SUMMARY
The renal corpuscle of the kidney comprises a glomerular vasculature embraced by podocytes and supported by mesangial myofibroblasts, which ensure plasma filtration at the podocyte-generated slit diaphragm. With a spectrum of podocyte-expressed gene mutations causing chronic disease, an enhanced understanding of podocyte development and function to create relevant in vitro podocyte models is a clinical imperative. To characterize podocyte development, scRNA-seq was performed on human fetal kidneys, identifying distinct transcriptional signatures accompanying the differentiation of functional podocytes from progenitors. Interestingly, organoid-generated podocytes exhibited highly similar, progressive transcriptional profiles despites an absence of the vasculature, although abnormal gene expression was pinpointed in late podocytes. On transplantation into mice, organoid-derived podocytes recruited the host vasculature and partially corrected transcriptional profiles. Thus, human podocyte development is mostly intrinsically regulated and vascular interactions refine maturation. These studies support the application of organoid-derived podocytes to model disease and to restore or replace normal kidney functions.
eTOC Blurb
Tran et al. performed single-cell RNA sequencing to provide an understanding of human podocyte development. Insights from the in vivo analysis was applied to extensively evaluate the formation of podocytes in vitro, highlighting autonomous programs of development and those requiring an interplay with adjacent cell types.
INTRODUCTION
The functional unit of the kidney, the nephron, is made up of more than 14 distinct cell types (Lee et al., 2015). The most proximal component, the renal corpuscle, contains the glomerular filtration apparatus with the vascular endothelium ensheathed by podocytes in a three-dimensional network supported by mesangial myofibroblasts, encased by an outer capsule of parietal epithelium (McMahon, 2016). Within the renal corpuscle, podocytes extend foot processes wrapping the glomerular vasculature thus forming an interface for blood filtration. Podocytes are highly susceptible to damage during kidney injury, and genetic defects to podocytes frequently result in severe kidney disease (Greka and Mundel, 2012).
Recent progress has seen the publication of several directed differentiation protocols that generate complex associations of kidney-like cell types – kidney organoids – from pluripotent human stem cells (Freedman et al., 2015; Morizane et al., 2015; Taguchi et al., 2014; Takasato et al., 2014, 2015; Yamaguchi et al., 2016). Podocyte-like cells (PLCs) have been independently described several times in these systems (Kim et al., 2017; Sharmin et al., 2016). Data suggests PLCs develop along a process akin to that occurring in vivo. When transplanted either under the mouse kidney capsule or subcutaneously, PLCs attract and interact with mouse endothelial cells (ECs) (Bantounas et al., 2018; van den Berg et al., 2018; Sharmin et al., 2016). Transcriptionally, PLCs, human glomeruli, and mouse podocytes partially overlap (Sharmin et al., 2016), and structurally, PLCs establish an apical-basal polarity resembling human capillary-loop stage podocytes (Bantounas et al., 2018; Kim et al., 2017; Sharmin et al., 2016).
These findings suggest that pluripotent stem cell-derived PLCs have potential for regenerative therapeutic approaches, disease modeling and drug discovery. However, realizing this potential requires a strong understanding of normal podocyte development to optimize in vitro programs and to accurately assess PLC-derived cell types. To this end, we employed scRNA-seq to obtain a detailed picture of the in vivo program of human podocyte development. Comparison with in vitro PLC production demonstrates an extensive but not complete, autonomous maturation in the absence of normal glomerular formation with PLCs displaying, vascular and mesangial organizing properties on transplantation beneath the mouse renal capsule. These data inform and support translational strategies with PLCs while highlighting areas where improvement is required to normalize PLC actions and properties.
RESULTS
Single cell transcriptomic analysis of human nephrogenesis
Analysis of kidney organoids generated by the directed differentiation of pluripotent stem cells suggests organoid nephron-like structures resemble fetal and not mature nephrons (Freedman et al., 2015; Morizane et al., 2015; Taguchi et al., 2014; Takasato et al., 2016). Given the lack of a comprehensive molecular frame-work for the formation of kidney cell-types in the human fetal kidney, conclusions were limited and primarily founded on analysis of a few specific podocyte markers and selected morphological criteria.
We have begun to assemble a frame-work for the earliest stages of human nephrogenesis, within the cortical nephrogenic zone (Lindström et al., 2018a, 2018b, 2018c, 2018d; O’Brien et al., 2016). To extend these analyses to include more mature cell types, we performed scRNA-Seq analyses on both the nephrogenic zone and the inner cortex. To preserve spatial information, we cut 300-μm thick vibratome sections of week 15-17 fetal kidney samples, manually dissected the outer nephrogenic cortex (Zone 1) and the inner cortex (Zone 2), and dissociated each region to enable scRNAseq (using the 10x Chromium platform, as described previously in Lindström et al., 2018c) (Figure 1A).
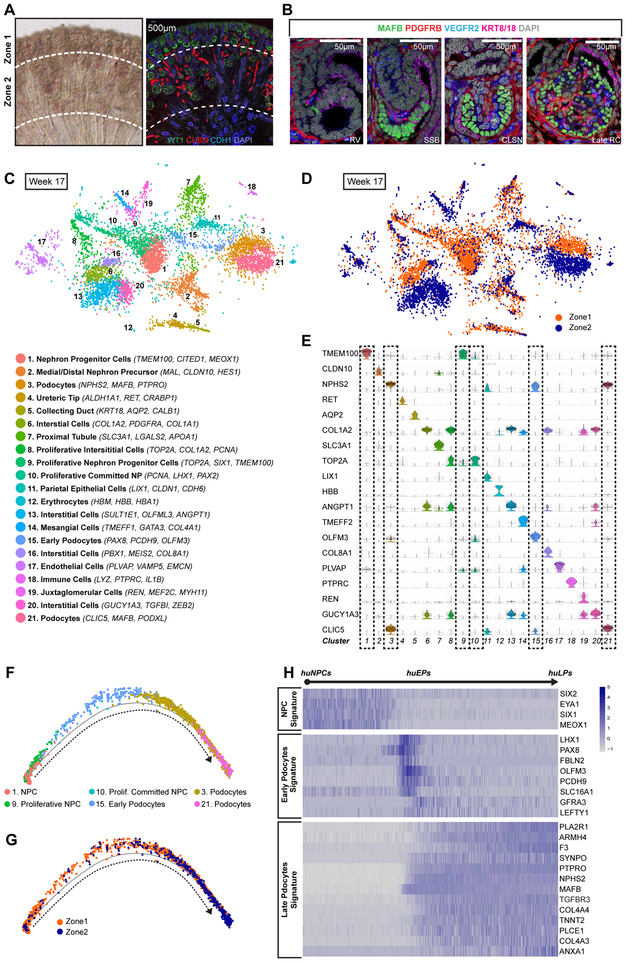
Figure 1: Single-Cell RNA-Seq Analyses Showing Transcriptional Changes during Differentiation of human NPCs to Podocytes.
(A) Left: Vibratome section of week 15-17 human fetal kidney containing Zone 1 and Zone 2 cells. Right: IF stain of a week 15-17 kidney cryosection highlighting mesenchymal progenitor cells, ureteric epithelial cells, early and late nephrons. Dotted lines indicated sites of micro-dissection to separate Zone 1 and Zone 2.
(B) IF stain showing morphogenesis of the renal corpuscle through RV, SSB, CLSN and late RC stages.
(C) Unsupervised clustering of Week 17 kidney cells from both Zone 1 and Zone 2 displayed in a tSNE plot with annotation of cluster identities. In parentheses are differentially expressed genes used for cluster identification).
(D) tSNE plot of Week 17 kidney cells colored by their original zonal identities.
(E) Violin plots of differentially expressed genes used to classify 21 clusters. Dotted-lined boxes mark uncommitted/committed NPC and podocyte clusters subjected to secondary analyses.
(F) Pseudotime trajectory from NPC to podocytes after removal of cells with strong cell-cycling signature. Cells are colored by their cluster identities.
(G) Pseudotime trajectory from NPCs to podocytes with cells colored by their original zonal identities.
(H) Heatmaps of selected genes whose expression changes along the differentiation trajectory from NPCs to podocytes.
RV: renal vesicle, SSB: S-shaped body nephron, CLSN: capillary loop stage nephron, Late RC: late renal corpuscle, huNPC: human nephron progenitor cell, huEP: human early podocyte, huLP: human late podocyte.
Zone 1 and Zone 2 cells enrich for early and late nephron cell-types, respectively, and display contiguous transcriptomic signatures of nephron development. To visualize renal corpuscle developmental programs with a focus on podocyte development, we performed immunofluorescent analysis against specific proteins in Zone 1 and 2 to distinguish podocytes (MAFB), ECs (VEGFR2), interstitial cells (PDGFRB), and epithelial nephron cell types (KRT8/18) in human fetal kidneys (Figure 1B). MAFB+ podocyte precursors emerged first in the proximal segment of the renal vesicle (RV). As the RV matured to comma-shaped body (CSB) nephrons and the glomerular cleft formed and expanded, cuboidal podocyte precursors lie on the proximal side of the glomerular cleft. Endothelial and interstitial cells invaded the glomerular cleft at the CSB stage and were likely progressively recruited during the transition to the S-shaped body (SSB) stage, as podocyte precursor cells extended along the length of the glomerular cleft (Figure 1B). The RV to SSB progression was limited to Zone 1. In zone 2, nephrons matured to the capillary loop stage (CLSN) and functional filtering nephrons. The progressive development of the renal corpuscle is highlighted by the interplay amongst podocytes (MAFB+), ECs (VEGFR2+), and mesangial cell population (PDGFRB+) as these cell types expand, rearrange and mature to form the renal filtration unit (Figure 1B).
Zone 1 and 2 scRNA-seq profiles from a week 17 human kidney were subjected to quality control filtering, and 7,518 single-cell transcriptional profiles were merged into one dataset. Cell groupings were identified by unsupervised clustering using a Gaussian mixture model (Lindström et al., 2018d; Scrucca et al., 2016) . Twenty-one clusters emerged from this analysis (Figure 1C). Each cluster was identified by the expression of known human kidney cell type markers (Lindström et al., 2018c, 2018a, 2018b, 2018d) (Figure 1C and 1E) (Supplemental Table 1). Following expectations, Zone 1 was enriched for early progenitor and differentiated cell types from the nephrogenic, interstitial, and ureteric lineages, whereas Zone 2 consisted of more mature cell-types (Figures 1C, 1D and S1E). ECs were identified in both Zone 1 and Zone 2, while mesangial cells (MCs) predominated in Zone 2 (Figure 1C, 1D and S1E). Cell cluster identification suggested a continuum of nephrogenic lineage cell progression from nephron progenitor cells (NPCs) to maturing nephron cell types.
As we also aimed to examine the presence of glomerular vascular and mesangial cells in the in vitro culture, signatures of these two cell types were identified from the scRNA-Seq datasets. We confirmed that co-expression of PDGFRB and GATA3 marked the pericytes recruited to renal corpuscles, and committed mesangial cells in late RC co-expressed GATA3 and TMEFF2 (Figure S4A, 5G and 5H). Human glomerular vascular cells activated PECAM1, EHD3 and GATA5 (Figure 5I and S4A), and GATA5 expression was also detected in the mouse glomerular vasculature (Messaoudi et al., 2015) (Figure 5K).
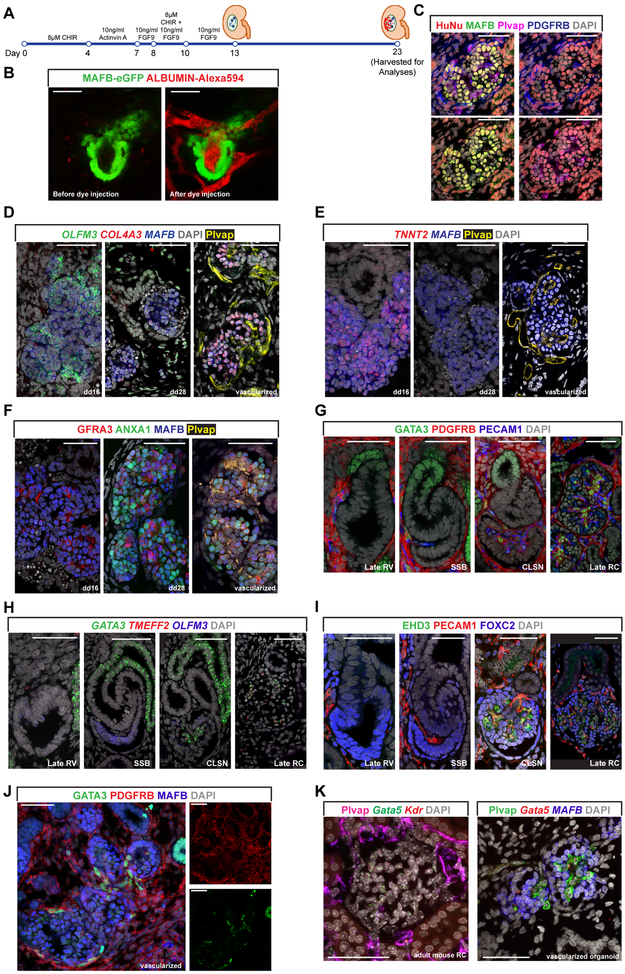
Figure 5: Examination of vasculature’s contribution to glomerular construction.
(A) Schematic diagram summarizing the experimental design to vascularize in vitro derived podocytes
(B) Multiphoton imaging snapshots of vascularized in vitro derived podocytes before and after Alexa 594-conjugated Albumin dye injection. Scale bars denote 50μm.
(C) IF analysis of vascularized organoids showing human or mouse origins of podocytes, interstitial and vascular components. Scale bars denote 50μm.
(D) and (E) Fluorescent in situ hybridization showing expression of EP gene (OLFM3) and LP genes (COL4A3 and TNNT2) in dd16, dd28 and vascularized in vitro derived MAFB+ podocytes. Scale bars denote 50μm.
(F) IF stains of in dd16, dd28 and vascularized in vitro derived podocytes. Scale bars denote 50μm.
(G) and (H) IF stain (G) and fluorescent in situ hybridization (H) highlighting PDGFRB+ GATA3+ pericytes and GATA3+ TMEFF2+ mesangial cells in human developing nephrons. Scale bars denote 50μm.
(I) IF analysis showing EHD3+ PECAM1+ glomerular endothelial cells in human developing nephrons. Scale bars denote 50μm.
(J) IF stain showing expression of GATA3 in interstitial cells recruited to podocyte clusters. Scale bars denote 50μm.
(K) IF stain combined with in situ hybridization (italicized gene names) showing mouse Gata5+ glomerular vascular cells in adult mouse renal corpuscle (LEFT panel) and vascularized organoid (RIGHT panel). Scale bars denote 50μm.
To validate key predictions from the week 17 kidney analysis, we performed scRNA-seq on a week 15 kidney sample. The 6,129 Zone 1 and 2 scRNA-Seq profiles formed 21 clusters (Figures S1A-D). Though the proportion of cells varied within clusters at week 15 and week 17, as expected, the cell type diversity was similar. Overall, the scRNA-seq quality was stronger in the week 17 sample, and to avoid a loss of resolution in more detailed studies of the podocyte, we focused on this sample for further analysis.
Establishing the developmental trajectory from nephron progenitor to human podocyte
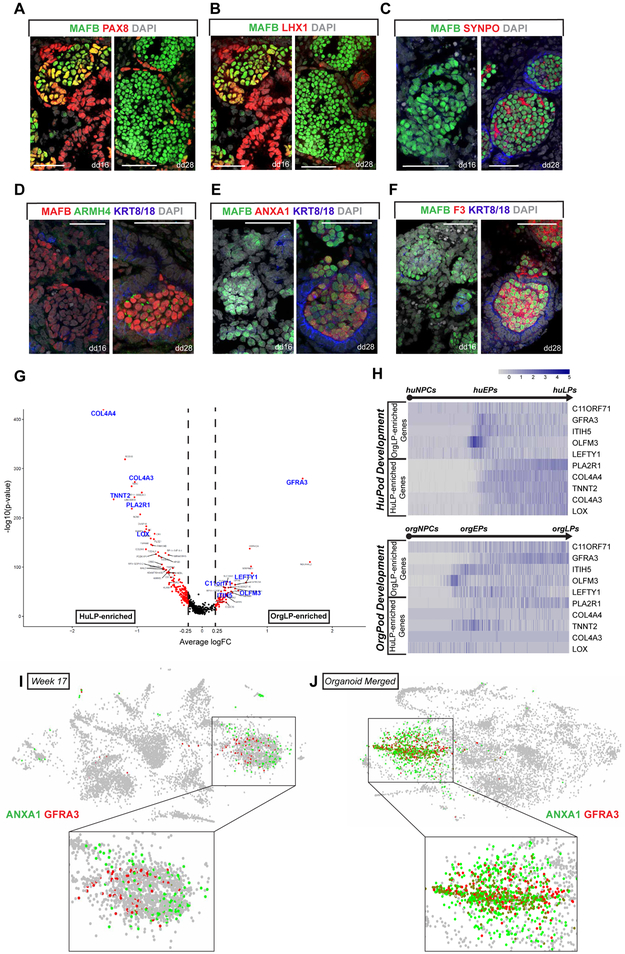
To identify normal transcriptional changes in the podocyte forming program, we selected 3,318 cells including: NPCs (cluster 1 and 9), proliferative developing nephron cells (cluster 10), early podocytes (cluster 15), and maturing podocytes (cluster 3 and 21). Monocle2.0 was used to assemble data into a predicted developmental trajectory (Figure S2A, S2B, 1F and 1G). One end-point of the trajectory comprised nephron progenitors (B1), and the other podocytes (B2) with a side branch of proliferating cells, collectively grouped by a strong cell cycle signature (branch B3), (Figure S2C). A second iteration of pseudotime analysis was performed from NPCs to podocytes (2,705 cells of branches B1 and B2). As expected, Zone 1 and Zone 2 cells concentrated at either ends of the trajectory (Figure 1F). NPC genes such as SIX1, SIX2, EYA1 and MEOX1 were present at the start of the trajectory and downregulated along the predicted developmental progression. The loss of an NPC signature corresponded to the onset of a podocyte signature. A cohort of genes predicted to display a distinct transient early podocyte (EP)-restricted activity (including LHX1, PAX8, FBLN2, OLFM3, PCDH9, SLC16A1, GFRA3, and LEFTY1) gave way to a late podocyte (LP) gene expression signature (including PLA2R1, ARMH4, F3, SYNPO, NPHS2, MAFB, TGFBR3, COL4A3, COL4A4, TNNT2, PLCE1 and ANXA1) (Figure 1H). Analysis of week 15 and 17 scRNA-seq confirmed EP and LP signatures within Zones 1 and 2, respectively (Figures S1F and S1G).
We generated comprehensive human EP (huEP) and LP (huLP) signatures by identifying genes whose expression correlated with representative EP and LP signatures (Figure S2D), and examined their expression along the human podocyte developmental trajectory (Figures S5) to generate curated lists of 158 EP and 104 LP signature genes. Among the EP genes, about half were activated in NPCs. Many of these were enriched in uncommitted and committed NPCs (red text, underlined – Figures S5), and were upregulated in the early period of podocyte development (Figure S2E). Seventy-five genes were transiently expressed in a narrow window of early podocyte development including a number of genes known to control or demarcate nephron induction (green text, underlined – Figures S5) (Figure S2E). Interestingly, 40 EP genes encoded ribosomal proteins (“RPL” or “RPS” genes) downregulated at the initiation of podocyte development. The LP signature list included genes encoding transcription factors known to regulate podocyte development, cytoskeletal components and extracellular matrix proteins linked to glomerular function (Figure S2F). Many EP (CCND1) and LP genes (CD151, GSN, COL4A4, THSD7A, NPHS2, CLIC5, VEGFA, PLA2R1, COL4A3, CTGF, and MAFB) have been associated with genetic disorders of the glomerulus or kidney (Figure S2K) (Online Mendelian Inheritance in Man, 2018). We confirmed the expression of 72/104 LP genes in adult podocytes using Human Protein Atlas, highlight their relevance as mature podocyte signature (Uhlén et al., 2010; Uhlén et al., 2015) (Supplemental Table 2).
To determine whether human podocyte differentiation is conserved in the mouse, we compared the human EP and LP signatures with the mouse microarray analyses of Mafb-GFP+ podocytes from E13.5 embryos and adult kidneys (Brunskill et al., 2011). Our analyses suggested a modest overlap between the two species, with 13 EP and 40 LP genes shared between mouse and human (Figure S2G and S2H), many of which have been described to be important for mouse podocyte morphogenesis (discussed later).
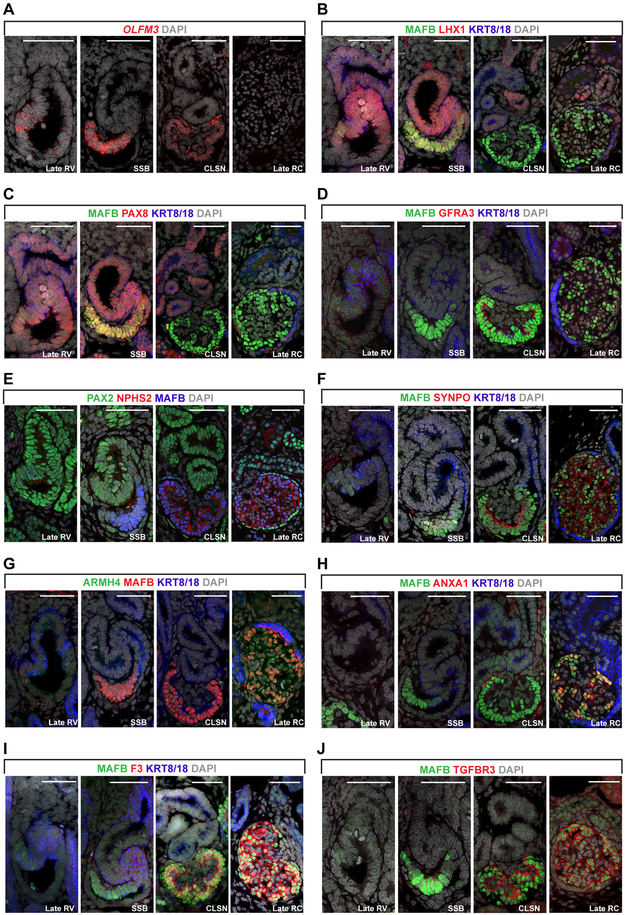
We pinpointed when in podocyte development EP and LP gene profiles were established by combining immunofluorescent detection of target protein, or RNAscope fluorescent in situ hybridization to a target mRNA, on sections of 15-17 week human fetal kidneys. RV and SSB podocyte precursors expressed EP markers (OLFM3, PAX8, and LHX1) but expression ceased prior to or at CLSN stages (Figure 2A-C). GFRA3 expression was restricted to CLSN podocytes (Figure 2D). We also examined the expression of RPS21, encoding Ribosomal Protein S21, using in situ hybridization and observed lower activity in mature renal corpuscle podocytes (Figures S2I and S2J). LP markers (NPHS2, SYNPO, ARMH4, ANXA1, F3, TGFBR3, PLA2R1, COL4A3, and TNNT2) were first detected in SSB or CLSNs, and the highest levels were observed in mature renal corpuscles (Figures 2E-J, S4E and S4F). Interestingly, ANXA1, which is only expressed in the late RC stage, displayed a unique distribution with markedly varying levels in individual podocytes within a single renal corpuscle (Figure 2J). Based on these observations, we estimated the expression windows of EP and LP genes, and categorized them into smaller groups. EP signature genes can be separated into those activated in NPCs and early committed NPCs (group EP1), those activated in RV and highly expressed in SSB-stage podocyte precursors (group EP2), and those transiently expressed in the CLSN podocytes (group EP3) (Figure S5). LP genes maintained high expression levels in late RC-stage podocytes, but can be divided into the early- and late-activated groups (groups LP1 and LP2, respectively) (Figure S6).
Figure 2: In vivo Validation of Early and Late Podocyte Signatures.
(A) Fluorescent in situ hybridization showing expression of EP gene OLFM3 in RV, SSB, CLSN and late RC stages. Scale bars denote 50μm.
(B) to (J) IF stains of EP markers (LHX1, PAX8 and GFRA3), and LP markers (NPHS2, SYNPO, ARMH4, ANXA1, F3, TGFBR3, and MAFB) showing their expression in RV, SSB, CLSN and late RC stages. Scale bars denote 50μm.
RV: renal vesicle, SSB: S-shaped body nephron, CLSN: capillary loop stage nephron, Late RC: late renal corpuscle, EP: early podocyte, LP: late podocyte.
In summary, the single-cell transcriptome profiles define the molecular development of the human podocyte lineage providing a molecular frame-work for analyzing development of PLCs in vitro.
Transcriptomic profiling of podocyte-like cells generated from MAFB-reporting hESCs
To facilitate analysis of in vitro derived PLCs, we generated a MAFB-P2A-eGFP H9 hESC line that places eGFP under the control of the MAFB gene (Figure S3A). MAFB encodes a key transcriptional regulator of podocyte gene expression that has been shown to play a critical role in mouse podocyte development (Sadl et al., 2002). Expression of the mouse and human gene can be detected in podocyte precursors as early as the RV stage (Figure 1B, 2A-J) and MAFB activity continues into mature podocytes in both the adult mouse and human fetal kidney (Lindström et al., 2018a).
MAFB-P2A-eGFP H9 hESCs were differentiated into kidney organoids using existing protocols (Morizane et al., 2015) with minor modifications (Figure 3A). Whole organoid transcriptional profiling at differentiation day (dd) 0, 8, 10, 16, 22 and 28, guided by parallel immunofluorescent analysis with informative markers, showed induction and differentiation of characteristic nephron cell types in the kidney organoids (Figure S3B and S3C). eGFP+ cells first appeared at dd13-14 with a subsequent increase in both the numbers of eGFP+ cells and the level of eGFP fluorescence to dd28 (Figure 3B). As expected, the eGFP signal was tightly correlated with endogenous MAFB activity and colocalized with WT1, a known determinant of podocyte programs (Berry et al., 2015; Chau et al., 2011; Gebeshuber et al., 2013; Hammes et al., 2001; Kann et al., 2015; Moore et al., 1999) (Figure S3D). RNAseq profiling of FACS-isolated MAFB-eGFP+ cells from day 28 organoids confirmed significant enrichment of MAFB and other podocyte marker genes (Figure 3C). Further, hierarchical clustering of eGFP+ cells with human fetal renal corpuscles, immortalized podocytes (imPod) before and after thermoswitch emphasized improved resemblance of hESC-derived podocytes to the in vivo cells, supporting their use as a markedly enhanced tool for in vitro studies (Figure S3F). Additionally, gene ontology (GO) analysis highlighted podocyte-related terms (Figure S3E). Collectively, these data demonstrate the eGFP+ labelling strategy specifically identifies PLCs in kidney organoid culture.
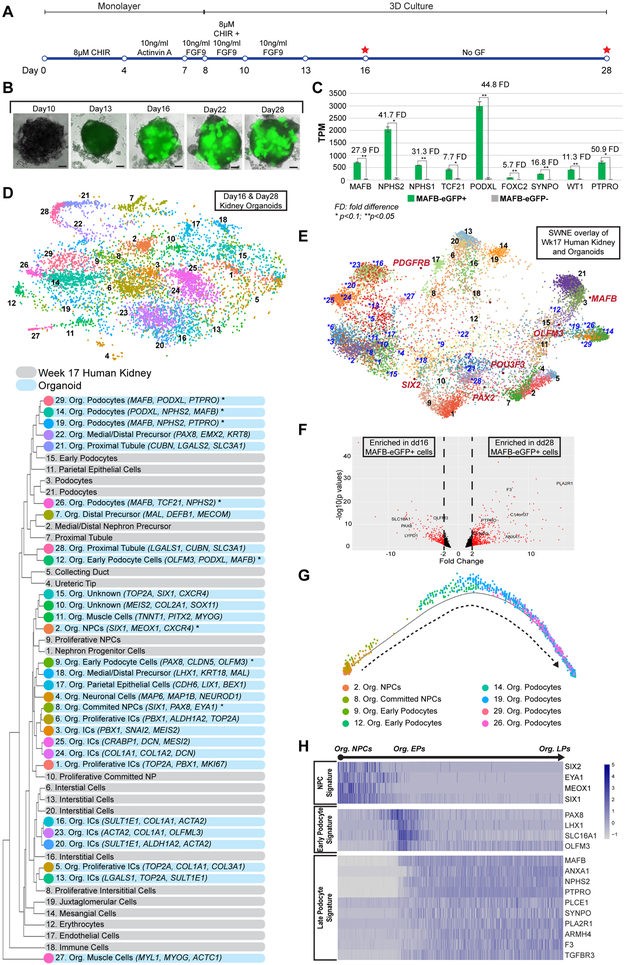
Figure 3: Single-cell RNA-Seq Analyses Showing Transcriptional Changes during in vitro derivation of podocytes.
(A) Schematic diagram summarizing the 28-day protocol to generate kidney organoids from human ESC. Red stars denote the time points selected for further analyses using scRNA-Seq.
(B) Bright-field and (gray scale) endogenous eGFP (green) imaging of the kidney organoids after transition to 3D cultures. Scale bars denote 200μm.
(C) RNA expression levels of known podocyte markers in MAFB-eGFP+ cells versus MAFB-eGFP− cells. Statistical significance is determined using student t-test.
(D) Top: Unsupervised clustering of cells from dd16 and dd28 presented in a tSNE plot. Bottom: Annotation of organoid cluster identities and differentially expressed genes used for cluster identification. Hierarchical clustering suggested relationship among organoid cell clusters and human week 17 cell types. Asterisks (*) mark organoid NPC and podocyte clusters selected for secondary analyses using Monocle.
(E) SWNE overlay of Week 17 human kidney and organoid clusters. Black numbers: Week 17 human kidney clusters; Blue numbers (with asterisks): organoid clusters. “Factor genes” in red are known kidney cell type markers.
(F) Volcano plot with annotations of selected genes that were differentially expressed between dd16 and dd28 MAFB-eGFP+ cells.
(G) Pseudotime trajectory from organoid NPCs to podocytes with cells colored by their cluster identities.
(H) Heatmaps of selected genes whose expression changes along the differentiation trajectory from organoid NPCs to podocytes.
Org: organoid, NPC: nephron progenitor cell, IC: interstitial cell, dd: differentiation day, EP: early podocyte, LP: late podocyte
To investigate whether PLCs develop along a similar trajectory to human fetal podocytes, we performed scRNAseq on organoids at dd16 and dd28 (2,153 cells from dd16, and 5,818 cells from dd28). These two time points were selected as organoids strongly expressed EP signatures (including PAX8, OLFM3, LYPD1, SLC16A1) at dd16, and mature nephron signatures (including F3, PTPRO, C14orf37 or ARMH4, SYNPO, ANXA1) at dd28. (Figure S3C). After quality control filtering, unsupervised clustering of scRNA profiles for 7,200 organoid cells assigned this population to 29 groups (Figure 3D, Figure S3G). Each of these was identified through expression of marker genes, and hierarchical clustering was performed to match these groupings with the most closely related clusters of the week 17 human kidney scRNA-seq (Figure 3D). Similarity Weighted Nonnegative Embedding (SWNE) analysis (Wu et al., 2018) of the in vivo and in vitro cells merged object concurred with the hierarchical clustering and confirmed our identifications of in vitro cell types (Figure 3E).
Organoids comprised cells with transcriptional signatures indicative of interstitial cells (COL1A1+, ALDH1A2+), nephron progenitors (SIX1+, MEOX1+), early developing nephron cells (PAX8+, LHX1+), precursors of the medial/distal segment (SOX9+, MECOM+, EMX2+), proximal tubule (LRP2+, SLC3A1+), parietal epithelial cells (LIX1+, CDH6+), and early and late podocytes (OLFM3+, MAFB+, NPHS2+). Two clusters unexpectedly expressed muscle markers (MYOG+, MYL1+) and another cluster displayed a prominent neuronal signature (MAP6+, NEUROD1+) supporting recent observations of lineage heterogeneity in kidney organoid protocols (Czerniecki et al., 2018; Wu et al., 2018). Two clusters (10 and 15) with mixed but incomplete NPC and IC signatures were left “unidentified”. Of note, organoids are devoid of vascular cells precluding vascular podocyte interactions in podocyte development in vitro.
To examine the PLC lineage more specifically, we identified 2,188 cells with NPC-like or PLC signatures and predicted developmental trajectories between the cell types (Figure S3H, S3I). As branch 3 (B3) contained mostly mitotic cells, we focused on the differentiation trajectory from organoid NPC to PLCs of 1,941 B1 and B2 cells (Figures S3H, S3J, 3G and S3I). We observed similar transcriptional changes to those identified in human podocyte development. EP signature genes (including OLFM3, PAX8, LHX1, SLC16A1) were transiently upregulated as NPC signature genes were deactivated, and downregulated when LP signature genes (including ANXA1, NPHS2, PTPRO, PLCE1, SYNPO, PLA2R1, ARMH4, F3, TGFBR3) were upregulated (Figure 3H). EP and LP genes were also enriched in dd16 and dd28 MAFB-eGFP+ cells, respectively, in bulk RNA-seq analyses (Figure 3F). Immunofluorescent analysis of EP and LP-associated proteins within PLCs at dd16 and dd28 confirmed the EP to LP gene expression transition (Figure 4A-F). In summary, PLCs follow a developmental trajectory similar to that observed in human podocyte development: dd16 PLCs resemble podocytes in SSBs while dd28 PLCs are similar to podocytes of the CLSN or later stages.
Figure 4: Comparison of in vitro derived podocytes to human podocytes.
(A) to (F) IF stains to validate expression of EP markers (PAX8 and LHX1) and LP markers (SYNPO, ARMH4, ANXA1, and F3) in dd16 and dd28 organoid podocytes. Scale bars denote 50μm.
(G) Volcano plot displaying genes either enriched in human LPs or organoid LPs. Blue text highlighting genes plotted in Figure 4H.
(H) Heatmaps showing expression of selected OrgLP-enriched genes (C11orf71, GFRA3, ITIH5, OLFM3, and LEFTY1) and HuLP-enriched genes (PLA2R1, COL4A4, TNNT2, COL4A3 and LOX) along the in vivo or in vitro podocyte differentiation trajectory.
(I) and (J) Feature plots showing expression of ANXA1 and GFRA3 analyzed using scRNA-Seq of Week 17 human fetal kidney (I), or kidney organoids (J).
Comparison of podocyte-like cells and human podocytes
To determine the overlap between PLCs and human podocytes, we performed differential expression gene test between their transcriptomes (Figure 4G) (Supplemental table 1). Of the previously identified EP and LP signature genes in the fetal kidney, most of these (137 EP genes and 84 LP genes) followed a similar early-late developmental time course in vitro (Figures S6). LP genes were absent, or expressed at low level, including genes encoding key extracellular matrix components linked to kidney disease (including COL4A3, COL4A4) and angiogenic factors (including CXCL12, EFNB1) (Figures 4G, 4H and S6) (Supplemental Table 2). Among the EP genes, we validated the expression of OLFM3 and GFRA3 in dd28 podocytes by in situ hybridization and immunofluorescence, respectively (Figure 5D and 5F). We also observed co-expression of EP (GFRA3) and LP genes (ANXA1) in dd28 MAFB+ cells (Figures 4J and 5F). While GFRA3 and ANXA1 were detected at distinct stages of in vivo podocyte development - CLSN and late RC respectively (Figures 2D, 2H and 4I), co-expression in dd28 organoids showed the down-regulation of EP signature genes was incomplete. Dd28 organoid were a mosaic of EP-like (e.g. GFRA3+ANXA1−) and LP-like (e.g. GFRA3-ANXA1+) cell types (Figures 4J and 5F). Further, some LP genes including TNNT2, TNNI1, CAPN2, DCN, IGFBP7, CTGF, CXCL12 and CYR61 were weakly active in dd16 podocytes, but subsequently downregulated in dd28 PLCs (see TNNT2 in Figure 5E). An examination of published scRNA-Seq profiling of kidney organoids (Czerniecki et al., 2018; Wu et al., 2018) also suggests similar mis-expression of EP and LP genes in other organoid datasets in the literature indicating the observations here are likely of broad relevance to in vitro organoid models (Figure S4C and S4D).
Collectively, these findings show a surprising level of maturation of in vitro derived podocytes that is independent of the vascular and mesangial components, and pinpoint aspects of in vitro podocyte programming for further improvement.
Examination of glomerular disease-relevant genes
To examine the applicability of in vitro derived podocytes to glomerular disease studies, we examined expression of genes associated with human glomerulosclerosis and glomerulopathy in the human glomerular cell types (Lepori et al., 2018, Online Mendelian Inheritance in Man, 2018). Forty-three genes were expressed in all three glomerular components: LMNA, COL4A1, MPV17, KANK2 and MYO1E were more highly expressed in the mesangial cells, while DGKE, KHDRBS3, SOX18, SLC7A7, KANK4 and TAL1 were enriched in the endothelial component. Of the twelve genes activated in podocytes, PAX2 and FAH were upregulated only in EPs, while PTPRO, COL4A3, COL4A4, PLCE1, CRB2, NPHS1, TCF21, and ARHGAP24 were first detected in EPs and showed increased activity in LPs (Figure S2K). We surveyed the expression of these disease-associated genes in in vitro EP and LP populations, and highlighted podocyte genes that were detected in the in vitro derived cells to inform future glomerular disease modeling studies (highlighted in red, Figure S2K).
Vasculature interactions in renal corpuscle development
Podocytes produce VEGFA, which promotes concurrent development of the vascular network and mesangial support system (Eremina et al., 2003, 2006). Organoids generated via the Morizane protocol adopted here are largely avascular (Figure S4B). To determine whether co-development with vascular cell types may normalize podocyte maturation programs, dd13-14 organoids were implanted beneath the kidney capsule of NOD/SCID mice (Figure 5A). Live imaging of blood flow facilitated by plasma labeling showed organoids were well vascularized by the host mouse circulatory system (Plvap+) 10 days after implantation (Figure 5B and Supplemental video 1). OLFM3 and GFRA3, EP signature genes which display persistent activity in vitro, continued to be expressed in late vascularized podocytes post transplantation (Figure 5D and 5F), Similarly, we failed to observed normalization of the LP signature gene TNNT2 which remained undetectable in PLCs post transplantation (Figure 5E). However, COL4A3, mutations in which result in glomerular degeneration in Alport’s syndrome was now expressed in vascularized podocytes (Figure 5D). Thus, vascular interactions partially normalized the expression of a critical component of the glomerular basement membrane.
Our scRNA-Seq analyses indicated that pericytes and mesangial cells were absent in the avascular organoids (Figure S4B). In vascularized organoids, MAFB+ HuNu+ in vitro derived podocytes formed renal corpuscle-like structure with PDGFRB+ HuNu+ organoid interstitial cells and PECAM1+ HuNu− mouse ECs (HuNu: Human Nucleus) (Figure 5C). Though TMEFF2 was not detected in PDGFRB+ cells recruited to podocyte clusters (data not showed), expression of GATA3 lends support to human organoid derived interstitial cells adopting an early pericyte identity (Figure 5J). Additionally, the expression of GATA5 in mouse endothelial cells attracted to podocyte clusters supports the activation of an early glomerular-specific vascular signature (Figure 5K).
In summary, in vitro derived podocytes nucleate vascular (host-derived) and mesangial (graft-derived) developmental programs, suggesting full maturation will require an interplay amongst all these cell types.
DISCUSSION
In this study, we extended recent reports from our group and others of human fetal kidney development, with a focus on the programs underlying human podocyte development (Lindström et al., 2018d, Menon et al., 2018). These data provide a benchmark for an unbiased assessment of human podocyte development as well as a resource for mining of regulatory mechanism to improve kidney organoid-directed programming of the kidney’s filtration system.
Development of the human podocyte
Deep profiling identified transcriptomic changes highlighting temporally distinct gene expression profiles in the staged progression of podocyte maturation. The transition from NPC to EP is associated with the down-regulation of a large set of ribosomal protein-encoding genes that remain active in other cell types of the human fetal kidney (Figures S4I and S4J), and the activation of EP signature genes profiled in our previous studies (Lindström et al., 2018c), several of which are critical for the formation of the mouse kidney (Müller et al., 1997; Ohuchi et al., 2000; Xu et al., 1999, 2003). NPCs are highly proliferative. However, proliferation drops as NPC commit to EP precursors, which undergo terminal cell divisions prior to forming mitotically quiescent mature podocytes (Hiromura et al., 2001). Genes of group EP2 are expressed transiently in the RV, SSB, and CLSN podocyte precursors, among which the expression of HEY1, CDH6, PAX8, JAG1, OLFM3, PCDH9 and LHX1 in human podocyte precursors has been documented (Lindström et al., 2018a, 2018d). Though their specific roles in podocyte development have not been examined, Cdh6, Pax8 and Lhx1 have been reported to play important roles in the mouse kidney formation or nephron induction (Bouchard et al., 2002; Kobayashi et al., 2005; Mah et al., 2000). Group EP3 comprises genes upregulated only in the CLSN, possibly as a transient response to endothelial/mesangial invasion and early establishment of signaling center at slit diaphragm. Among these, the transient expression of GFRA3, encoding GDNF receptor α3, hints at the possible importance of GDNF signaling in this specific time window of podocyte and glomerular formation.
Among LP signature genes, group LP1 are detected as early as RV-stage podocyte precursors and maintained expression in late podocyte development, while LP2 genes are highly expressed in late RC podocytes. A large portion of them are disease-relevant ECM protein-encoding genes, some with known functions in mouse podocyte development (including ACTN4, NPHS1, NPHS2, PODXL, SYNPO, TJP1, PLCE1, COL4A3, COL4A4) (Ahvenainen et al., 1956; Asanuma et al., 2006; Boute et al., 2000; Invest., 1970; Itoh et al., 2014; Kaplan et al., 2000; Lepori et al., 2018; Online Mendelian Inheritance in Man, 2018). Our comparative analyses with the mouse podocyte transcriptional profiles highlight novel cross-species shared LP genes. For instance, PHACTR4 (Phosphatase and Actin Regulator 4) has been shown to regulate directional migration of enteric neural crest cells (Zhang et al., 2012), but its importance in podocyte cytoskeletal architecture and migration has not been studied. Additionally, the anti-inflammatory function of ANXA1 (Annexin A1) has been highlighted in diseases of various organ systems (Jong et al., 2016; Ries et al., 2016) and may function in podocyte antigen presenting activities (Goldwich et al., 2013). Notably, we also identified novel human LP-specific genes (including S100A6, MYL9, TPPP6, SNCA, TNNI1, TNNT2, MYL12, PDLIM2, PALLD, GSN) that are associated with cytoskeletal proteins, suggesting species-specific functions in maintaining podocyte architecture.
Temporal expression of EP and LP genes is likely to be orchestrated by temporal programs of transcription factor activity. SIX1, SIX2, EYA1 and MEOX1, which are linked to the maintenance of mouse and human NPCs (Kobayashi et al., 2008; O’Brien et al., 2016; Self et al., 2006; Xu et al., 1999, 2003), could play a role in EP1 gene activation, while PAX8, LHX1, HEY1 activity would be more likely to play a role in EP2 and EP3 programs. Clearly, transient EP-restricted activity of these transcription factors rules out a progressive role in specification of mature podocytes. Transcription factors known to play essential role specifically in podocyte development (e.g. TCF21 and MAFB) were first detected in RV-stage podocyte precursors overlapping with NPC genes (SIX1 and TMEM100). Evidence points to conserved functions for several podocyte specifying transcriptional regulators between mouse and human, with activity continuing into LP programs regulating podocyte morphogenesis (Maezawa et al., 2014; Moriguchi et al., 2006). Signaling inputs from the slit diaphragm established at the capillary loop stage likely regulate LP gene activity (Greka and Mundel, 2012; Schell et al., 2014).
The renal corpuscle is composed of three tightly interacting cellular components: podocytes, vascular and mesangial cells. Disrupting their interactions results in defective renal corpuscle development in the mouse kidney (Eremina et al., 2003, 2006; Kikkawa et al., 2003; Levéen et al., 1994; Lindahl et al., 1998; Soriano, 1994). Our analysis is consistent with a conserved role of VEGF signaling in recruitment and organization of the vasculature. A transient activation of SEMA3A was observed in EPs; Sema3a regulates morphogenesis of podocytes and recruitment of ECs in the mouse kidney (Reidy et al., 2009). We also observed FBLN2 expression in podocytes; Fbln2 is required for vessel wall integrity (Chapman et al., 2010; Xu and Shi, 2014), and recent studies link Fbln2 to promoting vascular invasion (Kim et al., in preparation).
Development of in vitro derived podocytes in comparison with the in vivo counterpart and roles of the vasculature in renal corpuscle development
Recent studies have employed scRNA-Seq technology to identify kidney-like cell types in organoids through common markers (Table 1) (Czerniecki et al., 2018; Hale et al., 2018; Sharmin et al., 2016; Wu et al., 2018; Yoshimura et al., 2017). Our developmental biology-driven analyses deepen an understanding of the in vitro system, highlighting a differentiation trajectory similar to human podocyte development in the fetal kidney. The in vivo data will facilitate efforts to improve cell culture-directed methods of podocyte production and the prediction of regulatory mechanisms in the podocyte program. Of note, despite the absence of vascular or mesangial cell types, comparison of in vitro derived MAFB-eGFP+ cells with the in vivo podocyte developmental program indicated a striking overlap: 137 of 158 EP genes and 84 of 104 LP genes were expressed as expected. Podocytes in organoid transplants attract and interact with host vasculature and interstitial cells in the vicinity upregulate a pericyte signature. Further, there is a normalization of donor human podocyte programs in transplants. Together these results suggest further improvements can be made in vitro through the optimization of mesangial and vascular compartments in the organoid models.
Table 1:
Review recent approaches to assess in vitro derived podocytes.
| Study | Reporter Cell Line for Podocyte Identification |
Comparison with in vivo Podocytes | ||
|---|---|---|---|---|
|
In vivo Sample Type |
Method | Findings | ||
| Sharmin et al., 2016 | NPHS1-GFP iPSC | mouse adult podocytes, human glomeruli | Microarray | Shared genes among in vitro podocytes, human glomeruli and mouse adult podocytes are enriched for podocyte signature genes |
| Wu et al., 2018 | N/A | adult kidney cells | single-cell RNA Seq of organoids, and single-nucleus RNA-Seq of human adult kidney biopsy | Identified podocyte-like cells in organoids by comparing average gene expression of organoid and human fetal kidney cell clusters |
| Hale et al., 2018 | MAFB-BFPiPSC | human matrisome database | mass spectrometry | in vitro 3D-cultured podocytes and human matrisome have shared extracellular matrix proteins |
| Czerniecki et al., 2018 | N/A | Human Fetal Kidney (Menon et al., 2018) |
single-cell RNA Seq of organoids and human fetal kidney | Identified podocyte-like cells in organoids by comparing average gene expression of organoid and human fetal kidney cell clusters |
| Combes et al., 2019 | N/A | Human Fetal Kidney (Lindström et al., 2018) |
single-cell RNA Seq of organoids and human fetal kidney | Identified podocyte-like cells in organoids by comparing average gene expression of organoid and human fetal kidney cell clusters |
| Yoshimura et al., 2019 | NPHS1-GFP iPSC | Human Adult Podocytes | RNA Seq | Identified shared and different gene expression profiles between in vitro and in vivo podocytes |
| Tran et al. (our study) |
MAFB-eGFP H9 hESC | Human Fetal Kidney | single-cell RNA Seq of organoids and human fetal kidney | Identifed shared transcriptional signatures between in vitro and in vivo podocyte development, pinpointed abnormal gene expression in vitro, and highlighted the potential of in vitro derived podocytes as a model for human podocyte development and disease studies |
Our organoid approach was centered around generating the metanephric component of the kidney (Morizane and Bonventre, 2016; Morizane et al., 2015), and the absence of vascular cells can be due to distinct spatial and temporal developmental origin. Additionally, vascular development is heavily dependent on VEGF and a low physiological oxygen level (Kusuma et al., 2014); current organoid growth conditions may not favor growth of low numbers of vascular cells. Recently, Czerniecki and colleagues described an approach to generate kidney organoids with endothelial cells (Czerniecki et al., 2018). Our studies can facilitate assessment of glomerular endothelial specification as well as effects of endothelial cells on podocyte maturation and mesangial cell commitment in vitro. Interestingly, flow in bioengineered kidney organoid chips enhances endothelial growth though the effects on the maturation of renal corpuscle cell-types is not clear (Homan et al., 2019). Predictions of ligand-receptor interactions among the RC cell types may help in these efforts (Supplementary Table 2).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Dr. Andrew P. McMahon (amcmahon@med.usc.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Kidney Studies
De-identified human fetal tissue was obtained from elective terminations with informed consent with the approval of Institutional Review Boards of both Children’s Hospital of Los Angeles and the Keck School of Medicine of the University of Southern California. Gestational age was determined using guidelines from the American College of Obstetricians and Gynecologists using a combination of ultrasound and last menstrual measurements (O’Rahilly and Müller, 2010; O’Rahilly et al., 1987; Strachan et al., 1997). Samples were delivered on ice at 4°C in high glucose DMEM (Gibco) supplemented with 10% fetal bovine serum (Sigma Aldrich) and 25mM HEPES (Gibco).
Human embryonic stem cell line (hESC)
H9 hESC line (female) was obtained from WiCell (WA09). The MAFB-P2A-eGFP hESC line was generated by knocking-in the P2A-eGFP cassette into the 3’ end of MAFB locus by employing CRISPR-Cas9 technology. The donor plasmid, gRNA and Cas9 protein were introduced into H9 cells via electroporation (Thermofisher, MPK5000S). For single-cell cloning, the targeted H9 cells were dissociated into single cells using Accutase (StemCell Technologies), and 200 cells were seeded in a Geltrex-coated 100-mm dish with 10ml of mTeSR (supplemented with 10μm Y27632). Single cell-derived colonies were picked up and expanded for Southern blot validations. Key sequences important for cell line generation are listed in Supplemental Table 2 under “DNA Sequences (cell line)”.
METHOD DETAILS
hESC Maintenance and Differentiation to Generate Kidney Organoids
MAFB-P2A-eGFP H9 hESC cells were cultured in StemFit media (Ajinomoto, ASB01-R) supplemented with 10ng/ml of FGF2 (R&D, 273-F9) on Geltrex-coated plates (Geltrex from ThermoFisher, A1413302). When cells reach 60% confluent, differentiation was started following the Bonventre protocol with minor modification: 4 days of 8μM CHIR99021 (Sigma Aldrich, SML1046) treatment, followed by 3 days of 10ng/ml ActivinA (R&D, 338-AC-050) incubation, and 1 day of 10ng/ml FGF9 incubation. On day8, the cells were dissociated using TrypLE dissociation enzyme (Gibco, 12563011), and cell number was acquired. 75,000 cells/well were seeded on a 96-well ultralow-attachment plate (Corning, CLS7007) to form aggregates. The aggregates were formed in 3uM CHIR and 10ng/ml FGF9. On Day10, the media was changed to basal media with 10ng/ml FGF9. From Day13 to Day28, the aggregates were cultured in growth factor-free basal media. All differentiation steps were done using basal differentiation media (Advanced RPMI 1640 (Gibco, 12633020) + 1X Glutamax (Gibco, 35050079) + 1% Penicillin-Streptomycin (Invitrogen, 15070063).
Immunofluorescent analyses
Before the immuno-detection was performed, frozen sections tissue was thawed at room temp for 10 minutes. Antigen retrieval was done in 1X Citrate Buffer pH 6.0 (Sigma) in a pressure cooker. The slides were washed with water and air dried for 5 min. The slides were incubated with 1.5% Seablock (ThermoFisher) in PBS + 0.25%TritonX block buffer for 1 hour at room temperature, and in primary antibody mixture (diluted in block buffer) at 4°C o vernight. Primary antibodies used in the study are listed as follow: LHX1 (R&D, MAB2725,1:300), MAFB (R&D, MAB3810, 1:500), MAFB (Santa Cruz, sc-10022, 1:100), PAX8 (abcam, ab189249, 1:1000), KRT8/18 (DSHB, troma-1; 1:50), NPHS2 (abcam, ab50339, 1:10000), SYNPO (R&D, MAB8977, 1:300), TGFBR3 (R&D, AF-242-PB, 1:300), ANXA1 (Cell Signaling, 32934, 1:200), ARMH4/C14orf37 (Sigma Aldrich, HPA001789, 1:300), WT1 (abcam, ab89901, 1:1000), CUBN (Santa Cruz, sc-20607, 1:300), CDH1(Biosciences, 610182, 1:300 ), PDGFRB (abcam, ab32570, 1:500), VEGFR2 (Cell Signaling, 2479,1:150), GFRA3 (R&D, AF670, 1:300), PAX2 (R&D, AF3364, 1:500), F3 (R&D, AF2339, 1:500), PLVAP ( BioRad, MCA2539GA,1:300), GATA3 (R&D, AF2605, 1:300), PECAM1 (BD Pharmingen, BDB550274, 1:300), EHD3 (Novus Biologicals, NBP2-31896, 1:300), FOXC2 (R&D, AF6989, 1:300). Secondary antibodies conjugated with AlexaFluor 488, 555, 594, and 647 were purchased from Molecular Probes. Slides were incubated with 1 μg/ml Hoechst 33342 (Molecular Probes) in PBS for 5 min to stain the nuclei. Sections were mounted in ProLong Gold Antifade Reagent (Life technologies) and imaged at 10X or 63X using the Leica SP8 confocal microscope.
In situ hybridization
We utilized RNAscope® Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, Inc.) to perform fluorescent in situ hybridization. The slides were prepared as described above. Briefly, the tissues were treated with hydrogen peroxide and protease, then hybridized with RNAscope probes for 2 hours at 40°C using the HybEZ oven (Advanced Cell D iagnostics, Inc.). The probes were then amplified and detected with tyramide signal amplification fluorescence. To detect nuclei, the slides were incubated with 1 μg/ml Hoechst 33342 (Molecular Probes). The tissue was imaged at 63X using the Leica SP8 confocal microscope. The catalog numbers of probes from Advanced Cell Diagnostics, Inc. used in this work are listed as follow: MAFB (400801-C2), RPS21 (511381-C3), OLFM3 (549051-C3), PLA2R1 (524581), COL4A3 (461861) and TNNT2 (518991-C3).
Single-Cell RNA-Sequencing and Analyses of Human Fetal Kidneys
Human kidney samples were embedded in 4% agarose block, and were sectioned to 300-μm thick sections using the vibratome (Leica VT1000S). The vibratome sections were further dissected manually to separate the outer cortex from the inner cortex. The specimens were enzymatically digested using collagenase A/pancreatin. Live single cells (DAPI− DRAQ5+) were selected using fluorescence-activated cell sorting (FACS) and were captured by microfluidic droplets using the Chromium 10x genomics platform as described previously (Lindström et al., 2018c). Quality control, mapping to reference genome and count table assembly of the libraries were performed using CellRanger 2.1 (10x Genomics) (Lindström et al., 2018c). The data is available at GEO accession number GSE124472.
The datasets from Zone 1 and Zone 2 cells were merged into one for initial analysis using the MergeSeurat function in Seurat package (Satija et al., 2015). The merged dataset was log-normalized using NormalizeData function. The ScaleData function was used to scale and center genes in the dataset, regressing the following variables against each gene: nUMI, nGene, saturation, and orig.ident. Quality control filtering was performed on the cells using the following criteria: saturation ≥70%, ≤5% mitochondrial genes. Principle components (PCs) were calculated using the RunPCA function, and statistically significant PCs (p < 0.01, Jackstraw criteria - Chung and Storey, 2015) were selected to use for clustering. We performed clustering using mclust package (Scrucca et al., 2016) with 41 PCs. Cluster assignments were initialized by hierarchical clustering (“hcVVV” initialization in mclust). The number of clusters were decided based on Bayesien Information Criterion (BIC). Cell types were identified using known markers as well as GO term analyses of top differentially expressed genes.
Single-Cell RNA-Sequencing and Analyses of Human Kidney Organoids
Two dd16 and two dd28 kidney organoids of the same differentiation batch were dissociated by incubating with the Accumax™ Cell Dissociation Solution (Innovative Cell Technologies, Inc.) for 20min with gentle pipetting using P1000 tips. The dissociation enzyme was neutralized by autoMACS™ Running Buffer (Milteny Biotec), and the cells were centrifuged at 300 g for 5min. The pellet was resuspended in autoMACS™ Running Buffer (Milteny Biotec). The cell solution was run through a 40-μm cell strainer (Falcon) before live single cells (DAPI− DRAQ5+) were selected using FACS. The single cells were captured and subjected to single-cell sequencing using the Chromium 10x genomics platform (Lindström et al., 2018c). The two dd16 datasets and two dd28 datasets were merged with the Seurat package using MergeSeurat function. Quality control and cluster finding were performed as described above for the human kidney datasets.
Pseudotime Reconstruction of Lineages
NPCs, early induced NPCs, early podocytes and podocytes were subset from the merged Zone1/Zone2 dataset using the Seurat package. We used the Monocle2 package to reconstruct differentiation trajectory (Qiu et al., 2017). Cells assigned by Monocle to the “proliferative” branch were selected against as described in (Lindström et al., 2018d). Non-proliferative cells were re-analyzed to build a developmental trajectory.
Differentially Expressed Gene Test between Two Podocyte Clusters
Differentially expressed genes from podocyte clusters in each dataset were obtained using the FindMarkers function in Seurat (Bimod test) (Satija et al., 2015). The following comparisons were performed: Week 15 (clusters 5 and 7) Zone 1 versus Zone2, Week 17 (clusters 3, 15 and 21) Zone 1 versus Zone 2, or Week 17 Late Podocyte (clusters 3 and 21) versus Organoid Podocyte (clusters 14, 19, 26 and 29) (representing “Podocyte Differences”). To account for “batch” variation due to tissue handling and origins of tissues, we selected non-epithelial clusters from the datasets (non-podocytes and PAX2-negative clusters – listed in table below), and performed differentially expressed gene test to obtain gene lists representing “Background Differences” (Supplemental Table 2). Differentially expressed genes specific to podocyte were identified as “Podocyte Differences” genes absent from the “Background Differences” list, and were presented in volcano plots.
| Samples | Clusters selected for “Podocyte Differences” |
Clusters selected for “Background Differences” |
|---|---|---|
| Week 15 | 5, 7 | 6, 11, 14, 15, 16, 17, 18, 19, 20, 21 |
| Week 17 | 3, 15, 21 | 6, 8, 12, 13, 14, 16, 17, 18, 19, 20 |
| Organoid | 14, 19, 26, 29 | 1,3, 4, 5, 6, 11, 13, 16, 20, 23, 24, 25, 27 |
EP and LP gene list generation
To obtain the list of representative EP and LP genes (Figure S2C), we first inferred a set of genes that changed significantly across the differentiation trajectory using the differentialGeneTest function in Monocle2 (q-value < 0.05) (Qiu et al., 2017). We then filtered this list by selecting only genes correlated with known EP and LP markers: For a set of previously profiled markers (Figure S2D) we obtained the 50 genes with highest Pearson correlation with each one and selected, among the union of all correlated genes, the ones that were significant in the trajectory. Each resulting gene was visualized in a trajectory heatmap and manually curated to generate the lists of 158 EP genes and 104 LP genes.
Receptor-Ligand analysis
To create a list of candidate receptor-ligand pairs between all clusters we used the list of receptor-ligand pairs proposed by Ramilowski et al., 2015. Given two clusters i and j, we calculated a score Sk(i → j) for each receptor-ligand pair (rK, lk) given by:
where p(i, g) is the proportion of single-cells in cluster i whose expression of gene g is nonzero. We set a significance cutoff S > 0.25 by visual inspection of the score distribution.
Hierarchical clustering of human week 17 clusters and organoid cell clusters
Using the Seurat package, the Week 17 merged dataset was combined with the Kidney Organoid merged dataset (using MergeSeurat function) after identification of all clusters, and 70 PCs were calculated. To account for transcriptional differences due to origins of tissue, we examined all PCs using boxplots for distributions of cell loadings of each cluster; we removed PC2, which was homogeneous within one origin, but heterogeneous between human and organoid origins. Hierarchical clustering was then performed using the BuildClusterTree function in Seurat with PCs 1 to 70 excluding PC2.
SWNE analysis of the merged human week 17 and organoid cell clusters
The Seurat object of merged human week 17 and organoid cells generated above was used as the input for SWNE (Wu et al., 2018). To select genes as input for SWNE (var.genes), we extracted the genes with highest loadings in each significant principal component of our joint analysis (PCs 1 to 70, excluding PC2). We sorted the squared loadings of each gene in decreasing order and kept the genes whose cumulative squared sum is smaller than 0.5. Our candidate gene list is the union of all significant genes across all principal components. Using var.genes, SWNE embedding was generated with 20 k factors. The plot was generated with alpha.exp = 1.6, snn.exp = 1, n_pull = 8, and 6 embedded factor genes (SIX2, PDGFRB, POU3F3, PAX2, OLFM3, and MAFB).
mRNA-Seq of MAFB-eGFP+ cells from kidney organoids
Dd16 and dd28 kidney organoids from at least 3 differentiation batches were dissociated using the Accumax™ Cell Dissociation Solution (Innovative Cell Technologies, Inc.) and resuspended in autoMACS™ Running Buffer (Milteny Biotec) for FACS as described above. Live eGFP+ single cells were selected using stringent gate to separate the brightest eGFP+ DAPI− DRAQ5+ cells from eGFP− DAPI− DRAQ5+ cells. 50,0000 – 100,0000 cells collected from FACS were pelleted at 4°C, 2000 g for 10min, and resuspended in RLT buffer (RNeasy MicroKit, Qiagen). The RNeasy kit was used to purify RNA from the cells following the manufacturer’s protocol. mRNA-Seq libraries were prepared with KAPA Stranded mRNA-Seq Kit (Kapa Biosystems, #KK8420). The libraries were subsequently sequenced with Illumina NextSeq500 model with pair-end 75 bp setting.
mRNA-Seq of human fetal renal corpuscles
Human fetal kidneys of week 16-17 were minced and ground using the seal end of a 3-ml syringe plunger (BD Medical) on a 100-μm cell strainer (Falcon). Dissociated tissue passing through the strainer was collected in DMEM, and the flow-through was filtered using a 70-μm cell strainer (Falcon). Renal corpuscles trapped on the 70-μm strainer were collected in DMEM, concentrated by centrifuging at 1000 g for 5 min, and lysed in RLT buffer for RNA extraction (RNeasy MicroKit, Qiagen). The 70-μm strainer flow-through portion was also collected in RLT buffer for mRNA profiling. mRNA-Seq libraries were prepared as described above.
mRNA-Seq of immortalized podocytes
Human immortalized podocytes were obtained from Dr. Moin Saleem’s group (University of Bristol) and cultured following the existing protocols (Saleem et al., 2002; Lan et al., 2012). Cells were collected before thermoswitch (cultured at 33°C) an d after thermoswitch (cultured at 37°C) in RLT buff er (RNeasy MicroKit, Qiagen) for RNA extraction. mRNA-Seq libraries were prepared as described above.
mRNA-Seq data analysis
mRNA-Seq reads were aligned with TopHat2 (Kim et al., 2013; Trapnell et al., 2009) to hg38 assembly. The aligned reads were quantified with Partek Genomics Suite software, version 6.6 (St. Louis, MO, USA) to obtain both RPKM and counts at gene level. TPM was calculated by dividing the RPKM by the mapping ratio of the library to exon regions of the genome. To identify differentially expressed genes, count tables of the two groups of data being compared were first processed through DESeq2 (Love et al., 2014) to obtain the p values from negative binomial tests which evaluates the significance of difference by read counts. Next, the TPM tables of the same samples were used to calculate the two-sample independent t-test p values. Both p values were used in screening for differentially expressed genes (p < 0.05 in both cases). Only genes with TPM > 5 in the more highly expressed samples were selected. Top 2000 most variably expressed genes of all samples were selected and their TPM values were plotted with 'heatmap2' in the 'gplot' package of R.
Gene-list GO-term Queries
Differentially expressed genes or gene module lists were queried by ToppGene (Chen et al., 2009) identifying Biological Processes and Coexpression Atlas.
Glomerular disease-related gene expression analysis
Genes associated with human “glomerulosclerosis” and “glomerulopathy” were obtained from the Online Mendelian Inheritance in Man database and Lepori et al., 2018. The average expression levels of OMIM genes were calculated using the AverageExpression function in Seurat for the following cell populations: Week 17 early and late podocyte, Week 17 mesangial cells, Week 17 glomerular endothelial cells, Organoid early and late podocytes. Genes with average expression higher than 0 were presented using the DotPlot function in Seurat.
Analysis of published scRNASeq dataset
Datasets from the Czerniecki et al. (2018) and Wu et al. (2018) studies were acquired through GEO (GSE109718 and GSE118184, respectively). The Seurat package was used to obtain feature plots displaying expression of genes of interest.
Analysis of microarray datasets
Microarray data was acquired through GEO (GSE17142, GSE17143, GSE17145). The Oligo package (Carvalho et al., 2018) was used to process the CEL files. The Limma package (Ritchie et al., 2015) was used to normalize the data sets and perform statistical inference. To find differentially expressed genes between two given groups, we used the following threshold: DEseq value > 6, fold change > 3 and p-value < 0.01.
Renal capsule transplant
The procedure was adapted from (Yoshimura et al., 2017). Week 8-12 NOD.CB17-Prkdc<SCID>/J mice were anesthetized with Ketamine/Xylazine. Surgery site at the dorsal flank was shaved and swabbed with Proviodine/alcohol. An 8-10 mm incision was made, and the fascia was incised before the kidney was externalized. The kidney capsule was kept moistened with sterile saline during the procedure. A small incision was made in the outer membrane of the renal capsule at the caudal end, using a sharp 24g needle and the sub capsular space is flushed with 1ml of basal differentiation media using a blunted 24g needle 30g needle (B30-50, Strategic Applications, Inc.) attached to a 1 ml syringe. Two agarose rods (2mm long, 0.5mm diameter) were pushed into the sub capsular space in the shape of an open V using forceps. A 20g indwelling needle (SURFLO® PTFE I.V. Catheter needle, TESR-OX2025CA, VWR) attached to a 1 ml syringe and draw up 3-4 dd13-14 organoids basal differentiation media into the needle. The indwelling needle was inserted under the renal capsule to place organoids between the agarose rods. The capsule incision was cauterized, and the kidney was replaced into the retroperitoneum. The muscle layer was sutured, and the skin was closed with wound clips.
Intravital multiphoton microscopy
NOD SCID mice were anesthetized with isoflurane (3% in induction chamber and 1.5-1.8% for maintenance through nasal cone). Alexa594-conjugated bovine serum albumin (ThermoFisher) was administered iv. by retro-orbital injections to label the circulating plasma (30μL iv. bolus from 10 μg/ml stock solution). Subcapsular organoid implants in left kidney were exteriorized through a 8-10 mm incision under sterile conditions. In all procedures, body temperature was maintained by using a homeothermic blanket (Harvard Apparatus, Holliston, MA). The mouse was placed on the stage of an inverted microscope with the exposed kidney placed in a coverslip-bottomed chamber bathed in normal saline.
Image acquisition of intact organoids was performed in vivo using a Leica SP8 DIVE multiphoton confocal fluorescence imaging system (Leica Microsystems) powered by a Coherent Discovery laser system at 860 nm excitation (Coherent) and a DMI8 inverted microscope’s external Leica 4Tune spectral hybrid detectors (emission at 530±10nm for eGFP and 600nm for Alexa594-Albumin). Images were acquired in 12-bit, 512×512 pixel using a 40X Leica water immersion objective (NA 1.1). (Hackl et al., 2013; Kang et al., 2006). All animal protocols were approved by the Institutional Animal Care and Use of Committee at the University of Southern California.
DATA AND SOFTWARE AVAILABILITY
The bulk and single-cell RNA sequencing datasets are available under GEO accession numbers: GSE127344, GSE124392, and GSE124472.
Supplementary Material
SUPPLEMENTAL TABLE 2
A. Summary of RNA seq samples
B. Differentially Expressed Genes related to Figures 1C, S1A, 3D
C. GO analyses related to Figures 1C, S1A, 3D
SUPPLEMENTAL TABLE 2
A. Links to protein expression data of LP genes on Human Protein Atlas (related to Figure S5)
B. Mouse Podocyte MicroArray analyses (related to Figures S2G and S2H)
C. Tables of TPM values from mRNA-seq analyses of: Organoid differentiation progression, MAFB-eGFP+ and MAFB-eGFP− cells (Related to Figures 3 and S3)
D. Differentially Expressed Genes of Podocytes comparing: Zone1 vs Zone2 (Related to Figures S1F and S1G), in vivo vs in vitro (Related to Figure 4G)
E. Ligand-Receptor Predictions of Glomerular components (Related to Discussion)
F. DNA Sequences important for MAFB-P2A-eGFP H9 hESC generation (Related to Figure S3 and STAR Method – section Human embryonic stem cell line (hESC).
SUPPLEMENTAL VIDEO 1 (Related to Figure 5B)
Multiphoton live imaging of in vitro derived podocytes (green) transplanted under the kidney capsule of an immunocompromised mouse after Alexa 594-conjugated Albumin dye (red) injection. Scale bars denote 10μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LHX1 | R&D | MAB2725, RRID:AB_2135636 |
| MAFB | R&D | MAB3810, RRID:AB_2137675 |
| PAX8 | Abcam | ab189249, |
| KRT8/18 | DSHB | Troma-1 |
| NPHS2 | abcam | ab50339, RRID:AB_882097 |
| SYNPO | R&D | MAB8977 |
| TGFBR3 | R&D | AF-242-PB, RRID:AB_354417 |
| ANXA1 | Cell Signaling | 32934 |
| ARMH4/C14orf37 | Sigma Aldrich | HPA001789, RRID:AB_1078328 |
| WT1 | abcam | ab89901, RRID:AB_2043201 |
| CUBN | Santa Cruz | sc-20607, RRID:AB_2230037 |
| CDH1 | Biosciences | 610182, RRID:AB_397581 |
| PDGFRB | Abcam | ab32570, RRID:AB_777165 |
| VEGFR2 | Cell Signaling | 2479, RRID:AB_2212507 |
| GFRA3 | R&D | AF670, RRID:AB_2110447 |
| PAX2 | R&D | AF3364, RRID:AB_10889828 |
| F3 | R&D | AF2339, RRID:AB_442150) |
| Plvap | BioRad | MCA2539GA, RRID:AB_931734 |
| GATA3 | R&D | AF2605, RRID:AB_2108571 |
| PECAM1 | BD Pharmingen | BDB550274 |
| EHD3 | Novus Biologicals | NBP2-31896 |
| FOXC2 | R&D | AF6989, RRID:AB_10973139 |
| Fluorescent in situ hybridization probes | ||
| MAFB | Advanced Cell Diagnostics, Inc. | 400801-C2 |
| RPS21 | Advanced Cell Diagnostics, Inc. | 511381-C3 |
| OLFM3 | Advanced Cell Diagnostics, Inc. | 549051-C3 |
| PLA2R1 | Advanced Cell Diagnostics, Inc. | 524581 |
| TNNT2 | Advanced Cell Diagnostics, Inc. | 518991-C3 |
| COL4A3 | Advanced Cell Diagnostics, Inc. | 461861 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CHIR99021 | Sigma Aldrich | SML1046-25MG |
| Recombinant Human/Mouse/Rat Activin A Protein | R&D | 338-AC-050 |
| Recombinant Human FGF-basic (154 a.a.) | PeproTech | 10771-940 |
| Recombinant Human FGF-9 Protein | R&D | 273-F9 |
| Recombinant Human VEGF 165 Protein | R&D | 293-VE-010 |
| StemFit Basic02 | Ajinomoto | Basic02 |
| Y27632 | StemCell Technologies | 72302 |
| Accutase | StemCell Technologies | 07920 |
| Cas9 Protein with NLS | PNA Bio | CP02 |
| mTeSR | StemCell Technologies | 85850 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | 10X Genomics | 120237 |
| Chromium Single Cell A Chip Kit | 10X Genomics | 120236 |
| Chromium i7 Multiplex Kit | 10X Genomics | 120262 |
| RNeasy | QIAGEN | 74104 |
| RNAscope® Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics, Inc. | 323100 |
| KAPA Stranded mRNA-Seq Kit | Kapa Biosystems | KK8420 |
| Deposited Data | ||
| Bulk RNA-Seq of human fetal renal corpuscles and whole kidney (minus renal corpuscles) | Gene Expression Omnibus | GSE127344 |
| Bulk RNA-Seq of organoids collected at various differentiation timepoints | Gene Expression Omnibus | GSE124392 |
| scRNA-Seq of human fetal kidneys and organoids (day 16 and day 28) | Gene Expression Omnibus | GSE124472 |
| Experimental Models: Cell Lines | ||
| H9hESC | WiCell | WA09 |
| Experimental Models: Organisms/Strains | ||
| Immunocompromised mice | The Jackson Laboratory | NOD.CB17-Prkdc<SCID>/J, RRID:MGI:5652139 |
| Software and Algorithms | ||
| Seurat | Satija et al., 2015 | http://satijalab.org/seurat/ |
| Similarity Weighted Nonnegative Embedding (SWNE) | Wu et al., 2018 | https://github.com/yanwu2014/swne |
| Monocle 2.0 | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release/ |
| Cell Ranger 2.1 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
HIGHLIGHTS.
scRNA-seq identifies distinct transcriptional profiles of human podocyte development
Comparative analyses show similarities and differences of in vitro-derived podocytes
Implantation study suggests vascular interactions improve in vitro podocyte maturation
ACKNOWLEDGEMENTS
We thank the McMahon lab members for insightful discussions. Work in A.P.M.'s laboratory was supported by grants from the NIH (DK054364 and DK110792).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahvenainen EK, Hallman N, and Hjelt L (1956). Nephrotic syndrome in newborn and young infants. 2, 227–241. [PubMed] [Google Scholar]
- Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, and Mundel P (2006). Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol 8, 485–491. [DOI] [PubMed] [Google Scholar]
- Bantounas I, Ranjzad P, Tengku F, Silajdžić E, Forster D, Asselin M-CC, Lewis P, Lennon R, Plagge A, Wang Q, et al. (2018). Generation of Functioning Nephrons by Implanting Human Pluripotent Stem Cell-Derived Kidney Progenitors. Stem Cell Reports 10, 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg C, Ritsma L, Avramut C, Wiersma L, van den Berg B, Leuning D, Lievers E, Koning M, Vanslambrouck J, Koster A, et al. (2018). Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports 10, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R, Ozdemir D, Aronow B, Lindström N, Dudnakova T, Thornburn A, Perry P, Baldock R, Armit C, Joshi A, et al. (2015). Deducing the stage of origin of Wilms’ tumours from a developmental series of Wt1-mutant mice. Dis Model Mech 8, 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubüser A, and Busslinger M (2002). Nephric lineage specification by Pax2 and Pax8. Gene Dev 16, 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, and Antignac C (2000). NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet 24, 349–354 [DOI] [PubMed] [Google Scholar]
- Brunskill E, Georgas K, Rumballe B, Little M, and Potter S (2011). Defining the Molecular Character of the Developing and Adult Kidney Podocyte. Plos One 6, e24640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Sicot F-X, Davis E, Huang J, Sasaki T, Chu M-L, and Yanagisawa H (2010). Fibulin-2 and Fibulin-5 Cooperatively Function to Form the Internal Elastic Lamina and Protect From Vascular Injury. Arteriosclerosis Thrombosis Vasc Biology 30, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y-Y, Brownstein D, Mjoseng H, Lee W-C, Buza-Vidas N, Nerlov C, Jacobsen S, Perry P, Berry R, Thornburn A, et al. (2011). Acute Multiple Organ Failure in Adult Mice Deleted for the Developmental Regulator Wt1. Plos Genet 7, e1002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, and Storey J (2015). Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki S, Cruz N, Harder J, Menon R, Annis J, Otto E, Gulieva R, Islas L, Kim Y, Tran L, et al. (2018). High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 22, 929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina V, Sood M, Haigh J, Nagy A, Lajoie G, Ferrara N, Gerber H-P, Kikkawa Y, Miner J, and Quaggin S (2003). Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. Journal of Clinical Investigation 111, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina V, Cui S, Gerber H, Ferrara N, Haigh J, Nagy A, Ema M, Rossant J, Jothy S, Miner J, et al. (2006). Vascular Endothelial Growth Factor A Signaling in the Podocyte-Endothelial Compartment Is Required for Mesangial Cell Migration and Survival. J Am Soc Nephrol 17, 724–735. [DOI] [PubMed] [Google Scholar]
- Freedman B, Brooks C, Lam A, Fu H, Morizane R, Agrawal V, Saad A, Li M, Hughes M, Werff R, et al. (2015). Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nature Communications 6, 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeshuber C, Kornauth C, Dong L, Sierig R, Seibler J, Reiss M, Tauber S, Bilban M, Wang S, Kain R, et al. (2013). Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med 19, 481. [DOI] [PubMed] [Google Scholar]
- Goldwich A, Burkard M, Ölke M, Daniel C, Amann K, Hugo C, Kurts C, Steinkasserer A, and Gessner A (2013). Podocytes Are Nonhematopoietic Professional Antigen-Presenting Cells. J Am Soc Nephrol 24, 906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greka A, and Mundel P (2012). Cell biology and pathology of podocytes. Annu. Rev. Physiol 74, 299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl MJ, Burford JL, Villanueva K, Lam L, Suszták K, Schermer B, Benzing T, and Peti-Peterdi J (2013). Tracking the fate of glomerular epithelial cells in vivo using serial multiphoton imaging in new mouse models with fluorescent lineage tags. Nat. Med 19, 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes A, Guo J-K, Lutsch G, Leheste J-R, Landrock D, Ziegler U, Gubler M-C, and Schedl A (2001). Two Splice Variants of the Wilms’ Tumor 1 Gene Have Distinct Functions during Sex Determination and Nephron Formation. Cell 106, 319–329. [DOI] [PubMed] [Google Scholar]
- Hiromura K, Haseley LA, Zhang P, Monkawa T, Durvasula R, Petermann AT, Alpers CE, Mundel P, and Shankland SJ (2001). Podocyte expression of the CDK-inhibitor p57 during development and disease. Kidney Int. 60, 2235–2246. [DOI] [PubMed] [Google Scholar]
- Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, et al. (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invest. M-A (1970). Glomerular polyanion. Alteration in aminoucleoside nephrosis. Lab. Invest [PubMed] [Google Scholar]
- Itoh M, Nakadate K, Horibata Y, Matsusaka T, Xu J, Hunziker W, and Sugimoto H (2014). The structural and functional organization of the podocyte filtration slits is regulated by Tjp1/ZO-1. PLoS ONE 9, e106621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong R, Leoni G, Drechsler M, and Soehnlein O (2016). The advantageous role of annexin A1 in cardiovascular disease. Cell Adhes Migr. 11, 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JJ, Toma I, Sipos A, McCulloch F, and Peti-Peterdi J (2006). Quantitative imaging of basic functions in renal (patho)physiology. Am. J. Physiol. Renal Physiol. 291, F495–502 [DOI] [PubMed] [Google Scholar]
- Kann M, Ettou S, Jung Y, Lenz M, Taglienti M, Park P, Schermer B, Benzing T, and Kreidberg J (2015). Genome-Wide Analysis of Wilms’ Tumor 1-Controlled Gene Expression in Podocytes Reveals Key Regulatory Mechanisms. J Am Soc Nephrol 26, 2097–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodríguez-Pérez JC, Allen PG, Beggs AH, et al. (2000). Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet 24, 251–256. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Virtanen I, and Miner JH (2003). Mesangial cells organize the glomerular capillaries by adhering to the G domain of laminin alpha5 in the glomerular basement membrane. J. Cell Biol 161, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Refaeli I, Brooks C, Jing P, Gulieva R, Hughes M, Cruz N, Liu Y, Churchill A, Wang Y, et al. (2017). Gene-Edited Human Kidney Organoids Reveal Mechanisms of Disease in Podocyte Development. STEM CELLS 35, 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kwan K-M, Carroll T, McMahon A, Mendelsohn C, and Behringer R (2005). Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 132, 2809–2823. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Valerius, Mugford J, Carroll T, Self M, Oliver G, and McMahon A (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma S, Peijnenburg E, Patel P, and Gerecht S (2014). Low oxygen tension enhances endothelial fate of human pluripotent stem cells. Arterioscler. Thromb. Vasc. Biol 34, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan N, Saleem M, and Mathieson P (2012). Podocyte culture: Tricks of the trade. Nephrology 17, 525–531. [DOI] [PubMed] [Google Scholar]
- Lee JW, Chou C-LL, and Knepper MA (2015). Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J. Am. Soc. Nephrol 26, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepori N, Zand L, Sethi S, Fernandez-Juarez G, and Fervenza F (2018). Clinical and pathological phenotype of genetic causes of focal segmental glomerulosclerosis in adults. Clin Kidney J 11, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, and Betsholtz C (1994). Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 8, 1875–1887. [DOI] [PubMed] [Google Scholar]
- Lindahl P, Hellström M, Kalén M, Karlsson L, Pekny M, Pekna M, Soriano P, and Betsholtz C (1998). Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development 125, 3313–3322. [DOI] [PubMed] [Google Scholar]
- Lindström N, Tran T, Guo J, Rutledge E, Parvez R, Thornton M, Grubbs B, McMahon J, and McMahon A (2018). Conserved and Divergent Molecular and Anatomic Features of Human and Mouse Nephron Patterning. J Am Soc Nephrol Jasn 29, 825–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström N, McMahon J, Guo J, Tran T, Guo Q, Rutledge E, Parvez R, Saribekyan G, Schuler R, Liao C, et al. (2018). Conserved and Divergent Features of Human and Mouse Kidney Organogenesis. J Am Soc Nephrol Jasn 29, 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström N, Guo J, Kim A, Tran T, Guo Q, Brandine G, Ransick A, Parvez R, Thornton M, Basking L, et al. (2018). Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol Jasn 29, 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström NO, De Sena Brandine G, Tran T, Ransick A, Suh G, Guo J, Kim AD, Parvez RK, Ruffins SW, Rutledge EA, et al. (2018). Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev. Cell 45, 651–660.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa Y, Onay T, Scott R, Keir L, Dimke H, Li C, Eremina V, Maezawa Y, Jeansson M, Shan J, et al. (2014). Loss of the Podocyte-Expressed Transcription Factor Tcf21/Pod1 Results in Podocyte Differentiation Defects and FSGS. J Am Soc Nephrol 25, 2459–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah S, Saueressig H, Goulding M, Kintner C, and Dressler G (2000). Kidney Development in Cadherin-6 Mutants: Delayed Mesenchyme-to-Epithelial Conversion and Loss of Nephrons. Dev Biol 223, 38–53. [DOI] [PubMed] [Google Scholar]
- McMahon A (2016). Development of the Mammalian Kidney. Curr Top Dev Bio. 117, 31–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R, Otto EA, Kokoruda A, Zhou J, Zhang Z, Yoon E, Chen Y-CC, Troyanskaya O, Spence JR, Kretzler M, et al. (2018). Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi S, He Y, Gutsol A, Wight A, Hébert R, Vilmundarson R, Makrigiannis A, Chalmers J, Hamet P, Tremblay J, et al. (2015). Endothelial Gata5 transcription factor regulates blood pressure. Nat Commun 6, 8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AW, McInnes L, Kreidberg J, Hastie ND, and Schedl A (1999). YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 126, 1845–1857. [DOI] [PubMed] [Google Scholar]
- Moriguchi T, Hamada M, Morito N, Terunuma T, Hasegawa K, Zhang C, Yokomizo T, Esaki R, Kuroda E, Yoh K, et al. (2006). MafB Is Essential for Renal Development and F4/80 Expression in Macrophages. Mol Cell Biol 26, 5715–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R, Lam A, Freedman B, Kishi S, Valerius T, and Bonventre J (2015). Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 33, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U, Wang D, Denda S, Meneses JJ, Pedersen RA, and Reichardt LF (1997). Integrin alpha8beta1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell 88, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, and Itoh N (2000). FGF10 Acts as a Major Ligand for FGF Receptor 2 IIIb in Mouse Multi-Organ Development. Biochem Bioph Res Co 277, 643–649. [DOI] [PubMed] [Google Scholar]
- O’Brien L, Guo Q, Lee Y, Tran T, Benazet J-D, Whitney P, Valouev A, and McMahon A (2016). Differential regulation of mouse and human nephron progenitors by the Six family of transcriptional regulators. Development (Cambridge, England) 143, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, MD: ), 2018. World Wide Web URL: https://omim.org/ [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner H, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy K, Villegas G, Teichman J, Veron D, Shen W, Jimenez J, Thomas D, and Tufro A (2009). Semaphorin3a regulates endothelial cell number and podocyte differentiation during glomerular development. Development 136, 3979–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries M, Loiola R, Shah U, Gentleman S, Solito E, and Sastre M (2016). The anti-inflammatory Annexin A1 induces the clearance and degradation of the amyloid-β peptide. J Neuroinflamm 13, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, Xing CY, Ni L, Mathieson PW & Mundel P (2002) A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol 13, 630–8. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell Wanner, and Huber TB (2014). Glomerular development – Shaping the multi-cellular filtration unit. Seminars in Cell & Developmental Biology 36, 39–49. [DOI] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, and Raftery AE (2016). mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J 8, 289–317. [PMC free article] [PubMed] [Google Scholar]
- Self M, Lagutin O, Bowling B, Hendrix J, Cai Y, Dressler G, and Oliver G (2006). Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. The EMBO Journal 25, 5214–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin S, Taguchi A, Kaku Y, Yoshimura Y, Ohmori T, Sakuma T, Mukoyama M, Yamamoto T, Kurihara H, and Nishinakamura R (2016). Human Induced Pluripotent Stem Cell-Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation. Journal of the American Society of Nephrology 27, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P (1994). Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 8, 1888–1896. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, and Nishinakamura R (2014). Redefining the In Vivo Origin of Metanephric Nephron Progenitors Enables Generation of Complex Kidney Structures from Pluripotent Stem Cells. Cell Stem Cell 14, 53–67. [DOI] [PubMed] [Google Scholar]
- Takasato, Er, Becroft, Vanslambrouck, Stanley, Elefanty, and Little (2014). Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nature Cell Biology 16, 118–126. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er P, Chiu H, Maier B, Baillie G, Ferguson C, Parton R, Wolvetang E, Roost M, de Lopes S, et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er P, Chiu H, Maier B, Baillie G, Ferguson C, Parton R, Wolvetang E, Roost M, Lopes S, et al. (2016). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Bioinformatics S-S (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. (2010). Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol 28, 1248–1250. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BMM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, and Humphreys BD (2018). Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell 23, 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tamayo P, and Zhang K (2018). Visualizing and Interpreting Single-Cell Gene Expression Datasets with Similarity Weighted Nonnegative Embedding. Cell Syst 7, 656–666.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, and Shi G-PP (2014). Vascular wall extracellular matrix proteins and vascular diseases. Biochim. Biophys. Acta 1842, 2106–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P-X, Adams J, Peters H, Brown C, Heaney S, and Maas R (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nature Genetics 23, 113–117. [DOI] [PubMed] [Google Scholar]
- Xu P-X, Zheng W, Huang L, Maire P, Laclef C, and Silvius D (2003). Six1 is required for the early organogenesis of mammalian kidney. Development 130, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Morizane R, Homma K, Monkawa T, Suzuki S, Fujii S, Koda M, Hiratsuka K, Yamashita M, Yoshida T, et al. (2016). Generation of kidney tubular organoids from human pluripotent stem cells. Scientific Reports 6, 38353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Taguchi A, and Nishinakamura R (2017). Generation of a Three-Dimensional Kidney Structure from Pluripotent Stem Cells. Methods Mol. Biol 1597, 179–193. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim T-H, and Niswander L (2012). Phactr4 regulates directional migration of enteric neural crest through PP1, integrin signaling, and cofilin activity. Gene Dev 26, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL TABLE 2
A. Summary of RNA seq samples
B. Differentially Expressed Genes related to Figures 1C, S1A, 3D
C. GO analyses related to Figures 1C, S1A, 3D
SUPPLEMENTAL TABLE 2
A. Links to protein expression data of LP genes on Human Protein Atlas (related to Figure S5)
B. Mouse Podocyte MicroArray analyses (related to Figures S2G and S2H)
C. Tables of TPM values from mRNA-seq analyses of: Organoid differentiation progression, MAFB-eGFP+ and MAFB-eGFP− cells (Related to Figures 3 and S3)
D. Differentially Expressed Genes of Podocytes comparing: Zone1 vs Zone2 (Related to Figures S1F and S1G), in vivo vs in vitro (Related to Figure 4G)
E. Ligand-Receptor Predictions of Glomerular components (Related to Discussion)
F. DNA Sequences important for MAFB-P2A-eGFP H9 hESC generation (Related to Figure S3 and STAR Method – section Human embryonic stem cell line (hESC).
SUPPLEMENTAL VIDEO 1 (Related to Figure 5B)
Multiphoton live imaging of in vitro derived podocytes (green) transplanted under the kidney capsule of an immunocompromised mouse after Alexa 594-conjugated Albumin dye (red) injection. Scale bars denote 10μm.
Data Availability Statement
The bulk and single-cell RNA sequencing datasets are available under GEO accession numbers: GSE127344, GSE124392, and GSE124472.