2) | 0.761 | 3 (23.1) | 0.696 |
| M1b | 0 (0.0) | 18 (14.9) | 0.127 | 14 (15.4) | 0.120 | 1 (7.7) | 0.433 |
| M1c | 5 (29.4) | 42 (34.7) | 0.666 | 25 (27.5) | 0.870 | 4 (30.8) | 0.936 |
| Localization | |||||||
| Right upper lobe | 0 (0.0) | 31 (25.6) | 0.013 | 24 (26.4) | 0.012 | 0 (0.0) | - |
| Right lower lobe | 2 (11.8) | 12 (9.9) | 0.683 | 11 (12.1) | 0.970 | 3 (23.1) | 0.628 |
| Middle lobe | 2 (11.8) | 6 (5.0) | 0.256 | 3 (3.3) | 0.175 | 1 (7.7) | 0.709 |
| Left upper lobe | 4 (23.5) | 23 (19.0) | 0.744 | 17 (18.7) | 0.739 | 3 (23.1) | 0.977 |
| Left lower lobe | 3 (17.6) | 19 (15.7) | 0.735 | 10 (11.0) | 0.427 | 2 (15.4) | 0.869 |
| Lingula | 1 (5.9) | 1 (0.8) | 0.232 | 1 (1.1) | 0.291 | 0 (0.0) | 0.281 |
| Mixed | 5 (29.4) | 29 (24.0) | 0.764 | 25 (27.5) | 0.870 | 4 (30.8) | 0.936 |
| Distribution | |||||||
| Central | 6 (35.3) | 26 (21.5) | 0.225 | 19 (20.9) | 0.217 | 3 (23.1) | 0.691 |
| Peripheral | 7 (41.2) | 76 (62.8) | 0.088 | 59 (64.8) | 0.066 | 6 (46.2) | 0.785 |
| Central and peripheral | 4 (23.5) | 19 (15.7) | 0.485 | 13 (14.3) | 0.466 | 4 (30.8) | 0.698 |
| Malignant PE | 4 (23.5) | 36 (29.8) | 0.778 | 21 (23.1) | 0.968 | 4 (30.8) | 0.698 |
| Size (mm) | 50.4 ± 36.9 | 45.7 ± 22.4 | 0.461 | 46.4 ± 22.3 | 0.543 | 55.0 ± 28.9 | 0.714 |
| Brain meta at diagnosis | 2 (11.8) | 19 (15.7) | 0.663 | 11 (12.1) | 0.970 | 2 (15.4) | 0.773 |
| Brain meta at diagnosis and during follow-up |
6 (35.3) | 32 (26.4) | 0.562 | 21 (23.1) | 0.360 | 4 (30.8) | 0.794 |
Data are mean values ± standard deviations for continuous variables and number of patients with percentages in parentheses for categorical variables.
†Patients with EGFR or KRAS mutation and unknown ALK mutation status included.
‡Analysis restricted to patients with known ALK mutation status. Bold numbers indicate significant p-values (< 0.05).
Fig 1. ALK rearranged vs. ALK wildtype lung adenocarcinoma in never smokers.
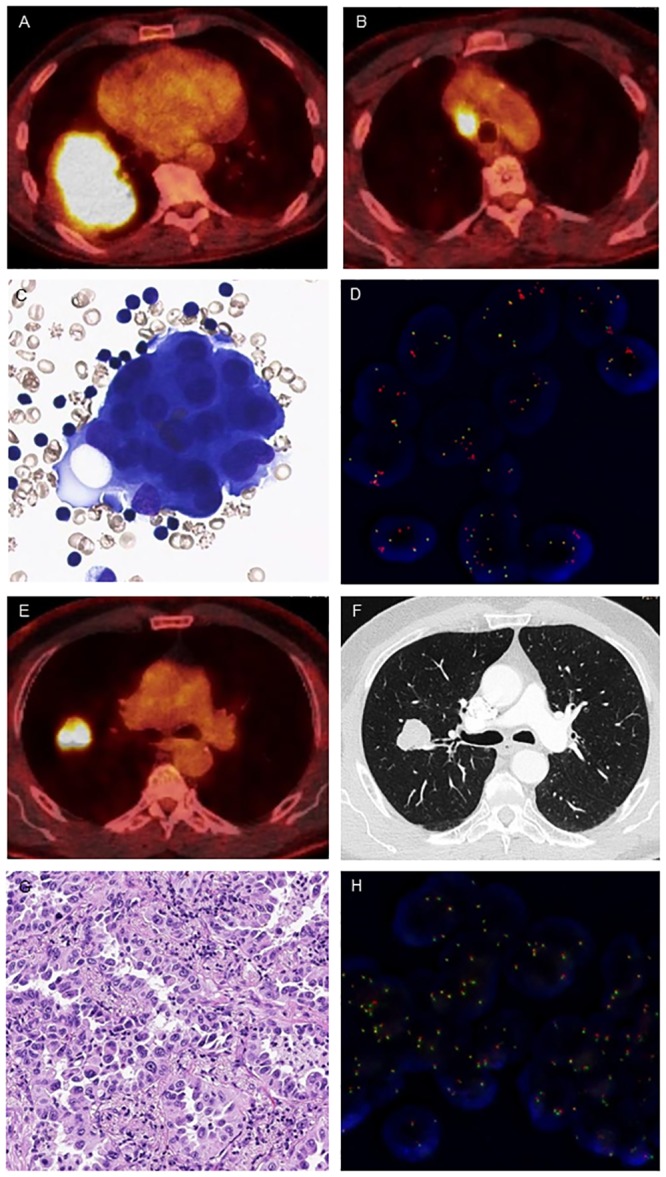
(A,B) PET-CT scans showing a large tumor mass in the right lower lobe together with ipsilateral mediastinal lymph node metastasis. (C) Adenocarcinoma morphology on cytological sample (May-Grünwald stain, x 400). (D) Positive ALK fluorescence in situ hybridization (FISH) with split signals and mainly isolated red signals. (E) PET-CT scan and (F) axial CT image showing a tumor nodule in the right upper lobe without mediastinal lymph node metastases. (G) Adenocarcinoma histology on resection specimen (hematoxylin & eosin stain, x 200). (H) ALK FISH negativity with fusion signals.
Correlation with outcome
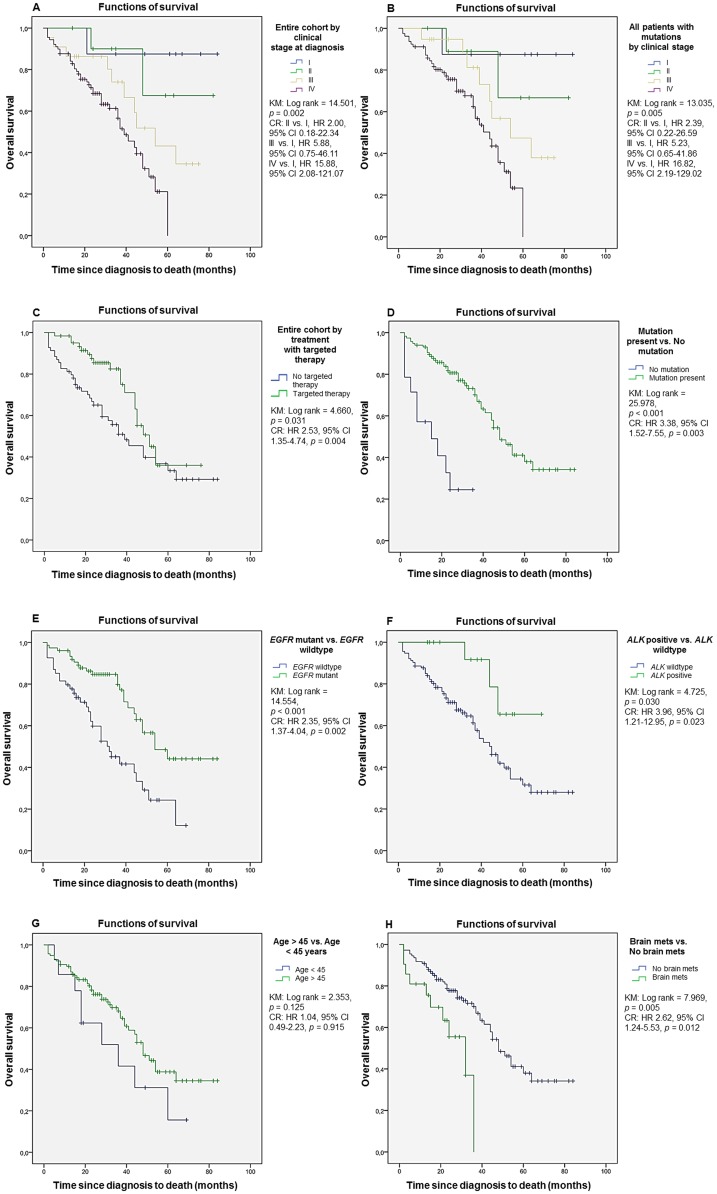
Early clinical stage (in the entire study cohort–p = 0.002, Fig 2a; in all patients with mutations in their tumors–p = 0.005, Fig 2b; in all patients with EGFR mutant and ALK rearranged tumors–p = 0.005) and treatment with targeted therapy (in the entire study cohort–HR 2.53, 95% CI 1.35–4.74, p = 0.004, Fig 2c; in all patients with mutations in their tumors–HR 2.61, 95% CI 1.39–4.90, p = 0.003; and in the cohort with EGFR mutation or ALK rearrangement–HR 2.22, 95% CI 0.99–4.99, p = 0.043) were independently associated with longer mOS. Patients with mutations in their tumors had significantly longer mOS compared to patients without mutations (mOS 52.2 vs. 16.9 mo; HR 3.38; 95% CI 1.52–7.55; p = 0.003, Fig 2d), and patients with EGFR mutated tumors had significantly better outcome compared to patients with EGFR wildtype and pan-negative tumors (mOS 57.8 vs. 35.0 mo, HR 2.35, 95% CI 1.37–4.04, p = 0.002, Fig 2e, and mOS 57.8 vs. 16.9 mo, HR 7.80, 95% CI 3.28–18.55, p < 0.001). Among the covariates included in the multivariable model (age, gender, clinical stage, treatment) only treatment with targeted therapy and clinical stage were found significant. In the subset of patients with EGFR mutant lung adenocarcinoma, no significant survival difference was found between patients with tumors carrying exon 19 deletions and patients harboring exon 21 p.L858R point mutation (mOS 47.0 vs. 58.7 mo, HR 1.02, 95% CI 0.34–3.08, p = 0.977). In contrast, patients with ALK rearranged tumors had significantly longer OS compared to the ALK wildtype and pan-negative cohorts (mOS 59.9 vs. 46.5 mo, HR 3.96, 95% CI 1.21–12.95, p = 0.023, Fig 2f and mOS 59.9 vs.16.9 mo, HR 34.78, 95% CI 3.48–34.65, p = 0.003). Age, gender, clinical stage and treatment with targeted therapy proved to be significant covariates in the multivariable regression model, and statistical significance for all variables was preserved when the analysis was restricted to the subset of patients for whom ALK mutation status was known. Patients > 45 years had insignificantly longer OS compared to patients < 45 years (mOS 50.7 vs. 36.2 mo, HR 1.04, 95% CI 0.49–2.23, p = 0.915, Fig 2g), while patients with brain metastases at diagnosis had significantly shorter OS compared to patients without brain metastases at diagnosis (mOS 24.6 vs. 51.5 mo, HR 2.62, 95% CI 1.24–5.53, p = 0.012, Fig 2h).
Fig 2. Overall survival in never smokers with lung adenocarcinoma.
(A) Entire cohort by clinical stage at diagnosis. (B) All patients with mutations by clinical stage at diagnosis. (C) Entire cohort by treatment with targeted therapy. (D) All patients by presence of oncogenic driver mutations. (E) All patients by EGFR mutation status. (F) All patients by ALK rearrangement status. (G) Entire cohort by age at diagnosis. (H) All patients by presence of brain metastases at diagnosis. KM, Kaplan Meier; CR, Cox regression.
Discussion
The study presents the molecular, demographic and clinicopathologic features of never smoker patients with lung adenocarcinoma, diagnosed at a single institution in Switzerland. We comprehensively analyzed associations between mutation status and clinicopathologic characteristics and outcome.
To the best of our knowledge, only two previous studies have thoroughly investigated the genomic characteristics of LCINS in Western populations [1,6]. In the first study, a large multicenter series from France, comprising 384 never smokers with NSCLC (adenocarcinoma histology– 85%), Couraud et al. [6] reported ALK rearrangement and EGFR, KRAS, BRAF, ERBB2 and PIK3CA mutation in 13%, 43%, 7%, 5%, 4% and 2% of tested patients, respectively, using different genotyping methods in overall 75 participating centers. The authors did not comprehensively analyze associations between mutation status and clinicopathologic and demographic patient characteristics, instead the study focused on epidemiologic data including information on exposure to occupational carcinogens and passive smoker exposure [6]. In the second study, a large retrospective single center series from Canada, comprising 712 never smoking lung cancer patients (adenocarcinoma histology– 87%), 515 of whom had tumor tissue available for molecular analysis, Korpanty et al. [1] reported EGFR, KRAS, TP53, ERBB2, BRAF and PIK3CA mutation and ALK rearrangement in 52.2%, 2.3%, 1.4%, 1,0%, 0.4%, 0.4% and 7.6% of patients, respectively, using MassARRAY technology (Sequenom, San Diego, CA) or MiSeq (Illumina, San Diego, CA) NGS personal genomics platforms as testing methods. In accordance with the above-mentioned studies [1,6] and the results of investigations from Asian populations [7,18,19] EGFR mutation was the most commonly encountered mutation type in our study population (58.7%), and the majority (55.6%) of EGFR mutations were deletions in exon 19. It is well known that the reported EGFR mutation rates in patients with lung adenocarcinoma vary widely among different populations worldwide (ranging from 10–20% in European and North American cohorts [20–23] to more than 50% in Asian populations [24,25]), and that EGFR mutation status is significantly associated with female gender and never smoking status [20–25]. When we confined the analysis to female never smokers in our cohort, we achieved a high EGFR mutation rate of 63.0%, which is consistent with previous reports showing EGFR mutation rates reaching up to 60% when focusing on female never smoker patients [24–26]. ALK translocations, detected in 3–7% of non-selected NSCLC cohorts [27–29], are reported to occur more commonly in non-smokers, lung adenocarcinomas and non-Asian vs. Asian populations [30]. The frequency of ALK rearrangements in our study (12.3%) was similar to that reported for never smoker subgroups in previous investigations (range, 4.5%-16.4% [7,18,28,29,31]). In a recent study, we analyzed mutation frequencies and associations between mutational status and clinicopathologic patient characteristics in a non-selected representative cohort of Swiss patients with newly diagnosed lung adenocarcinoma the majority of whom (354/469, 75.5%) were ever smokers (current smokers or ex-smokers) [17]. Compared with the current study, we found a significantly higher percentage of KRAS mutation (159/469, 33.9%), which was the most common mutation in this non-selected patient cohort, and lower frequencies of EGFR mutation (90/469, 19.2%) and ALK rearrangement (28/469, 6.0%). Similar to the results of the current study, EGFR mutated tumors more frequently had (multiple) extrathoracic metastases at diagnosis and tended to occur more frequently in the right upper lobe compared to EGFR wildtype and ALK rearranged tumors, while ALK positive lung adenocarcinomas were less frequently located in the right upper lobe compared to ALK wildtype tumors. In both studies, we found that ALK-rearranged tumors were more commonly associated with ipsilateral mediastinal or subcarinal lymph node metastasis (N2) compared to ALK wildtype and EGFR mutated lung adenocarcinoma. In a non-selected stage I-III NSCLC population, Paik et al. [32] reported lower tumor stage (pT1) and significantly higher frequency of lymph node metastases in ALK FISH-positive NSCLC cases compared to ALK FISH-negative NSCLC cases. The authors suggested that ALK-rearranged lung cancer might have unique biological features with a tendency to early lymph node metastasis despite small primary tumor size, which could explain higher incidences of ALK rearrangement in advanced NSCLC when compared with surgically resectable lung cancer [32].
Recent studies have suggested that never smoker patients with tumors harboring mutations may have significantly longer mOS compared to patients without identifiable mutations [1]. Besides, never smokers with ALK rearranged NSCLC are reported to have significantly better outcome compared to the ALK wildtype and pan-negative cohorts [1,7]. In accordance with these reports, patients with oncogenic driver mutations in our study had significantly longer mOS compared to patients without identifiable mutations, and patients with ALK translocation had significantly better outcome compared to patients without ALK rearrangement and the pan-negative cohort. Similarly, the presence of EGFR mutation in our study was significantly associated with longer OS in univariable and multivariable analysis, while, consistent with the results reported by Korpanty et al. [1] and in contrast to previous reports [33,34], no significant survival difference was found between patients with tumors harboring EGFR exon 19 deletions and patients with tumors harboring EGFR exon 21 mutations.
The initial onset of brain metastases is generally considered an unfavorable prognostic factor that increases the risk of death [35]. In a recent study, Korpanty et al. [1] found no significant survival difference in patients with and those without brain metastases at diagnosis. The authors suggested that the lack of survival difference may be related to the high proportion (~80%) of tumors harboring EGFR mutation or ALK translocation among patients with brain metastases in their study [1], as these mutations are known to be associated with favorable response of CNS disease to targeted tyrosine kinase inhibitors and higher response rates to whole brain radiation therapy compared to wildtype lung cancer [36–40]. In our study, patients with brain metastases at diagnosis had significantly shorter mOS compared to patients without brain metastases at diagnosis despite the fact that 81.0% of patients with brain metastases at presentation had tumors with two most common targetable mutations (71.4%–EGFR, 9.5%–ALK).
NSCLC diagnosed in young patients is a rare entity, with incidences ranging between 1.3 and 5.3% among patients ≤ 45 years at diagnosis [4,41–43]. The genomics and clinical characteristics of this disease are poorly understood, and studying the relationship between age and genotype in young NSCLC patients is challenging due to multiple confounding factors such as gender, race and smoking status [44]. Previous reports have indicated that younger age may be associated with an increased likelihood of harboring oncogenic driver mutations in patients with NSCLC [44,45]. Besides, recent data suggest that ALK rearrangements occur more frequently in younger NSCLC patients compared to older patients with lung cancer [44–47], whereas KRAS mutations appear to be less frequent in the younger patient cohort [44,45]. Regarding EGFR mutation frequency, a study by Sacher et al. [44] comprising 2237 NSCLC patients found an increased likelihood of EGFR mutations in patients diagnosed with NSCLC at a younger age, while Tanaka et al. [45] reported a significantly lower frequency of EGFR mutations in 81 lung adenocarcinoma patients ≤ 40 years compared with 1665 lung adenocarcinoma patients > 40 years at initial diagnosis. In our study, focusing on never smoker patients with adenocarcinoma histology, we found no significant differences in the frequency of EGFR mutation, KRAS mutation or ALK translocation between patients < 45 years and patients ≥ 45 years. 92.9% of patients < 45 years in our study had potentially targetable genetic alterations in EGFR, ALK, ROS1 and MET, 78.6% had EGFR mutation or ALK rearrangement (71.4%–EGFR, 7.1%–ALK), and 14.3% had tumors harboring TP53 mutation. Consistent with previous results [44,45], patients < 45 years had shorter mOS compared to patients ≥ 45 years, although the difference in survival was not statistically significant in our cohort. It has been suggested that the worse prognosis in young NSCLC patients could be partly related to the significantly higher prevalence of TP53 mutations in young patients with lung adenocarcinoma [45,48]. However, further studies are needed to investigate the underlying mechanisms that explain the more aggressive biology of lung cancer in younger patients.
There are several limitations to this study. First, this is a retrospective analysis of highly selected patients diagnosed and treated from 2011–2018 at a single tertiary referral academic institution. During that time, different sequencing methods were used, and we are aware of the heterogeneity of molecular testing methods and the fact that molecular pathology data were not completely comprehensive for all patients, which might have influenced the results of the current study. In addition, due to rapidly changing treatment guidelines different treatment regimens and sequence of these therapies were applied during the study period, including the incorporation of targeted therapy in routine practice for selected patients, and we acknowledge that our analyses are limited by the heterogeneity of treatment modalities and different combinations of therapies. Second, we did not attempt to identify comorbidities or specific exposures that may contribute to lung cancer risk including exposure to other carcinogens such as asbestos, radon, radiation therapy, and various other exposures in environmental, medical and/or occupational settings. Given the retrospective nature of the study, our analyses are further limited by uncertainties about errors and incompleteness of information about smoking exposure. Last, we did not analyze associations between programmed death-ligand 1 (PD-L1) expression and molecular features or outcome, although recent studies have shown that the predictive value of PD-L1 in NSCLC patients may be influenced by oncogenic driver mutation status [49]. Future prospective studies are needed to comprehensively investigate PD-L1 expression and the efficacy of PD-1/PD-L1 inhibitor therapy in different subsets of never smoker patients with oncogenic driver mutations.
Conclusion
This is the first comprehensive analysis of molecular, clinicopathologic and survival data of never smokers with lung adenocarcinoma diagnosed at a tertiary referral academic hospital in Switzerland. There was a high incidence of oncogenic driver mutations in our study population, and EGFR mutation and ALK rearrangement were the most common genetic alterations. EGFR mutated tumors were more commonly associated with extrathoracic metastases at diagnosis compared to EGFR wildtype tumors, while ALK rearranged tumors more commonly had ipsilateral mediastinal or subcarinal lymph node metastasis compared to EGFR mutated and ALK wildtype tumors. Patients < 45 years more commonly showed lymph node metastasis and extrathoracic metastases compared to patients > 45 years. Consistent with previous reports never smokers with oncogenic driver mutations had significantly longer OS compared to patients without identifiable mutations. Likewise, there were significant differences in survival between patients with EGFR mutated vs. EGFR wildtype tumors, EGFR mutated vs. pan-negative tumors, ALK rearranged vs. ALK wildtype tumors and ALK rearranged vs. pan-negative tumors (Table 5).
Table 5. Most relevant study results.
| Most common mutations: |
| EGFR (81/138,58.7%), ALK (17/138, 12.3%), TP 53 (8/138, 5.8%), MET (8/138, 5.8%), KRAS (6/138, 4.3%), ERBB2 (6/138, 4.3%), PIK3CA (4/138, 2.9%), BRAF (3/138, 2.2%), ROS1 (2/138, 1.4%), RET (2/138, 1.4%) |
| Correlation with clinicopathologic characteristics: |
| EGFR mutated vs. EGFR wildtype tumors |
| - extrathoracic metastases at diagnosis (55.6% vs. 35.1%, p = 0.018) |
| ALK rearranged vs. ALK wildtype tumors |
| - ipsilateral mediastinal or subcarinal lymph node metastasis (52.9% vs. 24.8%, p = 0.022) |
| ALK rearranged vs. EGFR mutated tumors |
| - ipsilateral mediastinal or subcarinal lymph node metastasis (52.9% vs. 22.2%, p = 0.016) |
| Patients < 45 years vs. patients > 45 years |
| - lymph node metastasis (100.0% vs. 73.4%, p = 0.022) |
| - extrathoracic metastases (78.6% vs. 43.5%, p = 0.013) |
| Correlation with outcome: |
| Patients with mutations vs. patients without mutations |
| - mOS 52.2 vs. 16.9 mo, p = 0.003 |
| Patients with EGFR mutated vs. EGFR wildtype tumors |
| - mOS 57.8 vs. 35.0 mo, p = 0.002 |
| Patients with EGFR mutated vs. pan-negative tumors |
| - mOS 57.8 vs. 16.9 mo, p < 0.001 |
| Patients with ALK rearranged vs. ALK wildtype tumors |
| - mOS 59.9 vs. 46.5 mo, p = 0.023 |
| Patients with ALK rearranged vs. pan-negative tumors |
| - mOS 59.9 vs. 16.9 mo, p = 0.003 |
| Patients with brain metastasis at diagnosis vs. patients without brain metastasis at diagnosis |
| - mOS 24.6 vs. 51.5 mo, p = 0.012 |
mOS: median overall survival.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Abbreviations
- ALK
Anaplastic lymphoma kinase
- BRAF
B rapidly accelerated fibrosarcoma
- EGFR
Epidermal growth factor receptor
- ERBB2
Erb-b2 tyrosine kinase 2
- FISH
Fluorescence in situ hybridization
- ICC
Immunocytochemistry
- IHC
Immunohistochemistry
- KRAS
Kirsten rat sarcoma
- LCINS
Lung cancer in never smokers
- MET
Mesenchymal epithelial transition proto-oncogene
- NGS
Next-generation sequencing
- NSCLC
Non-small cell lung cancer
- OS
Overall survival
- PCR
polymerase chain reaction
- PD-L1
Programmed death-ligand 1
- PIK3CA
Phosphatidylinositol-3 kinase catalytic subunit alpha
- RET
Rearranged during transfection proto-oncogene
- ROS1
ROS proto-oncogene 1
- SS
Sanger sequencing
- TP53
Tumor suppressor protein 53
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Korpanty GJ, Kamel-Reid S, Pintilie M, Hwang DM, Zer A, Liu G, et al. Lung cancer in never smokers from the Princess Margaret Cancer Centre. Oncotarget. 2018;9: 22559–22570. 10.18632/oncotarget.25176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7: 778–790. 10.1038/nrc2190 [DOI] [PubMed] [Google Scholar]
- 3.Lee YJ, Kim JH, Kim SK, Ha SJ, Mok TS, Mitsudomi T, et al. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer. 2011;72: 9–15. 10.1016/j.lungcan.2010.12.013 [DOI] [PubMed] [Google Scholar]
- 4.Luo W, Tian P, Wang Y, Xu H, Chen L, Tang C, et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int J Cancer. 2018;143: 1696–1705. 10.1002/ijc.31542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couraud S, Zalcman G, Milleron B, Morin F, Souquet PJ. Lung cancer in never smokers–a review. Eur J Cancer. 2012;48: 1299–1311. 10.1016/j.ejca.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 6.Couraud S, Souquet PJ, Paris C, Dô P, Doubre H, Pichon E, et al. BioCAST/IFCT-1002: epidemiological and molecular features of lung cancer in never-smokers. Eur Respir J. 2015;45: 1403–1414. 10.1183/09031936.00097214 [DOI] [PubMed] [Google Scholar]
- 7.Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, Rha SY, et al. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2012;118: 729–739. 10.1002/cncr.26311 [DOI] [PubMed] [Google Scholar]
- 8.Parente Lamelas I, Abal Arca J, Blanco Cid N, Alves Pérez MT, Dacal Quintas R, Gómez Márquez H, et al. Clinical characteristics and survival in never smokers with lung cancer. Arch Bronconeumol. 2014;50: 62–66. 10.1016/j.arbres.2013.09.015 [DOI] [PubMed] [Google Scholar]
- 9.Thun MJ, Hannan LM, Adams-Campbell LL, Boffetta P, Buring JE, Feskanich D, et al. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 2008;5: e185 10.1371/journal.pmed.0050185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toh CK, Gao F, Lim WT, Leong SS, Fong KW, Yap SP, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol. 2006;24: 2245–2251. 10.1200/JCO.2005.04.8033 [DOI] [PubMed] [Google Scholar]
- 11.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150: 1107–1120. 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511: 543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mounawar M, Mukeria A, Le Calvez F, Hung RJ, Renard H, Cortot A, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67: 5667–5672. 10.1158/0008-5472.CAN-06-4229 [DOI] [PubMed] [Google Scholar]
- 14.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29: 2046–2051. 10.1200/JCO.2010.33.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 8th ed UK: John Wiley & Sons, Ltd; 2017. [Google Scholar]
- 16.Travis WD, Brambilla E, Burka AP, Marx A, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. Lyon: IARC; 2014. [DOI] [PubMed] [Google Scholar]
- 17.Grosse A, Grosse C, Rechsteiner M, Soltermann A. Analysis of the frequency of oncogenic driver mutations and correlation with clinicopathological characteristics in patients with lung adenocarcinoma from Northeastern Switzerland. Diagn Pathol. 2019;14: 18 10.1186/s13000-019-0789-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Ren Y, Fang Z, Li C, Fang R, Gao B, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28: 4616–4620. 10.1200/JCO.2010.29.6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YJ, Cho BC, Jee SH, Moon JW, Kim SK, Chang J, et al. Impact of environmental tobacco smoke on the incidence of mutations in epidermal growth factor receptor gene in never-smoker patients with non-small-cell lung cancer. J Clin Oncol. 2010;28: 487–492. 10.1200/JCO.2009.24.5480 [DOI] [PubMed] [Google Scholar]
- 20.Ramlau R, Cufer T, Berzinec P, Dziadziuszko R, Olszewski W, Popper H, et al. Epidermal growth factor receptor mutation-positive non-small-cell lung cancer in the real-world setting in Central Europe: The INSIGHT Study. J Thorac Oncol. 2015;10: 1370–1374. 10.1097/JTO.0000000000000621 [DOI] [PubMed] [Google Scholar]
- 21.Vallee A, Sagan C, Le Loupp AG, Bach K, Dejoie T, Denis MG. Detection of EGFR gene mutations in non-small cell lung cancer: lessons from a single-institution routine analysis of 1,403 tumor samples. Int J Oncol. 2013;43: 1045–1051. 10.3892/ijo.2013.2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5: 2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 23.Bauml J, Mick R, Zhang Y, Watt CD, Vachani A, Aggarwal C, et al. Frequency of EGFR and KRAS mutations in patients with non small cell lung cancer by racial background: do disparities exist? Lung Cancer. 2013;81: 347–353. 10.1016/j.lungcan.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia N, An J, Jiang QQ, Li M, Tan J, Hu CP. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res. 2013;39: 328–335. 10.3109/01902148.2013.819535 [DOI] [PubMed] [Google Scholar]
- 25.Gao B, Sun Y, Zhang J, Ren Y, Fang R, Han X, et al. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol. 2010;5: 1130–1135. 10.1097/JTO.0b013e3181e05016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9: 154–162. 10.1097/JTO.0000000000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363: 1693–1703. 10.1056/NEJMoa1006448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115: 1723–1733. 10.1002/cncr.24181 [DOI] [PubMed] [Google Scholar]
- 29.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448: 561–566. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 30.Zhao F, Xu M, Lei H, Zhou Z, Wang L, Li P, et al. Clinicopathological characteristics of patients with non-small-cell lung cancer who harbor EML4-ALK fusion gene: a meta-analysis. PLoS One. 2015;10: e0117333 10.1371/journal.pone.0117333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27: 4247–4253. 10.1200/JCO.2009.22.6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paik JH, Choi CM, Kim H, Jang SJ, Choe G, Kim DK, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 2012;76: 403–409. 10.1016/j.lungcan.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 33.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16: 141–151. 10.1016/S1470-2045(14)71173-8 [DOI] [PubMed] [Google Scholar]
- 34.Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A, Zhou C, et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus chemotherapy in EGFR mutant lung cancer: a meta-analysis. J Clin Oncol. 2015;33: 1985–1965. 10.1200/JCO.2014.60.4769 [DOI] [PubMed] [Google Scholar]
- 35.She C, Wang R, Lu C, Sun Z, Li P, Yin Q, et al. Prognostic factors and outcome of surgically treated patients with brain metastases of non-small cell lung cancer. Thorac Cancer. 2019;10: 137–142. 10.1111/1759-7714.12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gounant V, Wislez M, Poulot V, Khalil A, Lavole A, Cadranel J, et al. Subsequent brain metastasis responses to epidermal growth factor receptor tyrosine kinase inhibitors in a patient with non-small-cell lung cancer. Lung Cancer. 2007;58: 425–428. 10.1016/j.lungcan.2007.07.010 [DOI] [PubMed] [Google Scholar]
- 37.Fekrazad MH, Ravindranathan M, Jones DV Jr. Response of intracranial metastases to erlotinib therapy. J Clin Oncol. 2007;25: 5024–5026. 10.1200/JCO.2007.13.3751 [DOI] [PubMed] [Google Scholar]
- 38.Eichler AF, Kahle KT, Wang DL, Joshi VA, Willers H, Engelman JA, et al. EGFR mutation status and survival after diagnosis of brain metastasis in nonsmall cell lung cancer. Neuro Oncol. 2010:12: 1193–1199. 10.1093/neuonc/noq076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HL, Chung TS, Ting LL, Tsai JT, Chen SW, Chiou JF, et al. EGFR mutations are associated with favorable intracranial response and progression-free survival following brain irradiation in non-small cell lung cancer patients with brain metastases. Radiat Oncol. 2012;7: 181 10.1186/1748-717X-7-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa DB, Shaw AT, Ou SH, Solomon BJ, Riely GJ, Ahn MJ, et al. Clinical experience with crizotinib in patients with advanced ALK-rearranged non-small-cell lung cancer and brain metastases. J Clin Oncol. 2015;33: 1881–1888. 10.1200/JCO.2014.59.0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu CL, Chen KY, Shih JY, Ho CC, Yang CH, Yu CJ, et al. Advanced non-small cell lung cancer in patients aged 45 years or younger: outcomes and prognostic factors. BMC Cancer. 2012;12: 241 10.1186/1471-2407-12-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidler MM, Gupta S, Soerjomataram I, Ferlay J, Steliarova-Foucher E, Bray F. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: a population-based study. Lancet Oncol. 2017;18: 1579–1589. 10.1016/S1470-2045(17)30677-0 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Chen SF, Zhen Y, Xiang J, Wu C, Bao P, et al. Multicenter analysis of lung cancer patients younger than 45 years in Shanghai. Cancer. 2010;116: 3656–3662. 10.1002/cncr.25100 [DOI] [PubMed] [Google Scholar]
- 44.Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016;2: 313–320. 10.1001/jamaoncol.2015.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K, Hida T, Oya Y, Yoshida T, Shimizu J, Mizuno T, et al. Unique prevalence of oncogenic genetic alterations in young patients with lung adenocarcinoma. Cancer. 2017;123: 1731–1740. 10.1002/cncr.30539 [DOI] [PubMed] [Google Scholar]
- 46.Nagashima O, Ohashi R, Yoshioka Y, Inagaki A, Tajima M, Koinuma Y, et al. High prevalence of gene abnormalities in young patients with lung cancer. J Thorac Dis. 2013;5: 27–30. 10.3978/j.issn.2072-1439.2012.12.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VandenBussche CJ, Illei PB, Lin MT, Ettinger DS, Maleki Z. Molecular alterations in non-small cell lung carcinomas of the young. Hum Pathol. 2014;45: 2379–2387. 10.1016/j.humpath.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 48.Ye T, Pan Y, Wang R, Hu H, Zhang Y, Li H, et al. Analysis of the molecular and clinicopathologic features of surgically resected lung adenocarcinoma in patients under 40 years old. J Thorac Dis. 2014;6: 1396–1402. 10.3978/j.issn.2072-1439.2014.08.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng TL, Liu Y, Dimou A, Patil T, Aisner DL, Dong Z, et al. Predictive value of oncogenic driver subtype, programmed death-1 ligand (PD-L1) score, and smoking status on the efficacy of PD-1/PD-L1 inhibitors in patients with oncogene-driven non-small cell lung cancer. Cancer. 2018. 10.1002/cncr.31871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.