Abstract
Chromatin is a significant barrier to many DNA damage response (DDR) factors, such as DNA repair enzymes, that process DNA lesions to reduce mutations and prevent cell death; yet, paradoxically, chromatin also has a critical role in many signaling pathways that regulate the DDR. The primary level of DNA packaging in chromatin is the nucleosome core particle (NCP), consisting of DNA wrapped around an octamer of the core histones H2A, H2B, H3 and H4. Here, we review recent studies characterizing how the packaging of DNA into nucleosomes modulates the activity of the base excision repair (BER) pathway and dictates BER subpathway choice. We also review new evidence indicating that the histone amino-terminal tails coordinately regulate multiple DDR pathways during the repair of alkylation damage in the budding yeast Saccharomyces cerevisiae.
1. Introduction
It has been estimated that endogenous sources of DNA damage cause more than 75,000 DNA lesions per day in each human cell, ranging from depurination/depyrimidination of bases, oxidative damage, nonenzymatic methylation by S-adenosylmethionine, cytosine deamination, to single-strand DNA breaks (Table 1) [1, 2]. In addition, cells acquire DNA damage from exogenous sources, such as UV radiation, chemical carcinogens, and ionizing radiation. Many of these lesions are non-helix distorting modifications to DNA bases (i.e., oxidation, methylation, etc.), which are primarily repaired by the base excision repair (BER) pathway. While unrepaired DNA base lesions can lead to mutations, the driving force behind human aging and cancer [3], the accumulation of repair ‘intermediates’, such as abasic sites during BER, is often more toxic and mutagenic then the original DNA base lesion [4], particularly during DNA replication. The postreplication repair (PRR) pathway functions as an alternative mechanism to allow proficient replication past DNA lesions, including BER intermediates, but it is not known to what extent the activity of the BER and PRR pathways are coordinated in eukaryotic cells. To add another layer of complexity, most repair (and lesion bypass) occurs on DNA that is packaged by histones, which create a DNA landscape that occludes excision repair enzymes [5–15], creating a barrier to DNA lesion detection, removal, and repair. In this review, we focus on recent studies characterizing how the packaging of DNA with histones fundamentally shape how base lesions are repaired, and propose a model in which histones actively coordinate the activities of BER and PRR pathways during the repair of alkylation damage in the model eukaryote Saccharomyces cerevisiae.
Table 1.
DNA lesion occurrence from natural endogenous damage
| DNA damage | Rate of occurrence (per cell per day)a |
|---|---|
| Oxidative | 74,000–100,000 in rats 10,000–11,500 in humans |
| Depurinations | 2,000–14,000 |
| Depyrimidinations | ~500 |
| Single-strand breaks | ~55,000 |
| Double strand breaks | 10–50 in humans |
| 7-methylguanine & 3-methyladenine | 7,200 |
| O6-methylguanine | 3,120 |
| Cytosine deamination | 192 |
1.1. DNA packaging in chromatin
Packaging of DNA in chromatin begins with binding of the four core histones H2A, H2B, H3, and H4. These small proteins (~100–150 amino acids) contain a structured histone fold domain, an unstructured amino-terminal tail (N-tail), and, in some cases, a carboxyl-terminal tail. The histone fold domain makes up ~70% of the total mass of each protein and allows for a “handshake motif” interaction between histones H2A and H2B, as well as histones H3 and H4, to form the histone octamer [16, 17]. This octamer complex is wrapped by ~147 bp of DNA to make up the nucleosome core particle (NCP), the basic structural subunit for DNA compaction in the nucleus (Figure 1). The DNA molecule is wrapped ~1.67 times around the histone octamer, and at each DNA superhelical location facing the octamer surface is an arginine residue that inserts into the minor groove [18, 19]. Furthermore, DNA-histone interactions in NCPs are not uniform, and the DNA ends show increased unwrapping-rewrapping dynamics compared to DNA more centrally positioned in the NCP [20].
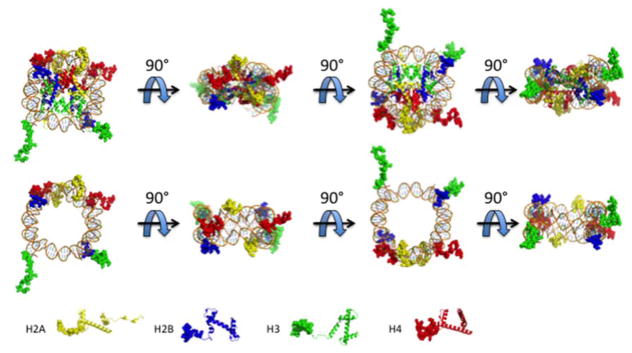
Figure 1.
The NCP and histone N-tails. The NCP is rotated at 90° intervals to show the positions of the histone N-tails (top panel). To better view the locations of the histone N-tails in the context of nucleosomal DNA, the histone core domains have been omitted and rotated 90° (bottom panel). H2A (yellow) H2B (blue) H3 (green) H4 (red). Nucleosome images are derived from the Protein Data Bank (PDB) using PDB ID 3LZ0.
The histone N-tails extend away from the NCP to create eight “appendages” (Figure 1) [21]. Specifically, all four histone N-tails have been shown to make intranucleosomal contacts with the DNA [22–26]. In oligonucleosomes, there are internucleosomal contacts such as interactions of H3/H4 N-tails to neighboring nucleosome DNA [26] and interactions between an acidic patch on the surface of H2A/H2B to the N-tail of H4 of a neighboring nucleosome [27, 28].
Humans and mice contain over 50 histone genes [29], which make genetic analysis of specific histone residues or domains in mammals technically challenging with current molecular tools. To identify cellular functions of the histone N-tails, the budding yeast S. cerevisiae has served as a useful genetic model since it contains only two copies of each histone gene (e.g., HTA1 and HTA2 for histone H2A). Mutational analyses have shown that the N-tails of certain pairs of histones have redundant functions for viability since the simultaneous deletion of the H2A/H2B or H3/H4 N-tails render yeast cells inviable [30–32]. More recently, it was found that combinatorial deletion of H2A and H4 N-tails also leads to yeast cell lethality, most likely due to shared functions between the two N-tail sequences [32–34]. Analysis of viable single and double N-tail deletion mutants (H2A/H3, H2B/H3, and H2B/H4) have shown that these tails are important for a variety of different processes including chromatin compaction, DDR, mitosis, and transcription [32].
The N-tails are highly solvent exposed and, therefore, easily accessible to soluble enzymes such as posttranslational modifying enzymes. Unsurprisingly, these tails are highly posttranslationally modified as compared to the histone fold domain [35]. The various histone posttranslational modifications (PTMs) and their roles in the cell have been extensively reviewed in this issue and elsewhere [36–38] and will not be discussed here.
1.2. DNA damage response
Given the broad spectrum of DNA damage, the cell has a variety of mechanisms to mitigate the abundance of lesions and optimize survival, which are termed the DNA damage response or DDR [39, 40]. When the cell detects a threshold of DNA lesions, it stops cell cycle progression at specific stages—G1/S, intra-S, or G2/M checkpoints—to allow for DNA repair. In S. cerevisiae, a complicated network of signaling complexes, including Mec1/Ddc2, Rad24/Rfc, MRX (Mre11, Rad50, Xrs2) with Tel1, and 9-1-1 (Rad17, Mec3, and Ddc1), are involved in sensing DNA damage and damage-associated DNA replication stress and promoting the phosphorylation of the key kinase Mec1 [41]. Mec1 then phosphorylates Mrc1 or Rad9 depending on whether there is replication stress (e.g., stalled replication forks during S phase) or DNA damage, respectively. Both Mrc1 and Rad9 activate Rad53 kinase to modulate other cellular processes (e.g., gene expression) to arrest cell cycle progression [42, 43]. However, if there is overwhelming cellular damage, the cell can activate programmed cell death in the form of autophagy, apoptosis, or necrosis [44–46]. Importantly, there are pathways, such as BER, NER, and DNA damage tolerance, that allow for removal or bypass of most DNA lesions so cells can resume cell-cycle progression and avoid commitment to programmed cell death.
1.3. Base excision repair
BER recognizes and repairs non-helix distorting lesions caused by oxidation, alkylation, and methylation of DNA bases. Repair occurs through four general steps: 1) recognition and cleavage of the damaged nucleotide base, 2) creation of a nick 5′ of the lesion, 3) DNA synthesis, and 4) DNA ligation [47] (Figure 2). The initial detection and cleavage of DNA lesions by BER is performed by DNA glycosylases. It has been proposed that a DNA glycosylase binds to random sites on the DNA and ‘slides’ along the DNA through one-dimensional diffusion until it either falls off or recognizes a lesion [48]. The DNA base containing the lesion is “flipped” into the active site of the glycosylase and the N-glycosidic bond is cleaved to create an apurinic/apyrimidinic (AP) or abasic site [49]. Among the DNA glycosylases of the cell, some have an additional DNA lyase activity that allows for DNA incision 3′ of the lesion by β-elimination to yield a 3′ α,β-unsaturated aldehyde and a 5′ phosphate or further conversion of the 3′ aldehyde to a 3′ phosphate via a -elimination [50]. These bifunctional glycosylases are distinct from the more common monofunctional glycosylases and are not discussed further, but have been extensively reviewed elsewhere [50–52].
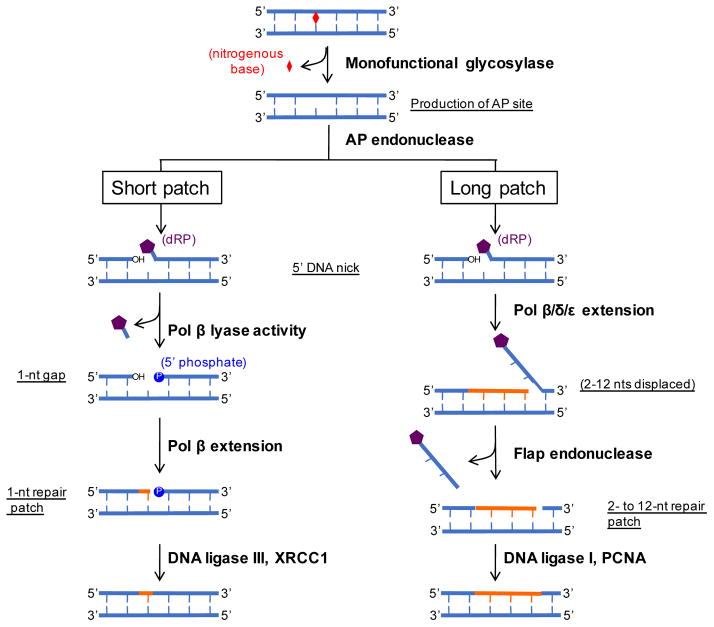
Figure 2.
BER pathway. A non-helix distorting lesion is recognized by a monofunctional glycosylase that cleaves at the N-glycosydic bond, which releases the base. This creates an abasic site that is recognized by an AP endonuclease that creates a 5′ nick. This substrate is then processed by SP or LP repair. SP repair uses the lyase activity of pol to remove the dRP moiety. Pol extends 1 nt followed by DNA ligation by the DNA ligase III/XRCC1 complex. LP repair proceeds by pol,, to extend •2 nts. The displaced DNA is cleaved by flap endonuclease followed by DNA ligation via the coordinated efforts of DNA ligase I and PCNA. Adapted from Figure 1 of Liu & Wilson [47].
Two types of DNA lesions that are recognized and cleaved by DNA glycosylases are 3-methyladenine and 7-methylguanine, which are the major DNA lesions created by the alkylating agent methyl methanesulfonate (MMS). These alkylated bases are recognized and cleaved by the yeast 3-methyladenine DNA glycosylase Mag1 and the human alkyl-adenine DNA glycosylase AAG [53, 54]. Approximately 10% of MMS-induced lesions are 3-methyladenine, which can inhibit DNA replication [55], while ~80% of lesions are 7-methylguanine. This latter adduct does not directly impact replication or transcription [56], but can promote mutagenesis and cytotoxicity upon conversion to an abasic site during BER or through spontaneous depurination [57].
Removal of a modified base by a monofunctional DNA glycosylase creates an abasic site, which, as mentioned earlier, is generally more mutagenic and cytotoxic than the original DNA base lesion. An AP endonuclease recognizes the abasic site and cleaves 5′ of the lesion to create a 3′ hydroxyl and a 5′ deoxyribose phosphate. At this point, the process can progress in one of two subpathways: short patch (SP; also termed single-nucleotide BER) or long patch (LP) repair (Figure 2). SP repair requires Pol β to remove the 5′ deoxyribose phosphate moiety to create a 1-nucleotide (nt) gap and a 5′ phosphate. One nt is then extended from the gap and the DNA ligase III and XRCC1 complex ligates the DNA strands [58]. Alternatively, LP repair creates a longer repair patch that consists of approximately 2–12 nts [59]. LP BER is initiated by pol β,, and/or extending the cleaved strand, using the intact strand as template, and displacing the 5′ deoxyribose phosphate-containing strand (Figure 2). This creates a “flap” of DNA that is then cleaved by a flap endonuclease that leaves a 5′ phosphate and a 3′ hydroxyl for the coordinated action of PCNA and DNA ligase I. A number of different mechanisms have been suggested to regulate BER pathway choice, including the type of DNA glycosylase used to initiate BER (i.e., monofunctional versus bifunctional) [60], the cell-cycle stage in which repair occurs [61], and the chromatin status of the lesion-containing DNA sequences (see below).
2. BER activity and subpathway choice is dictated by the chromatin context of the DNA lesion
Studies from our lab [62–64] and others [65, 66] have shown that the translational position (nucleotide position relative to the dyad axis) and rotational position (orientation of the DNA phosphate backbone relative to the histone octamer) of lesions within a nucleosome affects the accessibility of most BER enzymes to the lesion. The enzymatic activities of DNA glycosylase, AP endonuclease, pol β, and DNA ligase I are inhibited in NCPs when a DNA lesion is located close to the nucleosome dyad (center) or oriented toward the core histones (at an ‘In’ rotational setting) [63, 64, 67–69]. A recent study has shown that a rotational effect on initiation of repair may be DNA glycosylase-specific, since certain DNA glycosylases (e.g., human 8-oxoguanine DNA glycosylase [hOGG1]) were unable to cleave DNA lesions at either ‘Out’ or ‘In’ rotational settings in nucleosomes in vitro [70]. However, this study only analyzed DNA glycosylase activity for lesions at the nucleosome dyad axis [70]; future studies using lesions at different translational positions will be required to verify if these DNA glycosylases are influenced by the rotational position of DNA lesions elsewhere in the nucleosome.
Interestingly, the enzyme that is specific for LP BER, flap endonuclease, exhibits either similar or greater activity in nucleosomes in vitro as compared to naked DNA [71]. Intriguingly, a recent study suggests that particular variants in human BER enzymes are more strongly inhibited by nucleosomes than their ‘wild type’ counterparts [72]. This study analyzed ~8 different cancer-associated or naturally occurring variants in human AP endonuclease-1 (APE1), which processes abasic site intermediates in the BER pathway (see above). They found that two of these variants (APE1 R237C and G241R) were more strongly inhibited than wild type APE1 when their substrate, an abasic site analog known as tetrahydrofuran, was located in a reconstituted NCP in vitro at either an ‘in’ or ‘out’ rotational setting. In contrast, these same variants showed wild type levels of activity on naked DNA substrates, indicating that they had a specific defect in cleaving abasic sites located in nucleosomes (or potentially other DNA-bound proteins) [72]. While the mechanism underlying this effect has yet to be fully elucidated, these data suggest that cells containing these or similar APE1 variants may accumulate BER-associated abasic site intermediates in nucleosomes. As abasic sites are considerably more mutagenic than many primary base lesions, it would be interesting to test whether there is elevated mutagenesis associated with nucleosome DNA in individuals or tumors expressing these APE1 variants.
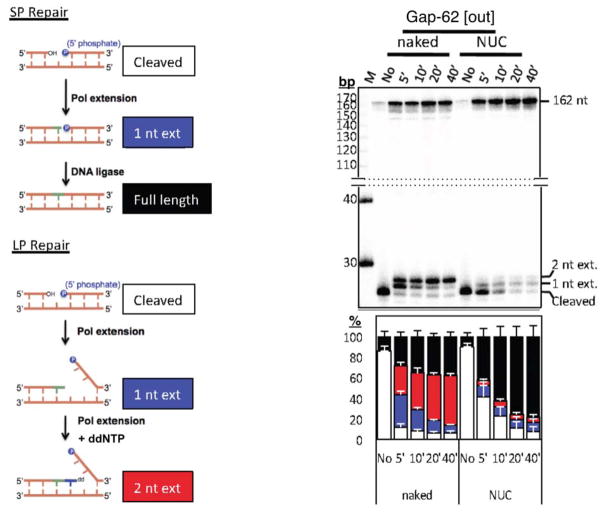
In addition to generally reducing the activity of BER enzymes, nucleosomes have also been recently implicated in regulating BER subpathway choice. To understand the ‘mechanism of choice’ between SP and LP BER in chromatin, we developed an assay to quantitatively measure repair patch sizes in nucleosomes following BER in cell extracts (Figure 3, left panel)[73]. We ‘designed’ nucleosomes (i.e. NCP + linker DNA) that contained either a uracil base or a 1-nt gap positioned at different translational and rotational settings within the NCP region or within adjacent linker DNA (Figure 4). Using bovine testis nuclear extracts or purified BER enzymes, we found a novel nucleosome-dependent bias for SP BER. With uracil substrates, BER polymerase extension is almost exclusively limited to 1 nt in NCP DNA (Figure 3, right panel). Furthermore, we observed that LP BER occurred in linker DNA, but its extension was halted once the DNA polymerase reached the edge of a positioned NCP. When pol β was immunodepleted from the nuclear extracts, repair in NCP DNA was significantly reduced, indicating that pol β plays an important role during BER in NCPs.
Figure 3.
The schematic of the assay to analyze repair in naked or nucleosome substrates is shown on the left. A representative gel of a gap substrate located at position −62 from the nucleosome dyad axis is shown after the indicated repair times. “M” is the marker lane and “No” is the no treatment lane. The composite bar graph is shown below the gel (white, cleaved; blue, 1 nt extension; red, 2 nt extension; black, full-length). Figure was adapted from Figs. 3a and 3d of ref. 73.
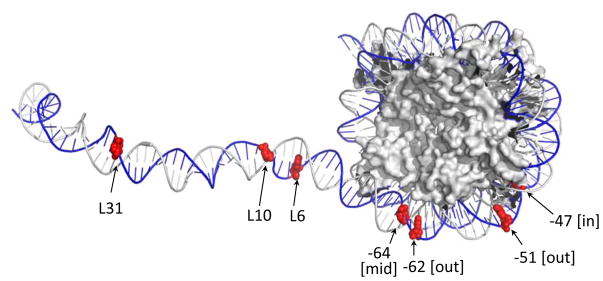
Figure 4.
Map of the various template lesions used in experiments such as those described in Figure 3. The locations of uracils (or gaps) are shown in red. The “L” indicates position of lesions in linker DNA from the edge of the NCP. The number at each lesion in the NCP represents its distance from the dyad center and brackets indicate its rotational position relative to the histone surface (as described in ref. 73). Nucleosome image from PDB ID 1ZBB.
In sperm, the genome is more compacted than somatic cells because of DNA-associated protamines, arginine-rich nuclear proteins that replace histones during spermatogenesis. Interestingly, pol β is highly expressed in zygotene and pachytene spermatocytes and is important for DNA repair in sperm cells [74, 75]. Given that pol β is the smallest mammalian DNA polymerase at 38 kDa [76], we hypothesize that pol β is a major contributor to repair of highly compact DNA (i.e., NCP DNA and spermatocyte DNA) because there is minimal occlusion via steric hindrance as compared to bulkier polymerases. This hypothesis is supported by a recent study that showed the activity of DNA glycosylases on DNA lesion-containing nucleosome substrates is generally more inhibited for large DNA glycosylases as compared to small DNA glycosylases. Specifically, the glycosylase activity of hOGG1 (38 kDa) and Fpg (30.2 kDa) is almost completely inhibited in nucleosome substrates as compared to the smaller UDG (25.67 kDa) and hAAG (24.3 kDa) glycosylases [70].
Since pol β is more intrinsically error-prone than replicative DNA polymerases [77], these results suggest that mutations introduced during repair synthesis might be enriched within nucleosomes. However, it has been recently reported that the activity of pol β within nucleosomes is significantly modulated by histone post-translational modifications [78]. Acetylation of lysine residues in histone H3 (H3K14 and H3K56) significantly inhibited pol β activity at specific locations within nucleosomes [78, 79]. It will be important to determine whether histone acetylation similarly modulates BER subpathway choice in nucleosomes, perhaps favoring LP BER by replicative DNA polymerases.
Recently, a novel subpathway of LP BER has been characterized in mammalian cells, which involves incision of an additional 8 nucleotides 5′ of the DNA lesion by the concerted activities of RECQ1 and XPF-ERCC1, yielding a 9-nucleotide 5′ gap [80]. This results in a patch size of ~20 nucleotides: 8 nucleotides 5′ of the DNA lesion, and up to 12 nucleotides 3′ of the lesion. Since this pathway required replicative DNA polymerases, but was independent of pol β, it was suggested that formation of an expanded 5′ gap may be important for efficiently loading replicative DNA polymerases during the DNA synthesis step of BER [80]. However, it is currently unclear how DNA packaging into nucleosomes might modulate this BER subpathway.
3. Repair of MMS-induced DNA alkylation damage in chromatin in vivo
Much less is known about the impact of chromatin on BER in vivo. While it appears that at least some strongly positioned nucleosomes impact BER efficiency in vivo [81, 82], it is not clear to what extent the translational or rotational positioning of the lesion influences BER. Recently, we reported the genome-wide map of UV radiation damage and repair in chromatin of yeast [83]. We have extended these studies to include the genome-wide map of MMS-induced damage, repair, and mutagenesis at single nucleotide resolution across the yeast genome [84]. We find that DNA packaging in chromatin significantly modulates BER of alkylation damage, and both the translational and rotational settings of lesions within nucleosomes significantly influence BER efficiency in intact cells. Importantly, slow repair near the center of strongly positioned nucleosomes correlates with increased MMS-induced mutations in yeast, indicating that chromatin-associated variations in BER efficiency impacts mutation rates in vivo [84].
4. N-tails of histones H2A and H3 coordinately regulate BER and PRR
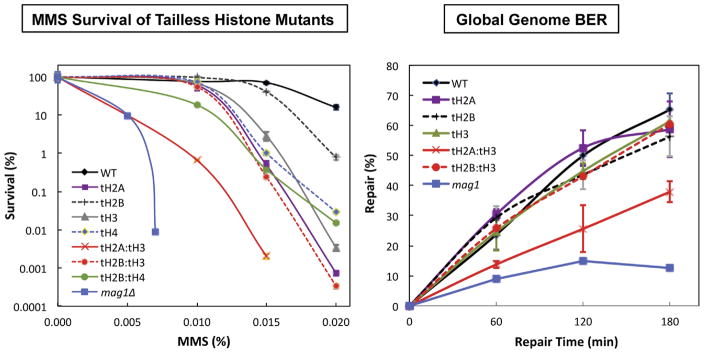
It has been shown that the histone N-tails are required for cellular resistance to DNA damaging agents [32]. Recently, we examined several DNA damage responses in the budding yeast S. cerevisiae with the N-tails deleted from all four canonical histones, both individually and in combination [85]. It was found that combinatorial N-tail deletions of histones H2A and H3 (tH2A:tH3) sensitize yeast cells to the DNA alkylating agent MMS (Figure 5). The tH2A:tH3 mutant cells were found to be deficient in the ‘global removal’ of MMS-induced DNA lesions when using an assay that analyzed DNA lesions in bulk DNA (i.e. ‘global genome BER’; Figure 5). This deficiency was due to the role of H2A and H3 N-tails in regulating both basal and MMS-induced expression of DNA glycosylase Mag1, as MAG1 mRNA expression was significantly decreased in the tH2A:tH3 mutant. Curiously, overexpression of Mag1 in a mutant lacking the H2A and H3 N-tails rescued BER activity, but exacerbated MMS sensitivity in these mutant cells, indicating that the BER defect in the tH2A:tH3 mutant was not primarily responsible for the MMS hypersensitivity phenotype [85]. We also found that the H3 N-tail functions in the Rad9/Rad53 DDR pathway, but this did not appear to be the primary cause of MMS sensitivity of the tH2A:tH3 mutants. Instead, multiple lines of evidence indicated that the H2A and H3 N-tails regulate post-replication repair (PRR) (Figure 6). Genetic experiments revealed that the tailless H2A/H3 mutants are in the RAD18 epistasis group, which regulates post-replication repair (PRR), and we observed increased levels of ubiquitylated PCNA and significantly lower mutation frequency in the tH2A:tH3 mutant, which are both indicative of a defect in PRR.
Figure 5.
Survival curves (left) and global genome BER (right) for the different N-tail deleted mutants as compared to the wild type and the BER-defective mag1Δ mutant is shown. Adapted from Figures 1 and 2 of ref. 85.
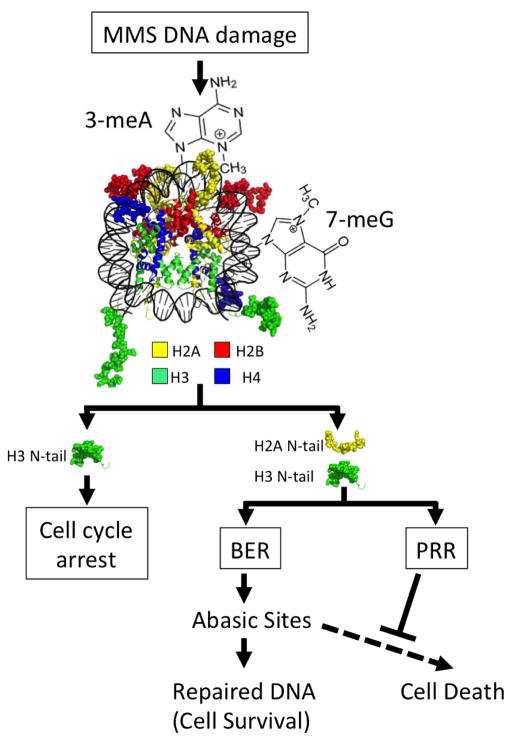
Figure 6.
The N-tails of H2A and H3 coordinate base excision repair (BER) and postreplication repair (PRR) to mitigate the cytotoxic effects of BER intermediates, such as abasic sites. The N-tails of H2A and H3 are highlighted in the model to show their role in different branches of the DNA damage response to methyl methanesulfonate (MMS). The N-tails of H2A and H3 are important for coordinately regulating BER, by modulating expression of Mag1, and PRR. Stimulating the PRR pathway could help suppress the cytotoxic effects of BER intermediates, such as abasic sites, during DNA replication. Nucleosome image from PDB ID IKX5.
These results identified novel roles of the histone H2A and H3 N-tails in regulating the expression of a critical BER enzyme (Mag1) and stimulating PRR in chromatin. Interestingly, our data indicated that the same histone tails are responsible for coordinating both BER and PRR pathways (Figure 6), which may have important implications for mutation avoidance and cell survival. As mentioned above, BER intermediates, such as abasic sites, are typically more toxic and mutagenic than the primary base lesion but can be efficiently bypassed during DNA replication by the PRR pathway. By coordinately stimulating both BER activity (via MAG1 expression) and PRR, the H2A and H3 N-tails may promote the repair of DNA base lesions while at the same time mitigating the cytotoxic effects of the resulting transient BER intermediates (Figure 6). This would explain why higher BER activity in the tH2A:tH3 mutant strain (due to MAG1 overexpression) significantly enhanced MMS sensitivity, since its additional defect in PRR made it unable to efficiently bypass the resulting BER intermediates. In summary, we propose that the coordinate regulation of the BER and PRR pathways by the H2A and H3 N-tails may function as a general strategy for mutation avoidance and cell survival in response to high levels of DNA base lesions.
5. Concluding remarks
During the past decade and a half, a series of studies have led to significant progress in our understanding of DNA base damage and BER at the nucleosome level in chromatin [60–69]. The translational and rotational position of lesions within a nucleosome can strongly affect the accessibility of BER enzymes in vitro. Recent studies in the yeast S. cerevisiae have demonstrated a strong modulation of MMS damage formation by DNA packaging into chromatin [84]. Thus, we are ‘on the cusp’ of revealing the influence of chromatin structure and function on BER and mutagenesis of single-base lesions in cells. These revelations will have important implications for understanding the molecular basis of mutational heterogeneity found in many human cancers [86–88].
Acknowledgments
We would like to thank Dr. Peng Mao for his critical reading of the manuscript. This work was supported by NIH grant ES002614 (to MJS and JJW) and an internal grant from the College of Veterinary Medicine at Washington State University (to JJW). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 2.Bernstein C, Prassad AR, Nfonsam V, Bernstein H. DNA Damage, DNA Repair and Cancer. In: Chen CC, editor. New Research Directions. Intech, open access; 2013. pp. 413–465. [Google Scholar]
- 3.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133:118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinz JM, Czaja W. Facilitation of base excision repair by chromatin remodeling. DNA repair. 2015 doi: 10.1016/j.dnarep.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balliano AJ, Hayes JJ. Base excision repair in chromatin: Insights from reconstituted systems. DNA repair. 2015 doi: 10.1016/j.dnarep.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guintini L, Charton R, Peyresaubes F, Thoma F, Conconi A. Nucleosome positioning, nucleotide excision repair and photoreactivation in Saccharomyces cerevisiae. DNA repair. 2015 doi: 10.1016/j.dnarep.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Meas R, Mao P. Histone ubiquitylation and its roles in transcription and DNA damage response. DNA repair. 2015;36:36–42. doi: 10.1016/j.dnarep.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakkenist CJ, Kastan MB. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA repair. 2015;36:8–12. doi: 10.1016/j.dnarep.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor JS. Design, synthesis, and characterization of nucleosomes containing site-specific DNA damage. DNA repair. 2015;36:59–67. doi: 10.1016/j.dnarep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X. In vitro chromatin templates to study nucleotide excision repair. DNA repair. 2015;36:68–76. doi: 10.1016/j.dnarep.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 13.Waters R, van Eijk P, Reed S. Histone modification and chromatin remodeling during NER. DNA repair. 2015;36:105–113. doi: 10.1016/j.dnarep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Polo SE, Almouzni G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA repair. 2015;36:114–121. doi: 10.1016/j.dnarep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA repair. 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2013;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 18.Hodges AJ, Gallegos IJ, Laughery MF, Meas R, Tran L, Wyrick JJ. Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae. Genetics. 2015;200:795–806. doi: 10.1534/genetics.115.175885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature structural & molecular biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Hayes JJ. Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J Biol Chem. 2007;282:32867–32876. doi: 10.1074/jbc.M706035200. [DOI] [PubMed] [Google Scholar]
- 22.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 angstrom resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 23.Ebralidse KK, Grachev SA, Mirzabekov AD. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature. 1988;331:365–367. doi: 10.1038/331365a0. [DOI] [PubMed] [Google Scholar]
- 24.Lee KM, Hayes JJ. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8959–8964. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausio J, Dong F, Vanholde KE. Use of Selectively Trypsinized Nucleosome Core Particles to Analyze the Role of the Histone Tails in the Stabilization of the Nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 26.Zheng CY, Lu X, Hansen JC, Jeffrey JH. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 27.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 28.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 29.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 30.Schuster T, Han M, Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986;45:445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- 31.Ling X, Harkness TA, Schultz MC, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes & development. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 32.Kim JA, Hsu JY, Smith MM, Allis CD. Mutagenesis of pairwise combinations of histone amino-terminal tails reveals functional redundancy in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5779–5784. doi: 10.1073/pnas.1203453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes & development. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Cote J. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 37.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 38.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 39.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanawalt PC. Historical perspective on the DNA damage response. DNA repair. 2015 doi: 10.1016/j.dnarep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldo V, Liang J, Wang G, Zhou H. Preserving Yeast Genetic Heritage through DNA Damage Checkpoint Regulation and Telomere Maintenance. Biomolecules. 2012;2:505–523. doi: 10.3390/biom2040505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y, Pastushok L, Xiao W. DNA damage-induced gene expression in Saccharomyces cerevisiae. FEMS microbiology reviews. 2008;32:908–926. doi: 10.1111/j.1574-6976.2008.00126.x. [DOI] [PubMed] [Google Scholar]
- 43.Jossen R, Bermejo R. The DNA damage checkpoint response to replication stress: A Game of Forks. Frontiers in genetics. 2013;4:26. doi: 10.3389/fgene.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisenberg T, Carmona-Gutierrez D, Buttner S, Tavernarakis N, Madeo F. Necrosis in yeast. Apoptosis. 2010;15:257–268. doi: 10.1007/s10495-009-0453-4. [DOI] [PubMed] [Google Scholar]
- 45.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci. 2012;37:162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prakash A, Doublie S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog Mol Biol Transl Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drohat AC, Maiti A. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. Org Biomol Chem. 2014;12:8367–8378. doi: 10.1039/c4ob01063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochimica et biophysica acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res. 2000;460:201–210. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Derfler B, Maskati A, Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2′-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic acids research. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rinne ML, He Y, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic acids research. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J Cell Physiol. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sattler U, Frit P, Salles B, Calsou P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003;4:363–367. doi: 10.1038/sj.embor.embor796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic acids research. 2015;43:10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mjelle R, Hegre SA, Aas PA, Slupphaug G, Drablos F, Saetrom P, Krokan HE. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA repair. 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 62.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. The Journal of biological chemistry. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. The Journal of biological chemistry. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Molecular and cellular biology. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic acids research. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. Embo J. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hinz JM. Impact of abasic site orientation within nucleosomes on human APE1 endonuclease activity. Mutation research. 2014;766–767:19–24. doi: 10.1016/j.mrfmmm.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 70.Olmon ED, Delaney S. Differential Ability of Five DNA Glycosylases to Recognize and Repair Damage on Nucleosomal DNA. ACS Chem Biol. 2017;12:692–701. doi: 10.1021/acschembio.6b00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huggins CF, Chafin DR, Aoyagi S, Henricksen LA, Bambara RA, Hayes JJ. Flap endonuclease 1 efficiently cleaves base excision repair and DNA replication intermediates assembled into nucleosomes. Molecular cell. 2002;10:1201–1211. doi: 10.1016/s1097-2765(02)00736-0. [DOI] [PubMed] [Google Scholar]
- 72.Hinz JM, Mao P, McNeill DR, Wilson DM., 3rd Reduced Nuclease Activity of Apurinic/Apyrimidinic Endonuclease (APE1) Variants on Nucleosomes: IDENTIFICATION OF ACCESS RESIDUES. J Biol Chem. 2015;290:21067–21075. doi: 10.1074/jbc.M115.665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meas R, Smerdon MJ. Nucleosomes determine their own patch size in base excision repair. Scientific reports. 2016;6:27122. doi: 10.1038/srep27122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hirose F, Hotta Y, Yamaguchi M, Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Experimental cell research. 1989;181:169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- 75.Alcivar AA, Hake LE, Hecht NB. DNA polymerase-beta and poly(ADP)ribose polymerase mRNAs are differentially expressed during the development of male germinal cells. Biol Reprod. 1992;46:201–207. doi: 10.1095/biolreprod46.2.201. [DOI] [PubMed] [Google Scholar]
- 76.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gearhart PJ, Wood RD. Emerging links between hypermutation of antibody genes and DNA polymerases. Nat Rev Immunol. 2001;1:187–192. doi: 10.1038/35105009. [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, Smerdon MJ. Site-specific Acetylation of Histone H3 Decreases Polymerase beta Activity on Nucleosome Core Particles in Vitro. The Journal of biological chemistry. 2016;291:11434–11445. doi: 10.1074/jbc.M116.725788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mao P, Wyrick JJ. Emerging roles for histone modifications in DNA excision repair. FEMS yeast research. 2016;16 doi: 10.1093/femsyr/fow090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woodrick J, Gupta S, Camacho S, Parvathaneni S, Choudhury S, Cheema A, Bai Y, Khatkar P, Erkizan HV, Sami F, Su Y, Scharer OD, Sharma S, Roy R. A new sub-pathway of long-patch base excision repair involving 5′ gap formation. Embo J. 2017 doi: 10.15252/embj.201694920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. The Journal of biological chemistry. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 82.Li S, Smerdon MJ. Base excision repair of N-methylpurines in a yeast minichromosome. Effects of transcription, dna sequence, and nucleosome positioning. The Journal of biological chemistry. 1999;274:12201–12204. doi: 10.1074/jbc.274.18.12201. [DOI] [PubMed] [Google Scholar]
- 83.Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ. Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome research. 2017;27:1674–1684. doi: 10.1101/gr.225771.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meas R, Smerdon MJ, Wyrick JJ. The amino-terminal tails of histones H2A and H3 coordinate efficient base excision repair, DNA damage signaling and postreplication repair in Saccharomyces cerevisiae. Nucleic acids research. 2015;43:4990–5001. doi: 10.1093/nar/gkv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR Australian Pancreatic Cancer Genome I, Consortium IBC, Consortium IM-S, PedBrain I. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, Staaf J, Sieuwerts AM, Brinkman AB, Martin S, Ramakrishna M, Butler A, Kim HY, Borg A, Sotiriou C, Futreal PA, Campbell PJ, Span PN, Van Laere S, Lakhani SR, Eyfjord JE, Thompson AM, Stunnenberg HG, van de Vijver MJ, Martens JW, Borresen-Dale AL, Richardson AL, Kong G, Thomas G, Sale J, Rada C, Stratton MR, Birney E, Nik-Zainal S. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383. doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]