Abstract
Purpose:
Optical redox imaging (ORI), based on collecting the endogenous fluorescence of reduced nicotinamide adenine dinucleotide (NADH) and oxidized flavoproteins (Fp) containing a redox cofactor flavin adenine dinucleotide (FAD), provides sensitive indicators of cellular metabolism and redox status. ORI indices (such as NADH, FAD, and their ratio) have been under investigation as potential progression/prognosis biomarkers for cancer. Higher FAD redox ratio, i.e., (FAD/(FAD+NADH)) has been associated with higher invasive/metastatic potential in tumor xenografts and cultured cells. This study is to examine whether ORI indices can respond to the modulation of oncogene DEK activities that change cancer cell invasive/metastatic potential.
Procedures:
Using lentiviral shRNA, DEK gene expression was efficiently knocked down in MDA-MB-231 breast cancer cells (DEKsh). These DEKsh cells, along with scrambled shRNA transduced control cells (NTsh), were imaged with a fluorescence microscope. In vitro invasive potential of the DEKsh cells and NTsh cells were also measured in parallel using the transwell assay.
Results:
FAD and FAD redox ratio in polyclonal cells with DEKsh were significantly lower than that in NTsh control cells. Consistently, the DEKsh cells demonstrated decreased invasive potential than their non-knockdown counterparts NTsh cells.
Conclusions:
This study provides direct evidence that oncogene activities could mediate ORI-detected cellular redox state.
Keywords: Optical redox imaging, FAD, NADH, redox ratio, DEK gene, knock down, breast cancer, invasive potential
Introduction
Optical redox imaging (ORI) is based on imaging the intrinsic fluorescence mainly from mitochondrial NADH and oxidized flavoproteins (Fp containing FAD) [1–2]. NADH and FAD are important intermediates or cofactors of bioenergetics and biosynthesis pathways. When excited by UV (~ 325–380 nm) and blue (~ 430–460 nm) lights, NADH and FAD emit blue (~ 425–490 nm) and green (~ 530–570nm) fluorescence, respectively [3]. In contrast, their respective redox counterparts NAD and FADH2 do not have fluorescence. The optical redox ratio in various forms (e.g., FAD/NADH, NADH/FAD, normalized FAD redox ratio = FAD/(FAD+NADH), or normalized NADH redox ratio = NADH/(FAD+NADH); Fp used interchangeably with FAD in the literature) has been used as a surrogate indicator for the mitochondrial NAD-coupled redox state [4–5] and correlates with the NAD+/NADH redox potential determined by biochemical approaches [6–7]. ORI has been shown to provide useful information for cancer diagnosis and treatment response [5, 8–9].
While evaluating the ORI indices as potential biomarkers for diagnosis/prognosis of cancer, we found that ORI FAD redox ratios correlate linearly with the invasive potentials of five cell lines of human melanoma [10]. For breast cancer, we found that the clinical biopsies of breast cancer patients exhibit significantly higher FAD, NADH, and FAD redox ratio in the cancerous tissues than adjacent normal tissues [11]. In animal models, we found that the triple negative breast cancer (TNBC) cell line MDA-MB-231 induced mouse xenografts with higher metastatic potential and presented a more oxidized state (higher FAD redox ratio) in the localized tumor regions than estrogen-receptor (ER) positive MCF-7 xenografts, which have lower metastatic potential [12]. It also has been observed in cell cultures that not only the more invasive TNBC MDA-MB-231 cells have higher FAD/NADH ratio than ER+ or HER2+ lines , the rank order of redox ratio of MDA-MB-231 versus another TNBC line MDA-MB-468 is consistent with the rank order of invasive potential [8, 13–15]. All these results suggest a correlation or an association between ORI redox ratio and breast cancer aggressiveness as represented by invasive/metastatic potential [8].
It has been widely reported that overexpression of oncogenes or loss of tumor suppressor genes results in reprogramming metabolism in cancer development [16–17]. It has been found that ORI indices are sensitive to genetic status of P53, PTEN and Her2/neu receptor in (pre)cancer [13, 18–19]. In particular, we observed that the more oxidized region of the mouse xenografts of p53-null colon cancer HCT116 cell line has a higher FAD redox ratio than the p53 wild-type counterpart [18]. These results implicated that oncogene overexpression or loss of tumor suppressor genes may be associated with the more oxidized NAD-coupled redox state (higher redox ratio); and silencing the oncogene activity in tumor cells may result in decrease in aggressiveness and more reduced redox state (lower redox ratio). However, to our knowledge, there is no report directly demonstrating that genetically manipulating the expression of a single oncogene under isogenic conditions results in concordant alterations in the ORI indices and cancer cell aggressiveness.
In this report, through lentiviral-particle delivered shRNA we efficiently knocked down the expression of the DEK oncogene in TNBC MDA-MB-231 cells. DEK is a chromatin remodeling protein whose overexpression has been reported to reprogram cellular metabolism, stimulate cell proliferation, increase invasion and metastasis, etc [20–22]. In addition, high DEK expression is an independent prognostic factor associated with aggressive disease and poor outcome in solid tumors, including breast cancer [23–25]. The ORI results we acquired demonstrated that MDA-MB-231 cells with decreased DEK expression (DEKsh) had a significantly lower FAD redox ratio than the vector treated control cells (NTsh). Our report provides the first direct evidence that knockdown of oncogene gene expression affects ORI-sensitive redox status in breast cancer.
Materials and Methods
Cell lines and cell culture
DEK gene knockdown polyclonal cell line MDA-MB-231-DEKsh and control cell line MDAMB-231-NTsh were prepared as previously described [20]. Briefly, lentiviral particles were produced by transiently transfecting HEK-293T cells with Sigma MISSION pLKO.1 shRNA constructs. Non-targeting (NTsh) and DEK-targeting (pLKO.1_DEK832 “DEKsh”) constructs were used. MDA-MB-231 cells were transduced with lentivirus and underwent selection in 2 μg/ml puromycin for 4 days then maintained in 1 μg/ml puromycin. Cells were grown in RPMI 1640 medium (Gibco®, catalog number 1949915, phenol red free) containing 10% FBS, 11.1 mM glucose, 2.0 mM glutamine, 100 μg/ml penicillin and 0.25% streptomycin at 37°C and 5% CO2. RPMI 1640 medium was also supplemented with 1μg/ml puromycin.
Western blotting
Total protein was extracted from MDA-MB-231 DEKsh and NTsh cells with radioimmunoprecipitation assay (RIPA) lysis and extraction buffer and 50 μg was subjected to SDS-PAGE analysis prior to transfer to a PVDF membrane. Blots were probed with specific antibodies to DEK (1:1000, BD Bioscience, San Jose, CA catalog number 610948) and Actin C4 (1:10,000, gift of James Lessard, Cincinnati Children’s Hospital) followed by an anti-mouse secondary antibody. Membranes were visualized with the ChemiDoc Imaging System (BioRad) and analyzed with Image Lab Software (v5.2.1, BioRad).
Cell seeding and preparation for imaging
One day before imaging, 200 μl MDA-MB-231 DEKsh or NTsh cell suspension totaling 105 cells (in complete RPMI 1640 medium supplemented with 1μg/ml puromycin) were seeded to a 35mm glass-bottom petri dish coated with poly-d-lysine (MatTek Inc.). After about 4 h when cells have attached to the bottom of the dish, 800 μl complete RPMI 1640 medium supplemented with 1μg/ml puromycin was added to the dish which was then incubated overnight. On the day of imaging, in order to minimize background fluorescence, cells were rinsed twice with PBS (with Ca2+ and Mg2+) followed by adding 1 ml live cell image solution (LCIS, Invitrogen Inc.) supplemented with 11 mM glucose and 2 mM glutamine (same concentrations as in RPMI 1640 medium) approximately 1hr before imaging. LCIS can keep cells healthy for up to 4 h at ambient atmosphere and temperature.
To demonstrate ORI of MDA-MB-231 cells are sensitive to perturbation of mitochondrial metabolism, MDA-MB-231 cells cultured in RPMI 1640 medium were seeded in petri dishes (200 μl 8×104 cells per dish) and prepared using the same protocol as described above.
Fluorescence image acquisition
Using the same equipment and parameters as previously described [15], NADH and FAD fluorescence signals were obtained with a DeltaVision Deconvolution Microscope System which has a 300 W xenon lamp as an excitation light source, a 12-bit CCD, and an objective of 40X/0.95 NA (image size 512 × 512, bin 2 × 2, pixel size 0.32mm). The microscope is mounted in a thermally controlled chamber to maintain the samples at 37°C. The wavelengths and bandpass widths of excitation filters are 360/40 nm and 470/40 nm for NADH and FAD channels, respectively. The emission filter wavelengths and bandpass widths are 455/50 nm and 520/40 nm, respectively, for NADH and FAD channels. No photobleaching was observed under the lamp power used in the experiments. For each cell culture dish, five random-selected fields of view (FOVs) were imaged with an exposure time of 3 s for each channel. Each FOV contained 20–30 cells. Dishes with MDA-MB-231 DEKsh and NTsh cells were interleaved for imaging under the same conditions. Two to three dishes were imaged for each cell line in one experiment. The experiments were repeated 3 times.
For ORI under perturbation of mitochondrial metabolism, MDA-MB-231 cells were first imaged as control before any treatment, then treated with 2.5 μg/ml oligomycin, and imaged again approximately 3–5 min after the drug administration. Same cells were then treated with 0.5 μM FCCP (carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone) followed by another ORI 3–5 min later. Three randomly-selected FOVs were imaged for each dish during each image acquisition and in total 12 dishes were studied in 4 experiments.
Imaging data analysis
The acquired data were first processed using a customized Matlab® program in several steps following the flow chart in Electronic Supplemental Materials (ESM) (SFig. 1). First, the excitation light intensity fluctuation was taken into account using automatically recorded intensity readings for each channel by a photosensor when the image was acquired. The FAD and NADH signal from each FOV were extracted and normalized to photosensor readings for the corresponding channel. Second, each raw image was flattened by performing a third-order polynomial surface fit to remove inhomogeneous illumination. Thirdly, the background intensity of each channel was obtained from a randomly selected cell-free area. The net intensities of FAD and NADH were obtained by first subtracting their respective background intensities, then as the last step thresholding at three times of the standard deviation of the background. FAD/(FAD+NADH) image was then obtained pixel-by-pixel from the net intensity image of FAD and NADH. For individual FOVs, the mean values for FAD, NADH, and FAD/(FAD+NADH) were first calculated. These mean values were then averaged across all FOVs to obtain mean values for individual dishes, followed by averaging across multiple dishes to be reported as means of the group. The standard deviations of the means (SD) and the p-values of Student’s t test were reported based on the number of dishes. Cell nuclei may exhibit relatively low signals in image and may not be excluded completely by the image thresholding. To ensure nuclei being excluded completely, we also performed single-cell-based analysis for DEK knockdown experiments by drawing regions of interests on the NADH images using the customized Matlab® program to circle around single cells and exclude the regions of nuclei. For each group we analyzed ~45–50 well-separated cells with nuclei identifiable that were randomly selected from different FOVs in eight dishes. Cells without clear regions of nuclei were not included in the single-cell-based analysis.
Invasive potential assay
Invasive potential of cancer cells were measured in the Vinnedge lab using the transwell invasion assay. MDA-MB-231 DEKsh and NTsh cells were plated in serum-free RPMI media for 24 h then viable cell counts were acquired using Trypan Blue staining. Viable cells were then seeded at a density of 5×104 cells in each Matrigel-precoated upper invasion chamber (Corning™ BioCoat™ Matrigel™ Invasion Chamber with Corning™ Matrigel Matrix, product number 354480, 6.5 mm well, 8.0 μm pore) and the lower chamber was filled with 500 μL standard complete RPMI 1640 cell culture medium (containing 10% FBS). The chambers were incubated in a humidified 37-degree incubator for 20 h. Chambers were cleared of non-invading cells and invading cells were fixed in methanol prior to staining with Giemsa. Four representative, non-overlapping images (the invasion chamber was divided into quadrants and images were positioned at approximately the center of each quadrant) were captured in each well with a Leica DMIL microscope with SPOT software and cells were averaged across these images to obtain the mean number of invading cells for each well. Biologically triplicate wells seeded with cells of different passages were used in the measurement. Invasive potential was presented as fold change in invading cell numbers comparing the DEK knockdown line to the control line. An unpaired Student’s t test was used to determine statistical significance.
Results
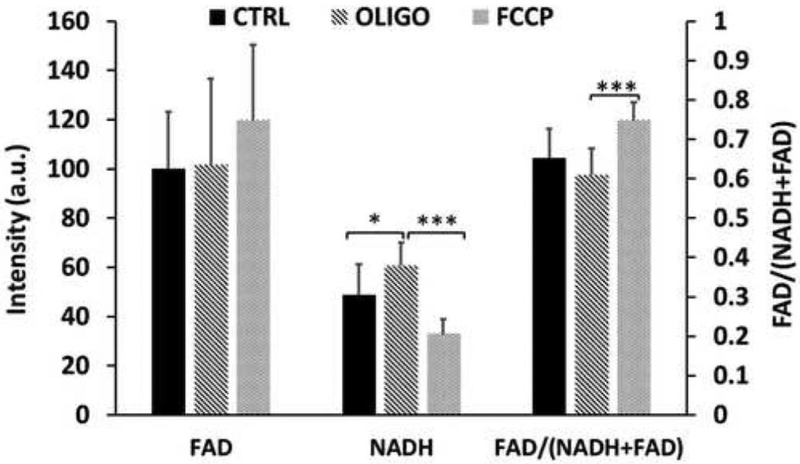
As shown in Fig. 1, we confirmed the redox indices (NADH, FAD and FAD redox ratio) of the MDA-MB-231 cells are sensitive to perturbation of mitochondrial metabolism. Administration of ATP synthase inhibitor oligomycin caused a significant NADH buildup and sequential administration of uncoupler FCCP lead to a significant decrease of NADH and an increase of the redox ratio.
Fig. 1.
Bar graph depicting the redox changes of MDA-MB-231 cells under sequential treatment of oligomycin (2.5 μg/ml) and FCCP (0.5 μM) (mean ± SD, n=12). Controls were not treated. * p<0.05, *** p<0.001.
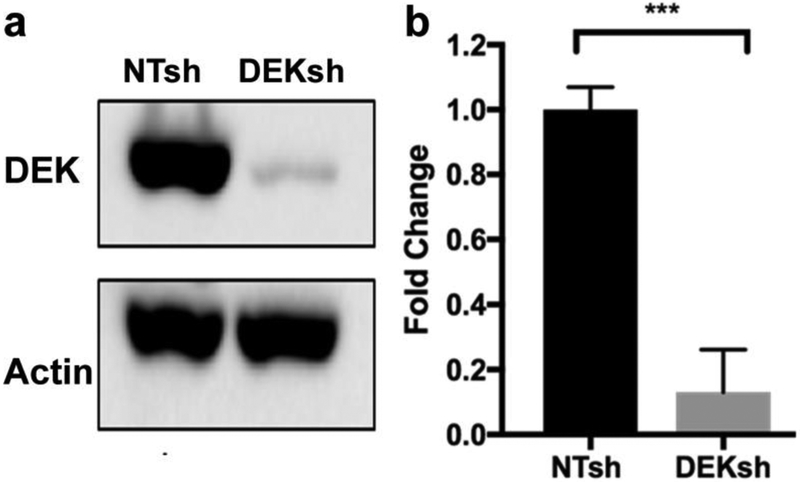
MDA-MB-231 cells were then transduced with lentivirus carrying shRNA targeting the DEK oncogene (231 DEKsh) or non-targeting control (231NTsh). DEK knockdown efficiency averaged 88% in early passage cells, as confirmed by Western blot (Fig. 2). Early passage DEK knockdown and control cells were imaged by ORI. Typical FOV images of FAD, NADH, and FAD/(FAD+NADH) of the two cell lines are shown in Fig. 3a. Compared to the control cells, 231DEKsh cells had 30.8% less FAD intensity (from 250 ± 44 arbitrary unit (a.u.) to 173 ± 28, p < 0.001, n = 8) and became significantly more reduced with the FAD redox ratio decreased by 8.1% (from 0.74 ± 0.03 to 0.68 ± 0.02, p < 0.001, n = 8). No significant difference in NADH was observed between the two cell lines (Fig. 3b). Consistent results, i.e., 25% decrease in FAD and 12% decrease in redox ratio (p<0.005) were obtained using the single-cell based analysis (Suppl. Fig. 2, see ESM) excluding the signal contribution from cell nuclei.
Fig. 2.
DEK western blot of cell lysates for MDA-MB-231 DEKsh and NTsh cells. a Representative image of gel bands. b Quantification of DEK activities. The band intensity ratio DEK/b-Actin of 231-DEKsh cells were normalized to that of 231-NTsh (5 independent experiments). *** p<0.001.
Fig. 3.
Optical redox imaging of MDA-MB-231 DEKsh and NTsh cells. a-b Typical pseudo-colored images of FAD, NADH and FAD redox ratio FAD/(FAD+NADH) for cultured human breast cancer cells MDA-MB-231 with a DEK knockdown (231-DEKsh) and b its control (231-NTsh). c Knockdown of DEK expression by shRNA in MDA-MB-231 resulted in a decrease in FAD fluorescence and FAD redox ratio, but no significant change in NADH fluorescence (n=8 dishes). Left y-axis is for NADH and FAD, and the right y-axis for FAD/(FAD+NADH). Bar height represents the mean and error bar represents the standard deviation (SD). *** p<0.001.
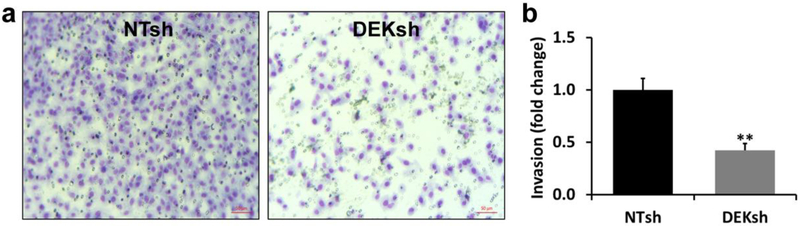
Previous work has demonstrated that loss of DEK expression in other breast cancer models results in decreased invasive potential in vitro and lung metastases in vivo [20, 22]. DEK knockdown in MDA-MB-231 cells resulted in significant decrease in invasive potential by 58% as measured by the Vinnedge lab, comparing to the NTsh control cells (p < 0.005) (Fig 4a and b). The significant decrease of invasive potential was also independently confirmed in the Li lab (Suppl. method and Suppl. Fig. 3 in ESM). Thus, we have observed that DEK oncogene knockdown in MDA-MB-231 cells results in a more reduced redox state (lower FAD redox ratio) and decreased invasive potential.
Fig. 4.
Results of invasive potential assays. a Pictures of typical field of views of invading MDA-MB-231 NTsh and DEKsh cells (lower-right corner scale bar 50 μm). Invaded cells that were stained with Giemsa dye. The blue stains indicate the nuclei. b Quantification of invasive potential shown as relative fold changes to the control MDA-MB-231 NTsh cells (3 biological triplicates), ** p<0.005.
Discussion
The ORI redox indices (FAD, NADH and redox ratios) are known to be sensitive to mitochondrial metabolism and NAD-coupled redox potential [1–2, 4, 6–7]. Therefore, we expect ORI is sensitive to genetic status of oncogenes and tumor suppressors that mediate cancer metabolic reprogramming. In addition to ORI’s sensitivity to the status of oncogenes and tumor suppressors including p53 and PTEN in (pre)cancer tissues [18–19] and the Her2/neu and ER among breast cancer cell lines [13–14], recently differential gene expression of PGC1α, a master regulator of mitochondrial biogenesis and function, was identified among ORI-detected redox subpopulations of MDA-MB-231 mouse xenografts [26].
This study is the first direct demonstration that genetic knockdown of an oncogene expression in cancer cells alters the optical redox indices. Varone, et al. has shown that overexpression of oncoprotein HPV E17 in engineered squamous epithelial tissues resulted in an increase of optical redox ratio (FAD/(NADH+FAD)) [7]. Interestingly high risk HPV E17 was shown to upregulate DEK in keratinocytes and fibroblasts [27]. Our study provides consistent and complementary results by showing that knock down of oncogene DEK resulted in decrease of redox ratio and invasive potential. Another study used fluorescence lifetime microscopy to evaluate how Wnt status and PDK-1 expression may change NADH lifetime and the balance between free NADH and bound NADH in colon cancer cells [28], but neither the change in total NADH nor the FAD and redox ratio were reported. It remains to be investigated whether the results of these studies will hold true for other oncogenes or tumor suppressors, e.g. cMyc, PIK3CA, VHL, known to mediate cancer metabolism.
In this study we demonstrated that the ORI-based redox indices can detect the changes directly induced by knock down of DEK gene expressions. DEK gene overexpression has been reported to reprogram metabolism in squamous cell carcinoma resulting in enhanced glycolysis and NAD+ [29]. Considering that FAD/(NADH+FAD) is a surrogate indicator of intracellular NAD+/(NAD++NADH) redox ratio [7], our data indicate that DEK knockdown changes intracellular NAD-coupled redox state. Furthermore, since ORI signals are mainly contributed by mitochondrial NADH and FAD, decrease of FAD redox ratio indicates that DEK knockdown likely resulted in a more reduced mitochondrial redox state. Since DEK promotes invasion and metastasis [22], correlated decreases of the redox ratio and invasive potential of DEK knockdown cells are consistent with the previous observations that associate higher or more oxidized redox ratio with cancer aggressiveness [10, 12, 15]. Note that when comparing malignant to normal cells or tissue, some studies indicate that breast cancer cells in culture have lower optical redox ratio than normal or untransformed cells [13–14] whereas other studies show that breast tumor tissues of clinical patients have higher redox ratio than adjacent normal breast tissue [11]. When comparing among breast cancers with different aggressiveness, different groups using ORI appear to reach largely consistent results that more oxidized redox ratio tends to be associated with higher aggressiveness [8, 30–31]. However, Santidrian, et al measured redox potential NADH/NAD+ using biochemical cycling assay and found that higher cellular NADH/NAD+ or more reduced redox state drives breast cancer cells to more metastasis [32]. Such biochemical cycling assay involves in vitro destructive processing of cell/tissue materials (e.g., homogenization, treatment with reagents, or isolation of mitochondria if mitochondrial redox potential is measured). On the other hand, ORI is noninvasive and the fluorescence signals are dominated by mitochondrial contributions. Future investigation on this apparent inconsistency between the biochemical cycling assay and ORI is needed.
We also observed a significant decrease of FAD signals due to DEK suppression. FAD fluorescence signals are contributed mainly by oxidized flavin prosthetic groups in 3 major metabolic enzymes, i.e., pyruvate dehydrogenase, α-ketoglutarate dehydrogenase and electron-transfer flavoprotein [33–34]. It remains unclear which component is responsible for the observed signal change. DEK overexpression has been reported to enhance glycolysis and mitochondrial respiration spared capacity, and knockdown of DEK decreased the transcription of metabolic enzymes regulating glycolysis and NAD metabolism [29]. It would be interesting to further investigate whether FAD signal decrease in our study might be associated with the suppression of the transcription of some of the redox enzymes.
We did not observe a significant change in NADH signals induced by DEK suppression in this study. Although NADPH is expected to be several fold or even nearly 10 fold less than NADH level in cells [35], NADPH may contribute to the total signals of NADH channel. Separate measurement of NADPH and NADH may illustrate how they change individually in response to DEK knockdown.
Other redox couples or systems including glutathione, ascorbate, thioredoxin and glutaredoxin [36–37] are directly or indirectly coupled to NAD(P)+/NAD(P)H redox potential, change of which may be detected by ORI. Thus, it remains to be studied whether ORI can be sensitive to or correlate with changes in other redox systems. Interestingly thioredoxin was reported to show a more oxidized state in prostate cancer but a more reduced state in breast cancer compared to benign tissue [38]. It remains to be investigated whether ORI-detected redox state may exhibit such dependence on specific cancer type.
Finally, note that we focused on studying a polyclone cell line generated from transduction of lentiviral shRNA instead of imaging single clone cell lines. It is common in cancer research for individual single clone subpopulations to have different gene expression levels of knock down. Thus the data obtained with this polyclone line likely reflect the “averaging effects” of the mixed cell subpopulations with various levels of DEK knockdown. In the future, multiple single clones of DEK knockdown may be imaged to demonstrate the correlation of changes of invasive potential and redox ratio. We may also validate such correlation in more breast cancer lines.
Supplementary Material
Acknowledgement:
This work was supported by the NIH Grant R01CA191207 (L.Z. Li) and NIH grant R37CA218072 (LMPV). The authors thank Ms. Jinxia Jiang for her assistance in part of the data acquisition, Dr. Zhenwu Lin for valuable discussion and support. The authors would also like to appreciate the valuable discussion with and support from Dr. Andrea Stout and Ms. Jasmine Zhao of Cell and Developmental Biology Microscopy Core, Perelman School of Medicine, University of Pennsylvania.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Li LZ (2012) Imaging mitochondrial redox potential and its possible link to tumor metastatic potential. J Bioenerg Biomembr 44:645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu HN, Li LZ (2014) Quantitative redox imaging biomarkers for studying tissue metabolic state and its heterogeneity. Journal of Innovative Optical Health Sciences 07:1430002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzbeta C, Dusan C (2014) Tissue fluorophores and their spectroscopic characteristics In Fluorescence Lifetime Spectroscopy and Imaging. CRC Press, pp 47–84. [Google Scholar]
- 4.Chance B, Schoener B, Oshino R, Itshak F, Nakase Y (1979) Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem 254:4764–4771. [PubMed] [Google Scholar]
- 5.Li LZ, Xu HN, Ranji M, Nioka S, Chance B (2009) Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Opt Health Sci 2:325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozawa K, Chance B, Tanaka A, et al. (1992) Linear correlation between acetoacetate/beta-hydroxybutyrate in arterial blood and oxidized flavoprotein/reduced pyridine nucleotide in freeze-trapped human liver tissue. Biochim Biophys Acta 1138:350–352. [DOI] [PubMed] [Google Scholar]
- 7.Varone A, Xylas J, Quinn KP, et al. (2014) Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues. Cancer Res 74:3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li LZ, Sun N (2014) Autofluorescence perspective of cancer diagnostics In Natural Biomarkers for Cellular Metabolism: Biology, Techniques, and Applications, Eds. Ghukasyan V, Heikal AA. New York: CRC Press, pp 273–297. [Google Scholar]
- 9.Kolenc OI, Quinn KP (2018) Evaluating cell metabolism through autofluorescence imaging of NAD(P)H and FAD. Antioxid Redox Signal. doi.org/ 10.1089/ars.2017.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li LZ, Zhou R, Xu HN, et al. (2009) Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci USA 106:6608–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu HN, Tchou J, Feng M, et al. (2016) Optical redox imaging indices discriminate human breast cancer from normal tissues. J Biomed Opt 21:114003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu HN, Nioka S, Glickson JD, et al. (2010) Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Opt 15:036010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh A, Cook RS, Rexer B, et al. (2012) Optical imaging of metabolism in HER2 overexpressing breast cancer cells. Biomed Optics Express 3:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrander JH, McMahon CM, Lem S, et al. (2010) Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res 70:4759–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun N, Xu HN, Luo Q, Li LZ (2016) Potential indexing of the invasiveness of breast cancer cells by mitochondrial redox ratios. Adv Exp Med Biol 923:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairns RA, Harris IS, Mak TW (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11:85–95. [DOI] [PubMed] [Google Scholar]
- 17.DeBerardinis RJ, Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2:e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu HN, Feng M, Moon L, et al. (2013) Redox imaging of the p53-dependent mitochondrial redox state in colon cancer ex vivo. J Innov Opt Health Sci 6:1350016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu HN, Nioka S, Li LZ (2013) Imaging heterogeneity in the mitochondrial redox state of premalignant pancreas in the pancreas-specific PTEN-null transgenic mouse model. Biomark Res 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Privette Vinnedge LM, McClaine R, Wagh PK, et al. (2011) The human DEK oncogene stimulates beta-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene 30:2741–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L, Huang X, Zhang W, et al. (2016) Critical role of DEK and its regulation in tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget 7:26844–26855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Privette Vinnedge LM, Benight NM, Wagh PK, et al. (2015) The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene 34:2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matrka MC, Cimperman KA, Haas SR, et al. (2018) Dek overexpression in murine epithelia increases overt esophageal squamous cell carcinoma incidence. PLoS Genet 14:e1007227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Xiong D, Zeng J, et al. (2017) Prognostic role of DEK in human solid tumors: a meta-analysis. Oncotarget 8:98985–98992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ying G, Wu Y (2015) DEK: A novel early screening and prognostic marker for breast cancer. Mol Med Rep 12:7491–7495. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z, Xu HN, Wang Y, Floros J, Li LZ (2018) Differential Expression of PGC1alpha in Intratumor Redox Subpopulations of Breast Cancer. Adv Exp Med Biol 1072:177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wise-Draper TM, Allen HV, Thobe MN, et al. (2005) The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol 79:14309–14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pate KT, Stringari C, Sprowl-Tanio S, et al. (2014) Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J 33:1454–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matrka MC, Watanabe M, Muraleedharan R, et al. (2017) Overexpression of the human DEK oncogene reprograms cellular metabolism and promotes glycolysis. PLoS One 12:e0177952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh AJ, Cook RS, Sanders ME, et al. (2014) Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res 74:5184–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alhallak K, Rebello LG, Muldoon TJ, et al. (2016) Optical redox ratio identifies metastatic potential-dependent changes in breast cancer cell metabolism. Biomed Opt Express 7:4364–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santidrian AF, Matsuno-Yagi A, Ritland M, et al. (2013) Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest 123:1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunz WS (1988) Evaluation of electron-transfer flavoprotein and alpha-lipoamide dehydrogenase redox states by two-channel fluorimetry and its application to the investigation of beta-oxidation. Biochim Biophys Acta 932:8–16. [DOI] [PubMed] [Google Scholar]
- 34.Kunz WS, Gellerich FN (1993) Quantification of the content of fluorescent flavoproteins in mitochondria from liver, kidney cortex, skeletal muscle, and brain. Biochem Med Metab Biol 50:103–110. [DOI] [PubMed] [Google Scholar]
- 35.Rehman AU, Anwer AG, Gosnell ME, Mahbub SB, Liu G, Goldys EM (2017) Fluorescence quenching of free and bound NADH in HeLa cells determined by hyperspectral imaging and unmixing of cell autofluorescence. Biomed Opt Express 8:1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee R (2008) Redox Biochemistry. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- 37.Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264:13963–13966. [PubMed] [Google Scholar]
- 38.Jorgenson TC, Zhong W, Oberley TD (2013) Redox imbalance and biochemical changes in cancer. Cancer Res 73:6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.