Abstract
Protocells, artificial cell-like particles, capable of autonomously synthesizing RNA and proteins based on a DNA template, are emerging platforms for studying cellular functions and for revealing the origins-of-life. Here, we show for the first time that artificial lipid-based vesicles, containing the molecular machinery necessary for transcription and translation, can be used to synthesize anti-cancer proteins inside tumors. The particles were engineered as stand-alone systems, sourcing nutrients from their biological microenvironment to trigger protein synthesis. When pre-loaded with template DNA, amino acids and energy-supplying molecules protocells synthesized, up to 2×107 copies of superfolder green fluorescent protein (sfGFP) were synthesized in each liposome. A variety of proteins, having molecular weights reaching 66 kDa and with diagnostic and therapeutic activities, were synthesized inside the particles. Incubating protein producing particles, encoded to secrete Pseudomonas exotoxin A (PE) with 4T1 breast cancer cells in culture, resulted in killing of most of the malignant cells. In mice bearing 4T1 tumors, histological evaluation of the tumor tissue after a local injection of PE-producing particles, indicating robust apoptosis. Protein producing particles are synthetic-biology platforms capable of synthesizing therapeutic proteins on-demand.
Keywords: protocells, nanoparticle, drug delivery, synthetic biology, liposome
1. Introduction
Drug delivery systems have greatly advanced over the past decade, enabling the targeted delivery of proteins and genes, as well as cell-based therapies.[1] In this study we propose to merge the fields of synthetic biology and drug delivery to form a platform that synthesizes therapeutic proteins inside the patient's body, specifically, inside a tumor.
Cell-free protein synthesis (CFPS) systems serve as an attractive tool in synthetic biology for different applications such as high-throughput protein expression and screening, production of toxic proteins, incorporation of non-natural amino acids, and in vitro selection and protein evolution. In addition, the potential of these systems in therapeutic protein development and production processes has been previously suggested;[2–4] for example, different CFPS systems were implemented to produce streptokinase, onconase and Pseudomonas exotoxin A.[4–6] The encapsulation of CFPS systems inside lipid vesicles has paved the way for improved and new applications; In vitro selection of membrane proteins can now be performed by protein cell-free production inside liposomes following by their integration to the liposome membrane.[7] In addition, these particles are utilized as a model for evolutionary research to explore the minimal cell components and processes required to enable cellular life.[8, 9] For example, de Souza et al. have explored the minimal size of a cell by using liposomes as an artificial model for the cellular compartment.[10] Furthermore, complex cellular processes such as a cascade expression of two proteins, DNA amplification and even communicational processes have been demonstrated using protocells.[7, 11] The interaction of artificial cells with natural cells has been promoted by engineering liposomes that release a small molecule in response to an external chemical trigger, which in turn induced Escherichia coli to produce a reporter protein.[12] Communication between artificial cells was accomplished by designing genetic circuits that control chemical signal exchange.[13]
Another application of artificial cell-like particles, which gains a large focus, is their use as biochemical micro-reactors – defined compartments in which protein synthesis is carried out.[14, 15] Recently, a complex protein production process has been demonstrated by the individual in vitro synthesis of two proteins in two-compartment lipid vesicles.[16] These bioreactors are mostly giant unilamellar vesicles (GUVs) with diameters in the range of 1-100 µm.[17] They possess high modularity due to the ability of the enclosed CFPS synthesis system to produce different proteins only by altering the DNA template.
Combining the fields of synthetic biology protocells with drug delivery offers a new therapeutic avenue – cell-free production of biologics inside the body. So far, little work has been done on implementing protein producing particles for therapeutic applications in vitro, and specifically in vivo.[18]
Here, we demonstrate the use of artificial cell-like particles as a tool for synthesizing therapeutic proteins at the disease site. A modified method for encapsulating CFPS inside liposomes is described (Figure 1A) and the resulted particles were characterized. The production of reporter and therapeutic proteins inside particles has been demonstrated both in vitro and in vivo.
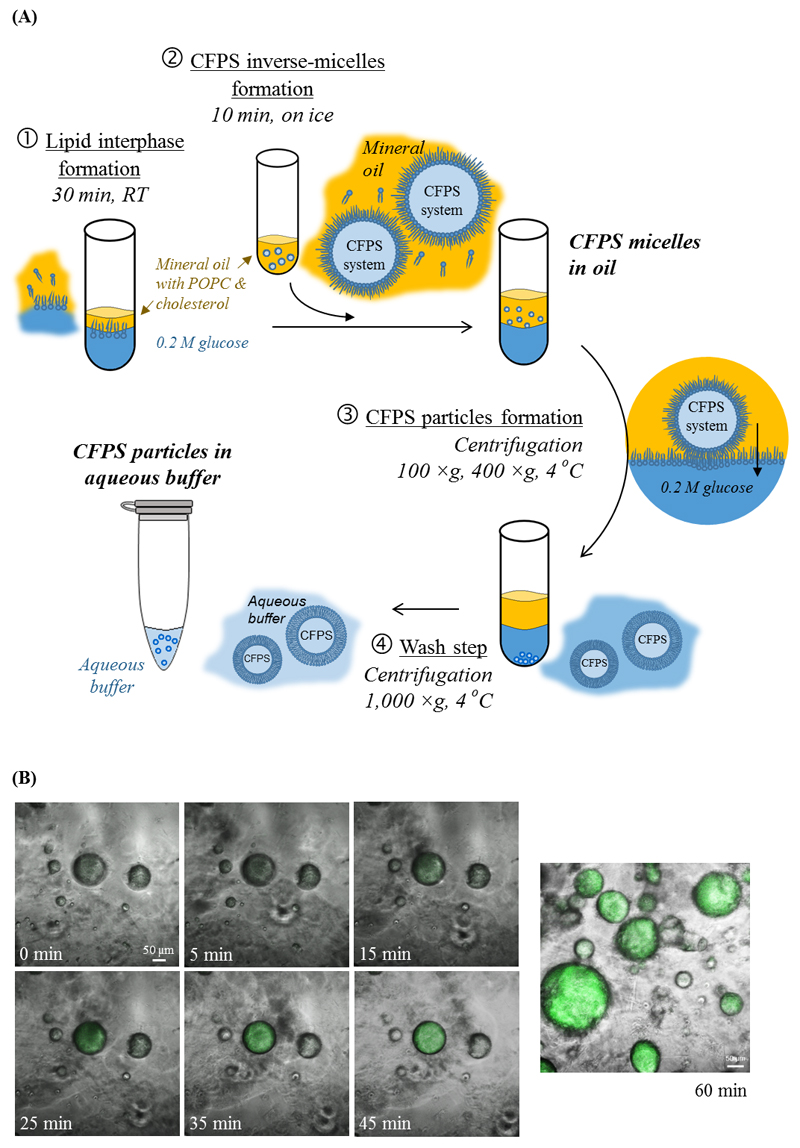
Figure 1. Protocells: Artificial, cell-like protein producing particles.
(A) A schematic representation of the process of producing protocells in the laboratory. A bacterial cell-free protein synthesis (CFPS) system is loaded into self-assembled lipid vesicles. At first, CFPS micelles are formed in a mineral oil (step 2). Centrifugation enables transforming the protein producing micelles into liposomes, by adding a lipid leaflet to the outer leaflet of the micelles (steps 3 and 4). (B) Protein synthesis in CFPS micelles. Confocal microscopy images of green fluorescent protein (sfGFP) as it is being produced inside CFPS micelles over a period of 60-minutes.
2. Results and Discussion
Cell-free protein synthesis (CFPS) systems are used for synthesizing proteins without the limitations of living cells. When enclosed inside a particle, these systems transform into a drug delivery system enabling on-demand protein supply. In this study we present a method for encapsulating the molecular machinery necessary for transcription and translation into artificial lipid vesicles. Physical and biochemical characterization of these cell-mimicking particles and of their capacity to produce proteins autonomously is presented. In addition, a therapeutic application both in vitro and in vivo is demonstrated.
Protein producing particles were constructed by encapsulating an E. coli-based CFPS system[4] inside lipid vesicles. The process, based on gentle agitation, centrifugation and liposome self-assembly is summarized in Figure 1A. In general, an aqueous solution containing the CFPS system is pipetted into a mineral-oil that contains a vesicle-forming lipid – POPC (having 16:0, 18:1 hydrocarbon tails) and cholesterol. Through several cycles of centrifugation (100, 400 and 1000 ×g) micelles, and thereafter liposomes containing the CFPS system are formed (Figure 1A). To inhibit unwanted protein production and to lengthen the activity life of the particles, the process is carried out at low temperatures, <4 °C. These mild conditions were found to be favorable for retaining the molecular machinery activity during the loading process, and are similar to the conditions used to precipitate live cells.[7, 15, 19, 20]
Superfolder green fluorescent protein (sfGFP, 27 kDa) was chosen as a model protein to examine protein synthesis inside the particles. This protein has improved folding kinetics, it can be fused to other proteins, and its fluorescence is proportional to the amount of expressed protein.[21] Time-lapse confocal images of protein-producing inverse-micelles in oil (Figure 1A-1) demonstrate that sfGFP can be observed inside the micelles already 15 minutes after initiating the protein production; the fluorescence increases over time until reaching signal saturation after 45 minutes (Figure 1B and Video S1).
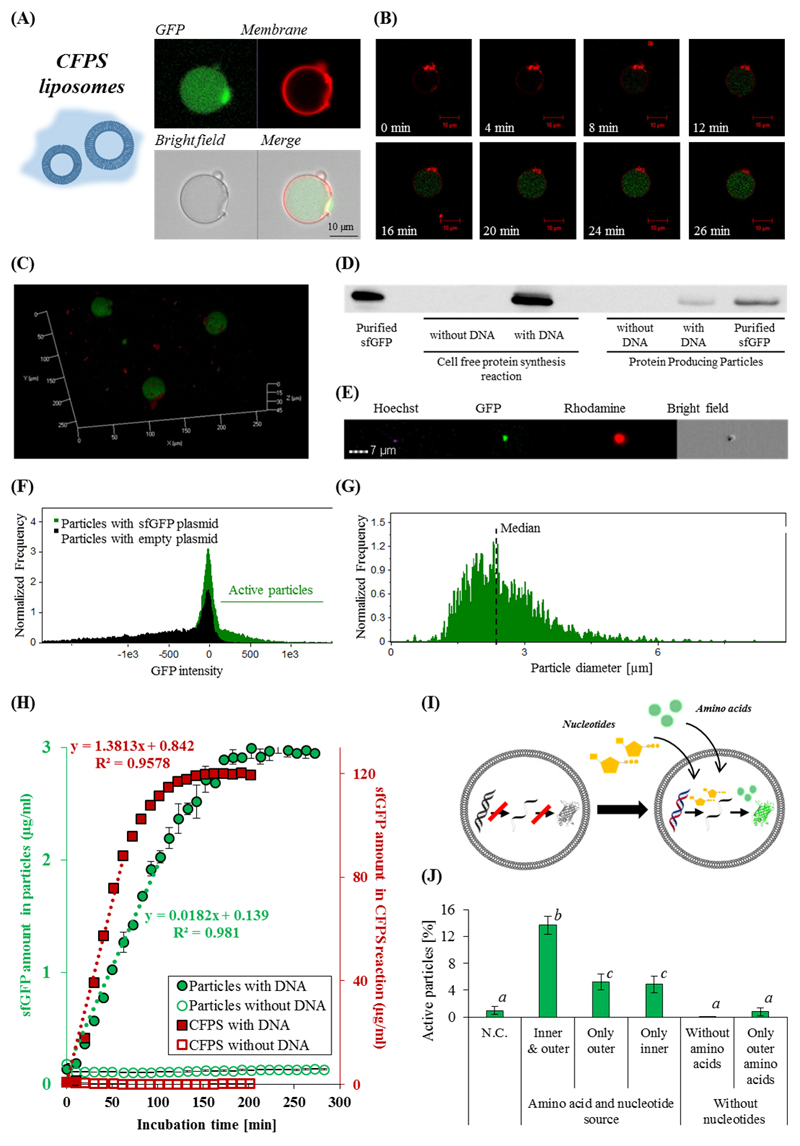
Next, we characterized the production of sfGFP inside liposomal particles. Liposomes are more suitable for CFPS drug delivery applications due to their improved stability, owed to the liposome' lipid bilayer, compared to the monolayer lipid leaflet in micelles (Figure 1A-1 and 1A-3).[22] After centrifugation, the liposomal artificial cell fraction was composed of 6.4±0.8 mM total lipids, of which 4.5±0.6 mM POPC and 1.9±0.6 mM cholesterol; having an average liposome concentration of (4±1)×108 particles mL-1 and occupying 3±1% of the volume of the dispersion. The liposome membrane was labeled with rhodamine (red) and sfGFP (green) was synthesized inside the liposome (Figure 2A, 2B and 2C, and Video S2). The rhodamine signal from the particle boundaries remained constant over time, while the sfGFP signal increased as more protein was produced inside the liposomes (Figure 2B). To confirm the production of proteins inside the particles, a 3D confocal reconstruction of the liposomes (Figure 2C and Video S3), and a Western blot analysis for sfGFP was performed (Figure 2D). Only when an appropriate DNA template was incorporated into the reaction, a band corresponding to sfGFP was detected. Taken together, these results confirmed autonomous protein production inside micelles and liposomes.
Figure 2. Protein production inside artificial cell-like particles.
Confocal microscopy images of sfGFP production inside protein producing liposomal particles after 60 minutes (A), and as a function of time (B), both at 37°C. sfGFP was imaged in the green channel, and the rhodamine-labeled liposome membrane was imaged in the red channel. (C) 3D image of the protein producing particles after 2 hours. (D) A Western blot analysis demonstrates that sfGFP (~27kDa) is produced both in bulk and in the particles. (E) Flow cytometry of sfGFP producing liposomes: Hoechst was used for nuclei staining, rhodamine was incorporated in the particle' membrane, and sfGFP was produced in the particles. The particles were analyzed for sfGFP synthesis (F), as a function of particle diameter (µm) (G). (H) sfGFP production kinetics inside artificial cell-like particles and in a cell-free bulk. A linear regression was fitted to the first third of each of the reactions incubation times. Error bars represent standard deviation of the mean from 2 independent repeats. (I)&(J) Artificial cell-like particles source molecular building blocks from their surrounding environment to trigger protein production. (I) An illustration of adding molecular building blocks that are necessary for transcription and translation through the particle membrane. (J) The effect of amino acids and nucleotide supplementation on sfGFP production inside particles. The fluorescence was measured after a 4 hour incubation. Error bars represent standard deviation of the mean from at least 3 independent repeats. Treatments that differ significantly by a two-tailed Student’s t test (p-value<0.05) are designated with different letters (a, b & c).
As long as the particles were held on ice, protein synthesis was retarded. To trigger production, the particles were warmed to physiological temperature (37 °C). Under these conditions, the CFPS system produced up to 380 μg/mL sfGFP in solution, compared to 5.3 μg/mL of sfGFP inside artificial cell-like particles, corresponding to 1 pico-gram-protein per particle after a two hour reaction.
When these reactions were monitored over time, faster protein production kinetics were observed in bulk compared to production inside the particles (Figure 2H). In both cases more than 80% of the protein was synthesized within 130 minutes. However, when considering the volumetric fractions of the active particles in the solution (Figure 2F), the resulted activity of the production, per active unit volume is 1.4-3.3 fold more efficient (in respect to the maximal activity of production in bulk and in particulate system).[23] Furthermore, protein synthesis in liposomes offers a self-confined system which can be harnessed for in vitro selection of membrane proteins, 'minimal cell' discovery, and for developing micro-bio-reactors.[7, 9, 13, 14, 20]
The fabrication process of protocells is imperfect, yielding both functional particles capable of synthesizing proteins alongside dysfunctional particles that do not produce proteins. To evaluate the efficiency of the process we analyzed the sfGFP expression particle-by-particle using flow cytometry. The particles were co-labeled and imaged using four different channels: Hoechst was used for DNA staining, GFP indicated the produced protein, rhodamine for membrane labeling and bright field (Figure 2E). Only 21-25% of the particles produced proteins efficiently (Figure 2F). This rather low efficiency may be explained by the need to encapsulate all the necessary components for carrying out transcription and translation at appropriate ratios inside the particle.[24] While further efforts need to be focused on increasing yield, the current method enables producing particles using common laboratory equipment.
Analyzing the effect of particle size on protein production showed that protein synthesis is confined to particles of sizes that resemble those of natural cells.[25] Protein synthesis was recorded for particles ranging in size from 376 nm up to 9±1 µm. The median size of active protein-producing particles was 2.9±0.5 µm, with the greatest production detected in particles ranging from 1.5 to 4 µm (Figure 2G).[26]
Therapeutic protein producing particles should have the capacity to stand-alone, sourcing nutrients from their surroundings. Molecular building blocks, such as amino acids and energy sources, must be able to cross the membrane to supplement for depleted materials.[27, 28] Therefore, we chose to construct the particles from POPC which forms a soft liquid-phase membrane at physiological temperature, which is permeable to small molecules.[28, 29] To test the ability of the particles to utilize building blocks present in their outer environment, particles lacking the main building blocks required for protein production (ATP, GTP, UTP and amino acids) were constructed. Then, the particles were incubated in solutions enriched with these molecular components, to allow for sfGFP synthesis (Figure 2I and 2J). A significant (p-value<0.05) protein production was measured in these particles, compared to particles incubated in nutrient-free buffers (Figure 2J). These results also indicate that both amino acids and nucleotides, which are necessary for the protein production process, can cross the membrane at a sufficient rate for facilitating continuous protein synthesis. Diffusion of low molecular weight molecules through the lipid membrane served as a trigger for initiating protein synthesis inside the particles.
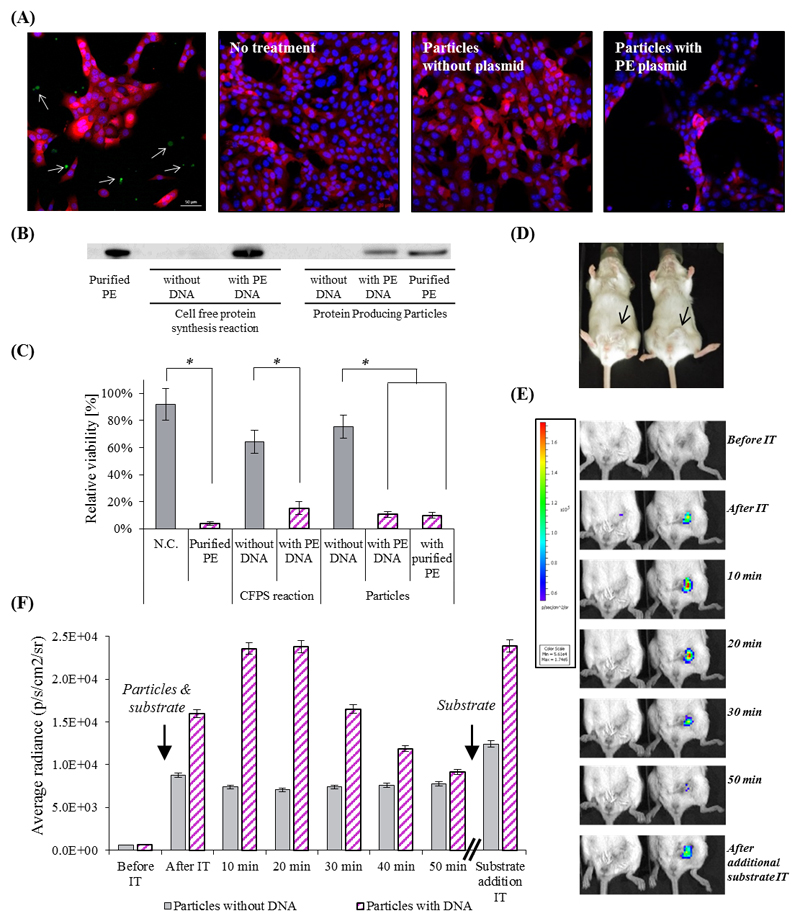
We evaluated the therapeutic capacity of these particles to produce Pseudomonas exotoxin A (PE) in the presence of triple-negative 4T1 breast cancer cells. PE, a 66 kDa bacterial toxin has been widely investigated for its use in cancer therapy, demonstrating that only several copies of the protein are capable of killing a target cell.[30, 31] PE has three domains: binding, translocation and catalytic domain. The translocation domain (II) guides the proteins across lipid membranes.[31] When PE-producing particles were added to 4T1 cells a toxic effect was observed within 24 hours, demonstrated by a decrease in cell confluency (Figure 3A). A Western blot assay confirmed the presence of PE in the particles (Figure 3B), and a quantitative MTT cell viability assay confirmed that more than 80% of the cells were killed by PE (Figure 3C). Our data suggest that after the protein is produced it is secreted from the particles into the media, possibly through the translocation domain, to enable its activity against the cancer cells.[32]
Figure 3. The therapeutic effect of artificial cell-like particles.
(A) Confocal microscopy analysis of the therapeutic potency of PE-producing particles on 4T1 breast cancer cells expressing a reporter mCherry gene. Upper left - Confocal microscopy image of mCherry cells with particles producing sfGFP. The cytoplasm of 4T1 cells is labeled red, the nucleus is labeled blue, and produced sfGFP is green (represented by arrows). Second and third to left - cells which were not treated or treated with the particles lacking a DNA template, respectively. Right – cells treated with particle containing PE-coding DNA template. (B) Western blot analysis verifies the production of PE inside the particles. (C) Cell viability was evaluated by an MTT assay. Error bars represent standard deviation of the mean from 3 independent repeats. *Significant difference between treatments, where p-value<0.05 according to a Student's t-test with a two-tailed distribution with equal variance. (D)&(E)&(F) Renilla Luciferase in vivo production inside particles injected to BALB/c mice bearing orthotopic 4T1 tumors in the mammary fat pad (D). (E) Protein producing particles DNA-encoded (right), or non-encoded (left), to synthesize luciferase, were injected intra-tumor with enzyme’s substrate. The luminescent signal from the particles was monitored using whole animal imaging for 50 min. At this point the luminescent signal decayed, and the substrate was refurbished, resuming the luminescent signal. The maximal instrument internal error is 3% of the obtained value.
After demonstrating the therapeutic potency of protein producing particles in vitro, we tested their capacity to serve as platforms for onsite production of therapeutic proteins in vivo.[3, 6, 18, 33] Specifically, we used the particles to synthesize proteins in orthotopic 4T1 tumors in the mammary fat pad of BALB/c mice (Figure 3D). At first, a reporter protein, Renilla luciferase, was encoded in the particles and injected intra-tumorally along with the enzyme’s substrate. The mice were monitored using a whole animal IVIS imaging system. An increase in the luminescent signal was observed over the first half hour, indicating the production of an enzymatically-active protein inside the tumor (Figure 3E). After which, a decrease in the signal was observed, possibly due to enzyme concentration depletion or to substrate decomposition. Sequential intra-tumor injections of the substrate re-ignited the luminescent signal, i.e. low substrate concentration was the cause for signal reduction and the produced enzyme was maintained in the tissue (Figure 3F).
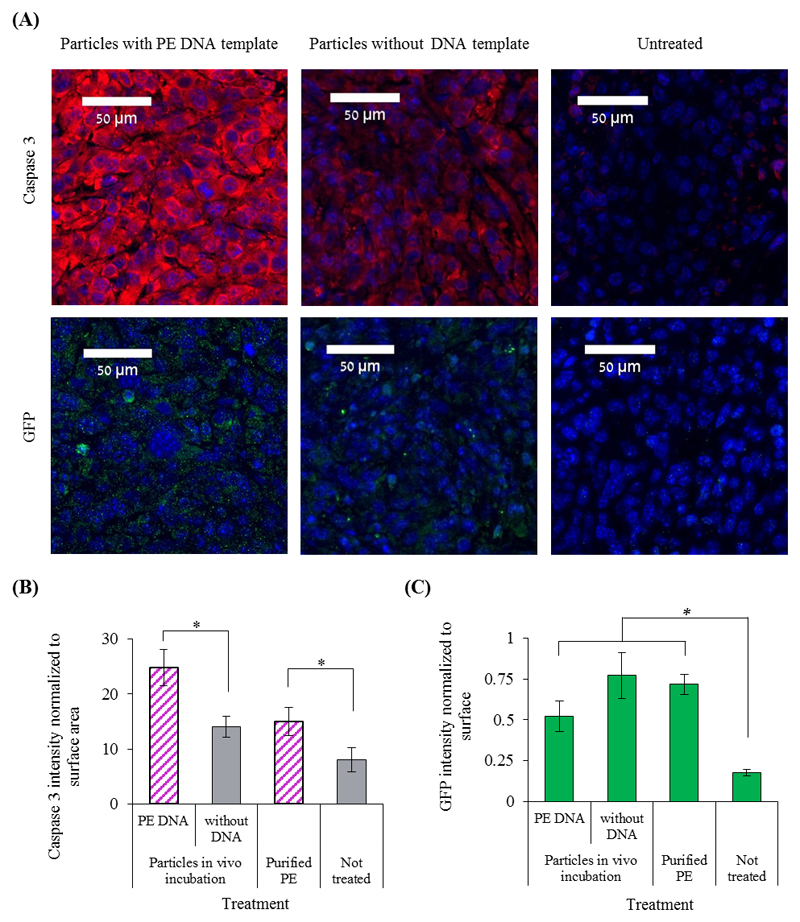
To assess the capacity of the particles to perform a therapeutic function in vivo, PE-encoded particles were injected into 4T1 tumors. All treated tumors were injected with particles encoding to sfGFP together with PE-particles, particles without DNA or purified PE (Figure 4A, 4C and S3). GFP was used as an indirect marker of PE levels in the tissue and enabled tracking the biodistribution of the particles in vivo. Histological evaluation of the tumors indicated an increase in the expression of the apoptotic marker caspase-3 compared to the controls (Figure 4A and 4B).[34] Interestingly, higher levels of the apoptotic marker were observed in tumors treated with the PE-producing particles compared to animals treated with the purified protein (Figure 4A and 4B). This toxicity may be attributed to the improved stability of the protein expressed in the particles,[35] an extended release profile of the protein in the diseased tissue,[36] as well as the inherent endotoxin toxicity of bacterial CFPS.[33] Other therapeutic applications may require an endotoxin-free bacterial extract or using an eukaryotic-based cell-free system.[37,13]
Figure 4. Pseudomonas exotoxin A (PE) production inside the tumor.
BALB/c mice bearing a 4T1 tumor in the mammary fat pad were injected intra-tumor with protein producing particles or purified PE toxin. Untreated tumors were used as a control group. (A) Immunofluorescence analysis of histology slides obtained from the tumors. Tissue slides were analyzed for the presence of both the apoptotic marker caspase-3 and GFP that was produced in the particles. In blue are the nuclei (DAPI), red indicates on caspase-3 expression and green indicates on GFP presence. (B)&(C) The levels of caspase-3 expression and of GFP presence, respectively, were evaluated by immunofluorescence analysis in tumors treated with PE-producing particles, or with protocells that lack a DNA template, or without treatment. Error bars represent standard deviation of the mean from 2-6 independent repeats. *Significant difference between treatments, where p-value<0.05 according to a Student's t-test with a one-tailed distribution with equal variance.
Taken together, these data indicate that artificial particles can autonomously synthesize a therapeutic protein inside an orthotopic tumor. In summary, utilizing synthetic particles as on-demand systems for supplying biologics may offer new treatment possibilities in cancer and other diseases; controlled drug production inside the body can enable their administration directly to the disease site, where and when they function is required.
3. Conclusions
In this study we demonstrate the self-assembly of lipid vesicles as carriers of a CFPS system for intra-tumoral drug production. The particles are prepared using a simple and inexpensive laboratory-scale apparatus. This platform was used to synthesize a variety of proteins with molecular weights of up to 66kDa, which possess different functionalities, including cytotoxic activity. The ability of the particles to interact with the environment and to exchange nutrients by diffusion is an advantage for the development of future applications and prolong the production of proteins in tissue. These artificial cell-like particles were used, for the first time, as internal bio-reactors to produce and treat tumors in vivo.
We believe such platforms present a new drug delivery approach – the production of therapeutic proteins directly at the disease site. In the future, these platforms may prove effective for synthesizing biologics encoded to address the patient's personalized needs.
4. Experimental Section
DNA templates
TargeTron® vector pAR1219 (Sigma-Aldrich, Rehovot, Israel) was used for T7 RNA polymerase over-expression in the preparation of the S30-T7 lysate. A superfolder GFP (sfGFP) template was purchased from Sandia BioTech (Albuquerque, New Mexico, USA) and was cloned into a pET9a vector using restriction sites NdeI and BamHI. A plasmid encoding to Renilla luciferase was obtained from the S30-T7 high yield protein expression system kit, purchased from Promega (Promega, Wisconsin, USA). A pVC45 f+t QQ∆ vector was used to produce Pseudomonas exotoxin A (PE). [38]
Lipids
The particle membrane included cholesterol (Sigma-Aldrich, Rehovot, Israel), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Lipoid, Ludwigshafen, Germany) and Rhodamine-labeled phospholipid - 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Avanti Lipids Polar, Alabaster, Alabama).
Preparation of S30-T7 lysate
S30-T7 lysate was prepared from Escherichia coli BL21(DE3) transformed with TargeTron® vector pAR1219 as we described previously.[4] The S30-T7 lysate aliquots were stored at -80 °C for further use.
In vitro protein synthesis using cell-free system based on S30-T7 lysate
The in vitro protein synthesis using the S30-T7 system was performed as we described previously.[4] The system was used to produce different proteins (sfGFP, Renilla luciferase, PE), by the incorporation of an appropriate DNA template and incubation at a constant temperature of 37 °C (unless mentioned otherwise) for 2 hours with vigorous shaking, 1200 RPM.
Superfolder GFP in vitro production
The amount of sfGFP produced by the CFPS system was evaluated according to previously prepared calibration curve at excitation wavelength of 488 nm and emission of 530 nm, using a 96 or 384 flat bottom black polystyrene plate by a plate reader (Tecan, Mannedorf, Switzerland).
In vitro PE cytotoxicity assay
The cytotoxic effect of PE was quantified based on a cell viability assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolinium bromide (MTT) reagent (Sigma-Aldrich, Rehovot, Israel), as we described previously.[4] The percentage of living cells was calculated with respect to the untreated wells.
Western blot analysis
Western blot analysis was used to confirm cell-free protein production, as we described previously.[4] Following electrophoresis (150 V), the gels were blotted onto nitrocellulose membranes and blocked with 5% nonfat milk powder in Tris-buffered saline. The membranes were probed for 1 hour at room temperature with anti-PE polyclonal antibody (Sigma-Aldrich, Rehovot, Israel) diluted by 1:15,000 or anti-His polyclonal antibody (GenScript, NJ, USA) diluted by 1:12,000 in Tris-buffered saline with 0.5% Tween-20 and 0.5% nonfat milk powder for the detection of PE and sfGFP respectively. After three washes, the blots were incubated with horseradish peroxidase-conjugated anti-rabbit (goat origin) secondary antibody (GenScript, NJ, USA) diluted to 1:20,000 and developed with Clarity™ Western ECL Blotting Substrate (BioRad, California, United States). The results were visualized using ImageQuant Las4000 (GE, Sweden).
Lipid vesicle preparation
Lipid vesicles (liposomes) were produced using the water/oil emulsion method as described previously with some modifications. [7, 15, 19, 20] Figure 1A demonstrates the modified protocol. A summarized protocol is presented in BOX 1.
BOX 1. Lipid vesicle preparation.
Step 1 - Lipid phase preparation:
Dissolve POPC and cholesterol in chloroform at a concentration of 50 mg mL-1 each.
Mix 100 μL of this lipid mixture with 500 μL of mineral oil in a glass vial, vortexed and heated to 80 °C for 1 hour to allow chloroform evaporation.
Store the resulted lipid phase at a room temperature for up to 2 weeks before usage.
Step 2 - Vesicle formation:
Layer 100 μL of the lipid phase over 300 µL of 200 mM glucose in a 1.5 mL plastic tube, and incubate it for 30 minutes. This will result in a biphasic solution.
Prepare 25 μL of a CFPS reaction mixture and 200 mM sucrose, and mix it with 150 µL of lipid phase in a glass vial by pipetting and moderate vortexing for 1 minute to obtain an emulsion.
Place the vial on ice for 10 minutes.
Layer the emulsion over the biphasic solution which was prepared at step 2.1.
Centrifuge at 100 xg for 9 minutes at 4 °C, and immediately afterwards at 400xg for 7 minutes.
Transfer the particle pellet to a new 1.5 mL plastic tube using a pipettor with 200 μL tip loaded with 200 mM glucose which is used to penetrate the oil/water interphase.
Centrifuge the collected particle suspension at 1,000 xg for 10 minutes at 4 °C.
Discard the supernatant was discarded, and resuspend the pellet in 25 μL of cold aqueous outer solution.
Incubate the particles at 37°C without shaking to enable protein production.
Lipid phase preparation - POPC and cholesterol were dissolved in chloroform at a concentration of 50 mg mL-1 each (1:2 molar ratio, respectively). Then, 100 µL of this lipid mixture were mixed with 500 µL of mineral oil (light oil) (Sigma-Aldrich, Rehovot, Israel) in a glass vial, vortexed and heated to 80 °C for 1 hour to allow chloroform evaporation. The resulted lipid phase was stored at a room temperature for up to 2 weeks before usage. When particle membrane labeling was required, a rhodamine-labeled phospholipid was incorporated by the addition of 6 μL of 1 mg mL-1 lipid stock solution to each 500 µL of mineral oil.
Vesicle formation - First, 100 µL of the lipid phase were layered over 300 µL of 200 mM glucose in a 1.5 mL plastic tube, resulted in a biphasic solution, and incubated for 30 minutes to form a stable lipid monolayer at the water/oil interphase (Figure 1A, step 1). Then, CFPS micelles were prepared (Figure 1A, step 2); 25 µL of an aqueous solution, designated as the inner solution of the micelles, were mixed with 150 µL of lipid phase in a glass vial by pipetting and moderate vortexing for 1 minute to obtain an emulsion. The inner solution of the micelles is comprised of the CFPS reaction mixture and 200 mM sucrose. Empty particles or particles encapsulating pure proteins were used as negative and positive controls, respectively. In these cases, the inner solution included only 200 mM sucrose and 3% (w/v) PEG. After the micelles were formed, the vial was placed on ice for 10 minutes to generate water/oil emulsion. Subsequently, the micelles solution was layered over the biphasic solution which was prepared earlier, and centrifuged at 100 xg for 9 minutes at 4 °C, and immediately afterwards at 400xg for 7 minutes (Figure 1A, step 3).
The particle pellet was transferred to a new 1.5 mL plastic tube as following described; a pipettor with 200 µL tip was loaded with 200 mM glucose and was used to penetrate the oil/water interphase. Then, the glucose solution was gently unloaded to remove oil remnants in the tip end, and the pellet was collected. Before unloading the pellet into a new tube, the tip end was wiped to remove any oil remnants. The collected particle suspension was centrifuged at 1,000 xg for 10 minutes at 4 °C (Figure 1A, step 4), the supernatant was discarded, and the pellet was resuspended in 25 µL of cold aqueous buffer solution, indicated as the outer solution. The aqueous outer solution was composed of 83 mM HEPES-KOH (pH=8), 21 mM magnesium acetate, 76 mM potassium acetate, 236.4 mM ammonium acetate, 4.5% (w/v) PEG, 61 mM D-(−)-3-Phosphoglyceric acid disodium salt, 3.8 mM of each natural amino acid (alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine), 1.8 mM ATP, 1.5 mM GTP, 1.2 mM UTP and 1.5 mM IPTG. The particles were incubated for 2 hours at 37ºC without shaking to enable protein production within the particles.
This protocol enabled the production of different volumes of particle suspensions, by maintaining the same volumetric ratios; for up to 50 µL particle suspension - 1.5 ml plastic tubes were used, otherwise 15 mL tubes were used. When the effect of amino acids and nucleotide presence was evaluated, one or both of these components were omitted from the inner or the outer solution. In this case, the particles were incubated for 4 hours to allow full protein synthesis to occur.
Lipid composition analysis of the particles
HPLC (1260 infinity, Agilent Technologies, Santa Clara, California, USA) equipped with an ELSD detector was employed to quantify the lipid composition of the liposomes. Lipid separation was achieved by using Luna C18 column, 5 mm, 100Å (Phenomenex LTP, Aschaffenburg, Germany) and an isocratic mobile phase compose of 87% 4mM ammonium acetate in methanol/10 % isopropanol/3% 4mM ammonium acetate in water. A constant flow rate of 1mL min-1 at 30 °C was applied. ELSD settings were adjusted to 3.5 bar of the inert gas flow and 40 °C as the nebulizer temperature. POPC and cholesterol calibration curve were obtained according to the peak areas in the obtained chromatograms. Liposomes were diluted 1:1 in methanol, centrifuged at 1,000 xg for 10 minutes to eliminate all non-lipid materials, and the supernatant was analyzed for its lipid composition (n=3).
Flow cytometry analysis
Particles were analyzed using flow cytometer instruments after the production of sfGFP inside them. 10,000 events were collected for each analyzed sample. Prior the analysis, the particles were diluted 25 fold in phosphate-buffered saline (PBS) and filtered with 70 µm cell strainer (BD Biosciences, San Jose, CA, USA) to eliminate aggregates.
In general, BD LSR-II analyzer (BD Biosciences, San Jose, CA, USA) with a 488 nm laser was used for excitation and a 530±30 nm filter for emission was used. Amnis ImageStream®X Mark II (Amnis, Seattle, WA, USA) with 405 nm, 488 nm and 561 nm lasers was used to analyze the particles that were membrane-labeled with rhodamine and stained with 1 µg mL-1 Hoechst 33342 (Life Technologies, USA). The emission spectrum was detected by 435-505 nm, 505-560 nm, 560-595 nm and 642-745 nm for Hoechst, GFP, rhodamine and bright field signals, respectively. Analysis was performed using the IDEAS analysis software (Amnis, Seattle, WA, USA), and the particles’ concentration and their diameter and volume distributions were calculated based on the rhodamine signal (n=3).
Microscopy analyses
Confocal microscope (LSM-710, Zeiss, Germany) was used to analyze sfGFP producing particles and to visualize the interaction and effect of the particles on cell culture. The PE cytotoxic effect was evaluated as follows: 4T1 over-expressing mCherry cells were seeded in an optical µ-slide 8 wells plate (ibdi, Madison, WI, USA), in an RPMI medium with 5 µg mL-1 Puromycin for 24 hours. Afterwards, the media were replaced with different treatments for an overnight incubation. Before imaging, the cells were stained with 1 µg mL-1 Hoechst for nuclei labelling. Acquisition was performed using the ZEN software and applying the 405, 488, 543, 639 nm lasers.
Establishment of a breast cancer tumor model
All animal studies were approved by, and complied with the institutional ethical committee at the Technion – Israel Institute of Technology. Fifty µL of 6×106 cells ml-1 of 4T1 over-expressing mCherry cell line were injected to 10-week-old BALB/c female mice (Harlan Laboratories Inc., Jerusalem, Israel); A BD Micro-Fine plus 29G insulin syringe (BD, New Jersey, United States) was used to inject the cells into each mouse’s 5th fat pad. Animal well-being was monitored daily by the research team and the Technion Pre-Clinical Research Authority veterinary staff.
Renilla luciferase production in vivo
Renilla luciferase production inside particles in vivo was monitored using IVIS 200 imaging system (PerkinElmer, Inc., Massachusetts, USA). Fifty µL of particles with luciferase-encoding DNA or without a DNA template were prepared and mixed with 10 µL of 170 µM of coelentrazine (Promega, Wisconsin, USA) - the enzyme’s substrate. The particles were kept on ice to eliminate in vitro protein production. The prepared mixtures of protein producing particles were injected intra-tumor to 10-week-old female BALB/c mice bearing a 4T1 cell-induced tumor. The experiment was carried 6 days after the tumor establishment. The mice were scanned in 10 minutes intervals for 1 hour, using the following IVIS scanning parameters: luminescence scan, working distance C, exposure time of 60 sec, large binning large and 1 f/s. During the experiment, the mice were anesthetized and their body temperature was kept using a red lamp.
In vivo PE effect
PE production and effect in vivo was evaluated by intra-tumor injection of different treatment mixtures to BALB/c mice bearing a 4T1 cell-induced tumor: 50 µL of particles with PE encoding DNA, particles without DNA, 120 µg mL-1 of purified PE and an untreated control group. All injected mixtures also included 25 µL of particles with sfGFP encoding DNA to verify the injection site in the following histology analysis. Each treatment group received the appropriate treatment for 2 subsequent days. At the third day, the mice were euthanized and the tumors were extracted and kept in 4% natural buffer formalin at room temperature for 24 hours before they were paraffin embedded.
To evaluate the levels of apoptotic markers expression in the tumor tissue, the slide from the tumors were analyzed by immunofluorescence. All slides were gone through antigen retrieval and blocking with 2.5% normal goat serum blocking solution (Vector Labratories inc., California, USA). Then, the slide were stained with polyclonal anti-caspase-3 antibody (rabbit source) (Abcam, Cambridge, United Kingdom), following a staining with a polyclonal secondary antibody to rabbit IgG H&L (Alexa Fluor® 647) (goat source) (Abcam, Cambridge, United Kingdom). Antibodies were diluted by 1:50 in a blocking serum. In addition, slides were stained with anti-GFP antibody – ChIP grade (rabbit source) (Abcam, Cambridge, United Kingdom) as a primary antibody for sfGFP detection, indicating the particles injection position.
The stained slides were scanned and analyzed using a 3D Histech Pannoramic MIDI scanner (3D Histech, Budapest, Hungary) using the following scanning parameters: a ×20 objective magnification, 30 msec DAPI exposure time, 700 msec EGFP and CY3 exposure times. The resulted images were analyzed using Fiji program to evaluate the caspase-3 and sfGFP levels in the tissue.
Statistical Analysis
Statistical analysis was performed using F test for variances to test if the different populations have the same variance. Accordingly, differences between experimental groups were evaluated using an unpaired, one or two-tailed disribution student’s T-test. Differences were considered significant at a p-value <0.05. Bar graphs and average values present data as means ± standard deviations of the mean.
Supplementary Material
Acknowledgements
This work was supported by ERC-STG-2015-680242.
Ms. Krinsky wishes to thank the Baroness Ariane de Rothschild Women Doctoral Program for its generous support.
Previous work related to this topic was supported by the Technion Integrated Cancer Center (TICC), the Russell Berrie Nanotechnology Institute, the Lorry I. Lokey Interdisciplinary Center for Life Sciences & Engineering, the Pre-Clinical Research Authority staff and the Biomedical Core Facility at the Rappaport Faculty of Medicine, as well as the Israel Ministry of Economy for a Kamin Grant (52752); the Israel Ministry of Science Technology and Space – Office of the Chief Scientist (3-11878); the Israel Science Foundation (1778/13); the Israel Cancer Association (2015-0116); the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); a Mallat Family Foundation Grant; A.S. acknowledges Alon and Taub Fellowships.
Contributor Information
Nitzan Krinsky, Laboratory for Targeted Drug Delivery and Personalized Medicine Technologies, Department of Chemical Engineering, Technion – Israel Institute of Technology, Haifa 32000, Israel; The Interdisciplinary Programs for Biotechnology, Technion – Israel Institute of Technology, Haifa 3200003, Israel.
Prof. Itai Benhar, Department of Molecular Microbiology and Biotechnology, Faculty of Life Sciences, Tel-Aviv University, Tel-Aviv 6997801, Israel
Dr. Dov Hershkovitz, Department of Pathology, Tel-Aviv Sourasky Medical Center, Tel Aviv 6423906, Israel
Dr. Avi Schroeder, Laboratory for Targeted Drug Delivery and Personalized Medicine Technologies, Department of Chemical Engineering, Technion – Israel Institute of Technology, Haifa 32000, Israel.
References
- [1].a) Torchilin VP. Nat Rev Drug Discovery. 2014;13:813. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bakh NA, Cortinas AB, Weiss MA, Langer RS, Anderson DG, Gu Z, Dutta S, Strano MS. Nat Chem. 2017;9:937. doi: 10.1038/nchem.2857. [DOI] [PubMed] [Google Scholar]; c) Chen Z, Wang J, Sun W, Archibong E, Kahkoska AR, Zhang X, Lu Y, Ligler FS, Buse JB, Gu Z. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He M. New Biotechnol. 2008;25:126. [Google Scholar]
- [3].Mohr BP, Retterer ST, Doktycz MJ. Expert Rev Proteomics. 2016;13:707. doi: 10.1080/14789450.2016.1209415. [DOI] [PubMed] [Google Scholar]
- [4].Krinsky N, Kaduri M, Shainsky-Roitman J, Goldfeder M, Ivanir E, Benhar I, Shoham Y, Schroeder A. PLoS One. 2016;11:e0165137. doi: 10.1371/journal.pone.0165137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Salehi AS, Smith MT, Bennett AM, Williams JB, Pitt WG, Bundy BC. Biotechnol J. 2016;11:274. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- [6].Tran K, Gurramkonda C, Cooper MA, Pilli M, Tarris J, Selock N, Han TC, Tolosa M, Zuber A, Penalber-Johnstone C, Dinkins C, et al. Biotechnol Bioeng. 2017 doi: 10.1002/bit.26439. [DOI] [PubMed] [Google Scholar]
- [7].Fujii S, Matsuura T, Sunami T, Nishikawa T, Kazuta Y, Yomo T. Nat Protoc. 2014;9:1578. doi: 10.1038/nprot.2014.107. [DOI] [PubMed] [Google Scholar]
- [8].Luisi PL, Ferri F, Stano P. Die Naturwissenschaften. 2006;93:1. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- [9].Blain JC, Szostak JW. Annu Rev Biochem. 2014;83:615. doi: 10.1146/annurev-biochem-080411-124036. [DOI] [PubMed] [Google Scholar]
- [10].Pereira de Souza T, Stano P, Luisi PL. Chembiochem. 2009;10:1056. doi: 10.1002/cbic.200800810. [DOI] [PubMed] [Google Scholar]
- [11].Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Nat Biotechnol. 2001;19:751. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- [12].Lentini R, Santero SP, Chizzolini F, Cecchi D, Fontana J, Marchioretto M, Del Bianco C, Terrell JL, Spencer AC, Martini L, Forlin M, et al. Nat Commun. 2014;5 doi: 10.1038/ncomms5012. 4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES. Nat Chem. 2017;9:431. doi: 10.1038/nchem.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nomura S, Tsumoto K, Hamada T, Akiyoshi K, Nakatani Y, Yoshikawa K. ChemBioChem. 2003;4:1172. doi: 10.1002/cbic.200300630. [DOI] [PubMed] [Google Scholar]
- [15].Noireaux V, Libchaber A. Proc Natl Acad Sci USA. 2004;101:17669. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Elani Y, Law RV, Ces O. Phys Chem Chem Phys. 2015;17:15534. doi: 10.1039/c4cp05933f. [DOI] [PubMed] [Google Scholar]
- [17].Walde P, Cosentino K, Engel H, Stano P. ChemBioChem. 2010;11:848. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- [18].Schroeder A, Goldberg MS, Kastrup C, Wang Y, Jiang S, Joseph BJ, Levins CG, Kannan ST, Langer R, Anderson DG. Nano Lett. 2012;12:2685. doi: 10.1021/nl2036047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Nishimura K, Matsuura T, Nishimura K, Sunami T, Suzuki H, Yomo T. Langmuir. 2012;28:8426. doi: 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]; b) Martini L, Mansy SS. Chem Commun. 2011;47:10734. doi: 10.1039/c1cc13930d. [DOI] [PubMed] [Google Scholar]; c) Pautot S, Frisken BJ, Weitz D. Proc Natl Acad Sci USA. 2003;100:10718. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Pontani LL, van der Gucht J, Salbreux G, Heuvingh J, Joanny JF, Sykes C. Biophys J. 2009;96:192. doi: 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Pautot S, Frisken BJ, Weitz DA. Langmuir. 2003;19:2870. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stano P, Kuruma Y, Souza TP, Luisi PL. In: Liposomes: Methods and Protocols, Volume 2: Biological Membrane Models. Weissig V, editor. Vol. 606. Humana Press; Totowa, NJ: 2010. p. 127. [Google Scholar]
- [21].Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat Biotechnol. 2006;24:79. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- [22].Torchilin VP. Nat Rev Drug Discovery. 2005;4:145. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- [23].Caschera F, Lee JW, Ho KK, Liu AP, Jewett MC. Chem Commun. 2016;52:5467. doi: 10.1039/c6cc00223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ivanir E. Master of Science in Biotechnology: Technion - Israel Institute of Technology. 2009 Feb [Google Scholar]
- [25].Kimmerling RJ, Lee Szeto G, Li JW, Genshaft AS, Kazer SW, Payer KR, de Riba Borrajo J, Blainey PC, Irvine DJ, Shalek AK, Manalis SR. Nat Commun. 2016;7 doi: 10.1038/ncomms10220. 10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tawfik DS, Griffiths AD. Nat Biotechnol. 1998;16:652. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]; Caschera F, Noireaux V. Artificial life. 2016;22:185. doi: 10.1162/ARTL_a_00198. [DOI] [PubMed] [Google Scholar]
- [27].a) Spirin AS, Swartz JR. In: Cell-free Protein Synthesis: Methods and Protocols. Swartz ASSaJR., editor. WILEY-VCH Verlag GmbH & Co. KGaA; Weinheim: 2008. p. 1. [Google Scholar]; Sawasaki T, Hasegawa Y, Tsuchimochi M, Kamura N, Ogasawara T, Kuroita T, Endo Y. FEBS Lett. 2002;514:102. doi: 10.1016/s0014-5793(02)02329-3. [DOI] [PubMed] [Google Scholar]; b) Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB. Science. 1988;242:1162. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- [28].Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Nature. 2000;407:599. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- [29].Mathai JC, Tristram-Nagle S, Nagle JF, Zeidel ML. J Gen Physiol. 2008;131:69. doi: 10.1085/jgp.200709848. [DOI] [PMC free article] [PubMed] [Google Scholar]; Israelachvili JN. Intermolecular and Surface Forces. Academic Press; London: 1992. [Google Scholar]
- [30].a) FitzGerald DJ, Willingham MC, Pastan I. Cancer Treat Res. 1988;37:161. doi: 10.1007/978-1-4613-1083-9_11. [DOI] [PubMed] [Google Scholar]; b) Borowiec M, Gorzkiewicz M, Grzesik J, Walczak-Drzewiecka A, Salkowska A, Rodakowska E, Steczkiewicz K, Rychlewski L, Dastych J, Ginalski K. Toxins (Basel) 2016;8 doi: 10.3390/toxins8110321. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ogata M, Chaudhary VK, Pastan I, FitzGerald DJ. J Biol Chem. 1990;265:20678. [PubMed] [Google Scholar]; d) Ogata M, Fryling CM, Pastan I, FitzGerald DJ. J Biol Chem. 1992;267:25396. [PubMed] [Google Scholar]
- [31].Shapira A, Benhar I. Toxins. 2010;2:2519. doi: 10.3390/toxins2112519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Menestrina G, Pederzolli C, Forti S, Gambale F. Biophys J. 1991;60:1388. doi: 10.1016/S0006-3495(91)82176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, et al. Cell. 2016;167:248. doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- [34].Elmore S. Toxicol Pathol. 2007;35:495. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pisal DS, Kosloski MP, Balu-Iyer SV. J Pharm Sci. 2010;99:2557. doi: 10.1002/jps.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- [37].a) Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA, Corchero JL, Villaverde A, Schaffer L, Head SR, Souvignier C, et al. Microb Cell Fact. 2015;14:57. doi: 10.1186/s12934-015-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stech M, Hust M, Schulze C, Dubel S, Kubick S. Eng Life Sci. 2014;14:387. doi: 10.1002/elsc.201400036. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Stech M, Kubick S. Antibodies. 2015;4:12. [Google Scholar]
- [38].Benhar I, Wang Q-c, FitzGerald D, Pastan I. J Biol Chem. 1994;269:13398. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.