Abstract
Previously three imaging methods, dynamic contrast enhanced magnetic resonance imaging (DCE-MRI), T1ρ-MRI, and low temperature NADH/Fp (reduced nicotinamide adenine dinucleotide/oxidized flavoprotein) fluorescence imaging (redox scanning) were reported to differentiate the mouse xenografts of a less metastatic human melanoma cell line A375P and a more metastatic line C8161. The more metastatic melanoma is characterized by less blood perfusion/permeability and more oxidized mitochondrial redox state in the tumor core and lower T1ρ relaxation time averaged across the tumor section. These features may be useful for identifying imaging biomarkers for cancer metastatic potential. Here, we have employed T1ρ - and T2-weighted MRI to image mouse xenografts of two human breast cancer lines (more metastatic MDA-MB-231 and less metastatic MDA-MB-468) on a vertical bore 9.4-T Varian MR system. The preliminary results indicated that the more metastatic MDA-MB-231 tumors had shorter T1ρ relaxation constants on average than the less metastatic MDA-MB-468 tumors, and T1ρ relaxation might be a potential biomarker of breast tumor metastatic potential. Distinct ring-like structures were observed on T1ρ-weighted MR images of the breast tumors, indicating tumor core and rim difference. This observation appears to be consistent with the tumor core-rim difference previously observed by DCE-MRI and redox scanning on aggressive melanoma xenografts.
1. Introduction
A biomarker of tumor aggressiveness or metastatic potential may help physicians to select proper treatment strategies and evaluate treatment response for cancer patients. Imaging biomarkers have the advantage of providing spatial and/or temporal distribution information about biological or physiological indices and have been increasingly utilized in the translational cancer research and clinical practice. The long-term objective of this study is to identify imaging biomarkers for tumor metastatic potential.
Previously, proton magnetic resonance imaging (MRI) and optical imaging techniques have been employed to characterize mouse xenografts of human melanoma cell lines with different levels of aggressiveness [1]. The aggressiveness of these lines had been characterized by the clinical data, the measurement of the tumor metastatic potential in mouse models, and the invasive potential of cell lines in vitro by the Boyden chamber method. DCE-MRI observation and histological assay of the vascular structure of tumor tissue indicated that a highly metastatic melanoma had lower blood perfusion in the tumor core compared to a poorly metastatic melanoma. T1ρ-MR indicated that the average T1ρ value in metastatic melanoma was shorter than in indolent melanoma. Although T1ρ-MRI is sensitive to pH, oxygenations, and protein concentrations, the underlying mechanism for this difference between the aggressive and indolent tumors awaits further investigation. Distinct tumor core-rim difference was also observed by low temperature NADH/Fp fluorescence imaging [2], which provides not only images of the relative intensity of NADH (reduced nicotinamide adenine dinucleotide) and Fp (oxidized flavoprotein) fluorescence in tissue, but also images of redox ratio. Higher Fp redox ratio, Fp/(Fp+NADH), indicates a more oxidized mitochondrial redox state. The melanoma tumor core exhibited more oxidized mitochondrial redox status compared to the tumor rim as indicated by the Fp redox ratio. The Fp redox ratios in the oxidized tumor regions were also found to correlate linearly with the aggressiveness of five melanoma xenografts spanning the full range of tumor progression to metastasis.
The low temperature NADH/Fp fluorescence imaging has also been applied to mouse xenografts of human breast cancer cell lines MCF-7 (poorly metastatic) and MDA-MB-231 (highly metastatic) [3]. The preliminary redox imaging data obtained were consistent with those of melanoma mouse xenografts, i.e., the more metastatic breast tumors exhibit more oxidized tumor cores with higher Fp redox ratios than those of indolent tumors. Several redox imaging biomarkers including Fp redox ratio, NADH, and Fp nominal concentrations have been shown to differentiate between MCF-7 and MDA-MB-231 tumors. Therefore, it would be interesting to explore whether DCE-MRI and T1ρ-MRI can distinguish between the poorly metastatic and highly metastatic breast tumors xenografted in mice, and whether the similar results consistent with those obtained on melanoma models can be obtained on breast tumor models as well.
In this paper, we report the preliminary T1ρ-MRI data obtained on a highly metastatic breast tumor xenograft MDA-MB-231 and a less metastatic breast tumor xenograft MDA-MB-468 [4]. Our data indicate that T1ρ-MRI can differentiate between these two lines with different metastatic potential and MDA-MB-231 breast tumors exhibited core-rim difference on T1ρ-MRI. Our study suggests that clinically translatable T1ρ-MRI may be a possible imaging biomarker for breast cancer metastatic potential.
2. Methods
Human breast cancer cell lines (MDA-MB-231 and MDA-MB-468) were grown in PRMI culture medium reaching about 100% confluency and then harvested to be subcutaneously implanted into the flanks or the upper parts of legs of athymic nude mice (NCI strain 01B74 NCr-nu/nu). Each inoculation consists of about 10 million breast cancer cells in 100 μl PBS buffer (pH=7). When tumors reached the size of 6~9 mm in diameter, the mice were subject to MRI scans under anaesthesia with continuous O2 flow doped with 1–2% isofluorane. Three MDA-MB-231 tumors and two MDA-MB-468 tumors were imaged by T1ρ- and T2-weighted MRI.
T1ρ-MRI of mouse tumors were conducted on a 9.4-T vertical bore Varian MR spectrometer. T1ρ-MR sequence [5] started with a spin-locking three pulse cluster (time of spin locking denoted as TSL and spin-locking frequency as SLF) to lock the proton magnetization in the transverse plane and then followed with a regular spin-echo data acquisition. A single slice spin echo T1ρ-MRI sequence (TR 3s, TE, 4 TSLs ranging from 15–125 ms, field of view (FOV) 2.0 cm) was used to acquire images of a plane sectioning the central portion of a tumor. Customized IDL (Interactive Data Language) programs were used to fit the data to a single exponential component decay to obtain the T1ρ relaxation time and T1ρ maps. Regular T2-MRI (TR 1.5s, 5 equal spaced TEs varying from 12–67 ms, thickness 1–2 mm, FOV 2 cm) was also conducted to obtain T2-weighted MR images of tumor slices and generate T2 maps with a single exponential fit of the signal changes versus TEs. For some tumors, a multi-slice T1ρ-MRI sequence (TR 3s, TE 12ms, 4 equally-spaced TSLs starting from 15–30 ms to about 120 ms, thickness 2 mm, matrix 64×64) and T2-MRI sequence were used to acquire T1ρ- and T2-weighted images, respectively, across the entire tumor volume.
3. Results and Discussion
Figure 1 shows preliminary results from single slice MRI. The mean T1ρ and T2 relaxation times of each tumor group were plotted in the graph. The more metastatic MDA-MB-231 tumors had shorter average T1ρ relaxation constants than the less metastatic MDA-MB-468 tumors, whereas T2 does not differentiate well between these two lines.
Fig. 1.
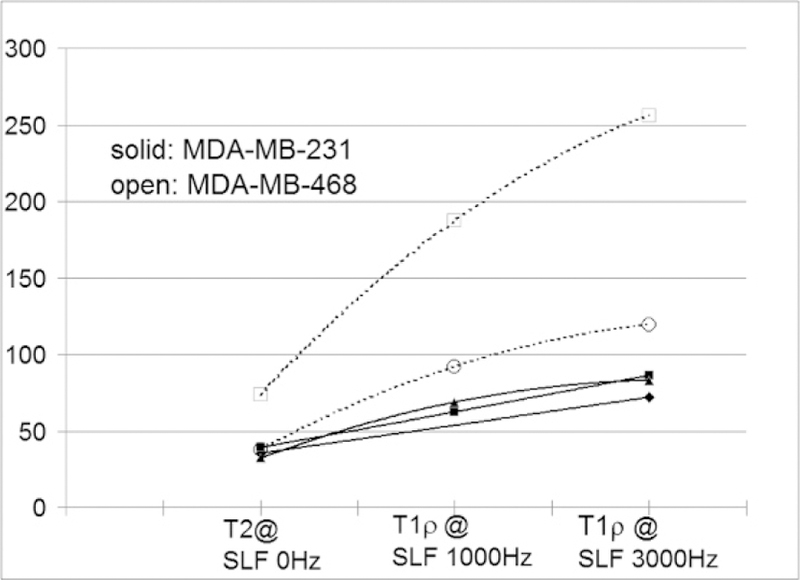
Comparison of the average T2 and T1ρ relaxation time constants (in ms) between MDA-MB-468 (N=2, open symbols and dashed lines) and more metastatic MDA-MB-231 tumors (N=3, solid symbols and solid lines).
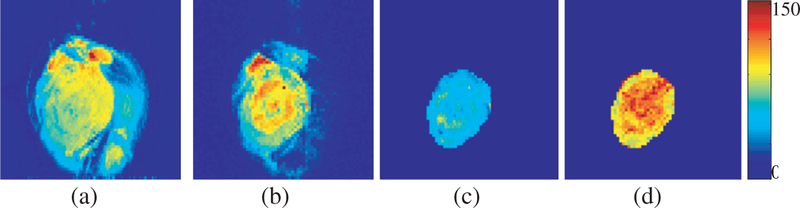
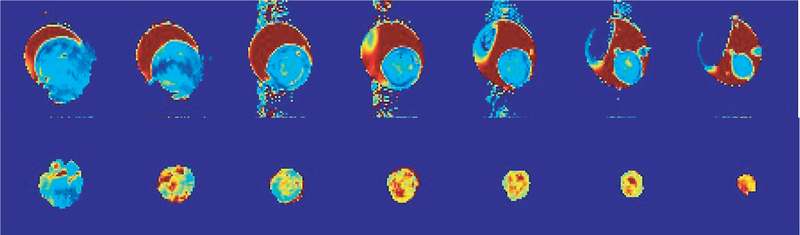
A distinct ring-like structure was observed on T1ρ weighted MR images of breast tumors, indicating tumor core and rim difference in MDA-MB-231 (Fig. 2). The tumor core-rim difference was also observed on T2-weighted images but appeared to be less distinct. Thus, T1ρ maps appear to show higher heterogeneity than T2 maps. This is further supported by multi-slice T1ρ and T2 maps indicating hot regions (higher relaxation values) in the tumor core areas, whereas T2 maps did not show a distinct difference (Fig. 3). This observation appears to be consistent with the tumor core-rim difference previously observed by DCE-MRI and redox scanning on aggressive melanoma xenografts.
Fig. 2.
Representative images of a slice sectioning the center of a MDA-MB-231 tumor. (a) T2-weighted image; (b) T1ρ-weighted image; (c) T2 map; (d) T1ρ map. The range of T2 or T1ρ relaxation time constants in ms is indicated by the colorbar.
Fig. 3.
Multi-slice T2 (upper row) and T1ρ (lower row) maps for a typical MAD-MB-231 tumor. Image orientation was in parallel with the body surface and the top of the tumor. The depth of the image slices (1mm thickness) ranged from about 1 mm (right) to 7mm (left). Field of view = 2.5 cm. The dark red regions in the 1st row were not tumors but gels used for enhancing B1 field homogeneity for more effective T1ρ-MR.
4. Discussion
Our studies on melanoma and breast cancer mouse xenografts support that T1ρ relaxation time constant can be a potential biomarker for cancer aggressiveness in the clinic. T1ρ relaxation rates reflect the interaction between water protons and macromolecules, and T1ρ relaxation is sensitive to macromolecular concentration and pH, oxygen saturation and cell death. Further study is needed to explore the underlying mechanism for the T1ρ differences.
Heterogeneities in relaxation constants were also observed for some MDA-MB-468 tumors. Work is in progress to characterize these heterogeneities in more tumors and compare with those of MDA-MB-231. Work is also in progress to study more breast tumors with MRI followed by redox scanning, and compare the MR images with the redox scanning images of the same breast tumor at various depths from the tumor surface. Since the higher Fp redox ratio in the tumor core but not in the rim has been shown as a potential biomarker for tumor metastatic potential, it will be interesting to test if T1ρ value in the tumor core and rim may differ in their power to predict tumor aggressiveness as well.
5. Conclusions
Clinically translatable T1ρ- and T2-MR imaging has been applied to mouse xenografts of two human breast tumor lines MDA-MB-231 (more metastatic) and MDA-MB-468 (less metastatic). Our preliminary data indicated that the average T1ρ value might distinguish between these two tumor lines but the average value of T2 did not. Multislice T1ρ images showed a distinct tumor core-rim difference with some core regions having higher T1ρ values. Our studies on melanoma and breast cancer mouse xenografts support that T1ρ relaxation time constant can be a potential biomarker for cancer aggressiveness in the clinic.
Acknowledgments
This work is supported by the Susan G. Komen Foundation Career Catalyst Research Grant (Lin Z. Li, KG080169) and Center for Magnetic Resonance and Optical Imaging, an NIH-supported research resource (Ravinder Reddy, RR02305). We would also like to acknowledge the technical assistance received from Dr. Steve Pickup and valuable discussions with Dr. Jerry Glickson.
References
- 1.Li LZ, Zhou R, Xu HN et al. (2009) Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proceedings of the National Academy of Sciences of the USA 106:6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li LZ, Xu HN, Ranji M et al. (2009) Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. Journal of Innovative Optical Health Sciences 2:325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu HN, Nioka S, Glickson JD et al. (2010) Quantitative mitochondrial redox imaging of breast cancer metastatic potential. Journal of Biomedical Optics, 15:036010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieuwerts AM, Klijn JG and Foekens JA (1997) Assessment of the invasive potential of human gynecological tumor cell lines with the in vitro Boyden chamber assay: influences of the ability of cells to migrate through the filter membrane. Clinical & Experimental Metastasis 15:53–62 [DOI] [PubMed] [Google Scholar]
- 5.Wheaton AJ et al. (2004) Pulse sequence for multislice T1ρ-weighted MRI. Magnetic Resonance in Medicine 51:362–369 [DOI] [PubMed] [Google Scholar]