Abstract
Natural killer (NK) cells are innate lymphocytes that play an integral role in tumor rejection and viral clearance. Unlike their other lymphocyte counterparts, NK cells have the unique ability to recognize and lyse target cells without prior exposure. However, there are no known NK cell-specific genes that are exclusively expressed by all NK cells. Therefore, identification of NK cell-specific genes would allow a better understanding of why NK cells are unique cytotoxic lymphocytes. From the Immunological Genome (ImmGen) Consortium studies, we identified kruppel-like factor 12 (Klf12), encoding a novel transcription factor, preferentially expressed in C57BL/6 mouse NK cells. KLF12 was dispensable for NK cell development, IFN-γ production, degranulation, and proliferation in Klf12 knockout mice. RNA-sequencing analysis revealed increased expression of Btg3, an anti-proliferative gene, in KLF12-deficient NK cells compared to wild-type (WT) NK cells. Interestingly, competitive mixed bone marrow (BM) chimeric mice exhibited reduced development of KLF12-deficient NK cells, altered IFN-γ production and degranulation, and impairment of NK cell proliferation in vitro and in vivo in response to mouse cytomegalovirus (MCMV) infection. KLF12-deficient NK cells from BM chimeric mice also expressed higher levels of the interleukin-21 receptor (IL-21R), which resulted in increased IL-21R signaling and correlated with greater inhibition of NK cell proliferation. Furthermore, IL-21 induced Btg3 expression, which correlated with arrested NK cell maturation and proliferation. In summary, we found that KLF12 regulates mouse NK cell proliferation potentially by regulating expression of Btg3 via IL-21.
Introduction
Patients with genetic mutations resulting in diminished natural killer (NK) cell numbers or function succumb to recurrent herpesvirus and papillomavirus infections (1–4), highlighting the importance of NK cells in controlling certain viral infections. NK cells are cytotoxic lymphocytes that have the unique ability to recognize and lyse target cells without prior exposure. NK cells also secrete cytokines, such as interferon-γ (IFN-γ), to activate other immune cells to coordinate appropriate immune responses against pathogens (5).
Mouse cytomegalovirus (MCMV) infection is an ideal model to study NK cell activation, expansion, and effector function. At the onset of infection, IL-12 production by dendritic cells is critical for early NK cell production of IFN-γ and control of viral load (6–8). A subset of NK cells expressing the activating Ly49H+ receptor in C57BL/6 mice specifically recognizes the MCMV-encoded glycoprotein, m157 (9, 10). Ly49H+ NK cells expand, contract, and persist after MCMV infection (11). These cells conferred specific protection against MCMV re-challenge and not other heterologous infections, indicating that these are MCMV-specific memory NK cells (12, 13).
NK cells share expression of many genes with their lymphocyte counterparts; therefore, we sought to find genes preferentially expressed by NK cells in the hematopoietic cell lineage to understand their unique activation and cytotoxic capabilities. From the Immunological Genome (ImmGen) Consortium, we identified kruppel-like factor 12 (KLF12), a novel transcription factor, to be preferentially expressed in mouse NK cells. KLF12 is a zinc finger transcription factor in the Kruppel-like factor family. Similar to KLF3 and KLF8, KLF12 has a conserved PVDLS domain at the N-terminus that binds to the corepressor, CtBP1 (14–16). Klf12 transcripts are found in the kidney, endometrial stromal cells, primary gastric tumors, and various cancer cell lines (15, 17–19). Prior studies have demonstrated that KLF12 binds to a conserved CACCC sequence and functions as a transcriptional repressor or activator, suggesting that the function of KLF12 is context- and cell type-specific (17, 20, 21). KLF12 target genes are largely unknown, but include NR4A1 (Nur77), TRAP2A, FOXO1, SLC14A2, and EGR1 (17, 20, 22–25).
In this study, we assessed the role of KLF12 in mouse NK cells as a potential transcriptional regulator of NK cell development and/or effector functions. To address this, we generated a mouse with floxed Klf12 loci and crossed these the mice expressing β-actin Cre recombinase to delete KLF12 expression. We assessed the development, proliferation, and effector functions of KLF12-deficient NK cells in response to in vitro stimulation and MCMV infection.
Materials and Methods
Mice
Mice were obtained from the following sources: wild-type C57BL/6 (WT) and C57BL/6 CD45.1 mice were purchased from the National Cancer Institute (Frederick, MD), Rosa26-Flippase C57BL/6 mice from Dr. R. Locksley and β-actin Cre transgenic C57BL/6 mice from Dr. M. McManus, UCSF, and Klra8−/− (Ly49H-deficient) C57BL/6 mice from Dr. S. Vidal, McGill University. The Klf12 targeting vector was purchased from the International Mouse Knockout Consortium and electroporated into E14–129/Ola embryonic stem cells. Selected clones were then microinjected into C57BL/6 females and heterozygotes were backcrossed at least nine generations onto the C57BL/6 background. Klf12-floxed mice were genotype by PCR using the following primer pairs: WT, forward, 5’-CACAGCGAGTTCCCCAAGAT-3’, reverse 1, 5’-GGACGCACATACAGCTTCCT-3’, reverse 2, 5’-AGGGAAAGGGTCGAGAGACA-3’, Flox, forward, 5’-CACAGCGAGTTCCCCAAGAT-3’, reverse 3, 5’-TAGGAGGTGTGGCTTTGCTG-3’, reverse 4, 5’-CCCTTTGTTGTCCGCCTACT-3’. Mice were bred and housed in a specific pathogen-free facility and experiments were performed according to UCSF Institutional Animal Care and Use Committee and National Institutes of Health guidelines.
Quantitative PCR
Total RNA from splenocytes or indicated cellar populations was prepared using the Ambion RNAqueous kit and converted into cDNA. Quantitative PCR analysis with SYBR green master mix (Roche) was performed on the cDNA following standard conditions with the following primer pairs: Klf12, forward, 5’-CTGGCGAACCACATAGGCCCAG-3’, reverse, 5’-CGGCGGCCTACATTTACGTGAT-3’, Btg3, forward, 5’-TGCTGCCGGTATGGAGAGAA-3’, reverse, 5’-GGTCACCTTATCCAGAGCCC-3’, Hprt, forward, 5’-CACAGGACTAGAACACCTGC-3’, reverse, 5’-GCTGGTGAAAAGGACCTCT-3’. Primer pairs to confirm disrupted Klf12: Klf12 exons 2–3, forward, 5’-GCTAATGCTTGATGGAATGCC-3’, reverse, 5’-AGTTGTGGACGTTTGGAGAC-3’, exons 5–6, forward, 5’-ACATCCATCCCCGGTATCCA-3’, reverse, 5’-TGGCGTCTTGTGCTCTCAAT-3’. Expressions were normalized to HPRT.
Southern blot and long range PCR
Genomic DNA (gDNA) from selected stem cell clones was processed using the Promega Wizard gDNA purification kit. gDNA was digested overnight with EcoRV, transferred onto a membrane, probed with α−32P dATP against the 5’ arm of the Klf12 targeting vector, and exposed to film. Probes were amplified using the following primer pair: forward, 5’-TCTCCCTCTTGGTGGTCACT-3’, reverse, 5’-GATGCCTGAAAACCGCACAG-3’. The 3’ arm of the targeting vector was amplified by PCR using Takara Primestar GXL DNA polymerase with the following primers: forward, 5’-GGATCTCATGCTGGAGTTCTTCGCC-3’, reverse 1, 5’-CCAAAGCCCCTATACCCTTCCCCGC-3’, and reverse 2, 5’-ATCTGGCGTGGGCGGCCAGCAGTTC-3’.
Ex vivo NK cell stimulations and proliferation assays
Splenocytes were resuspended in RPMI-1640 supplemented with 10% FCS, 50 μM β-mercaptoethanol, 100 μM non-essential amino acids, 1 mM sodium pyruvate, 50 mM HEPES, and 2 μM L-glutamine. One million cells were stimulated for 6 hr in the presence of BD GolgiStop with 20 ng/ml recombinant mouse (rm) IL-12 (R&D Systems #419-ML/CF) and 10 ng/ml rmIL-18 (R&D Systems #B0025), with 20 ng/ml PMA and 200 ng/ml ionomycin, or with 10 μg/ml of plate-bound anti-NK1.1 antibody (clone PK136), were fixed and permeabilized with BD Cytofix/Cytoperm, and then stained for IFN-γ and CD107a. For phosphoSTAT5 (BD #612599) and phosphoSTAT3 (BD #557814) staining, one million cells were stimulated for the indicated times and concentrations with recombinant human (rh) IL-15 (R&D Systems #247-ILB) or rmIL-21 (Zymogenetics), fixed with 1% paraformaldehyde (PFA) in PBS, permeabilized with methanol, and stained. For proliferation assays, one million cell trace violet (Invitrogen #C34557)-labeled splenocytes were cultured in 10 ng/ml rhIL-15, 1×105 PFA-fixed RMA lymphoma cells, or mouse cytomegalovirus m157-transduced RMA cells supplemented with 50 U/ml rhIL-2 (NCI Preclinical Repository) or 100 ng/ml rmIL-21.
Bone marrow chimeras
Donor bone marrow was isolated from femurs and tibias of 8–10 wk-old WT and Klf12F/F C57BL/6 mice, mixed 1:1 in sterile PBS, and 5×106 cells were injected i.v. into WT recipients lethally irradiated with 1200 Gy. Recipient mice were injected i.p. with 200 μg anti-CD4 (GK1.5), 200 μg anti-CD8 (2.43), and 100 μg anti-NK1.1 (PK136) depleting antibodies at the time of transfer and maintained on antibiotic pellets (Bio-Serv, S0443) for 2 wks. Recipients reconstituted for at least 8 wks were used for experiments.
NK cell enrichment and adoptive transfer
NK cells were enriched from spleens using mAbs against Ter119, Gr-1, CD4, CD8, CD19, and CD5. Qiagen anti-rat IgG magnetic beads were used to deplete the indicated populations. Enriched NK cells from WT or KLF12-deficient mice for adoptive transfer were mixed 1:1 and injected i.v. into Ly49H-deficient C57BL/6 recipients one day prior to MCMV infection.
MCMV infection and viral titers
Mice were injected i.p. with 1×104 PFU or 2.5×103 PFU of Smith strain MCMV for adoptive NK cell transfer experiments. For MCMV quantitation, oral lavage was collected by washing the sublingual cavity with sterile saline solution and used directly in qPCR analysis (26). DNA was isolated from blood using a ReliaPrep Blood gDNA Miniprep System kit (Promega) and 1 μl was used for qPCR analysis. MCMV primer pairs: forward, 5’-AGCCACCAACATTGACCACGCAC-3’ and reverse, 5’-GCCCCAACCAGGACACACAACTC-3’.
RNA-sequencing
Total RNA from 6 wk-old Klf12+/+ and Klf12F/F β-actin Cre− littermate males was isolated from CD3−NK1.1+ NK cells, CD4+TCRβ+CD25− T cells, CD8+TCRβ+CD25− T cells, and B220+IgM+IgD+ B cells following the ImmGen standard protocol (https://www.immgen.org/). One thousand cells (≥ 99% purity) were double-sorted directly into 96-well plates containing TCL buffer (Qiagen) and 1% β-mercaptoethanol, frozen, and analyzed by the Broad Technology Labs for SmartSeq2 library preparation and NextSeq500 sequencing. Transcripts were quantified using Cuffquant and then normalized using DEseq (27–31). Raw data were interpreted by Database for Annotations, Visualization and Integrated Discovery (DAVID), analyzing all differentially expressed genes using default parameters (32).
Abs and flow cytometry
Single cell suspensions were incubated with anti-CD16 + CD32 (2.4G2) monoclonal antibody (mAb) to block Fc receptors 15 min on ice and then stained with mAbs to cell surface or intracytoplasmic antigens of interest. mAbs were purchased from BD Biosciences, BioLegend, eBioscience, and Tonbo Biosciences. Samples were acquired on an LSR II or LSR Fortessa (BD Biosciences) and analyzed on FlowJo software (Tree Star).
Statistical analysis
The unpaired, two-tailed Student t test was used to analyze results in Prism GraphPad. A p-value of ≤0.05 was considered significant. Error bars represent standard deviation (SD).
Results
Klf12 is preferentially expressed in mature mouse NK cells
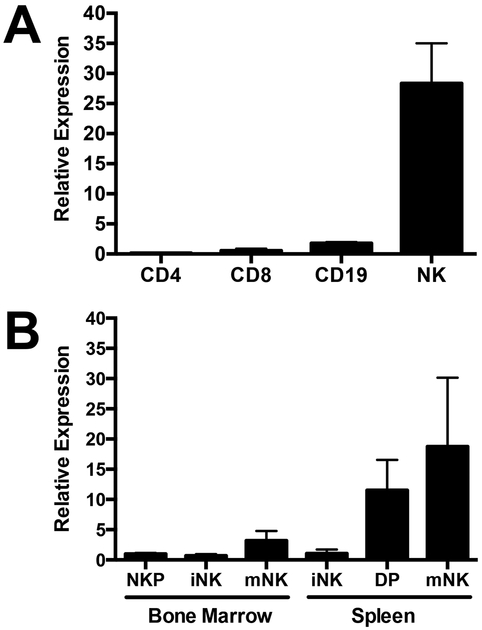
ImmGen results revealed that mouse NK cells preferentially expressed a novel transcription factor, kruppel-like factor 12, KLF12. Consistent with ImmGen data, quantitative PCR from highly purified lymphocyte populations from splenocytes of C57BL/6 mice confirmed that NK cells expressed 25-fold more Klf12 transcripts than CD4+, CD8+ T cells, and CD19+ B cells (Fig. 1A). Klf12 expression increased as splenic NK cells transitioned from semi-mature CD27+CD11b+ (DP) to fully mature CD27−CD11b+ (mNK) NK cells (Fig. 1B). Analysis of microarray and RNA-Seq data from ImmGen indicated absence or low transcription of Klf12 in all other hematopoietic cell types, including macrophages, monocytes, dendritic cells, granulocytes, mast cells, ILC2, ILC3, γδ T cells, thymocytes, iNKT cells, hematopoietic stem cells, and virus-activated CD8+ T cells (https://www.immgen.org). Furthermore, ILC1 do not express Klf12 (33). Taken together, mouse NK cells preferentially express Klf12.
FIGURE 1.
KLF12 is preferentially expressed in mature mouse NK cells. Quantitative PCR of Klf12 expression in (A) sorted splenic populations defined as CD4+ T cells: TCRβ+NK1.1−CD8−CD4+CD25−; CD8+ T cells: TCRβ+NK1.1−CD4−CD8+CD25−; CD19+ B cells: TCRβ−NK1.1−CD19+; NK cells: TCRβ−NK1.1+, and (B) sorted NK cell developmental stages in bone marrow defined as NKP: CD3−CD8−CD19−Ter119−Gr1−NK1.1−DX5−CD122+, iNK: CD122+NK1.1+DX5−, mNK: CD122+NK1.1+DX5+. Developmental stages in spleen defined as iNK: TCRβ−NK1.1+CD27+CD11b−, DP: TCRβ−NK1.1+CD27+CD11b+, mNK: TCRβ−NK1.1+CD27−CD11b+. Data are representative of 2 experiments (n = 3 mice/experiment).
NK cell development and function are normal in KLF12-deficient mice
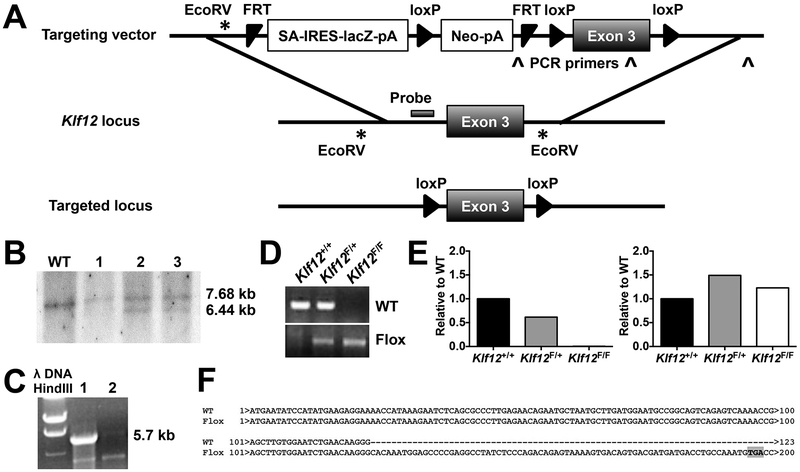
To investigate the role of KLF12 in NK cells, we generated Klf12 knockout mice using a targeted vector to excise exon 3 of Klf12 (Fig. 2A). Integration of the targeting vector was confirmed by the presence of a 6.44 kb band in the Southern blot of selected stem cell clones and a 5.7 kb band detected in mice by long range PCR (Fig. 2B, 2C). The lacZ and neomycin cassettes were removed by crossing to Rosa26-Flippase mice, resulting in progeny mice bearing loxP sites flanking exon 3 of Klf12 (Fig. 2A). β-actin Cre recombinase excised exon 3 of Klf12 and the progeny were genotyped by PCR (Fig. 2D). Klf12 transcripts without exon 3 were detected in whole splenocytes from Klf12F/F mice (Fig. 2E) that resulted in a pre-mature translational stop codon (Fig. 2F). The putative truncated protein encodes a 65 amino acid peptide lacking all functional KLF12 protein domains. However, as none of the antibodies currently available are KLF12-specific, we were unable to measure KLF12 protein in WT or knockout mice. The Klf12-null mice bred in predicted Mendelian ratios and there were no abnormalities observed in growth or weight compared to WT or heterozygous littermates. T cell development in the thymus and spleen and T cell development in bone marrow, spleen, lymph nodes, and the peritoneal cavity is normal in Klf2-deficient mice when compared with WT and heterozygous littermates (Supplemental Fig. 1). Additionally, stimulation of CD4+ and CD8+ T and B cells isolated from lymph nodes through their antigen receptors by immobilized anti-CD3 or anti-IgM, respectively, resulted in downstream signaling of ERK (Supplemental Fig. 2) and proliferation (Supplemental Fig. 3) equivalent to WT or heterozygous littermates.
FIGURE 2.
Targeted disruption of the Klf12 locus. (A) Schematic of the targeting strategy into the Klf12 locus to generate Klf12 conditional knockout mice after excision of the lacZ and neomycin cassettes by Flippase recombination. (B) Southern blot of gDNA from selected ES cell clones digested with EcoRV and hybridized to a probe indicated in (A). The 7.68 kb band is the WT allele and the 6.44 kb band is the targeted allele. (C) Long range PCR of genomic (g)DNA from heterozygous (lane 1) and WT (lane 2) mice amplifying the 3’ arm of the targeting vector with primers indicated in (A). The 5.7 kb band is the targeted allele. (D) PCR of mouse gDNA from indicated genotypes. (E) Quantitative PCR amplifying exons 2–3 (left) or exons 5–6 (right) of Klf12. (F) PCR sequence of Klf12 cDNA from WT or Klf12F/F β-actin Cre+ mice. The premature stop codon is highlighted in grey. (E-F) Data are representative of n = 3 mice/genotype.
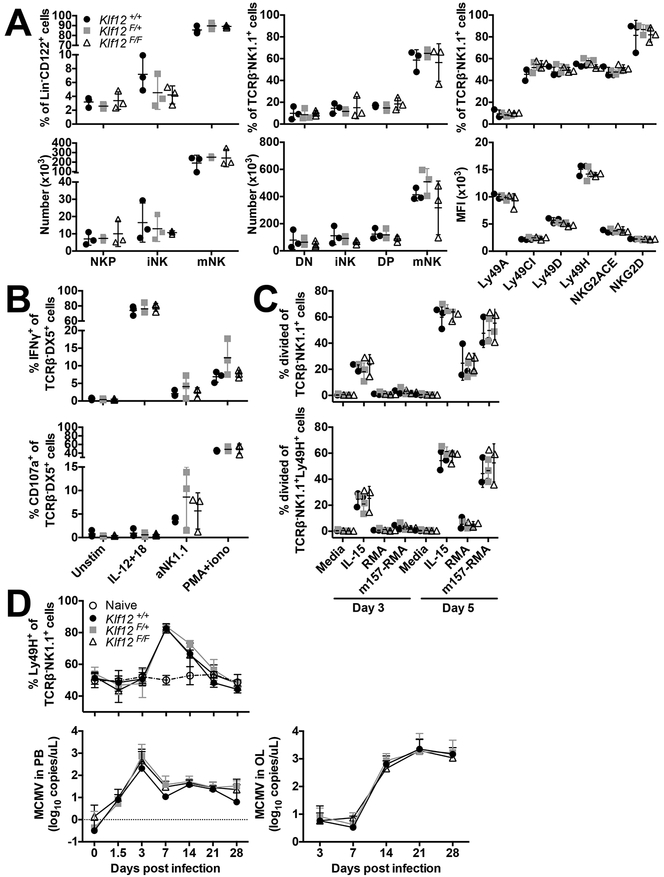
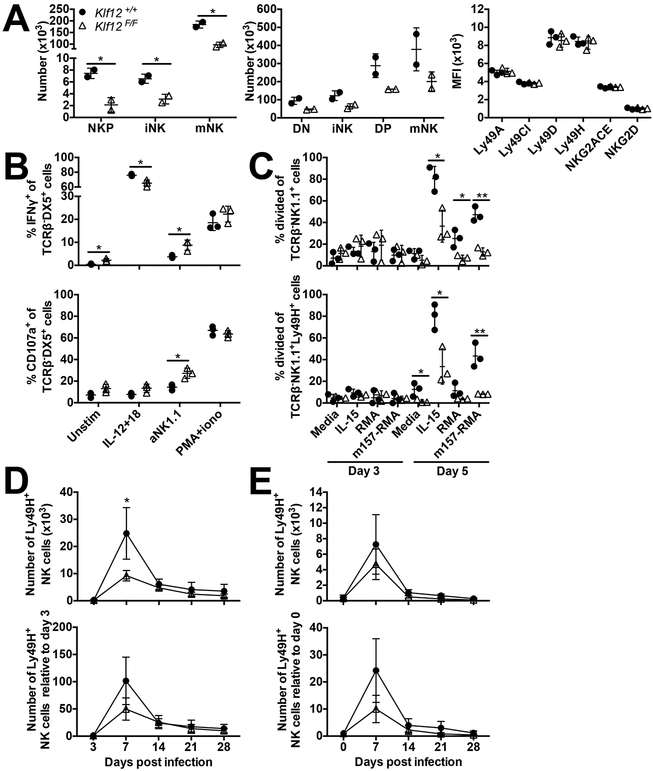
Phenotypic analysis of Klf12+/+, Klf12F/+, and Klf12F/F NK cells was performed to determine whether KLF12 is required for NK cell development. We examined NK developmental subsets as defined by expression of CD27 and CD11b and expression of Ly49 and NKG2 activating and inhibitory receptors in the bone marrow (BM) and spleen. Klf12F/+ and Klf12F/F mice had normal frequencies and numbers of all NK cell developmental subsets in the BM and spleen, equivalent to Klf12+/+ littermate controls. Expression of activating and inhibitory receptors was also similar to Klf12+/+ controls (Fig. 3A). Furthermore, the number and frequency of liver ILC1 and NK cells was equivalent in Klf12+/+ and Klf12F/F mice (data not shown). Therefore, KLF12 deficiency does not affect NK cell development or expression of NK receptors.
FIGURE 3.
NK cell development and function are normal in KLF12-deficient mice. (A) Percentage and total cell numbers of bone marrow (left panels) and splenic (middle panels) NK cell developmental subsets in Klf12+/+ (circle), Klf12F/+ (square), and Klf12F/F (triangle) mice. DN NK cell developmental subset defined as TCRβ−NK1.1+CD27−CD11b−. NK receptor expression (right panels) gated on splenic TCRβ−NK1.1+ NK cells. (B) IFN-γ and CD107a staining gated on TCRβ−DX5+ NK cells after in vitro stimulation for 6 hr. (A-B) Data are representative of 3 experiments (n = 3 mice/genotype/experiment). (C) Proliferation of TCRβ−NK1.1+ and TCRβ−NK1.1+Ly49H+ NK cells after in vitro stimulation. Data are representative of 2 experiments (n = 3 mice/genotype/experiment). (D) Percentage of Ly49H+ cells gated on TCRβ−NK1.1+ NK cells in the blood and viral titers in the blood and oral lavage following MCMV infection. Data are representative of 2 experiments (n = 2–4 mice/genotype/experiment).
We assessed whether KLF12 deficiency affected NK cell effector functions. Klf12F/+ and Klf12F/F NK cells produced interferon-γ (IFN-γ) and degranulated similar to Klf12+/+ NK cells upon stimulation with interleukin (IL)-12 and −18, anti-NK1.1, or phorbol 12-myristate 13-acetate (PMA) and ionomycin (Fig. 3B). Furthermore, the in vitro proliferative capacity of Klf12F/+ and Klf12F/F NK cells upon stimulation with IL-15, the RMA lymphoma cell line, and MCMV m157 (ligand of the activating Ly49H receptor)-transduced RMA cells was comparable to Klf12+/+ NK cells (Fig. 3C). Therefore, KLF12 deficiency does not affect the in vitro NK cell effector functions and cytokine- and antigen-driven proliferation in the knockout mice.
We also tested in vivo NK cell responses upon MCMV infection. Klf12+/+, Klf12F/+, and Klf12F/F mice were infected with Smith strain MCMV and expansion of Ly49H+ NK cells and viral titers were monitored over 28 days. Naïve, uninfected Klf12+/+ and Klf12F/F mice had a similar percentage of Ly49H+ NK cells in the blood. After MCMV infection, there was comparable expansion of Ly49H+ NK cells in Klf12+/+, Klf12F/+, and Klf12F/F mice. Viral titers in the blood and oral lavage were indistinguishable (Fig. 3D). Therefore, KLF12 deficiency does not alter NK cell effector functions in vitro and upon in vivo MCMV challenge.
KLF12-deficient NK cells intrinsically express more Btg3 transcripts
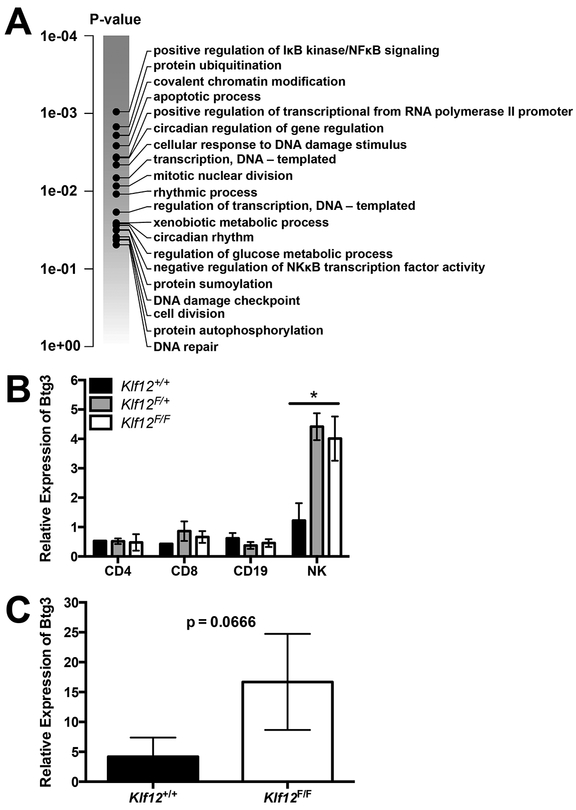
We hypothesized that redundancy or compensatory mechanisms with other KLF members may mask the effects of KLF12 deficiency in NK cell development and function. Therefore, we performed RNA-Seq to determine the effects of KLF12 deficiency on the transcriptome of Klf12F/F mice. Total RNA was collected from purified T cells, B cells, and NK cells from 6 wks old Klf12+/+ and Klf12F/F β-actin Cre− littermate male mice. Klf12F/F NK cells had increased transcripts involved in regulating cellular transcription, NFκB activity, and cellular division compared to Klf12+/+ NK cells (Fig. 4A). Of the differentially expressed genes, we confirmed increased expression of B cell translocation gene 3, Btg3, encoding an anti-proliferative protein, in Klf12F/F NK cells but not in Klf12F/F T cells, B cells, or Klf12+/+ NK cells (Fig. 4B). Furthermore, Klf12F/F NK cells isolated from mixed BM chimeric mice had more Btg3 transcripts than Klf12+/+ NK cells purified from the same mice (Fig. 4C). Altogether, KLF12-deficient NK cells intrinsically express more Btg3 transcripts. By contrast, no significant transcriptional differences were observed comparing Klf12+/+ and Klf12F/F T cells and B cells, indicating that the alterations in Klf12F/F NK cells were cell intrinsic.
FIGURE 4.
KLF12-deficient NK cells intrinsically express more Btg3 transcripts. (A) Gene set enrichment analysis of Klf12+/+ and Klf12F/F splenic NK cells defined as CD3−NK1.1+ (n = 3 mice/genotype). Quantitative PCR of Btg3 expression in (B) sorted splenic populations defined as CD4+ T cells: TCRβ+NK1.1−CD8−CD4+CD25−; CD8+ T cells: TCRβ+NK1.1−CD4−CD8+CD25−; CD19+ B cells: CD3−NK1.1−CD19+; NK cells: CD3−NK1.1+, and (C) sorted splenic TCRβ−NK1.1+ NK cells from Klf12+/+ and Klf12F/F BM chimeric mice. (B-C) Data are representative of 2 experiments (n = 2–3 mice/genotype/experiment for (B)) and (n = 3 mice/experiment for (C)). Full transcription datasets are available at gene expression omnibus GSE128962.
KLF12-deficient NK cells are competitively disadvantaged
Although in the intact KLF12-deficient mice we observed no marked alteration in NK cell phenotype, in many cases gene deficiencies in knockout mice are only revealed when the gene-deficient cells are in competition with WT cells. Therefore, we generated mixed BM chimeras of Klf12+/+ and Klf12F/F cells at a 1:1 ratio to assess the role of KLF12 in NK cells in a competitive setting. After reconstitution of the hematopoietic cells for about two months, we observed reduced absolute numbers of Klf12F/F NK cell developmental subsets in the BM and spleen (Fig. 5A). The levels of expression (as reflected by mean fluorescence intensity (MFI)) of activating and inhibitory NK receptors were comparable between Klf12+/+ and Klf12F/F cells (Fig. 5A). In a competitive setting, Klf12F/F NK cells produced less IFN-γ upon IL-12 + IL-18 stimulation, but more IFN-γ upon anti-NK1.1 stimulation. Klf12F/F NK cells also degranulated more than Klf12+/+ NK cells upon anti-NK1.1 stimulation (Fig. 5B). Thus, KLF12 deficiency alters NK cell development and effector function, but not receptor expression, when the NK cells are developed in a competitive setting. By contrast, Klf12F/F and WT CD4+ and CD8+ T cells responded equivalently to MCMV infection (Supplemental Fig. 4).
FIGURE 5.
KLF12-deficient NK cells are competitively disadvantaged. (A) Total cell numbers of bone marrow (left panel) and splenic (middle panel) NK cell subsets in bone marrow chimeras at a 1:1 ratio of Klf12+/+ (circle) and Klf12F/F (triangle) cells. NK receptor expression (right panel) gated on splenic TCRβ−NK1.1+ NK cells. Data are representative of 6 experiments (n = 3 mice/experiment). (B) IFN-γ and CD107a staining gated on TCRβ−DX5+ NK cells after in vitro stimulation for 6 hr. Data are representative of 3 experiments (n = 3 mice/experiment). (C) Proliferation of TCRβ−NK1.1+ and TCRβ−NK1.1+Ly49H+ NK cells after in vitro stimulation. Data are representative of 8 experiments (n = 2–3 mice/experiment). (D-E) Number (top panels) and relative change in number (bottom panels) of TCRβ−NK1.1+Ly49H+ NK cells in the blood during MCMV infection in (D) bone marrow chimeras and (E) adoptive transfer model where a 1:1 ratio of Klf12+/+ and Klf12F/F TCRβ−NK1.1+Ly49H+ NK cells were transferred into Ly49H-deficient hosts one day prior to MCMV infection. Data are representative of (D) 3 experiments (n = 3–6 mice/experiment) or (E) 6 experiments (n = 3–5 mice/experiment). *p < 0.05, ** p < 0.005.
Because we observed increased expression of the anti-proliferative gene, Btg3, in Klf12F/F NK cells, we directly assessed whether in vitro proliferation of Klf12F/F NK cells was preferentially affected in BM chimeras compared to Klf12+/+ NK cells. We observed a statistically significant proliferative impairment of Klf12F/F NK cells in BM chimeric mice in response to IL-15 or co-culture with m157-expressing RMA lymphoma cells compared to Klf12+/+ NK cells (Fig. 5C). Klf12F/F NK cells also had impaired proliferation in vivo when BM chimeric mice were infected with MCMV. At day 7 post-MCMV infection, we observed a statistically significant reduction in the numbers of Ly49H+ Klf12F/F NK cells compared to Ly49H+ Klf12+/+ NK cells (Fig. 5D). Furthermore, this observation was recapitulated when a mixture of mature Ly49H+ Klf12+/+ and Klf12F/F NK cells were adoptively transferred into Ly49H-deficient mice and infected with MCMV (Fig. 5E). Together, these findings reveal that KLF12 deficiency impairs NK cells to proliferate in response to cytokines and antigens when in competition with WT NK cells.
KLF12-deficient NK cells from bone marrow chimeric mice have altered common γ chain receptor expression but normal IL-15R signaling
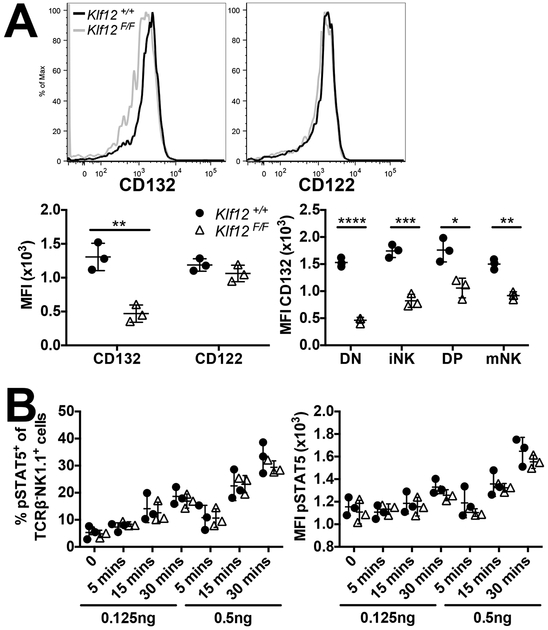
The common γ chain (CD132) cytokine receptor family is important for lymphocyte development during BM reconstitution and for lymphocyte activation. We observed slightly decreased CD132 expression on Klf12F/F NK cells compared to Klf12+/+ NK cells in BM chimeric mice, whereas expression of the β-chain (CD122) was identical. Furthermore, CD132 expression was significantly decreased on all Klf12F/F splenic NK cell developmental subsets compared to Klf12+/+ NK cells (Fig. 6A). Expression of CD132, but not CD122, was decreased on KLF12-deficient NK cells from mixed BM chimeric mice.
FIGURE 6.
KLF12-deficient NK cells from BM chimeric mice have decreased CD132 expression but normal responsiveness to IL-15. (A) Representative histogram of CD132 and CD122 expression on Klf12+/+ (black line) and Klf12F/F (grey line) splenic TCRβ−NK1.1+ NK cells. MFI of CD132 and CD122 expression on splenic TCRβ−NK1.1+ NK cells (left panel) and NK cell developmental subsets (right panel). Data are representative of 2 experiments (n = 3 mice/experiment). (B) Percentage and MFI of pSTAT5 in splenic TCRβ−NK1.1+ NK cells upon ex vivo IL-15 stimulation. Data are representative of 5 experiments (n = 2–3 mice/experiment). *p < 0.05, ** p < 0.005, *** p < 0.001, **** p < 0.0001.
We examined which members of the common γ chain cytokine receptor family might be affected by the decreased expression of CD132 on KLF12-deficient NK cells. We hypothesized that the responsiveness of IL-15 might be affected by KLF12 deficiency because it is important for NK cell development (34, 35). We examined the phosphorylation of STAT5 in NK cells from BM chimeric mice upon IL-15 stimulation as a functional measure of responsiveness to IL-15. The percentages of NK cells responding and MFI of pSTAT5 were similar between Klf12+/+ and Klf12F/F NK cells upon culture with low and high concentrations of IL-15. Thus, responsiveness to IL-15 remains intact and unaffected by KLF12 deficiency.
KLF12-deficient NK cells from bone marrow chimeric mice have increased IL-21R expression and signaling correlating with less NK cell proliferation
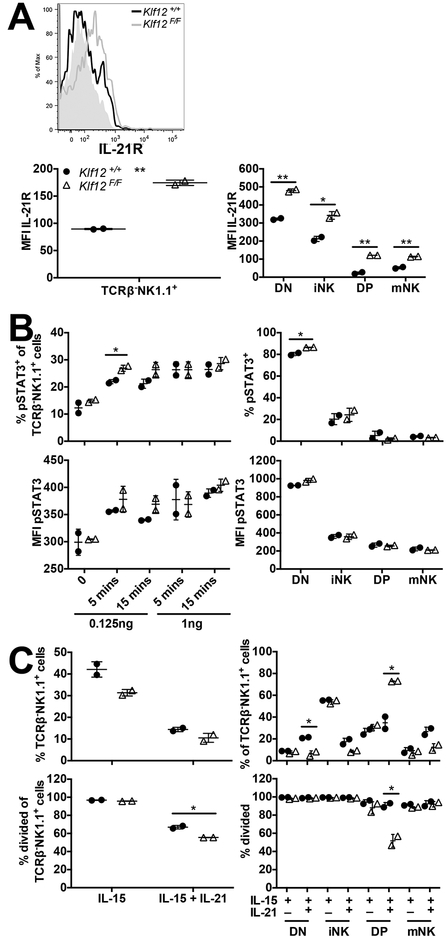
The IL-21R is another member of the common γ chain cytokine receptor family that has been implicated in NK cell maturation and proliferation (36–38). Although IL-21R is dispensable for mouse NK cell development, IL-21 induces their maturation but inhibits their proliferation (37). Furthermore, IL-21 enhances NK cell responses against tumors expressing ligands for the activating NK receptor, NKG2D (39). Given the inhibitory role of the IL-21R in NK cell proliferation, we assessed IL-21R expression on NK cells in BM chimeric mice. Interestingly, we observed higher levels of IL-21R expression on splenic Klf12F/F NK cells. This was also evident on all splenic Klf12F/F NK cell developmental subsets. In fact, the most immature subset of NK cells, lacking expression of CD27 and CD11b (termed DN), expressed the highest levels of the IL-21R (Fig. 7A). Thus, expression of the IL-21R is increased on mouse NK cells by KLF12 deficiency.
FIGURE 7.
IL-21R expression and signaling are increased in KLF12-deficient NK cells from mixed BM chimeric mice. (A) Representative histogram of IL-21R expression on Klf12+/+ (black line) and Klf12F/F (grey line) splenic TCRβ−NK1.1+ NK cells. The grey-filled histogram is the fluorescence minus one control. MFI of IL-21R expression on splenic TCRβ−NK1.1+ NK cells (left panel) and NK cell developmental subsets (right panel). Data are representative of 2 experiments (n = 2 mice/experiment). (B) Percentage and MFI of pSTAT3 in splenic TCRβ−NK1.1+ NK cells (left panels) upon IL-21 stimulation ex vivo and NK cell developmental subsets (right panels) upon 0.125 ng/ml IL-21 stimulation ex vivo for 15 min. Data are representative of 2 experiments (n = 2 mice/experiment). (C) Percentage of splenic TCRβ−NK1.1+ NK cells (left panels) and NK cell developmental subsets (right panels) after 7 days of in vitro culture in 10 ng/ml IL-15 in the absence or presence of 100 ng/ml IL-21. Data are representative of 2 experiments (n = 2 mice/experiment). *p < 0.05, ** p < 0.005.
We measured pSTAT3 expression upon IL-21 stimulation to determine whether increased IL-21R expression on KLF12-deficient NK cells resulted in increased IL-21-induced signaling. At a low concentration of IL-21, we detected increased percentages and levels of expression (MFI) of pSTAT3 in Klf12F/F NK cells compared to Klf12+/+ NK cells in the mixed BM chimeras (Fig. 7B). Furthermore, it was the most immature subset of NK cells (DN) that upregulated pSTAT3 the most compared to other developmental subsets. This is not surprising because the DN subset expressed the highest levels of IL-21R and therefore should have increased pSTAT3 upregulation.
Subsequently, we evaluated whether increased IL-21R expression and signaling in KLF12-deficient NK cells correlated with greater IL-21-mediated inhibition of proliferation. We cultured splenocytes from mixed BM chimeric mice with IL-15 in the presence or absence of IL-21 and monitored the maturation and percentage of NK cells for 7 days. We observed reduced percentages of Klf12F/F NK cells upon IL-15 culture compared to Klf12+/+ NK cells (Fig. 7C). By day 7 of IL-15 culture, all NK cells had proliferated (measured by the percentage of divided cells) and we were unable to observe differences in proliferation as we observed on day 5 of IL-15 culture (Fig. 5C). Addition of IL-21 to the culture reduced the overall percentages of NK cells and inhibited the proliferation of Klf12F/F NK cells more than Klf12+/+ NK cells. In fact, the most immature Klf12F/F NK cell DN subset was significantly reduced in percentage by IL-21, but there was no difference in proliferation of total NK cells, possibly because the Klf12F/F NK cell DN subset were driven to mature in response to the IL-21. Although there was a higher percentage of Klf12F/F semi-mature CD27+CD11b+ (DP) subset of NK cells, they did not proliferate as well as their Klf12+/+ counterparts. KLF12 deficiency in NK cells resulted in increased expression and signaling of the IL-21R. This enhanced IL-21R signaling correlated with greater inhibition of NK cell proliferation, particularly in the most immature DN NK cell subset.
IL-21 stimulation induces Btg3 expression in NK cells
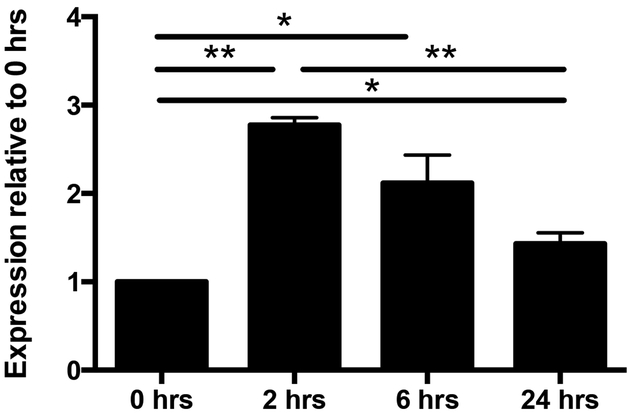
Given that KLF12-deficient NK cells have impaired proliferation, intrinsically express more Btg3 transcripts, and express higher levels of the IL-21R, we sought to determine whether IL-21 might inhibit NK cell proliferation via BTG3. To address this, we cultured enriched WT NK cells in IL-21 and assessed Btg3 expression. We observed a significant increase in Btg3 expression after 2 hr of IL-21 stimulation and then a progressive decrease in expression with time. Even after 24 hr of continual IL-21 stimulation, Btg3 expression remains significantly increased (Fig. 8). Thus, IL-21 directly induces Btg3 expression in NK cells and correlates with the decreased proliferative activity.
FIGURE 8.
IL-21 induced Btg3 expression in NK cells. Quantitative PCR of Btg3 expression in enriched C57BL/6 wild-type splenic TCRβ−NK1.1+ NK cells cultured with 100 ng/ml of IL-21 for the indicated times. Expression of Btg3 was normalized to 0 hr. Data are representative of 3 experiments (n = 2–3 mice/experiment). *p < 0.05, ** p < 0.005.
Discussion
Unlike their other lymphocyte counterparts, NK cells have the unique ability to recognize and lyse target cells without prior exposure via expression of their germline-encoded receptors. However, there are no NK cell-specific genes that are exclusively expressed by all NK cells. Transcription factors, signaling components downstream of NK activating or inhibitory receptors, and even expression of certain NK activating receptors are shared amongst NK cells and other lymphocytes. From the ImmGen studies, we identified Klf12, encoding a novel transcription factor, to be preferentially expressed in mouse NK cells and not in ILC1, ILC2, ILC3, T cells, or B cells. In this study, we generated Klf12 knockout mice to assess its role in NK cell development or effector function. We used β-actin Cre recombinase to delete KLF12 in all cells. We observed normal lymphocyte development, proliferation, and activation in KLF12-deficient mice (data not shown). We also generated mice in which KLF12 was conditionally deleted only in NK cells using Ncr1-Cre mice with similar results (data not shown). However, we found that in competitive mixed BM chimeras, KLF12-deficient NK cells demonstrated less robust proliferation that correlated with higher levels of expression of Btg3, an anti-proliferative gene, which is upregulated by IL-21. Notably, KLF12-deficient NK cells express higher levels of IL-21R and have elevated pSTAT3 signaling in response to IL-21 compared to WT NK cells.
In the absence of competition with WT NK cells, KLF12 is dispensable for NK cell lineage commitment, development, and effector functions. KLF12-deficient NK cells produced IFN-γ and degranulated equivalently as WT NK cells. Furthermore, NK cell proliferation in vitro and in vivo in the context of MCMV infection was also unperturbed by KLF12 deficiency in the Klf12F/F mice. The lack of a NK cell phenotype may be due to compensatory mechanisms or redundancy among other members of the KLF family, particularly KLF3 and KLF8. Both KLF3 and KLF8 share the conserved PVDLS domain with KLF12 (14, 16) and KLF3 is highly expressed in mouse NK cells (https://www.immgen.org/). Interestingly, KLF12-deficient NK cells have similar expression of KLF3 and KLF3 transcripts as WT NK cells (data not shown, GSE128962). However, we were unable to address whether compensatory mechanisms masked KLF12 deficiency in vivo because of embryonic lethality in C57BL/6 KLF3-deficient mice and KLF3, KLF8-double knockout mice (40, and personal communication with M. Crossley). Further studies generating NK cell-specific double- or triple- knockout mice would be required to uncover potential compensatory pathways in KLF12-deficient NK cells.
Recent studies have shown that KLF12 regulates proliferation of many cancer cell lines. Over-expression of KLF12 in endometrial and lung cancer cell lines correlated with decreased apoptosis, increased cellular proliferation, and increased in vivo tumor growth (41–43). Conversely, down regulation of KLF12 resulted in a proliferative defect in multiple cancer cell lines (19, 23, 44–47). These observations were recapitulated in primary human cancer cells and mouse kidney cells (48,49). Here, we observed a proliferative defect in KLF12-deficient NK cells in a competitive setting in BM chimeric mice upon antigen- and cytokine-mediated proliferation. Although we observed a proliferative impairment upon IL-15 stimulation, the expression of pSTAT5 was comparable between KLF12-deficient and WT NK cells. Interestingly, we detected increased pSTAT3 expression in KLF12-deficient NK cells upon IL-21 stimulation, which correlated with greater IL-21-mediated inhibition of proliferation. Similarly, over-expression of KLF12 in an endometrial adenocarcinoma cell line resulted in decreased pSTAT3 expression upon leukemia inhibitory factor (LIF) stimulation, which inhibits cellular differentiation (42). The proliferative defect in KLF12-deficient NK cells might be an intrinsic effect of pSTAT3 expression.
Alternatively, the proliferative impairment observed in KLF12-deficient NK cells may be due to the increased expression of an anti-proliferative transcription factor, Btg3. BTG3 is a member of the anti-proliferative BTG/Tob protein family that inhibits entry into the S-phase of cell cycle progression. It has been shown that upon DNA damage, p53 binds to the BTG3 promoter and induces its expression in order to regulate cell cycle checkpoints (50). Downregulation of BTG3 expression is associated with enhanced cell proliferation, growth, and migration (51, 52). Conversely, over-expression of BTG3 is associated with suppressed proliferation, reduced cancer invasiveness, and cellular apoptosis in primary cancers and cancer cell lines (53–55).
During MCMV infection, IL-21 induces mouse NK cells to produce IL-10, which affects dendritic cell activation and CD8+ T cell responses (38, 56–58). We also observed proliferative impairment of KLF12-deficient NK cells during MCMV infection in the mixed BM chimeric mice and in the adoptive transfer of mature NK cells. IL-21 present during MCMV infection may be inhibiting NK cell proliferation and inducing IL-10 production to modulate adaptive immune responses. However, we observed comparable numbers of KLF12-deficient and WT CD4+ AND CD8+ naïve, effector memory, and MCMV-specific NKG2D+ CD8+ T cells in MCMV-infected BM chimeric mice (data not shown), suggesting that the T cell responses against MCMV are normal.
Although the IL-21R is not required for mouse NK cell development, IL-21 induces NK cell maturation, inhibits their proliferation, and enhances their NKG2D-mediated anti-tumor response (36–39). In this study, we noted increased expression of the IL-21R on KLF12-deficient NK cells compared to WT NK cells in mixed BM chimeric mice. Increased IL-21R expression resulted in enhanced IL-21 signaling and correlated with inhibition of KLF12-deficient NK cell proliferation. It appears as though IL-21 is driving NK cell maturation, but in the absence of KLF12 NK cell maturation may be arrested at the semi-mature DP stage. This is consistent with the fact that KLF12 is upregulated at the semi-mature DP stage. Moreover, IL-21 alone induces Btg3 expression in NK cells, which may reinforce the proliferative defect in KLF12-deficient NK cells in competitive situations. Conditional deletion of Btg3 and Il21r in NK cells will be needed to definitively address this hypothesis.
Supplementary Material
Key Points.
KLF12 deficiency does not affect NK cell development and effector functions.
KLF12-deficient NK cells have increased expression of Btg3 and the IL-21 receptor.
IL-21 induced Btg3 and inhibited KLF12-deficient NK cell proliferation.
Acknowledgments
We thank the UCSF Cell and Genome Engineering Core for assistance and Dr. Hong-Erh Liang in the Dr. Richard Locksley lab for guidance in generating Klf12 conditional knockout mice, the ImmGen Consortium for performing the RNA-Seq experiment, Dr. Alice Chan in the Dr. Mark Anderson lab for technical guidance, and members of the Lanier Lab for helpful discussions.
This work was supported by National Science Foundation Grant GRFP 1650113, the National Institute of Health Grant T32AI007334 (to V.C.L.), and AI068129 (to L.L.L).
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Biron CA, Byron KS, and Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med 320: 1731–1735. [DOI] [PubMed] [Google Scholar]
- 2.Mace EM and Orange JS. 2019. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol. Rev 287: 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, Cohen JI, Frenkel EP, Bagwell JC, Sullivan JL, Biron CA, Spalding C, Zerbe CS, Uzel G, Holland SM, and S Orange J. 2013. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood 121: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Notarangelo LD, and Mazzolari E. 2006. Natural killer cell deficiencies and severe varicella infection. J. Pediatr 148: 563–564. [DOI] [PubMed] [Google Scholar]
- 5.Lam VC and Lanier LL. 2016. NK cells in host responses to viral infections. Curr. Opin. Immunol 44: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orange JS and Biron CA. 1996. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J. Immunol 156: 4746–4756. [PubMed] [Google Scholar]
- 7.Orange JS, Wang B, Terhorst C, and Biron CA. 1995. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J. Exp. Med 182: 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orange JS and Biron CA. 1996. An absolute and restricted requirement for IL-12 in natural killer cell IFN-gamma production and antiviral defense. Studies of natural killer and T cell responses in contrasting viral infections. J. Immunol 156: 1138–1142. [PubMed] [Google Scholar]
- 9.Arase H, Mocarski ES, Campbell AE, Hill AB, and Lanier LL. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296: 1323–1326. [DOI] [PubMed] [Google Scholar]
- 10.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, Fremont DH, and Yokoyama WM. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U. S. A : 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, and Yokoyama WM. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol 2: 951–956. [DOI] [PubMed] [Google Scholar]
- 12.Sun JC, Beilke JN, and Lanier LL. 2009. Adaptive immune features of natural killer cells. Nature 457: 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min-Oo G and Lanier LL. 2014. Cytomegalovirus generates long-lived antigen-specific NK cells with diminished bystander activation to heterologous infection. J. Exp. Med 211: 2669–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang DT, Pevsner J, and Yang VW. 2000. The biology of the mammalian Krüppel-like family of transcription factors. Int. J. Biochem. Cell Biol 32: 1103–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth C, Schuierer M, Günther K, and Buettner R. 2000. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12). Genomics 63: 384–390. [DOI] [PubMed] [Google Scholar]
- 16.Schuierer M, Hilger-Eversheim K, Dobner T, Bosserhoff AK, Moser M, Turner J, Crossley M, and Buettner R. 2001. Induction of AP-2alpha expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. J. Biol. Chem 276: 27944–27949. [DOI] [PubMed] [Google Scholar]
- 17.Suda S, Rai T, Sohara E, Sasaki S, and Uchida S. 2006. Postnatal expression of KLF12 in the inner medullary collecting ducts of kidney and its trans-activation of UT-A1 urea transporter promoter. Biochem. Biophys. Res. Commun 344: 246–252. [DOI] [PubMed] [Google Scholar]
- 18.Shen X, Hu Y, Jiang Y, Liu H, Zhu L, Jin X, Shan H, Zhen X, Sun L, Yan G, and Sun H. 2013. Krüppel-like factor 12 negatively regulates human endometrial stromal cell decidualization. Biochem. Biophys. Res. Commun 433: 11–17. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, Imoto I, Katai H, Yamaguchi T, Inazawa J, Hirohashi S, Ishikawa Y, and Shibata T. 2009. Krüppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int. J. Cancer 125: 1859–1867. [DOI] [PubMed] [Google Scholar]
- 20.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, and Buettner R. 1999. Transcriptional regulation of the AP-2alpha promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol. Cell. Biol 19: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, and Lloyd JA. 2005. A functional screen for Krüppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells. Mol. Dis 35: 227–235. [DOI] [PubMed] [Google Scholar]
- 22.Huang C, Jiang Y, Zhou J, Yan Q, Jiang R, Cheng X, Xing J, Ding L, Sun J, Yan G, and Sun H. 2017. Increased Krüppel-like factor 12 in recurrent implantation failure impairs endometrial decidualization by repressing Nur77 expression. Reprod. Biol. Endocrinol 10.1186/s12958-017-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SH, Park YY, Cho SN, Margalit O, Wang D, and DuBois RN. 2016. Krüppel-Like Factor 12 Promotes Colorectal Cancer Growth through Early Growth Response Protein 1. PloS One. 10.1371/journal.pone.0159899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Zhu X, Chen J, Jiang Y, Zhang Q, Kong C, Xing J, Ding L, Diao Z, Zhen X, Sun H, and Yan G. 2015. Krüppel-like factor 12 is a novel negative regulator of forkhead box O1 expression: a potential role in impaired decidualization. Reprod. Biol. Endocrinol 10.1186/s12958-015-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Zhang H, Jiang Y, Xue B, Diao Z, Ding L, Zhen X, Sun H, Yan G, and Hu Y. 2015. MicroRNA-181a is involved in the regulation of human endometrial stromal cell decidualization by inhibiting Krüppel-like factor 12. Reprod. Biol. Endocrinol 10.1186/s12958-015-0019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamimura Y and Lanier LL. 2014. Rapid and sequential quantitation of salivary gland-associated mouse cytomegalovirus in oral lavage. J. Virol. Methods 205: 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg S. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, and Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 30.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R. 2014. R.Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9: 171–181. [DOI] [PubMed] [Google Scholar]
- 31.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A. 2014. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol 107: 4.22.1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, and Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 33.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, Busslinger M, Smyth MJ, Belz GT, and Carotta S. 2014. Differential requirement for Nfil3 during NK cell development. J. Immunol 192: 2667–2676. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, and Peschon JJ. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med 191: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, and Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9: 669–676. [DOI] [PubMed] [Google Scholar]
- 36.Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, West J, Schrader S, Burkhead S, Heipel M, Brandt C, Kuijper JL, Kramer J, Conklin D, Presnell SR, Berry J, Shiota F, Bort S, Hambly K, Mudri S, Clegg C, Moore M, Grant FJ, Lofton-Day C, Gilbert T, Rayond F, Ching A, Yao L, Smith D, Webster P, Whitmore T, Maurer M, Kaushansky K, Holly RD, and Foster D. 2000. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature 408: 57–63. [DOI] [PubMed] [Google Scholar]
- 37.Kasaian MT, Whitters MJ, Carter LL, Lowe LD, Jussif JM, Deng B, Johnson KA, Witek JS, Senices M, Konz RF, Wurster AL, Donaldson DD, Collins M, Young DA, and Grusby MJ. 2002. IL-21 limits NK cell responses and promotes antigen-specific T cell activation: a mediator of the transition from innate to adaptive immunity. Immunity 16: 559–569. [DOI] [PubMed] [Google Scholar]
- 38.Brady J, Hayakawa Y, Smyth MJ, and Nutt SL. 2004. IL-21 induces the functional maturation of murine NK cells. J. Immunol 172: 2048–2058. [DOI] [PubMed] [Google Scholar]
- 39.Takaki R, Hayakawa Y, Nelson A, Sivakumar PV, Hughes S, Smyth MJ, and Lanier LL. 2005. IL-21 enhances tumor rejection through a NKG2D-dependent mechanism. J. Immunol 175: 2167–2173 [DOI] [PubMed] [Google Scholar]
- 40.Funnell AP, Mak KS, Twine NA, Pelka GJ, Norton LJ, Radziewic T, Power M, Wilkins MR, Bell-Anderson KS, Fraser ST, Perkins AC, Tam PP, Pearson RC, and Crossley M. 2013. Generation of mice deficient in both KLF3/BKLF and KLF8 reveals a genetic interaction and a role for these factors in embryonic globin gene silencing. Mol. Cell. Biol 33: 2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding L, Ding Y, Kong X, Wu J, Fu J, Yan G, and Zhou H. 2019. Dysregulation of Krüppel-like factor 12 in the development of endometrial cancer. Gynecol. Oncol 152: 177–184. [DOI] [PubMed] [Google Scholar]
- 42.Huang C, Sun H, Wang Z, Liu Y, Cheng X, Liu J, Jiang R, Zhang X, Zhen X, Zhou J, Chen L, Ding L, Yan G, and Jiang Y. 2018. Increased Krüppel-like factor 12 impairs embryo attachment via downregulation of leukemia inhibitory factor in women with recurrent implantation failure. Cell Death Discov. doi: 10.1038/s41420-018-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godin-Heymann N, Brabetz S, Murillo MM, Saponaro M, Santos CR, Lobley A, East P, Chakravarty P, Matthews N, Kelly G, Jordan S, Castellano E, and Downward J. 2016. Tumour-suppression function of KLF12 through regulation of anoikis. Oncogene 35: 3324–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mak CS, Yung MM, Hui LM, Leung LL, Liang R, Chen K, Liu SS, Qin Y, Leung TH, Lee KF, Chan KK, Ngan HY, and Chan DW. 2017. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol. Cancer doi.org/10.1186.s12943-017-0582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu M, Jin H, Xu C, Sun B, Mao Z, Bi W, and Wang Y. 2014. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget 5: 9472–9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan B, Li Q, Shen L, Rao Q, Wang Y, Zhu Y, Zhou XJ, and Li XH. 2016. MicroRNA-205 directly targets Krüppel-like factor 12 and is involved in invasion and apoptosis in basal-like breast carcinoma. Int. J. Oncol 49: 720–734. [DOI] [PubMed] [Google Scholar]
- 47.Du Y, Chen Y, Wang F, and Gu L. 2016. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med 37: 13557–13569. [DOI] [PubMed] [Google Scholar]
- 48.Song P and Yin SC. 2019. Long non-coding RNA 319 facilitates nasopharyngeal carcinoma carcinogenesis through regulation of miR-1207–5p/KLF12 axis. Gene 680: 51–58. [DOI] [PubMed] [Google Scholar]
- 49.Shin Y, Kim DY, Ko JY, Woo YM, and Park JH. 2018. Regulation of KLF12 by microRNA-20b and microRNA-106a in cystogenesis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 32: 3574–3582. [DOI] [PubMed] [Google Scholar]
- 50.Ou YH, Chung PH, Hsu FF, Sun TP, Chang WY, and Shieh SY. 2007. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 26: 3968–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren XL, Zhu XH, Li XM, Li YL, Wang JM, Wu PX, Lv ZB, Ma MH, Liao WT, Wang W, Ding YQ, and Liang L. 2015. Down-regulation of BTG3 promotes cell proliferation, migration and invasion and predicts survival in gastric cancer. J. Cancer Res. Clin. Oncol 141: 397–405. [DOI] [PubMed] [Google Scholar]
- 52.Liu L, Liu S, Duan Q, Chen L, Wu T, Qian H, Yang S, Xin D, He Z, and Guo Y. 2017. MicroRNA-142–5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am. J. Transl. Res 9: 2394–2402. [PMC free article] [PubMed] [Google Scholar]
- 53.Du Y, Liu P, Zang W, Wang Y, Chen X, Li M, and Zhao G. 2015. BTG3 upregulation induces cell apoptosis and suppresses invasion in esophageal adenocarcinoma. Mol. Cell. Biochem 404: 31–38. [DOI] [PubMed] [Google Scholar]
- 54.Mao D, Qiao L, Lu H, and Feng Y. 2016. B-cell translocation gene 3 overexpression inhibits proliferation and invasion of colorectal cancer SW480 cells via Wnt/β-catenin signaling pathway. Neoplasma 63: 705–716. [DOI] [PubMed] [Google Scholar]
- 55.An Q, Zhou Y, Han C, Zhou Y, Li F, and Li D. 2017. BTG3 Overexpression Suppresses the Proliferation and Invasion in Epithelial Ovarian Cancer Cell by Regulating AKT/GSK3β/β-Catenin Signaling. Reprod. Sci 24: 1462–1468. [DOI] [PubMed] [Google Scholar]
- 56.Biron CA and Tarrio ML. 2015. Immunoregulatory Cytokine Networks: 60 Years of Learning from Murine Cytomegalovirus. Med. Microbiol. Immunol 204: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen H, Chen SY, Folkersen L, Nolan GP, and Lanier LL. 2017. EBI3 regulates the NK cell response to mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A 114: 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, and Biron CA. 2009. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J. Exp. Med 206: 2235–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.