Abstract
Ossifying fibromyxoid tumor (OFMT) is an uncommon mesenchymal neoplasm of uncertain differentiation and intermediate malignant potential. Recurrent PHF1 gene rearrangements are detected in up to 80% of OFMTs. We describe the clinicopatho-logic features of five OFMTs harboring a novel PHF1-TFE3 fusion. In two cases, RNA sequencing identified a fusion transcript composed of PHF1 exon 11 fused to TFE3 exon 3, whereas in a third case PHF1 exon 12 was fused to TFE3 exon 7. A FISH break-apart assay revealed rearrangements in both PHF1 and TFE3 genes in all cases. The cohort included three males and two females with a median age of 64 years. One OFMT originated in the scapula, while four occurred in the deep soft tissues. Two OFMTs had typical features, whereas three were classified as malignant. Despite uniform cytologic features and fibromyxoid stroma compatible with an OFMT diagnosis, none showed a peripheral shell of lamellar bone. S100 expression was focally present in only one case, while desmin was positive in three cases. All tumors showed strong nuclear immunopositivity for TFE3. All three malignant OFMTs developed metastases, either regionally or to the lung. One patient died of disease 1 year after diagnosis, while the remaining two are alive with disease. In summary, we report novel recurrent PHF1-TFE3 fusions in a subset of OFMTs with aggressive clinical behavior. The PHF1-TFE3 fusions resulted in consistent protein TFE3 overexpression which can be used as a reliable screening tool, adding OFMT as another tumor driven by TFE3 oncogenic activation pathway.
Keywords: fusion, ossifying fibromyxoid tumor, PHF1, TFE3
1 |. INTRODUCTION
Ossifying fibromyxoid tumor (OFMT) is a rare soft tissue neoplasm, affecting middle-aged and older adults with a slight male predominance and presenting as slow growing, superficial or deep-seated masses in the extremities, head and neck area or trunk. The tumors have a local recurrence rate of 10%−20% and a very low risk of metastatic disease. Grossly, OFMT is a multinodular neoplasm surrounded by a thick fibrous capsule, often containing a thin shell of trabecular bone. Like most other translocation-positive soft tissue tumors, OFMT has a uniform cytomorphology and is composed of epithelioid, ovoid, short spindled, and sometimes rhabdoid cells arranged in nests, cords or lace-like architectural patterns, typically in a fibromyxoid or myxohyaline stroma. In typical OFMT mitoses are scarce. The appropriate terminology of lesions with worrisome histologic features, such as high cellularity and increased mitotic activity, is still debatable, in particular if such cases should be designated as atypical or malignant OFMT.1,2 Most OFMTs are positive for S100, while the remaining 10%−20% of negative cases may show only desmin reactivity or a nonspecific immunoprofile. The genetic hallmark of OFMTs are recurrent PHF1 gene rearrangements, detected in up to 80%, spanning both typical and diagnostically challenging tumors at the atypical or malignant end of the morphologic spectrum.3–5 The most common PHF1 fusion partner is EP400, reported in almost half of cases,5 with occasional cases showing variant fusions with MEAF6 and EPC1 genes.5 Other less common gene fusions include ZC3H7B-BCOR, CREBBP-BCORL1, and KDM2A-WWTR1 reported in a handful of cases.5,6 Prompted by an index case of malignant OFMT with an unusual immunoprofile and a novel PHF1-TFE3 fusion detected by RNA sequencing (RNA Seq), we sought to investigate this abnormality in a group of OFMTs lacking defined genetic alterations.
2 |. MATERIALS AND METHODS
2.1 |. Index case and patient selection
A 70-year-old male patient presented with a large, deep-seated soft tissue mass within the gluteal muscle. Grossly, the tumor measured 10 cm in largest diameter and showed an infiltrative growth within the surrounding soft tissues, with multiple satellite nodules extending along the fascia (Figure 1). Microscopically, the tumor was composed of encapsulated nodules of myxohyaline stroma with radiating cords of uniform epithelioid and ovoid spindle cells with atypical hyperchromatic nuclei, indistinct nucleoli and granular eosinophilic cytoplasm. The tumor showed overtly malignant features, including infiltrative growth within skeletal muscle, increased mitotic activity of up to 16 mitoses in 25 HPF corresponding to 5 mm2, areas of necrosis, and lymphovascular invasion (Figure 2). Immunohistochemically, the tumor diffusely expressed cytokeratins (including CK19), E-cadherin, CD56, and synaptophysin, in addition to focal EMA and S100. Desmin stain was negative. Given the immunophenotype of the tumor, the differential diagnosis included neuroendocrine carcinoma and myoepithelial carcinoma. Staging work-up at diagnosis detected two small lung metastases, measuring 6 and 13 mm. Subsequently, two inguinal lymph node metastases of 12 mm were identified 2 months after initial presentation, which were treated with local radiotherapy. The patient died of progressive disease (lung metastases) 1 year after diagnosis. By RNA sequencing a novel PHF1-TFE3 fusion gene was identified in keeping with a malignant OFMT.
FIGURE 1.
Gross examination of a resected malignant OFMT specimen (index case OFMT#1) arising in the deep gluteal muscle of a 70-year-old-male. The tumor had a nodular fibromyxoid cut-surface, being partly well-circumscribed and partly infiltrative, showing multiple small satellite lesions extending along the fascia [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 2.
Microscopic appearance of the index malignant OFMT case (OFMT#1) showing monomorphic ovoid spindle cells with an infiltrative growth within the muscle (A, 200×), arranged in solid sheets (A, 200×) and cords within a hyalinized stroma (B, 100×). Higher power shows cohesive epithelioid tumor cells with nuclear atypia and increased mitotic activity (C, 400×). Immunohistochemistry shows focal expression of S100 (D, 100×) and diffuse staining for cytokeratin (E, AE1:AE3, 100×) and TFE3 (F, 200×)
In order to establish its recurrent potential as well as estimate the frequency of this novel PHF1-TFE3 fusion, we searched our files for OFMT cases with either unknown genetic abnormalities or tumors with PHF1 rearrangements, but lacking a known gene partner. A combined molecular approach, including targeted RNA Seq and fluorescence in situ hybridization (FISH) was used to identify fusion candidates. Tumors were re-reviewed and morphologic findings were recorded, especially ones related to worrisome or atypical histologic features. Previously described diagnostic criteria for malignancy were applied.2,5 Briefly, malignant OFMT was defined as showing high nuclear grade or high cellularity and mitotic activity > 2/50 HPF. Atypical OFMT has been defined as a tumor with histologic findings deviating from typical OFMT, but fall short meeting the above criteria for malignancy. All cases were tested by IHC for S100, desmin, and TFE3; however, most cases had a more extensive immunopanel available, as described in the index case. Clinical follow-up was retrieved where available. The study was approved by the Institutional Review Board.
2.2 |. Targeted RNA sequencing
In three cases, targeted RNA sequencing was performed on samples of primary tumors. Total RNA was extracted from formalin-fixed paraffin-embedded tissue scrolls (3–4 per case) using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA); it was assessed using the RNA 6000 Nano Bioanalyzer Kit (Agilent, Mississauga, ON) and quantitated using the Qubit RNA HS Assay Kit (ThermoFisher Scientific, Mississauga, ON). An input of 20–100 ng total RNA and the TruSight RNA Fusion Panel were used to prepare the RNA-seq libraries (Illumina, San Diego, CA), following manufacturer’s instructions and as previously described.7,8 Sequencing of each sample was performed with 76 base-pair paired-end reads on an Illumina MiSeq at eight samples per flow cell (~3 million reads per sample). The results were then analyzed using the STAR and BOWTIE2 aligners, and Manta and JAFFA fusion callers, respectively.9,10
2.3 |. Fluorescence in situ hybridization (FISH)
In all five primary tumors, FISH was performed on interphase nuclei from paraffin-embedded 4-μm sections using bacterial artificial chromosomes (BAC) custom probes, flanking genes of interest. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI (4′,6-diamidino-2-phenylindole) in an antifade solution, as previously described.5,11 The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score. All cases were tested for ALK gene rearrangements.
3 |. RESULTS
A total of five OFMT cases, including the index case, displayed PHF1-TFE3 fusions, presenting in three males and two females, with a median age of 64 years (range 62–70) (Table 1). None of these cases were previously reported. All except one OFMT originated in the deep soft tissues, spanning a wide range of locations (gluteal muscle, thigh, abdominal wall and cervical, paraspinal), while one tumor appeared to be centered in the scapula. Median tumor size was 10 cm (range 5.2–19 cm). Follow-up was available in all but one recent case. Three patients diagnosed with malignant OFMT developed lung metastases or locoregional metastases (two at presentation, one after 4 years). One patient developed bone metastases 10 years after diagnosis. Of the four patients with available follow-up, one died of disease 1 year after diagnosis, while the remaining three are alive with disease 6 months, 2 years, and 10 years after diagnosis.
TABLE 1.
Clinical, pathologic, and molecular findings of the PHF1-TFE3 fusion-positive OFMTs
| OFMT | Age/sex | Location | Size (cm) | Histology | IHC pos | IHC neg | RNAseq | FU |
|---|---|---|---|---|---|---|---|---|
| 1 | 70/M | Gluteal | 10 | Epithelioid and spindle | TFE3, S100 (focal) | Desmin | PHF1-TFE3 | DOD, 12 months |
| 2 | 62/M | Thigh | 19 | Epithelioid | TFE3, desmin | S100 | NP | AWD, 6 months |
| 3 | 69/F | Abdominal wall | 5.2 | Spindle | TFE3, desmin | S100 | PHF1-TFE3 | NED, 24 months |
| 4 | 59/F | Cervical paraspinal | 8.9 | Epithelioid | TFE3, desmin | S100 | NP | Recent case |
| 5 | 64/M | Scapula | 13.5 | Epithelioid and spindle | TFE3 | S100, desmin | PHF1-TFE3 | AWD,10 years |
AWD, alive with disease; DOD, died of disease; FU, follow up; NED, no evidence of disease.
3.1 |. The histologic spectrum of OFMTs with PHF1-TFE3 fusions resembles that of other OFMTs, yet tumors with malignant features are overrepresented
All tumors had morphologic features compatible with OFMT, as illustrated in Figures 2 and 3. In two cases (OFMT #2&4) only needle biopsies were available, while the remaining three underwent surgical resection of the mass, allowing full assessment of histologic features. The tumors showed a multinodular growth surrounded by thin, fibrous capsules. Peripheral ossification was not found in any of the cases. The tumors were composed of uniform epithelioid to ovoid cells with scant light eosinophilic cytoplasm and round-ovoid nuclei with fine chromatin, embedded in a loose fibromyxoid stroma (Figures 2–4). By histologic criteria, two OFMTs had typical features, whereas three tumors (OFMT #3&5 and the index case OFMT #1, as described above) were overtly malignant, harboring areas with increased cellularity, nuclear atypia and mitotic activity (range 5–16 mitoses in 25 HPF equaling to 5 mm2). Malignant OFMTs with a PHF1-TFE3 fusion were morphologically similar to other PHF1-rearranged genetic subsets (Figure 3). One of the three malignant OFMTs (OFMT #5) presented in a 62-year-old male as a 13.5 cm bone tumor that was centered in the scapula and invaded surrounding soft tissue. The patient developed lung metastases 2 years after initial diagnosis, local tumor recurrences in the thoracic wall 4 years after diagnosis, and bone metastases in the femur 10 years after diagnosis and is alive with disease 10 years after initial diagnosis. Notably, the lung and bone metastases were composed of epithelioid cells arranged in trabeculae and pseudoglandular formations that expressed cytokeratins, CD56 and synaptophysin (Figure 4), suggestive of neuroendocrine differentiation, as also found in the malignant OFMT (index case OFMT #1, see Section 2). Focal areas of more conventional OFMT were detected in one of the recurrence.
FIGURE 3.
Microscopic findings of four additional OFMTs with PHF1-TFE3 fusions. Tumors displayed a typical morphology, such as OFMT#3 arising in the abdominal wall of a 69-year old female (A); OFMT#4 arising in the deep soft tissue of the posterior neck in a 59-year-old-female (B), a malignant OFMT, OFMT#2, arising in the deep soft tissue of the thigh in a 62-year-old male (C); and a malignant OFMT, OFMT#5 arising in the scapula of a 64-year-old male (D). These tumors were composed of ovoid to epithelioid tumor cells, arranged in reticular, fascicular, and trabecular patterns, with the malignant tumors showing increased cellularity. The microscopic features of these four OFMTs with PHF1-TFE3 fusions match that of a malignant OFMT with a PHF1-MEAF6 fusion as shown in Figures 3E–F for comparison
FIGURE 4.
Pathologic findings of a malignant OFMT arising in the scapula of a 64-year-old male (OFMT#5). Metastases in bone (A, 100×) and lung (B, 100×) have a trabecular and pseudoglandular morphology (B, 100×) with tumor cells expressing cytokeratins (C, 100×), CD56 (D, 200×), synaptophysin (E, 200×), and TFE3 (F, 100×)
By immunohistochemistry, all but one of the cases were negative for S100 protein. Desmin was focally expressed in three cases. After the molecular findings became available, all tumors were tested in retrospect for TFE3 and showed diffuse nuclear staining (Figures 2 and 4).
3.2 |. RNA sequencing and fluorescence in situ hybridization
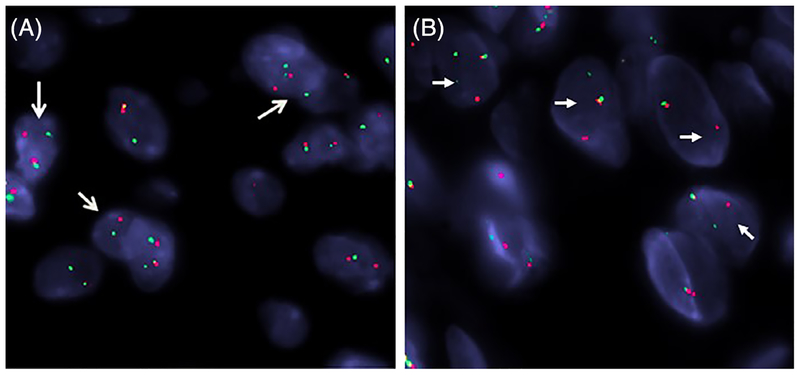
RNA sequencing was performed in the three cases with overtly malignant features (OFMT #1&3&5) (Figure 5). OFMT1 had exonic breakpoints resulting in a fusion between middle of PHF1 exon 11 and middle of TFE3 exon 3. OFMT3 showed intronic breakpoints resulting in a fusion between PHF1 exon 11 and TFE3 exon 3. OFMT5 showed a fusion between PHF1 exon 12 and TFE3 exon 7. These results were further validated by FISH in each case, showing a break-apart signal in both genes. In the remaining two cases with typical morphology (OFMT #2&4) (Figure 6), FISH revealed rearrangements in both PHF1 and TFE3 genes (6). Additionally, FISH was also performed confirming the TFE3 gene rearrangements in the lung metastasis and one local recurrence of case#5, which showed unusual neuroendocrine features.
FIGURE 5.
Diagrammatic representation of the PHF1-TFE3 fusion showing (A) PHF1 gene on 6p21.32 is fused to TFE3 on xp11.23 (thick arrows show the direction of transcription of each gene; delicate red arrows show the two fusion variants encountered. (B) RNAseq reads show the dominant transcript resulting in the fusion of PHF1 exon 11 to TFE3 exon 3. Protein domains for both PHF1 and TFE3 are also illustrated above [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 6.
FISH analysis showed the presence of break-apart signals (arrows) in both PHF1 (A) and TFE3 genes (B) (OFMT #2; red, centromeric; green, telomeric) [Color figure can be viewed at wileyonlinelibrary.com]
4 |. DISCUSSION
OFMT is a rare soft tissue neoplasm of uncertain histogenesis and intermediate malignant potential.12 Established diagnostic criteria in separating typical OFMT from atypical or malignant variants remain elusive. However, most studies define a malignant phenotype based on high cellularity and mitotic activity, which often correlates with lack of S100 reactivity.2,5 In contrast, atypical OFMT has been defined more loosely, as lesions which deviate from conventional OFMT morphology, typically lacking the peripheral shell of bone and S100 expression, but not meeting the criteria for malignancy.2 In the context of these challenging cases, molecular studies are quite helpful to confirm the diagnosis of OFMT.
The genetic hallmarks of OFMTs are recurrent gene fusions that involve PHF1 gene rearrangement in approximately 80% of cases, regardless of risk of malignancy. The most common gene fusion variant is EP400-PHF1 accounting for 44% of cases, with more than half displaying typical histology and co-expression of S100 and desmin. Other less common partners include EPC1 and MEAF6.5 In about 30% of tumors with PHF1 gene rearrangements no gene partner is identified, suggesting additional undetected fusion variants yet to be determined. Triggered by the RNA-Seq result of a novel PHF1-TFE3 fusion in an index case with unusual pathologic findings and overt features of malignancy, we investigated this genetic abnormality in a cohort of OFMTs lacking a known PHF1 gene partner. A total of 5 (10%) cases were identified of the 51 cases studied carrying PHF1 gene abnormalities. This small cohort was enriched by patients who followed a clinically aggressive course. By histologic criteria this genetic abnormality spanned two typical and three overtly malignant OFMT cases. Despite a uniform epithelioid to ovoid cytomorphology and fibromyxoid stroma compatible with an OFMT diagnosis, none of the cases showed evidence of a peripheral shell of lamellar bone and S100 expression was only focally present in one malignant OFMT. Desmin was positive in three of the cases and was negative in two malignant OFMTs. Malignant OFMTs with a PHF1-TFE3 fusions were morphologically similar to other PHF1-rearranged genetic subsets. As expected from its genetic abnormality resulting in TFE3 oncogenic activation through gene fusions, all tumors showed strong nuclear positivity for TFE3 immunohistochemically.
By RNA-Seq, the three PHF1-TFE3 fusion positive cases showed upregulation of TFE3 mRNA. The transcription factor E3 (TFE3) is a member of the microphthalmia (MiT) family, together with MITF, TFEB and TFEC, sharing a helix-loop-helix leucine zipper (bHLH-LZ) dimerization domain motif, a transactivation domain and basic region involved in DNA contact and binding.13 Because of their sequence homology, all MiT family members bind to identical DNA recognition sequences (CA [T/C]GTG) termed E-boxes. Similar to other Xp11 translocation associated tumors, the TFE3-fusion positive OFMTs retain the bHLH-LZ and transcriptional activation domains of TFE3.14 Chromosomal translocations involving TFE3 gene have been described in a number of neoplasms, including mesenchymal and epithelial malignancies. In alveolar soft part sarcoma, an unbalanced translocation, der(17)t(X;17)(p11;q25) results in the formation of an ASPSCR1-TFE3 fusion gene.13 TFE3 related fusions with various gene partners have also been described in the so-called Xp11 renal cell carcinoma as well as in a small subset of perivascular epithelioid cell tumors (PEComa), including SFPQ (PSF), PRCC, ASPSCR1, NONO, RBM10, and DVL2.11,15,16 In most of these various fusion variants, the TFE3 breakpoint resides in exons 3–5.15,17 More recently, recurrent YAP1-TFE3 fusions have been reported in a distinct subset of epithelioid hemangioendothelioma, showing vasoformative features and abundant eosinophilic cytoplasm.14 The fusion transcript in that study was defined as YAP1 exon 1 to TFE3 exon 4. In our three malignant OFMT cases, the RNA-seq showed that PHF1 exon 11 was fused to TFE3 exon 3 in two cases, whereas in the third case PHF1 exon 12 was fused to TFE3 exon 7 (Figure 4 Figure 5).
In about 5%−10% of cases, OFMTs are characterized by non-PHF1 type fusions, including ZC3H7B-BCOR, CREBBP-BCORL1, and KDM2A-WWTR1.5,6 Some of these rare alternative fusions have been associated with a malignant phenotype and aggressive clinical outcome. Moreover, in this current series, the index patient with malignant OFMT followed a fulminant course with lung and locoregional metastases, succumbing to disease 1 year after diagnosis. Two additional patients with malignant OFMT remain alive with disease after developing a similar pattern of local and distant metastases. It remains to be determined in larger series of patients if oncogenic TFE3 activation as the driver of OFMT pathogenesis is associated with a more unfavorable outcome compared to other PHF1 fusion variants.
In summary, we report novel recurrent PHF1-TFE3 fusions in a subset of OFMTs with aggressive clinical behavior, including local recurrences and distant lung metastases. Although morphologically the tumors had features in keeping with OFMT, none showed areas of peripheral ossification and all but one was negative for S100 protein. The PHF1-TFE3 fusions resulted in protein TFE3 overexpression, adding OFMT as an additional tumor driven by oncogenic activation of TFE3 pathway. Thus, in the setting of tumors with malignant phenotype and a nonspecific immunoprofile (S100/desmin negativity), TFE3 immunoreactivity might serve as a useful ancillary test in these challenging cases. Our results add OFMT to the list of tumors driven by TFE3 oncogenic activation through recurrent gene fusions, suggesting that TFE3 immunoreactivity can be thus used as a screening tool for this genetic abnormality.
Funding information
National Cancer Institute, Grant/Award Number: P30 CA008748; National Institue of Health, Grant/Award Number: P50 CA217694; National Institute of Health, Grant/Award Number: P50 CA 140146–01; Cycle for Survival, Slifka Foundation; St Baldrick Foundation
REFERENCES
- 1.Miettinen M, Finnell V, Fetsch JF. Ossifying fibromyxoid tumor of soft parts—a clinicopathologic and immunohistochemical study of 104 cases with long-term follow-up and a critical review of the literature. Am J Surg Pathol. 2008;32:996–1005. [DOI] [PubMed] [Google Scholar]
- 2.Folpe AL, Weiss SW. Ossifying fibromyxoid tumor of soft parts: a clinicopathologic study of 70 cases with emphasis on atypical and malignant variants. Am J Surg Pathol. 2003;27:421–431. [DOI] [PubMed] [Google Scholar]
- 3.Gebre-Medhin S, Nord KH, Moller E, et al. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012; 181:1069–1077. [DOI] [PubMed] [Google Scholar]
- 4.Graham RP, Weiss SW, Sukov WR, et al. PHF1 rearrangements in ossifying fibromyxoid tumors of soft parts: a fluorescence in situ hybridization study of 41 cases with emphasis on the malignant variant. Am J Surg Pathol. 2013;37:1751–1755. [DOI] [PubMed] [Google Scholar]
- 5.Antonescu CR, Sung YS, Chen CL, et al. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors—molecular characterization shows genetic overlap with endome-trial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao YC, Sung YS, Zhang L, Chen CL, Huang SC, Antonescu CR. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP-BCORL1 and KDM2A-WWTR1. Genes Chromosomes Cancer. 2017;56:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson BC, Hornick JL, Fletcher CDM, et al. Dermatofibrosarcoma protuberans with a novel COL6A3-PDGFD fusion gene and apparent predilection for breast. Genes Chromosomes Cancer. 2018;57:437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson BC, Sung YS, Rosenblum MK, et al. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018;42:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 11.Argani P, Zhong M, Reuter VE, et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol. 2016;40:723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher C, Bridge JA, Hogendoorn PC, et al. WHO Classification of Tumours of Soft Tissue and Bone. 4th ed. Lyon: IARC; 2013:281–295. [Google Scholar]
- 13.Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. [DOI] [PubMed] [Google Scholar]
- 14.Antonescu CR, Le Loarer F, Mosquera JM, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agaram NP, Sung YS, Zhang L, et al. Dichotomy of genetic abnormalities in PEComas with therapeutic implications. Am J Surg Pathol. 2015;39:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR. RBM10-TFE3 renal cell carcinoma: a potential diagnostic pitfall due to cryptic Intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am J Surg Pathol. 2017;41:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XT, Xia QY, Ni H, et al. Xp11 neoplasm with melanocytic differentiation of the prostate harbouring the novel NONO-TFE3 gene fusion: report of a unique case expanding the gene fusion spectrum. Histopathology. 2016;69:450–458. [DOI] [PubMed] [Google Scholar]