Abstract
Osteosarcoma is the most common primary bone tumor during childhood and adolescence. Several reports have presented data on serum biomarkers for osteosarcoma, but few reports have analysed circulating microRNAs (miRNAs). In this study, we used next generation miRNA sequencing to examine miRNAs isolated from microvesicle-depleted extracellular vesicles (EVs) derived from six different human osteosarcoma or osteoblastic cell lines with different degrees of metastatic potential (i.e., SAOS2, MG63, HOS, 143B, U2OS and hFOB1.19). EVs from each cell line contain on average ~300 miRNAs, and ~70 of these miRNAs are present at very high levels (i.e., more than 1,000 reads per million). The most prominent miRNAs are miR-21-5p, miR-143-3p, miR-148a-3p and 181a-5p, which are enriched between 3 to 100 fold and relatively abundant in EVs derived from metastatic SAOS2 cells compared to non-metastatic MG63 cells. Gene ontology analysis of predicted targets reveals that miRNAs present in EVs may regulate the metastatic potential of osteosarcoma cell lines by potentially inhibiting a network of genes (e.g., MAPK1, NRAS, FRS2, PRCKE, BCL2 and QKI) involved in apoptosis and/or cell adhesion. Our data indicate that osteosarcoma cell lines may selectively package miRNAs as molecular cargo of EVs that could function as paracrine agents to modulate the tumor micro-environment.
Keywords: Cancer, Osteosarcoma, Metastasis, Cell aggressiveness, Exosomes, MicroRNA
1. Introduction
Bone malignancies are the third leading cause of cancer-related death in children and adolescents (Siegel et al., 2018) . Osteosarcoma is the most frequent primary tumor in bone and is the fourth most frequently treated solid tumor in pediatric patients (Mirabello et al., 2009b; Ward et al., 2014). Worldwide osteosarcoma incidence rates have a bimodal age distribution with a primary peak in incidence occurring in children and adolescents ages 0 to 24 years, followed for a plateau of very low incidence (25 to 59 years) and a secondary peak in elderly (60 to 85 years) (Mirabello et al., 2009a; Mirabello et al., 2009b). The earlier incidence osteosarcoma peak observed in children and adolescent is affecting roughly between 200 to 2,000 patients on different continents across the globe (Mirabello et al., 2009a). Although osteosarcomas are not very common, osteosarcoma patients which do not received chemotherapy treatment will develop metastatic disease after surgical resection at rates close to 80%, thus showing the aggressiveness of this bone cancer (Harris et al., 1998; Marina et al., 2004).
One of the parameters predicting the clinical outcome of osteosarcomas is the presence of metastasis at diagnosis, which counts for about 10-20% of all patients, while 30-40% of patients without metastasis at diagnosis will relapse and develop metastasis during progression of disease independently of treatment (Bhattasali et al., 2015; Daw et al., 2015; Isakoff et al., 2015). Standard clinical care for the treatment of osteosarcomas is dose-intensive multi-agent chemotherapy (pre- and post-operatively) combined with limb-sparing surgery or amputation (Daw et al., 2015; Guillon et al., 2011). Osteosarcoma derived metastasis are mostly located within the lungs (85-90%) and are the leading cause of death from this disease (Bhattasali et al., 2015; Isakoff et al., 2015) . Metastatic osteosarcoma is associated with poor prognosis with overall survival rates remaining at ~20% after five years (Allison et al., 2012; Friebele et al., 2015; Saraf et al., 2018; Zhu et al., 2013). Therefore, it is of considerable interest to identify new molecular therapeutic targets that specifically interfere with osteosarcoma tumor progression within their tumor niche to reduce the incidence of lung metastasis.
Previously, our laboratories have analyzed several cellular and molecular processes related to normal osteoblast growth and differentiation, as well as osteosarcoma progression and metastasis. For example, we have investigated molecular mechanisms mediating cell cycle control and osteoblast phenotype retention in osteoblasts or osteosarcoma (Galindo et al., 2005; Lucero et al., 2013; Pratap et al., 2003; Varela et al., 2016), pathways controlling cell motility and migration (van der Deen et al., 2012), paracrine signalling and Wnt-related cell signalling (Araya et al., 2018; Bravo et al., 2018; Galindo et al., 2007; Vega et al., 2017), as well as DNA sensitivity to radiation (Mamo et al., 2017). We also have examined oncogenic deregulation of mechanisms controlling the activities of tumor suppressor (e.g., p53 and pRB) (Pereira et al., 2009; San Martin et al., 2009; van der Deen et al., 2013), as well as mechanisms of tumor cell adhesion and metastasis (Villanueva et al., 2019). We have also examined miRNA expression in primary osteosarcomas and osteoblastomas (Riester et al., 2017), similar to other studies that examined osteosarcoma related non-coding RNAs (e.g., lncRNAs, circRNAs and miRNAs (Lin et al., 2017; Otoukesh et al., 2018; Sun et al., 2014; Zhang et al., 2017). Recently, we investigated molecular components of the osteosarcoma secretome to understand paracrine effects of secreted proteins or extracellular vesicles in the microenvironment that are associated with osteosarcoma tumor progression and metastasis (Jerez et al., 2017; Villanueva et al., 2019).
In many cancer cell types, secreted factors such as soluble proteins, macromolecular complexes and extracellular vesicles (EVs) have been associated with tumor progression and poor prognosis (Makridakis and Vlahou, 2010; Paltridge et al., 2013). EVs may act as tumor messengers carrying proteomic, genomic and transcriptomic information from one cell to another within the same tumor or to other locations distal from the primary tumor (Lobb et al., 2017) to facilitate the preparation of new niches for future metastasis. Because EVs may convey information from the primary tumor, we performed proteomic analysis of osteosarcoma EVs (Jerez et al., 2017) and showed that EVs carry proteins involved in tumor progression process. In this work, we examined the small non-coding RNA content of EVs from five distinct osteosarcoma cell lines compared to the corresponding parent cells, because miRNAs may control protein expression through effects on mRNA translation and/or stability (Hammond, 2005; Romero-Cordoba et al., 2014; Ul Hussain, 2012) . MiRNAs have been implicated in cancer progression (Croce, 2009; Wang and Wang, 2012; Zhang et al., 2010) either by reduced expression of miRNAs that suppress oncogene expression (Negrini et al., 2009) or by elevated expression of oncomirs (Esquela-Kerscher and Slack, 2006) that inhibit the activity of tumor suppressor genes that normally block tumor suppression by preventing excessive cell proliferation or migration. Identification of EV specific miRNAs in osteosarcomas could lead to new miRNA based therapeutic strategies that increase or decrease miRNA levels in bone cancer patients (Bader et al., 2010; Nana-Sinkam and Croce, 2011).
To establish osteosarcoma specific gene expression patterns and identify new osteosarcoma biomarkers with diagnostic or prognostic potential, several studies have examined miRNA expression during normal osteoblast differentiation or in osteosarcomas focusing on intracellular or circulating miRNAs as part of macromolecular complex (Kushlinskii et al., 2016; Lulla et al., 2011; Maire et al., 2011; Ouyang et al., 2013; van Wijnen et al., 2013). Our study characterizes miRNAs from EVs derived from osteosarcomas to identify potential secreted/circulating miRNA molecules that may participate in osteosarcoma tumor progression or metastasis.
2. Methods
2.1. Cell lines
Human osteosarcoma cell lines (SAOS2, MG63, U2OS, HOS, and 143B) and an immortalized human fetal osteoblast cell line (hFOB1.19) were obtained from the American Type Culture Collection (ATCC) and maintained as recommended. SAOS2 and U2OS cells were cultured in McCoys 5A medium (Sigma–Aldrich, St Louis, MO, USA) supplemented with 15% and 10% of fetal bovine serum (FBS) (HyClone Laboratories Inc, Logan, UT, USA), respectively. MG63, HOS, and 143B cells were maintained in DMEM (Gibco, Grand Island, NY, USA) with 10% FBS. HFOB1.19 cells were maintained in DMEM/F12 with 10% FBS. All culture media were supplemented with L-glutamine (2mM) and penicillin-streptomycin cocktail (100 units/mL).
2.2. EVs and RNA Isolation
EVs were isolated from conditioned media by ultracentrifugation as previously described (Jerez et al., 2017) . Briefly, 48-hrs serum-free conditioned media were clarified by centrifugation to remove cell debris, filtered and concentrated through 0.22 μm, and ultra-centrifuged at 100,000g. RNA for RNA sequencing was isolated using Total Exosome RNA and Protein Isolation Kit from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA). Small RNA for qPCR was isolated using mirVana miRNA Isolation kit from Invitrogen (Thermo Fisher Scientific). As exogenous control, a heterologous microRNA (i.e., cel-miR-39-5p) was included before RNA isolation in all samples at 20 pM. RNA was quantified using a MaestroNano Micro-Volume Spectrophotometer (Maestro-Gen, Taiwan) and Bioanalyzer 2100 with 2100 expert software (Agilent Technologies, Santa Clara, CA, USA).
2.3. EVs size and concentration analysis
The size distribution and concentration of EV preparations were directly determined using a NanoSight N300 nanoparticle tracking analysis (NTA) device (Malvern Instruments, Malvern, UK). Before each session, NanoSight equipment was calibrated by determining the size and concentration of latex polystyrene beads (Malvern Instruments, Cat. No.: NTA4088, 100 nm). The camera level was set to 9, the temperature to 20°C and an arbitrary threshold of 5 was used. Serum-free culture medium (5μL) was diluted 500 times in PBS to obtain a solution free of extracellular microvesicles (ECMVs) as negative control. For each sample, 2 videos of 60s with more than 200 detected tracks per video were taken and analyzed using NTA software 2.3 with default settings that apply the Stokes-Einstein Equation to determine the size of the particles from their Brownian motion.
2.4. miRNA sequencing
MicroRNAs were sequenced using the NEBNext Small RNA library prep kit on an Illumina HiSeq 2000 as previously described (Martin, 2011) (Riester et al., 2015; Riester et al., 2017). Short reads were trimmed of adapters with Cutadapt. Trimmed miRNA sequences greater than 17 nucleotides in length were then aligned to the reference genome and miRBase reference sequences using Bowtie (Langmead et al., 2009). Expression of known miRNAs and prediction of novel miRNAs prediction was performed using miRDeep2 (Friedlander et al., 2008) with the CAP-miRSeq analysis pipeline (Sun et al., 2014).
2.5. Data Analysis
All in silico data analyses were carried out using miRNAs with expression of at least 10 normalized reads per million (RPM) for further analysis. Unsupervised hierarchical clustering was performed using the Pearson correlation method with the Morpheus matrix visualization platform (Broad Institute, Cambridge, MA, USA). Target prediction for individual miRNAs was carried out using TargetScan (Lewis et al., 2005). Prediction of target genes for miRNA sets for selected cell line was also performed using the computational online tool ComiR for combinatorial microRNA target prediction (Coronnello and Benos, 2013). Gene Ontology analysis for biological processes associated with miRNA target genes was performed using DAVID 6.8 (Huang da et al., 2009a; Huang da et al., 2009b), and gene networks of predicted miRNA targets were created with STRINGv11 (Szklarczyk et al., 2019).
2.6. Real-Time qPCR validation
Selected miRNAs that were differentially expressed based on RNA-seq data were validated using TaqMan® microRNA assays (Life Technologies, Carlsbad, CA, USA). Real-time qPCR reactions were performed using the stably expressed hsa-miR-103a-3p as endogenous reference (Riester et al., 2015; Riester et al., 2017). MiRNA expression levels were quantified using the 2ΔCt method. TaqMan® microRNA assays were used for hsa-miR-103, hsa-miR-21, hsa-miR-26a, hsa-miR-30, hsa-miR-143, hsa-miR-148a, and hsa-miR-181a and cel-miR-39 (Life Technologies, Carlsbad, CA, USA).
3. Results
3.1. Isolation and characterization of EVs derived from osteosarcoma cells
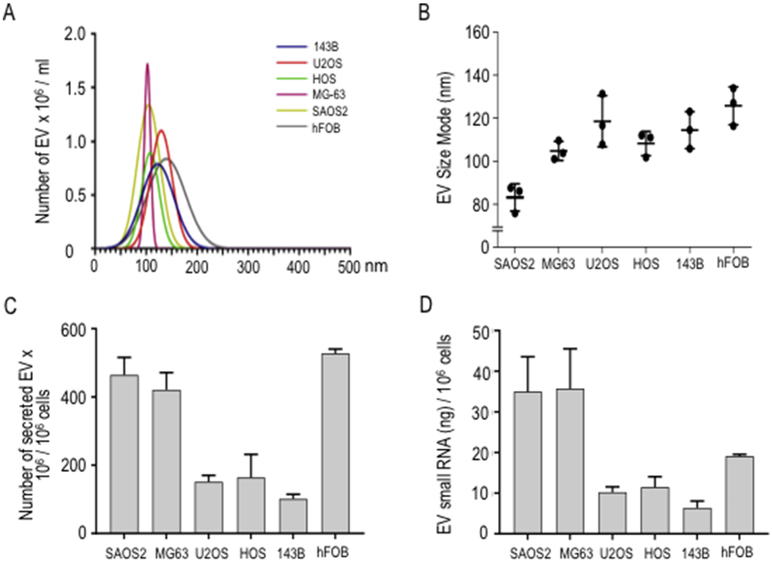
EVs were isolated from conditioned culture media of five human osteosarcoma cell lines. Two of these lines are considered metastatic (SAOS2 and 143B), whereas three are non-metastatic (MG63, U2OS and HOS) based on data from a recent study (Ren et al., 2015). We also examined the non-tumorigenic immortalized osteoblastic hFOB1.19 cell line. As previously reported, the EVs we derived from osteosarcoma cells resemble exosomes, because they present as cup-shaped by transmission electron microscopy and express characteristic exosomal markers (i.e., CD9, CD63 and CD81) (Jerez et al., 2017). We analyzed the diameter sizes and concentrations of isolated EVs by nanoparticle tracking analysis (NTA) using a NanoSight device. NTA shows that size distributions for EVs derived from all six cell lines are unimodal and generally range from 30 to 250nm (Fig. 1A). EVs preparations from each of the cell lines show differences in size distribution (MG63, 80-120nm; SAOS2, 40-120nm; HOS, 110-160nm; 143B, 30-220nm; U2OS, 60-200 nm; hFOB1.19; 30-250nm). The most frequently occurring diameter sizes (mode) of EVs derived from osteosarcoma cells (Fig. 1B) show less variation and ranges between 84 to 126 nm (i.e., SAOS2: 84nm; MG63: 105nm; HOS: 109 m; 143B: 115nm; U2OS: 119nm; hFOB1.19: 126nm). The amount of secreted EVs normalized per million cells shows that hFOB1.19, SAOS2 and MG63 produce significantly more EVs (respectively, 526 × 106 EVs, 465 × 106 and 418 × 106 EVs per million cells) than HOS, U2OS and 143B cells (respectively, 166 × 106, 153 × 106, and 100 × 106 EVs per million of cells) (Fig. 1C). In addition, the total amounts of RNA packaged into EVs ranges between 7 and 36ng RNA per cell line (per 106 cells, respectively, MG63: 36ng, SAOS2: 35ng, hFOB1.19: 19ng, HOS: 12ng, U2OS: 10ng, and 143B: 7ng) (Fig. 1D). The measured range and mode of diameter sizes for the EVs, the amount of secreted EVs per cell, as well as the amount of RNA packaged into EVs is in accordance with expected values for each of these descriptive parameters of EVs. We note that the five osteosarcoma cell lines produce a smaller number of EVs, which are typically smaller in size than EVs produced by non-tumor derived hFOB1.19 cells. These differences may perhaps reflect distinctions in biological properties between the various cell lines. Furthermore, neither the number and size of EVs, nor the amount of RNA packaged into EVs correlates with the biological status of osteosarcoma cell lines as metastatic or non-metastatic, similar to previous observations we made for the amount of protein delivered into EVs from osteosarcoma cells (Jerez et al., 2017)
Fig. 1.
Characterization of exosome-rich extracellular vesicles (EVs) derived from human osteosarcoma or osteoblastic cell lines. EVs were isolated from 24 hours conditioned culture medium obtained from osteosarcoma (SAOS2, MG63, U2OS, HOS, and 143B) and osteoblastic (hFOB1.19) cells. Diameter size (nm) distribution and concentration of purified EVs were determined by NTA (A), and mode size (nm) of three replicates were plotted (B). Relative number of secreted EVs contained in equal volume aliquots of purified EVs was normalized to 1 million of cells (C). Relative concentration of small RNA (ng) into EVs contained in equal volume aliquots of purified EVs was determined and normalized to amount of EVs secreted by 1 million cells (D).
3.2. Next generation RNA sequencing and identification of EV miRNAs
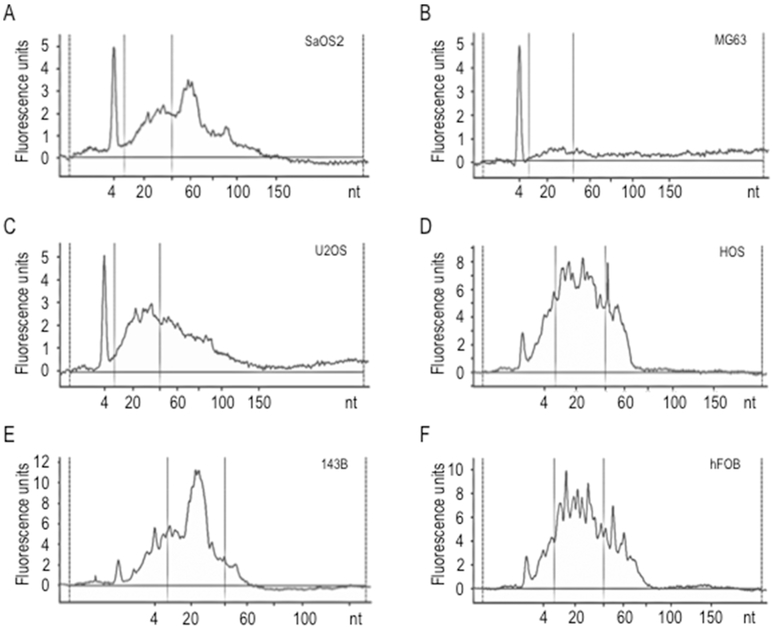
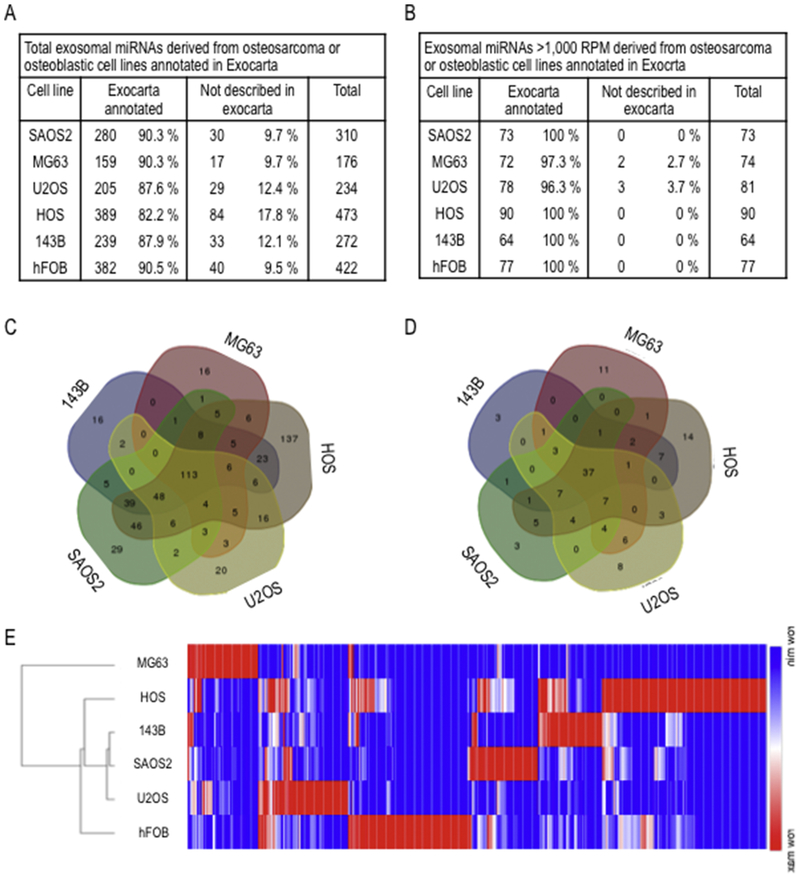
Analysis of RNA size distribution for RNA preparations from EVs revealed presence of small RNAs (~20 nucleotides) (Fig. 2). Therefore, we performed next generation sequencing to identify miRNAs enriched in EVs in our panel of six cell lines. Sequencing of small non-coding RNAs by miRNA sequencing revealed a total of 400 to 600 distinct miRNAs in EVs from each of the cells lines (listed in Supplementary file S1). We detected 237 miRNAs that were exclusively detected in EVs for any of the osteosarcoma cell lines and 77 miRNAs were unique for hFOB1.19 cells. The majority of these miRNAs have previously described in EXOCARTA as present in EVs from other cell types (Fig. 3A), and this is particularly evident if only the most abundant miRNAs are considered (i.e., RPM>1,000) (Fig. 3B). Thus, our analysis appears to detect EV miRNAs with a high degree of confidence. We examined the number of miRNAs exclusively detected in EVs from metastatic (SAOS2 and 143B) versus non-metastatic cells (MG63, U2OS and HOS). This analysis reveals that SAOS2 and 143B cells secrete 50 versus 209 exclusive miRNAs, and 7 versus 43 of these miRNAs are highly expressed (Figs. 3C and D). Comparison of miRNAs in EVs secreted by metastatic 143B cells (i.e., derived from non-metastatic HOS cells by vKi-ras transformation) versus HOS cells, shows that 143B and HOS cells secrete, respectively, 24 versus 225 exclusive miRNAs, and 8 versus 34 of these miRNAs are highly expressed. Thus, a trend emerges suggesting that metastatic osteosarcoma cells may perhaps secrete a more restricted set of cell line-specific miRNAs compared to non-metastatic osteosarcoma cells (Figs. 3C and D). Unsupervised hierarchical clustering of expression patterns of EV miRNAs detected in our panel of six cell lines revealed that metastatic SAOS2 and 143B cells cluster differently from non-metastatic MG63 and hFOB1.19 cells (Figs. 3A and E). The same results were found regardless of whether all expressed miRNAs were examined, or whether the analysis was limited to strongly expressed miRNAs (>1000 RPM) (Figs. 3B and 4A). Taken together, the detection of many cell line specific miRNAs in the six cell lines indicates considerable heterogeneity in the presence of miRNAs in EVs, consistent with the distinct biological phenotypes of each of these cell types.
Fig. 2.
Characterization of small RNAs purified from EVs derived from human osteosarcoma or osteoblastic cell lines. Bioanalyzer electropherograms for each cell line (A, SAOS2; B, MG63; C, U2OS; D, HOS; E, 143B; F, hFOB) show EVs small RNA molecules abundance, expressed as Fluorescence Units [FU] according to molecule nucleotide [nt] extension.
Fig. 3.
Analysis of EVs miRNAs derived from osteosarcoma or osteoblastic cell lines. Total miRNAs identified by NGS (A) and only robustly expressed miRNAs with more than 1000 RPM (B) were compared to those previously annotated in Exocarta data base (http://www.exocarta.org/), that include miRNAs previously identified on exosomes. 5-sets venn diagrams of total miRNAs (C) and robustly expressed miRNAs (D) show distribution of miRNAs among osteosarcoma cell lines. Unsupervised hierarchical clustering analysis (Pearson correlation method) of all EVs miRNAs detected in osteosarcoma or osteoblastic cell lines (E).
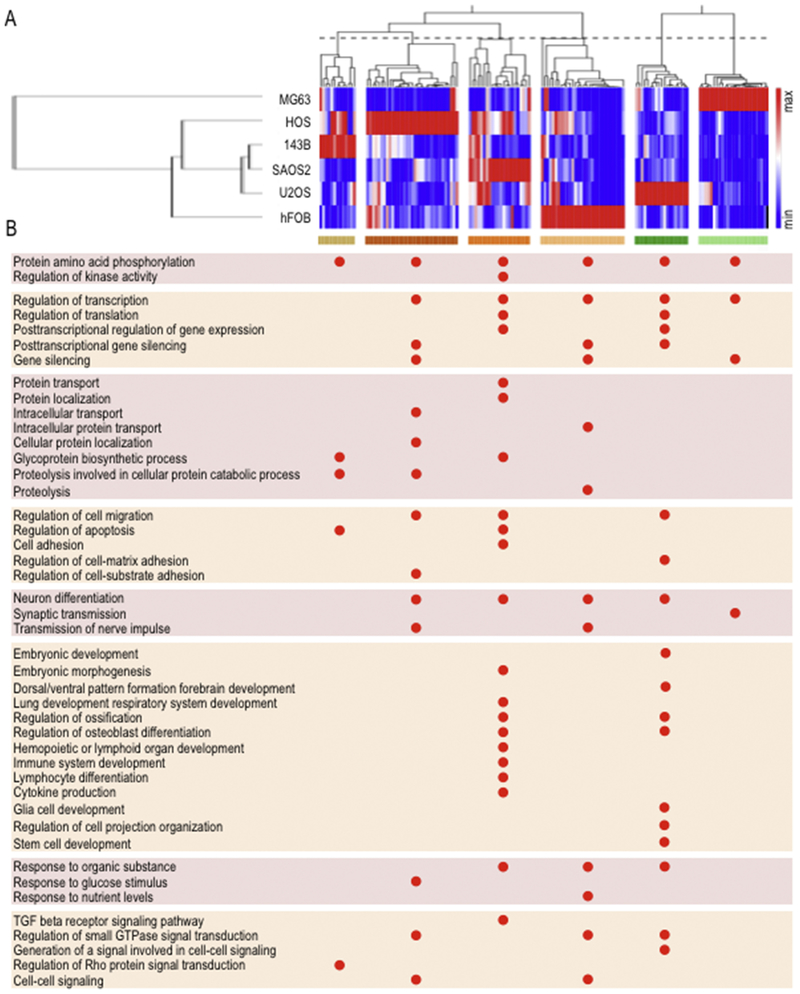
Fig. 4.
Gene ontology analysis of predicted target genes of EV miRNAs revealed biological categories related to tumor progression and metastasis. Unsupervised hierarchical clustering analysis (Pearson correlation method) of robustly expressed EVs miRNAs in osteosarcoma or osteoblastic cell lines show miRNA clusters according to their high relative expression on each cell line (A). Gene ontology analysis of predicted target genes, revealed by TargetScan (http://www.targetscan.org/), associated to miRNA clusters showed in A was assessed using DAVID 6.8 (http://david.ncifcrf.gov/) to identify pathways and biological processes functionally linked to tumor progression and metastasis (B).
3.3. Predicted target genes of EV miRNAs relate to tumor progression and metastasis
MiRNAs in cancer cells act as post-transcriptional regulators of their mRNA targets to control expression of genes involved in tumor progression. To understand the biological activities of highly abundant EV miRNAs for each cell line, we performed gene ontology analysis of potential target genes (Figs. 4A and B). We observed that the different sets of predicted target genes for each miRNA converge toward similar annotation clusters. Clusters with enrichment scores >1 for each of the miRNAs contain essentially the same biological gene ontology categories that are directly or indirectly related to tumorigenesis or metastasis (Fig. 4B). For example, putative target genes for miRNAs in EVs from metastatic SAOS2 cells form annotation clusters for a large number of biological categories, including glycoprotein biosynthesis and protein transport/localization process, cell adhesion, regulation of apoptosis, lung development, lymphoid organ and immune system development, lymphocyte differentiation and cytokine production, as well as the TGFβ receptor signalling pathway and regulation of kinase activity (Fig. 4B). These ontology categories are related to tumor progression properties such as immune evasion, survival and lung metastasis, consistent with the general idea that miRNAs in EVs may contribute to the metastatic potential of osteosarcoma cells.
3.4. Identification of specific EVs miRNAs relate to tumor progression and metastasis
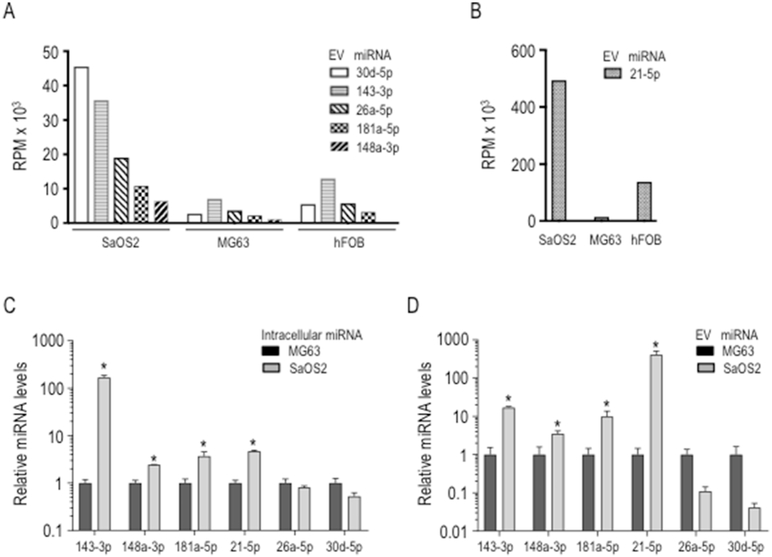
To identify specific miRNAs that may support tumor progression and metastasis, we compared the top 50 most abundant EV miRNAs from metastatic SAOS2 cells with those from non-metastatic MG63 cells. Six miRNAs with the highest fold changes (FC) of greater than 5 fold between these cell types were selected (Figs. 5A and B): miR-21-5p, miR-30d-5p, miR-26a-5p, miR-143-3p, miR-181a-5p and miR-148a-5p. We also monitored specific enrichment of each miRNA in EVs by comparing miRNA abundance in EVs with expression of these miRNAs in the corresponding cell type (Figs. 5C and D) using RT-qPCR with Taqman probes. Expression levels of intracellular miRNAs were normalized using miR-103-3p, which is a validated endogenous control for osteosarcoma samples (Riester et al., 2017). Spike-in methodology using cel-miR-39 at the moment of the RNA isolation was used to normalize EV-miRNA expression levels. The results show that SAOS2 cells have much higher intracellular expression levels of miR-143-3p, miR-21-5p, miR-181a-5p and miR-148-5p than MG63 cells (>3 fold). Similarly, these same miRNAs are also higher in EVs from SAOS2 cells compared to MG63 cells (>2 fold). Because these four miRNAs exhibit higher expression levels in metastatic SAOS2 cell line compared to non-metastatic MG63 cells, we focused our studies on the putative targets of this set of four microRNAs.
Fig. 5.
Analysis of EVs miRNAs differentially expressed in metastatic versus non-metastatic osteosarcoma cell lines. Expression levels of specific EVs miRNAS: miR-30d-5p, miR-143-3, miR-26a-5p, miR181a-5p, miR-148a-5p (A), and miR-21-5p (B) as determined by RNA sequencing in the metastatic (SAOS2) and non-metastatic (MG63) osteosarcoma cell lines as well as the osteoblastic cell line hFOB1.19. MiRNAs expression was validated by qPCR using Taqman probes for both intracellular (C) and EV (D) compartments.. QPCR values were normalized to endogenous expression of miR-103-3p. Expression levels in SAOS2 cells are shown in Log10 scale relative to MG63.
3.5. EV miRNAs in SAOS2 cells are predicted to target metastasis related genes.
We investigated the putative mechanisms by which miRNAs enriched in EVs could potentially support the metastatic potential of SAOS2 cells. We retrieved target genes for miR-143-3p, miR-21-5p, miR-181a-5p and miR-148-5p from the TargetScan database (Figs. 6 and 7). We first prioritized genes that are targeted by three or more of these four miRNAs (Fig. 6A). Remarkably, as many as 75 distinct predicted targets are shared by these four miRNAs group. This set of 75 genes was then subject to network analysis using STRINGv11, and 31 of these genes form a cohesive network (Fig. 6B) with a core of six very well connected genes (Fig. 6C). Gene ontology (GO) analysis using DAVID 6.8 revealed that this set of 31 networked genes is involved in cell adhesion and apoptosis (Fig. 6D). Examination of all putative targets for each of the four miRNAs is consistent with this conclusion (Fig. 6E). Hence, the four most abundant miRNAs enriched in metastatic SAOS2 cells are predicted to suppress genes required for cell adhesion and apoptosis.
Fig. 6.
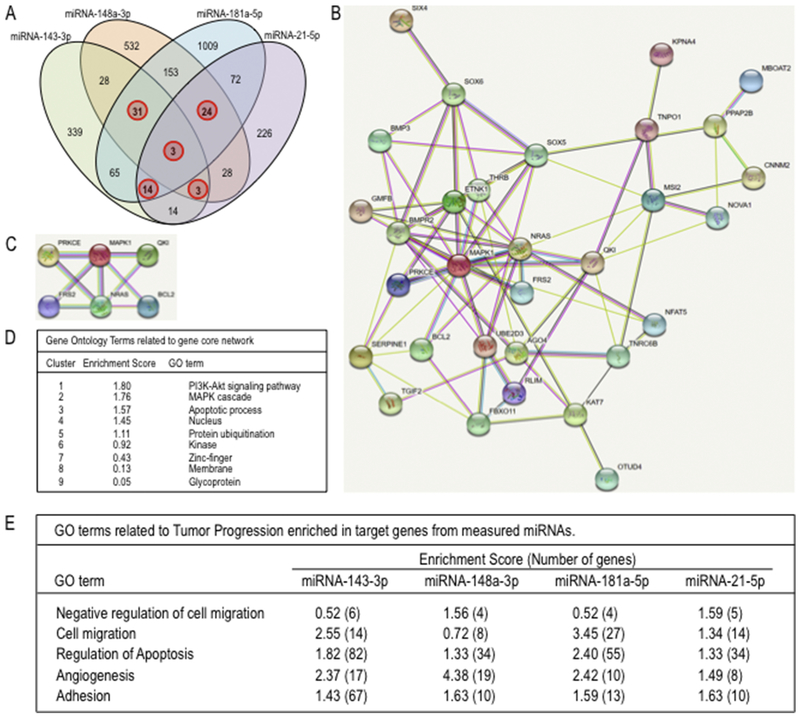
Identification of biological processes related to tumor progression and metastasis reveled by physical interaction network analysis of proteins encoding for genes targeted by EVs miRNAs differentially expressed for metastatic osteosarcoma cells. Target prediction for selected EVs miRNAs: miR-143-3, miR181a-5p, miR-148a-5p, and miR-21-5p, differentially overexpressed in the metastatic SAOS2 osteosarcoma cell line, was carried out using TargetScan. 4-Set venn diagram show number of genes (red circle) that are targeted for multiple EVs miRNAs (A). The resulting 75 multiple miRNA target genes were further analysed with STRINGv11 (http://string-db.org/) to identify functional interaction networks containing 31 of these target genes (B) and a high confidence core network of six target genes (C). The gene network components showed in C (D) as well as gene target for each EVs miRNA (E) were analyzed with David 6.8 to identify specific pathways and biological processes functionally linked to tumor progression and metastasis.
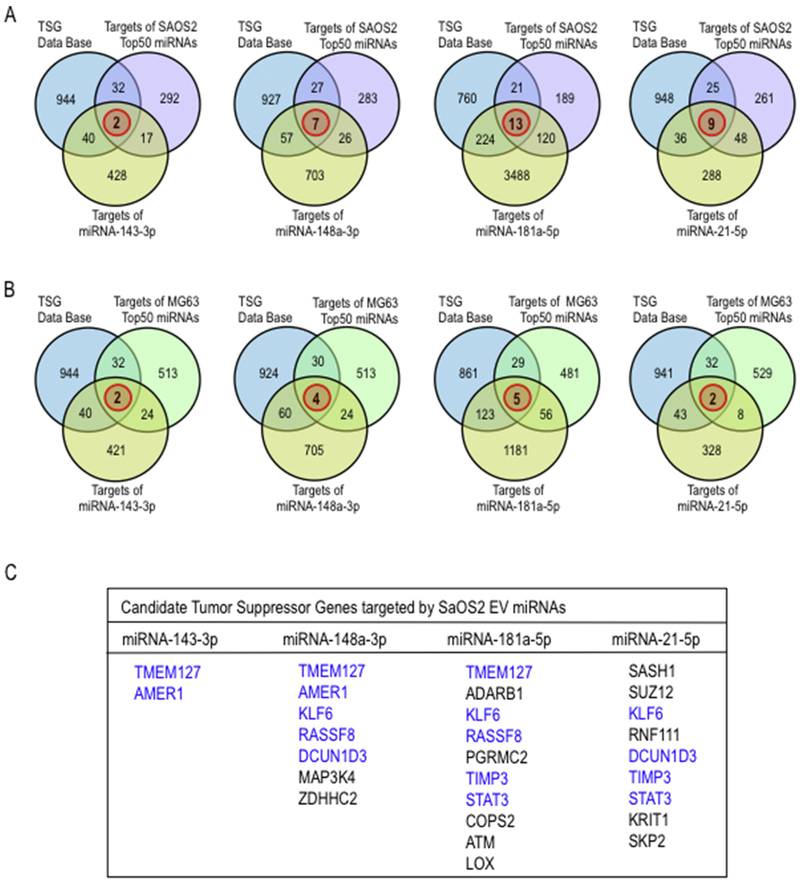
Fig. 7.
Identification of predicted tumor suppressor genes targeted by EVs miRNAs differentially expressed for metastatic osteosarcoma cells. Venn diagrams show the significant overlap (red circle) of the target genes of each selected EVs miRNAs: miR-143-3, miR181a-5p, miR-148a-5p, and miR-21-5p, and tumour suppressor genes listed in Tumor Supressor Gene database (TSGene) (http://bioinfo.uth.edu/TSGene/) as well as target genes predicted by ComiR (http://www.benoslab.pitt.edu/comir/) for combinatorial microRNA target prediction for the set of more expressed top 50 EVs miRNA derived from metastatic SAOS2 (A) and non-metastatic MG63 (B) cell lines, along with the list of tumor supressor genes commonly targeted by these EVs miRNAs in SaOS2 cells (C).
EVs may affect the tumorigenic properties of adjacent osteosarcoma cells through auto-paracrine mechanism. We tested the hypothesis whether miRNAs expressed in metastatic osteosarcoma cells and transmitted in EVs have the potential to control tumor suppressor proteins either within the parent cell or through transmission to adjacent cells via EVs. We investigated which of the predicted miRNA targets for the four selected EV miRNAs (mir-21-5p, mir-143-3p, mir-181a-5p and mir-148a-5p) may target known tumor suppressor genes that are actively expressed in osteosarcoma cells. First, we identified the most probable target genes for robustly expressed miRNAs in EVs from SAOS2 and MG63 using ComiR. This analysis yielded a list of 1,000 most probable targets based on miRNA expression levels. A similar analysis was made for the non-tumor cell line hFOB1.19 to permit elimination of miRNAs involved in normal cellular processes that are unrelated to tumorigenesis. This subtraction yielded a list of 343 target genes for SAOS2 EV-miRNAs and 571 target genes for MG63 EV-miRNAs. We filtered this gene list for Tumor Suppressor Genes (TSG) from the TSG Data base (https://bioinfo.uth.edu/TSGene/) (Zhao et al., 2016; Zhao et al., 2013) (Fig. 7A). This filtering revealed multiple predicted tumor suppressor genes that targeted by the four most abundant EV-miRNAs from SAOS2 cells (Figs. 7A and B). We note that one of the miRNAs target the collagen-crosslinking enzyme LOX, while two miRNAs target the MMP inhibitor TIMP3. The possibility arises that SAOS2 related miRNAs may influence metastatic potential of osteosarcomas in part by modulating ECM remodelling. The results presented in Figures 6 and 7 suggest that the miRNA cargo present in EVs from metastatic osteosarcoma cells may promote both tumorigenic and metastatic properties (Fig. 8).
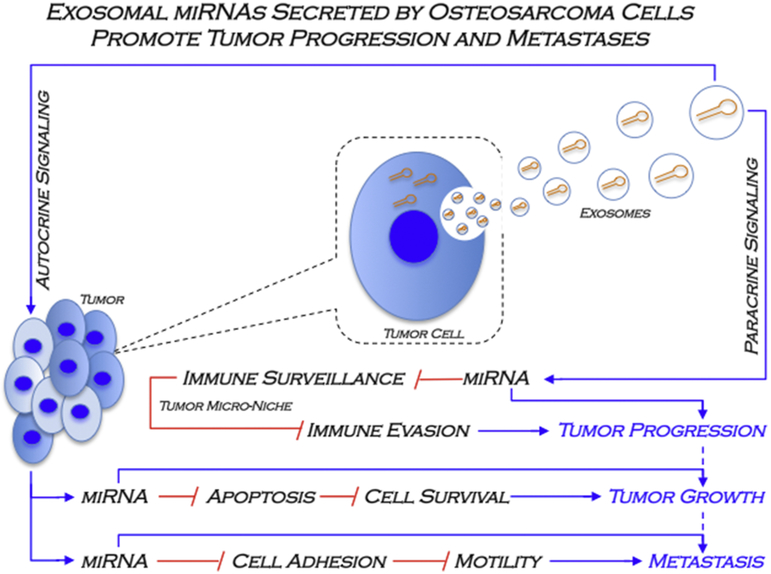
Fig. 8.
A model of the miRNAs secreted into EVs by metastatic osteosarcoma cells in tumor progression and metastasis. Previous studies provided evidence that SAOS2 cell, which exhibit metastatic properties in pre-clinical osteosarcoma animal models, secrete EVs that were characterized as exosomes. The current study shows that intracellular miRNAs can be packaged in EVs (exosomes), which are secreted to the extracellular milieu to establish cell-cell communication at the tumor microenvironment. These EVs miRNAs potentially modulate expression of target genes related to different gene ontology, defining putative pathways and biological categories related to the autocrine (e.g. apoptosis, cell adhesion, cell migration) and paracrine (e.g. immune evasion) control of tumor progression and metastasis in osteosarcoma.
4. Discussion
The content of tumor-derived EVs represents a potential source of possible tumor biomarkers (Duijvesz et al., 2011; Taylor and Gercel-Taylor, 2008) which may be used as diagnostic or prognostic markers, or perhaps represent a source of potential therapeutic targets (Hu et al., 2012; Viaud et al., 2010). Recent studies suggest that in EVs from tumors may modulate tumor growth and/or metastasis either positively or negatively (Aigner, 2011; Al-Nedawi et al., 2009; Trang et al., 2008). Proteomic analysis of osteosarcoma derived EVs revealed that these vesicles contain different proteins involved in tumor progression (Jerez et al., 2017). In this study, we characterized the miRNA content of derived EVs from six distinct osteosarcoma or osteoblastic cell lines. EV preparations from these lines differ to some degree in physical and biochemical properties (e.g., diameter and number of EVs per cell, RNA content), suggesting heterogeneity in the types of EVs that are produced by different cell types. Analysis of miRNAs from osteosarcoma derived EVs by next generation sequencing revealed that EVs from each cell type contains at least 300 distinct miRNAs. Unsupervised hierarchical clustering of the most abundant miRNAs (>1000 RPM) revealed qualitative differences in miRNAs that appear to be preferentially present in metastatic cell types (e.g., SAOS2 and 143B) as opposed to non-metastatic cell lines (e.g., MG63 and hFOB1.19).
Quantitative RT-qPCR of the four most relevant miRNAs (miR-181a-5p, miR-143-3p, miR-21-5p and miR-148a-3p) showed that those are highly abundant in EVs from SAOS2 cells compared to MG63 cells. Of these, miR-21 represents a classical oncomir and is present at elevated levels in many different tumors compared to normal tissue (Ren et al., 2016). MiR-181a-5p and miR-148a-3p are very abundant in EVs from metastatic SAOS2 cells and have been characterized as circulating miRNAs in serum samples from gastro-intestinal cancer patients (Ghaedi et al., 2019; Jacob et al., 2017). MiR-181a-5p has been reported to play a role in tumor progression, because it is overexpressed in gastric cancer (Kim et al., 2011; Zhang et al., 2012) , breast cancer (Mansueto et al., 2010; Taylor et al., 2013), and colorectal cancer (Takahashi et al., 2012). In addition, miR-181a-5p may represent a biomarker for endometrial cancer (Yanokura et al., 2010). MiR-148a-3p is overexpressed in osteosarcoma biopsies (Ma et al., 2014; Maire et al., 2011) and is a potential serum biomarker for thymic ephitelial cancer (Bellissimo et al., 2016). Less is known about miR-143-3p, but its elevated expression in SAOS2 cells is incongruent with recent studies suggesting that this miRNA may counter metastatic properties of squamous cell carcinoma (Han et al., 2018). Our gene ontology analysis of predicted target genes for miR-181a-5p, miR-143-3p, miR-21-5p and miR-148a-3p suggests that these miRNAs control proteins that regulate apoptosis, angiogenesis, cell adhesion and regulation of cell migration. Taken together, the four most abundant miRNAs in EVs from metastatic SAOS2 cells have previously established biological roles in different cancer types.
5. Conclusions
Characterization of metastatic and non-metastatic osteosarcoma cell lines reveals that they each produce EVs with distinct miRNA cargo. These miRNA are predicted to target a network of genes that may suppress cell adhesion and apoptosis, thus perhaps increasing the metastatic potential of osteosarcoma cells. Because miR-21-5p and miR-143-3p show higher levels in the metastatic osteosarcoma cell lines, it is possible that these miRNAs could find use as prognostic biomarkers for osteosarcoma. We conclude that miRNAs from osteosarcoma cell lines that are packaged into EVs may be transmitted to cells within the osteosarcoma micro-niche as part of as paracrine mechanism to promote osteosarcoma growth or metastasis.
Supplementary Material
Highlights: Extracellular vesicles from osteosarcoma cell lines contain multiple microRNAs, including miR-21-5p, miR-143-3p, miR-148a-3p and 181a-5p that are abundant in SAOS2 cells. Common target genes for these miRNAs may form a gene network that could control the metastatic potential of osteosarcoma cells. The data suggest that osteosarcomas produce extracellular vesicles with molecular cargos that are predicted to modulate the osteosarcoma microenvironment.
Acknowledgments
We thank the members of our research groups for stimulating discussions, including Francisco Villanueva and Nelson Varela (University of Chile).
Funding
This study was supported by National Fund for Scientific and Technological Development (FONDECYT Chile), grant numbers 1130931 (to MG) and 1151157 (to CEI), Millennium Science Initiative from Ministry for the Economy, Development and Tourism, Chile, grant number P09/016-F (to MG), Chilean Economic Development Agency (CORFO, Chile) grant number 14IDL2-30168 (to CEI). Additional support was provided by PhD fellowship of the National Commission for Scientific and Technological Research (CONICYT Chile) (to SJ and HA). This work was also supported by National Institutes of Health (NIH) grant R01 AR049069 (to AJvW).
Abbreviations:
- miRNA
microRNA
- EV
extracellular vesicle
- RPM
Reads Per Million mapped reads
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aigner A 2011. MicroRNAs (miRNAs) in cancer invasion and metastasis: therapeutic approaches based on metastasis-related miRNAs. Journal of molecular medicine (Berlin, Germany). 89:445–457. [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, and Rak J. 2009. Microvesicles: messengers and mediators of tumor progression. Cell cycle (Georgetown, Tex.). 8:2014–2018. [DOI] [PubMed] [Google Scholar]
- Allison DC, Carney SC, Ahlmann ER, Hendifar A, Chawla S, Fedenko A, Angeles C, and Menendez LR. 2012. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012:704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya HF, Sepulveda H, Lizama CO, Vega OA, Jerez S, Briceno PF, Thaler R, Riester SM, Antonelli M, Salazar-Onfray F, Rodriguez JP, Moreno RD, Montecino M, Charbonneau M, Dubois CM, Stein GS, van Wijnen AJ, and Galindo MA. 2018. Expression of the ectodomain-releasing protease ADAM17 is directly regulated by the osteosarcoma and bone-related transcription factor RUNX2. Journal of cellular biochemistry. 119:8204–8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, and Winkler M. 2010. The promise of microRNA replacement therapy. Cancer research. 70:7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimo T, Russo E, Ganci F, Vico C, Sacconi A, Longo F, Vitolo D, Anile M, Disio D, Marino M, Blandino G, Venuta F, and Fazi F. 2016. Circulating miR-21-5p and miR-148a-3p as emerging non-invasive biomarkers in thymic epithelial tumors. Cancer biology & therapy. 17:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattasali O, Vo AT, Roth M, Geller D, Randall RL, Gorlick R, and Gill J. 2015. Variability in the reported management of pulmonary metastases in osteosarcoma. Cancer medicine. 4:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo D, Salduz A, Shogren KL, Okuno MN, Herrick JL, Okuno SH, Galindo M, van Wijnen AJ, Yaszemski MJ, and Maran A. 2018. Decreased local and systemic levels of sFRP3 protein in osteosarcoma patients. Gene. 674:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronnello C, and Benos PV. 2013. ComiR: Combinatorial microRNA target prediction tool. Nucleic acids research. 41:W159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM 2009. Causes and consequences of microRNA dysregulation in cancer. Nature reviews. Genetics. 10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw NC, Chou AJ, Jaffe N, Rao BN, Billups CA, Rodriguez-Galindo C, Meyers PA, and Huh WW. 2015. Recurrent osteosarcoma with a single pulmonary metastasis: a multi-institutional review. British journal of cancer. 112:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijvesz D, Luider T, Bangma CH, and Jenster G. 2011. Exosomes as biomarker treasure chests for prostate cancer. European urology. 59:823–831. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, and Slack FJ. 2006. Oncomirs - microRNAs with a role in cancer. Nature reviews. Cancer. 6:259–269. [DOI] [PubMed] [Google Scholar]
- Friebele JC, Peck J, Pan X, Abdel-Rasoul M, and Mayerson JL. 2015. Osteosarcoma: A Meta-Analysis and Review of the Literature. American journal of orthopedics (Belle Mead, N.J.). 44:547–553. [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, and Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nature biotechnology. 26:407–415. [DOI] [PubMed] [Google Scholar]
- Galindo M, Kahler RA, Teplyuk NM, Stein JL, Lian JB, Stein GS, Westendorf JJ, and van Wijnen AJ. 2007. Cell cycle related modulations in Runx2 protein levels are independent of lymphocyte enhancer-binding factor 1 (Lef1) in proliferating osteoblasts. Journal of molecular histology. 38:501–506. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, and van Wijnen AJ. 2005. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. The Journal of biological chemistry. 280:20274–20285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi H, Zadian SS, Alipoor S, Hadianpour S, and Alipoor B. 2019. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene. 687:135–142. [DOI] [PubMed] [Google Scholar]
- Guillon MA, Mary PM, Brugiere L, Marec-Berard P, Pacquement HD, Schmitt C, Guinebretiere JM, and Tabone MD. 2011. Clinical characteristics and prognosis of osteosarcoma in young children: a retrospective series of 15 cases. BMC cancer. 11:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM 2005. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS letters. 579:5822–5829. [DOI] [PubMed] [Google Scholar]
- Han L, Tang M, Xu X, Jiang B, Wei Y, Qian H, and Lu X. 2018. MiR-143-3p suppresses cell proliferation, migration, and invasion by targeting Melanoma-Associated Antigen A9 in laryngeal squamous cell carcinoma. Journal of cellular biochemistry. [DOI] [PubMed] [Google Scholar]
- Harris MB, Gieser P, Goorin AM, Ayala A, Shochat SJ, Ferguson WS, Holbrook T, and Link MP. 1998. Treatment of metastatic osteosarcoma at diagnosis: a Pediatric Oncology Group Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 16:3641–3648. [DOI] [PubMed] [Google Scholar]
- Hu G, Drescher KM, and Chen XM. 2012. Exosomal miRNAs: Biological Properties and Therapeutic Potential. Frontiers in genetics. 3:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Isakoff MS, Bielack SS, Meltzer P, and Gorlick R. 2015. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 33:3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, and Myklebust MP. 2017. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 8:87837–87847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerez S, Araya H, Thaler R, Charlesworth MC, Lopez-Solis R, Kalergis AM, Cespedes PF, Dudakovic A, Stein GS, van Wijnen AJ, and Galindo M. 2017. Proteomic Analysis of Exosomes and Exosome-Free Conditioned Media From Human Osteosarcoma Cell Lines Reveals Secretion of Proteins Related to Tumor Progression. Journal of cellular biochemistry. 118:351–360. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, Choi IJ, Munroe DJ, and Green JE. 2011. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC medical genomics. 4:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushlinskii NE, Fridman MV, and Braga EA. 2016. Molecular Mechanisms and microRNAs in Osteosarcoma Pathogenesis. Biochemistry. Biokhimiia. 81:315–328. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, and Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20. [DOI] [PubMed] [Google Scholar]
- Lin YH, Jewell BE, Gingold J, Lu L, Zhao R, Wang LL, and Lee DF. 2017. Osteosarcoma: Molecular Pathogenesis and iPSC Modeling. Trends in molecular medicine. 23:737–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb RJ, Lima LG, and Moller A. 2017. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Seminars in cell & developmental biology. 67:3–10. [DOI] [PubMed] [Google Scholar]
- Lucero CM, Vega OA, Osorio MM, Tapia JC, Antonelli M, Stein GS, van Wijnen AJ, and Galindo MA. 2013. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. Journal of cellular physiology. 228:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla RR, Costa FF, Bischof JM, Chou PM, de FBM, Vanin EF, and Soares MB. 2011. Identification of Differentially Expressed MicroRNAs in Osteosarcoma. Sarcoma. 2011:732690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Zhang X, Chai J, Chen P, Ren P, and Gong M. 2014. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 35:12467–12472. [DOI] [PubMed] [Google Scholar]
- Maire G, Martin JW, Yoshimoto M, Chilton-MacNeill S, Zielenska M, and Squire JA. 2011. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer genetics. 204:138–146. [DOI] [PubMed] [Google Scholar]
- Makridakis M, and Vlahou A. 2010. Secretome proteomics for discovery of cancer biomarkers. Journal of proteomics. 73:2291–2305. [DOI] [PubMed] [Google Scholar]
- Mamo T, Mladek AC, Shogren KL, Gustafson C, Gupta SK, Riester SM, Maran A, Galindo M, van Wijnen AJ, Sarkaria JN, and Yaszemski MJ. 2017. Inhibiting DNA-PKCS radiosensitizes human osteosarcoma cells. Biochemical and biophysical research communications. 486:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, Iaccarino A, Troncone G, and Fusco A. 2010. Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes & cancer. 1:210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina N, Gebhardt M, Teot L, and Gorlick R. 2004. Biology and therapeutic advances for pediatric osteosarcoma. The oncologist. 9:422–441. [DOI] [PubMed] [Google Scholar]
- Martin M 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 17:10–12. [Google Scholar]
- Mirabello L, Troisi RJ, and Savage SA. 2009a. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. International journal of cancer. 125:229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, and Savage SA. 2009b. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 115:1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, and Croce CM. 2011. MicroRNAs as therapeutic targets in cancer. Translational research : the journal of laboratory and clinical medicine. 157:216–225. [DOI] [PubMed] [Google Scholar]
- Negrini M, Nicoloso MS, and Calin GA. 2009. MicroRNAs and cancer--new paradigms in molecular oncology. Current opinion in cell biology. 21:470–479. [DOI] [PubMed] [Google Scholar]
- Otoukesh B, Boddouhi B, Moghtadaei M, Kaghazian P, and Kaghazian M. 2018. Novel molecular insights and new therapeutic strategies in osteosarcoma. Cancer cell international. 18:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang L, Liu P, Yang S, Ye S, Xu W, and Liu X. 2013. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Medical oncology (Northwood, London, England). 30:340. [DOI] [PubMed] [Google Scholar]
- Paltridge JL, Belle L, and Khew-Goodall Y. 2013. The secretome in cancer progression. Biochimica et biophysica acta. 1834:2233–2241. [DOI] [PubMed] [Google Scholar]
- Pereira BP, Zhou Y, Gupta A, Leong DT, Aung KZ, Ling L, Pho RW, Galindo M, Salto-Tellez M, Stein GS, Cool SM, van Wijnen AJ, and Nathan SS. 2009. Runx2, p53, and pRB status as diagnostic parameters for deregulation of osteoblast growth and differentiation in a new pre-chemotherapeutic osteosarcoma cell line (OS1). Journal of cellular physiology. 221:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, and van Wijnen AJ. 2003. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer research. 63:5357–5362. [PubMed] [Google Scholar]
- Ren L, Mendoza A, Zhu J, Briggs JW, Halsey C, Hong ES, Burkett SS, Morrow J, Lizardo MM, Osborne T, Li SQ, Luu HH, Meltzer P, and Khanna C. 2015. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 6:29469–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Shen Y, Zheng S, Liu J, and Jiang X. 2016. miR-21 predicts poor prognosis in patients with osteosarcoma. British journal of biomedical science. 73:158–162. [DOI] [PubMed] [Google Scholar]
- Riester SM, Arsoy D, Camilleri ET, Dudakovic A, Paradise CR, Evans JM, Torres-Mora J, Rizzo M, Kloen P, Julio MK, van Wijnen AJ, and Kakar S. 2015. RNA sequencing reveals a depletion of collagen targeting microRNAs in Dupuytren's disease. BMC medical genomics. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester SM, Torres-Mora J, Dudakovic A, Camilleri ET, Wang W, Xu F, Thaler RR, Evans JM, Zwartbol R, Briaire-de Bruijn IH, Maran A, Folpe AL, Inwards CY, Rose PS, Shives TC, Yaszemski MJ, Sim FH, Deyle DR, Larson AN, Galindo MA, Cleven AGH, Oliveira AM, Cleton-Jansen AM, Bovee J, and van Wijnen AJ. 2017. Hypoxia-related microRNA-210 is a diagnostic marker for discriminating osteoblastoma and osteosarcoma. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 35:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, and Hidalgo-Miranda A. 2014. miRNA biogenesis: biological impact in the development of cancer. Cancer biology & therapy. 15:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin IA, Varela N, Gaete M, Villegas K, Osorio M, Tapia JC, Antonelli M, Mancilla EE, Pereira BP, Nathan SS, Lian JB, Stein JL, Stein GS, van Wijnen AJ, and Galindo M. 2009. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfbeta in osteosarcoma cells. Journal of cellular physiology. 221:560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf AJ, Fenger JM, and Roberts RD. 2018. Osteosarcoma: Accelerating Progress Makes for a Hopeful Future. Frontiers in oncology. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A. 2018. Cancer statistics, 2018. CA: a cancer journal for clinicians. 68:7–30. [DOI] [PubMed] [Google Scholar]
- Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, and Kocher JP. 2014. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC genomics. 15:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, and Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research. 47:D607–d613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, Boland CR, and Goel A. 2012. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PloS one. 7:e46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, and Gercel-Taylor C. 2008. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic oncology. 110:13–21. [DOI] [PubMed] [Google Scholar]
- Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, and Schiemann WP. 2013. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. The Journal of clinical investigation. 123:150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Weidhaas JB, and Slack FJ. 2008. MicroRNAs as potential cancer therapeutics. Oncogene. 27 Suppl 2:S52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Hussain M 2012. Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell and tissue research. 349:405–413. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Lapointe D, Gupta S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS, and van Wijnen AJ. 2012. Genomic promoter occupancy of runt-related transcription factor RUNX2 in Osteosarcoma cells identifies genes involved in cell adhesion and motility. The Journal of biological chemistry. 287:4503–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deen M, Taipaleenmaki H, Zhang Y, Teplyuk NM, Gupta A, Cinghu S, Shogren K, Maran A, Yaszemski MJ, Ling L, Cool SM, Leong DT, Dierkes C, Zustin J, Salto-Tellez M, Ito Y, Bae SC, Zielenska M, Squire JA, Lian JB, Stein JL, Zambetti GP, Jones SN, Galindo M, Hesse E, Stein GS, and van Wijnen AJ. 2013. MicroRNA-34c inversely couples the biological functions of the runt-related transcription factor RUNX2 and the tumor suppressor p53 in osteosarcoma. The Journal of biological chemistry. 288:21307–21319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ, Taipaleenmaki H, Hesse E, Riester S, and Kakar S. 2013. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Current osteoporosis reports. 11:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela N, Aranguiz A, Lizama C, Sepulveda H, Antonelli M, Thaler R, Moreno RD, Montecino M, Stein GS, van Wijnen AJ, and Galindo M. 2016. Mitotic Inheritance of mRNA Facilitates Translational Activation of the Osteogenic-Lineage Commitment Factor Runx2 in Progeny of Osteoblastic Cells. Journal of cellular physiology. 231:1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega OA, Lucero CMJ, Araya HF, Jerez S, Tapia JC, Antonelli M, Salazar-Onfray F, Las Heras F, Thaler R, Riester SM, Stein GS, van Wijnen AJ, and Galindo MA. 2017. Wnt/beta-Catenin Signaling Activates Expression of the Bone-Related Transcription Factor RUNX2 in Select Human Osteosarcoma Cell Types. Journal of cellular biochemistry. 118:3662–3674. [DOI] [PubMed] [Google Scholar]
- Viaud S, Thery C, Ploix S, Tursz T, Lapierre V, Lantz O, Zitvogel L, and Chaput N. 2010. Dendritic cell-derived exosomes for cancer immunotherapy: what's next? Cancer research. 70:1281–1285. [DOI] [PubMed] [Google Scholar]
- Villanueva F, Araya H, Briceno P, Varela N, Stevenson A, Jerez S, Tempio F, Chnaiderman J, Perez C, Villarroel M, Concha E, Khani F, Thaler R, Salazar-Onfray F, Stein GS, van Wijnen AJ, and Galindo M. 2019. The cancer-related transcription factor RUNX2 modulates expression and secretion of the matricellular protein osteopontin in osteosarcoma cells to promote adhesion to endothelial pulmonary cells and lung metastasis. Journal of cellular physiology. [DOI] [PubMed] [Google Scholar]
- Wang L, and Wang J. 2012. MicroRNA-mediated breast cancer metastasis: from primary site to distant organs. Oncogene. 31:2499–2511. [DOI] [PubMed] [Google Scholar]
- Ward E, DeSantis C, Robbins A, Kohler B, and Jemal A. 2014. Childhood and adolescent cancer statistics, 2014. CA: a cancer journal for clinicians. 64:83–103. [DOI] [PubMed] [Google Scholar]
- Yanokura M, Banno K, Kobayashi Y, Kisu I, Ueki A, Ono A, Masuda K, Nomura H, Hirasawa A, Susumu N, and Aoki D. 2010. MicroRNA and endometrial cancer: Roles of small RNAs in human tumors and clinical applications (Review). Oncology letters. 1:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, and Lai M. 2010. The microRNA network and tumor metastasis. Oncogene. 29:937–948. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, Tong D, Wu D, Li C, Wei Q, Zhang X, Li D, Liu P, Cui H, Tang H, and Ji F. 2017. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 8:61687–61697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Nie Y, Du Y, Cao J, Shen B, and Li Y. 2012. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 33:1589–1597. [DOI] [PubMed] [Google Scholar]
- Zhao M, Kim P, Mitra R, Zhao J, and Zhao Z. 2016. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic acids research. 44:D1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Sun J, and Zhao Z. 2013. TSGene: a web resource for tumor suppressor genes. Nucleic acids research. 41:D970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, McManus MM, and Hughes DP. 2013. Understanding the Biology of Bone Sarcoma from Early Initiating Events through Late Events in Metastasis and Disease Progression. Frontiers in oncology. 3:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.