Abstract
The ability to regenerate the entire retina and restore lost sight after injury is found in some species and relies mostly on the epigenetic plasticity of Müller glia. To understand the role of mammalian Müller glia as a source of progenitors for retinal regeneration, we investigated changes in gene expression during differentiation of retinal progenitor cells (RPCs) into Müller glia. We also analyzed the global epigenetic profile of adult Müller glia. We observed significant changes in gene expression during differentiation of RPCs into Müller glia in only a small group of genes. We found a high similarity between RPCs and Müller glia on the transcriptomic and epigenomic levels. Our findings also indicate that Müller glia are epigenetically very close to late-born retinal neurons, but not early-born retinal neurons. Importantly, we found that key genes required for phototransduction were highly methylated. Thus, our data suggest that Müller glia are epigenetically very similar to late RPCs. Meanwhile, obstacles for regeneration of the entire mammalian retina from Müller glia may consist of repressive chromatin and highly methylated DNA in the promoter regions of many genes required for the development of early-born retinal neurons. In addition, DNA demethylation may be required for proper reprogramming and differentiation of Müller glia into rod photoreceptors.
Keywords: retina, Müller glia, development, epigenetics, histone modifications, DNA methylation
Introduction
Vision loss, resulting from retinal diseases or injuries, has a strong influence on normal human life [1–3]. Current treatment paradigms are all essentially oriented on slowing the rate of neurodegenerative changes; the time has come to look beyond this model and seek ways to regenerate lost retinal tissue – a strategy that is utilized by some species, but is not found mammals. Teleost fish, such as zebrafish (Danio rerio), have a remarkable self-healing ability to regenerate the damaged retina[4–10]. The regeneration of the entire retina and restoration of lost sight after injury relies mostly on the epigenetic plasticity of Müller glia in these animals [4–10]. The epigenetic plasticity of zebrafish’s Müller glia allows them to undergo a reprogramming process in response to injury, producing a proliferating population of retinal progenitor cells (RPCs), which later differentiate into all retinal cell types and restore vision (a self-healing retina) [4–10]. However, mammalian Müller glia have reduced reprogramming abilities due to restricted epigenetic plasticity [11–14]. Even when mammalian Müller glia are genetically and/or pharmacologically forced to be reprogramed, these cells generate low numbers of late-born retinal neurons, such as bipolar cells and rod photoreceptors [11–14]. Thus, a comprehensive characterization of the Müller glia epigenetic state is required to understand the restricted epigenetic plasticity of these cells.
To understand the role and place of Müller glia as a source of progenitors for retinal regeneration, we need to look into Müller glia origins. In the developing retina, RPCs generate all types of retinal neurons and Müller glia in a temporal order: retinal ganglion cells (RGCs) are the first-born and Müller glia are the last-born retinal cell type [15–17]. During retinal development, RPCs undergo a transition from early RPCs to late RPCs [15–17]. While early RPCs give rise to late RPCs and early-born retinal neurons (RGCs, cone photoreceptors, horizontal cells, and early-born amacrine cells), late RPCs generate only late-born retinal neurons (bipolar cells, rod photoreceptors, and late-born amacrine cells) and Müller glia [15–17]. Notch signaling is the key signaling cascade that regulates retinal development and is responsible for maintaining the RPC phenotype and, later, the Müller glia phenotype[15, 16]. The differentiation of RPCs into retinal neurons requires reduced Notch signaling activity following reduced expression of Hes1 and Hes5 transcription factors, which primarily inhibit the expression of pro-neuronal transcription factors (i.e. reduced Hes1 and Hes5 expression allows neuronal phenotypes in retina) [15, 16]. In differentiating Müller glia, Hes1 and Hes5 are expressed in a high and sustained manner, preventing all possible neuronal phenotypes [15, 16]. Thus, all of these data and the ability of adult mammalian Müller glia to be reprogramed into progenitors and differentiate only into late-born retinal neurons suggest that mammalian Müller glia might only be reprogrammed into late RPCs. However, the reason for the very low efficiency of reprogramming Müller glia into late RPCs in mammals and the mechanism by which zebrafish Müller glia are able to reprogram into all types of RPCs and generate the entire retina after injury remains unclear. Investigation of the Müller glia epigenetic state in mammals could lead to significant understanding of this mystery. In this study, we compared RPC and Müller glia transcriptomes during retinal development and performed an epigenetic study of adult murine Müller glia in order to identify possible mechanisms that prevent mammalian Müller glia from regenerating the entire retina.
Material and Methods
Animals
All experiments were performed in compliance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals, the Association for Research in Vision and Ophthalmology (ARVO) statement for use of animals in ophthalmic and vision research, and the University of Miami Institutional Animal Care and Use Committee’s (IACUC) approved protocol. C57BL/6 J (stock number 000664) mice were obtained from the Jackson Laboratory (Bar Harbor, Maine, United States). Mice were housed under standard conditions of temperature and humidity, with a 12-hour light to dark cycle and free access to food and water.
Isolation of Notch1+ cells, Glast+ cells, and cortical astrocytes
We used the same protocol for immunomagnetic cell separation described in Dvoriantchikova et al. to isolate Notch1+ cells with monoclonal biotin-conjugated anti-Notch1 mouse antibodies (130–096-557, Miltenyi Biotec, Auburn, CA) [18]. To isolate Glast+ cells from retinas, we used The Neural Tissue Dissociation Kit (130–093-231, Miltenyi Biotec, Auburn, CA), which was specially optimized for use with anti-Glast (ACSA-1) antibodies (130–118-984, Miltenyi Biotec, Auburn, CA) in immunomagnetic cell separation. Isolated P0, P3, P7, and P14 Notch1+ cells and P7, P14, P21, and P28 Glast+ cells were used for RNA purification (qRT-PCR, microarray study). P28 Glast+ cells were used for the ChIP-seq study and the purified DNA was used for whole genome bisulfite sequencing (WGBS). Primary cortical astrocytes were obtained using the ‘shaking method’ as previously described [19].
Immunohistochemistry
Immunohistochemistry for Glast (Slc1a3) and Gfap was performed as described previously [20]. Briefly, the fixed retinas were sectioned to a thickness of 100 μm with a Vibratome (Leica Microsystems) and immunostained with the anti-Glast (ACSA-1) antibody (130–118-984, Miltenyi Biotec, Auburn, CA), the anti-Gfap antibody (Table 1), and species-specific secondary fluorescent antibodies (Thermo Fisher Scientific, Carlsbad, CA). Control sections were incubated without primary antibodies. Imaging was performed with a Leica TSL AOBS SP5 confocal laser microscope (Leica Microsystems).
Table 1.
List of oligonucleotides and antibodies used in this study
| Gene | PCR primers |
|---|---|
| Prm1 | F: 5’ – ACGCAGGAGTTTTGATGGAC - 3’ R: 5’ – CCCTCTCACCACTTTTCTTACC – 3’ |
| Gas2l1 | F: 5’ – ACAAGCAAACGTAGCACCAC – 3’ R: 5’ – GACCAGCCAGACAGCAAAC – 3’ |
| Gapdh | F: 5’ – GGTGCTGAGTATGTCGTGGA – 3’ R: 5’ – GTCTTCTGGGTGGCAGTGAT – 3’ |
| Ascl1 | F: 5’ – CATCTCCCCCAACTACTCCA – 3’ R: 5’ – GGTTGGCTGTCTGGTTTGTT – 3’ |
| Hes5 | F: 5’ – CAAGGAGAAAAACCGACTGC – 3’ R: 5’ – GTGCAGGGTCAGGAACTGTA – 3’ |
| Notch1 | F: 5’ – TGTTGTGCTCCTGAAGAACG – 3’ R: 5’ – GTGGGAGACAGAGTGGGTGT – 3’ |
| Sox9 | F: 5’ – TGCAGCACAAGAAAGACCAC – 3’ R: 5’ – CCCTCTCGCTTCAGATCAAC – 3’ |
| Otx2 | F: 5’ – GGGCTGAGTCTGACCACTTC – 3’ R: 5’ – GGCCTCACTTTGTTCTGACC – 3’ |
| Il10ra | F: 5’ – AACTGCCAAGCCCTTCCTAT – 3’ R: 5’ – AAGCGAGTCTCAGTGGTGGT – 3’ |
| Il10rb | F: 5’ – CGTGGAAGACACCATCATTG – 3’ R: 5’ – TGGTCGAGAAGAAACCCTTG – 3’ |
| Foxn4 | F: 5’ – CACAGACCCCACCTCTTCAT – 3’ R: 5’ – TCACTGTTGCCAGAGACAGG – 3’ |
| Slc1a3 | F: 5’ – GCCTATCCAGTCCAACGAAA – 3’ |
| R: 5’ – CAAGAAGAGGATGCCCAGAG – 3’ | |
| Glul | F: 5’ – GGACAAATGCGGAGGTTATG – 3’ R: 5’ – ACTGGTGCCTCTTGCTCAGT – 3’ |
| Gfap | F: 5’ – AGAAAGGTTGAATCGCTGGA −3’ R: 5’ – CGGCGATAGTCGTTAGCTTC – 3’ |
| Protein | Primary antibody |
| Slc1a3 | #130–095-822; Miltenyi Biotec |
| Gfap | # C9205; MilliporeSigma |
| Glul | # MAB302; MilliporeSigma |
Immunocytochemistry
After immunomagnetic cell separation, 5 × 104 Glast+ cells from P7 and P28 were placed on PDL- and laminin-pretreated cover slips in a drop (70 μl) of Neurobasal media and kept in a CO2 incubator for 30 min to allow the cells to attach. After this, the cells were fixed in 4% PFA and blocked with 5% normal donkey serum with 0.15% Tween-20 in PBS at pH 7.4. Cells were then incubated with the anti-Glul antibody and anti-Gfap antibody (Table 1) followed by species-specific fluorescent secondary antibodies (Thermo Fisher Scientific, Carlsbad, CA). Negative controls were incubated with the secondary antibody only. DAPI was used to visualize the nucleus of the cells. Imaging was performed with a confocal laser microscope (Leica TSL AOBS SP5; Leica Microsystems). Individual cover slips were sampled randomly to collect a total of 10 images using a 20X objective lens. The Glul (glutamine synthetase, GS; Müller glia marker), Gfap (astrocyte marker), and DAPI positive cells (total cell number) were counted using ImageJ software. The percentage of Glul and Gfap positive cells, relative to the total number of cells, was calculated.
Quantitative RT-PCR analysis
Quantitative RT-PCR analysis was executed using gene-specific primers as described before (Table 1) [18, 20]. Briefly, RNA was purified from samples using the Absolutely RNA® Nanoprep kit (Agilent Technologies, Santa Clara, CA) and reverse transcribed with SuperScript III Reverse Transcriptase (ThermoFisher Scientific, Grand Island, NY) to produce cDNA. Quantitative PCR was then performed (Rotor-Gene Q, Qiagen, Valencia, CA) using a kit (SYBR GREEN PCR MasterMix; Qiagen, Valencia, CA). Relative expression was calculated by comparison with a standard curve following normalization to expression of the housekeeping gene Gapdh. Data are presented as average ± SEM. Quantitative RT-PCR data were analyzed with the Student’s t-test. Values of P < 0.05 were designated as statistically significant.
RNA extraction, probe preparation, and array hybridization
Three independent biological replicates were obtained for comparative profiling of purified P0 Notch1+, P3 Notch1+, P7 Notch1+, P14 Notch1+, and P14 Glast+ cells. Two independent biological replicates were obtained for comparative profiling of purified P7 Glast+, P21 Glast+ and P28 Glast+ cells. RNA was purified from samples using the Absolutely RNA® Nanoprep kit (Agilent Technologies, Santa Clara, CA). RNA samples were sent to Ocean Ridge Biosciences (Palm Beach Gardens, FL, USA) for processing using MEEBO microarrays. Biotin-labeled complementary RNA (cRNA) was prepared from the total RNA according to Van Gelder’s protocol [21]. Biotinylated cRNA samples were fragmented, diluted in a hybridization buffer, and loaded on to the MEEBO microarray slides (for more information on the MEEBO oligonucleotide set please refer to http://alizadehlab.stanford.edu/). The slides were hybridized for 16–18 hours in a Model 400 hybridization oven (Scigene, Sunnyvale, CA). After hybridization, the microarray slides were washed, stained with Streptavidin-Alexa-647 (ThermoFisher Scientific, Grand Island, NY), and scanned using an Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA).
Microarray data analysis
Spot intensities for each probe were calculated by subtracting median local background from median local foreground for each spot and were then normalized. The mouse probes’ intensities were filtered to identify all probes with an intensity above a normalized threshold. For statistical analysis, microarray data were examined for differences using One-way ANOVA or the Student’s t-test. Values of P < 0.05 were designated as statistically significant. To execute hierarchical and k-means clustering, log2-transformed, significant (F > F crit = 2.832, signal threshold > 500) mouse probes were used for analysis by Gene Cluster 3.0 software according to the manual [22]. TreeView software was employed to visualize clustering results [23].
ChIP-Seq
Since some photoreceptor-related genes belong to the X chromosome, we used P28 Glast+ cells isolated from the retinas of only male mice to avoid exclusion of this chromosome from analysis in this study. All antibodies were validated by Diagenode Inc. Freshly isolated samples were cross-linked for 9 min at room temperature in 1% formaldehyde (Sigma-Aldrich, F8775–25ML) in 1X PBS. To stop the cross linking reaction, glycine (Sigma-Aldrich, G-7403) was added to a final concentration of 0.125 M. From this point onwards we worked on ice. The cells were centrifuged at 300× g for 10 minutes at 4°C and the supernatant was aspirated. The cross-linked cells were washed in 1 ml of ice cold HBSS containing a protease inhibitor cocktail (PIC, 200x; final concentration 1x; Sigma-Aldrich, P8340). The cells were centrifuged again at 300 × g for 10 minutes at 4°C, the supernatant was discarded, and the cell pellets were stored at −80°C. The ChIP-seq experiment was conducted by Diagenode’s ChIP-seq profiling service. The chromatin was prepared using the True MicroChIP Kit (Diagenode Cat# C01010130). Chromatin was sheared using the Bioruptor® Pico sonication device (Diagenode Cat# B01060001) combined with the Bioruptor® Water cooler for 7 cycles using 30” [ON] 30” [OFF] settings. Shearing was performed in 0.65 ml Bioruptor® Pico Microtubes (Diagenode Cat# C30010011) with 2.5 × 104 cells in 100μl. 25μl of this chromatin was used to assess the size of the DNA fragments obtained by a High Sensitivity NGS Fragment Analysis Kit (DNF-474) on a Fragment Analyzer™ (Advanced Analytical Technologies, Inc.). ChIP was performed using IP-Star® Compact Automated System (Diagenode Cat# B03000002) following the protocol of the aforementioned kit. Chromatin corresponding to 2.5 × 104 cells was immunoprecipitated using the following antibodies and amounts: H3K4me1 (0.5 μg; Diagenode Cat# C15410194), H3K4me3 (0.5 μg; Diagenode Cat# C15410003–50), H3K9me3 (0.5 μg; Diagenode Cat# C15410193), and H3K27me3 (0.5 μg; Diagenode Cat# C15410195). Chromatin corresponding to 10% was set apart as Input. After reverse cross-linking, the DNA is quantified using a Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, Q32854). Moreover, quantitative PCR analysis was performed to check ChIP efficiency using the following primers: Prm1 and Gas2l1 (Table 1). Libraries were made from the input and ChIP’d DNA (500 pg) using the MicroPlex Library Preparation Kit v2 (12 indices) (Diagenode Cat# C05010013). Library amplification was evaluated with a High Sensitivity NGS Fragment Analysis Kit (DNF-474) on a Fragment Analyzer™ (Advanced Analytical Technologies, Inc.). Libraries were then purified using Agencourt® AMPure® XP (Beckman Coulter) and quantified using a Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, Q32854).
ChIP-Seq data analysis
Libraries were pooled and sequenced on an Illumina HiSeq 4000 with single-end reads of 50bp in length, running HiSeq Control Software HD version 3.4.0.38. Quality control of sequencing reads was done by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned to the reference mouse genome (mm10) obtained from the UCSC genome browser using BWA software v.0.7.5a [24]. Samples were filtered for regions blacklisted by the ENCODE project. Subsequently, samples were deduplicated using SAMtools v1.3.1 [25]. Alignment coordinates were converted to BED format using BEDTools v.2.17 and peak calling was performed using SICER with customized parameters for each histone mark [26]. To integrate our ChIP-seq data and identify the major combinatorial and spatial patterns of marks (so-called chromatin states), we used ChromHMM software (http://compbio.mit.edu/ChromHMM/) according to the manual [27]. Annotation of the peaks and identified ChromHMM segments was carried out with the R Bioconductor package “Annotatr” [28].
Whole genome bisulfite sequencing (WGBS)
Since some photoreceptor-related genes belong to the X chromosome, we used samples isolated only from male mice for the methylation analysis to avoid exclusion of this chromosome from the study. Genomic DNA was purified from P28 Glast+ cells using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). The DNA concentration of the samples was measured using the Qubit® dsDNA BR Assay Kit (ThermoFisher Scientific, Grand Island, NY). DNA quality of the samples was evaluated with the Fragment AnalyzerTM and the DNF-487 Standard Sensitivity genomic DNA Analysis Kit (Advanced Analytical). WGBS was performed by Diagenode Inc. Genomic DNA was sheared using the Bioruptor® Pico sonication device (Diagenode Cat# B01060001) combined with the Bioruptor® Water cooler for 15 cycles using 30” [ON] 30” [OFF] settings. Shearing was executed in 0.2 ml Bioruptor® Pico Microtubes with Caps (Diagenode Cat# C30010020). 1μl of this sample was used to evaluate the size of the DNA fragments obtained by a High Sensitivity DNA chip for the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). DNA concentration of the sample was measured after shearing using the Qubit® dsDNA BR Assay Kit (ThermoFisher Scientific, Grand Island, NY). WGBS libraries were prepared using the Whole Genome Bisulfite Sequencing (RRBS) Kit (Diagenode Cat# C02030034) following the kit manual. 1μg of sheared genomic DNA was used to start library preparation for each sample. Following library preparation, samples were bisulfite converted and amplified by PCR using 9 amplification cycles. Final PCR clean-up was performed twice using a 1.1X beads:sample ratio of Agencourt® AMPure® XP (Beckman Coulter). DNA concentration of the libraries was evaluated using the Qubit® dsDNA HS Assay Kit (ThermoFisher Scientific, Grand Island, NY). The library profiles were checked using the High Sensitivity DNA chip for the 2100 Bioanalyzer (Agilent). WGBS libraries were sequenced on a HiSeq3000 (Illumina) using 150 bp single-end sequencing.
WGBS data analysis
The sequenced reads were tested for quality using the FastQC tool. Adapter removal was done using Trim Galore! v0.4.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The cleaned reads were then aligned to the Mus musculus reference genome (Genome Reference Consortium 37, mm10) using bismark v0.16.1 [29]. The average read coverage for our samples was 15. The cytosine2coverage and bismark_methylation_extractor modules of bismark were used to identify the methylation state of all cytosines in a CpG, CHH, or CHG context (for every single mappable read) and to calculate the percentage of methylation for each CpG, CHH, or CHG site. DNA methylation analysis from high-throughput bisulfite sequencing results were performed using the Bioconductor R packages “MethylSeekR” and “methylKit” according to software documentation [30, 31]. Annotation of identified segments and regions was carried out with the R Bioconductor package “Annotatr”.
Data Availability
The raw files from the microarray study, ChIP-Seq, and WGBS have been deposited in the NCBI Gene Expression Omnibus (GEO) database. They are accessible through GEO accession number GSE122337 (GSE122301, GSE122302, GSE122356).
Results
Molecular profiling of developing Müller glia
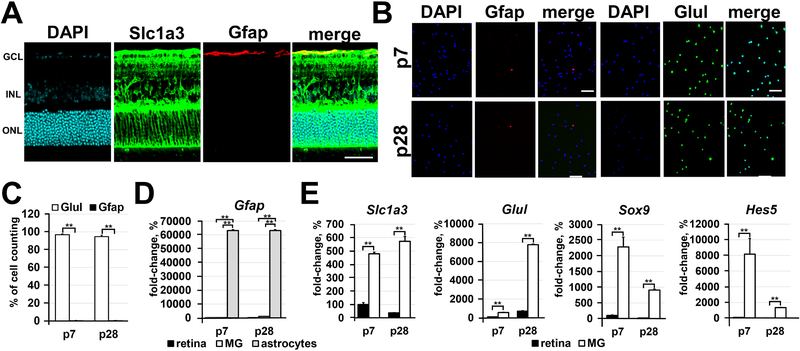
To study molecular changes during differentiation of RPCs into Müller glia, we isolated Notch1+ cells (Notch1 is a marker of RPCs) from postnatal-day (P) 0, 3, 7, and 14 retinas, and Glast + cells (Glast/Slc1a3 is a marker of Müller glia precursors and Müller glia in the retina) from P7, P14, P21, and P28 retinas. To isolate Notch1+ cells we used the protocol described in our study [18]. To purify Glast+ cells we utilized an immunomagnetic cell separation protocol using monoclonal biotin-conjugated antiGlast mouse antibodies and anti-biotin magnetic microbeads. Glast is mostly expressed in brain radial glia, and is also expressed in (mostly cortical) brain astrocytes. Since astrocytes are present in the retina (Figure 1A), we tested the contamination of isolated Glast+ cells with retinal astrocytes using immunocytochemistry and qRT-PCR. Using the astrocyte marker Gfap, and Müller glia marker Glul (glutamine synthetase; GS), we found that Glast+ cells isolated from P7 and P28 retinas were almost all Glul-positive (P7: 97±1% Glul+ vs. 0.4±0.3% Gfap+; P28: 95±2 Glul+ vs. 0.3±0.3 Gfap+; Figures 1B and 1C). Meanwhile, we found a significantly high expression of Gfap in isolated brain astrocytes compared to Gfap expression in the retina and Glast+ cells isolated from the retina (Figure 1D). We also found significant enrichment of Müller glia markers (Glast (Slc1a3), Glul, Sox9, Hes5) in Glast+ cells isolated from P7 and P28 retinas compared to the entire retina (Figure 1E). Thus, immunomagnetic separation of highly purified Müller glia using an anti-Glast antibody proved to be an effective technique for the purposes of our study.
Figure 1.
Immunomagnetic separation is an effective approach for the isolation of highly pure Müller glia. A) The spatial distribution of Glast (Slc1a3) proteins evaluated by immunohistochemistry in P28 retinas reveals the presence of Glast mostly in Müller glia and, to some extent, in retinal astrocytes (labeled by Gfap). DAPI was used to visualize the cell nucleus. Bar is 50 μm. (GCL - ganglion cell layer; INL - inner nuclear layer; ONL - outer nuclear layer) B) Since the anti-Glast (Slc1a3) antibody can recognize Müller glia and some retinal astrocytes, we tested the distribution of Glast+ cells isolated from the retina depending on cell type using the Müller glia marker (Glul; glutamine synthetase) and astrocyte marker (Gfap). Glast+ cells were isolated from retinas of P7 and P28 animals. Bar is 50 μm. C) The percentage of Müller glia (Glul; glutamine synthetase marker) and astrocytes relative to the total number (DAPI labeled cells) of Glast+ counted cells was calculated. D) and E) Expression of astrocyte and Müller glia markers in the entire retina and in isolated astrocytes, P7 and P28 Glast+ cells was evaluated using qRT-PCR. Astrocytes isolated from P3 brains were used as a positive control for Gfap expression. For each gene, the results are expressed as a fold-change of the corresponding value for P7 retina ± SE of the mean (n = 6).
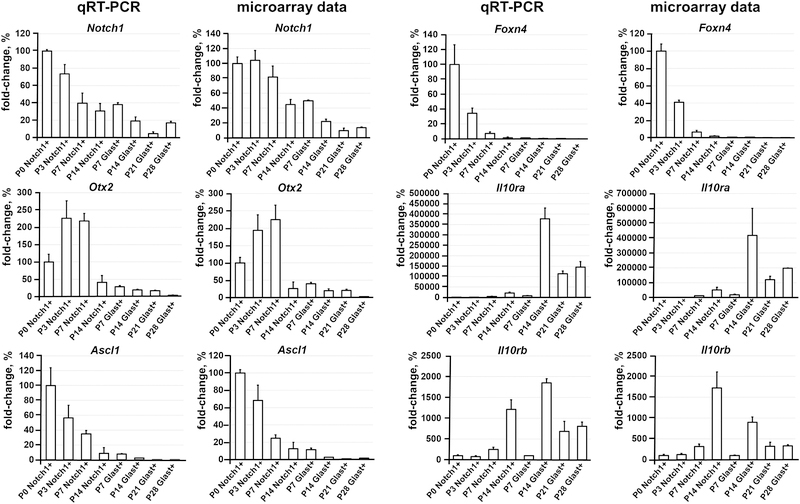
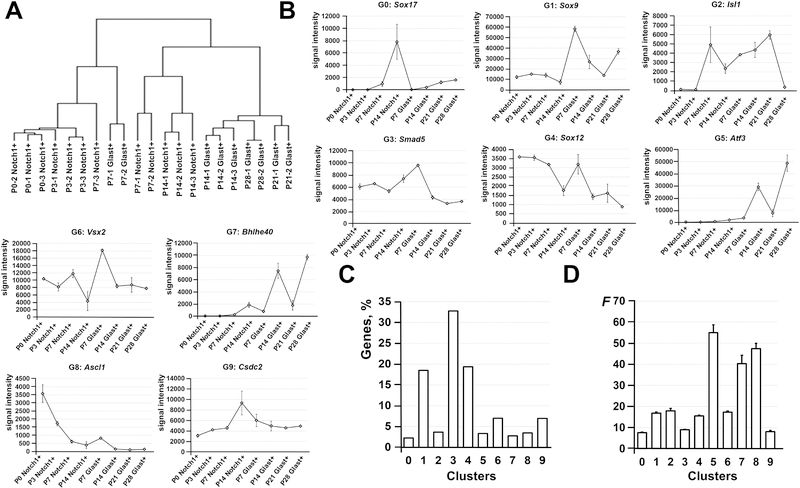
To characterize and compare P0, P3, P7, and P14 Notch1+ cells (RPCs) and P7, P14, P21, and P28 Glast+ cells (Müller glia precursors and adult Müller glia), RNA extracted from these cells was used for microarray analysis. The results of our statistical analysis demonstrated that a total of 10,047 genes passed the quality control criteria and One-Way ANOVA test (F > Fcrit. = 2.83) to identify genes that are regulated differently between the studied groups and time points (Supplementary Data S1). The changes in gene expression were verified using quantitative RT-PCR for a group of genes (Figure 2). For further analysis, we performed hierarchical clustering of differentially expressed genes to group samples of Notch1+ and Glast+ cells on the basis of similarities in their expression of these genes (Figure 3A). We found that our samples formed three subgroups (clustered together): 1) P0, P3, P7 Notch1+ cells, and P7 Glast+ cells; 2) P7 and P14 Notch1+ cells; 3) P14, P21, and P28 Glast+ cells (Figure 3A). The distance between subgroups 2 and 3 is less compared to subgroup 1 (Figure 3A). These data indicate that the gene expression profiles of P0, P3, and P7 Notch1+ cells shared more similarities (or lower distances between objects when forming the cluster/subgroup) with the gene expression profiles of P7 Glast+ cells (Figure 3A, Supplementary Data S1). Meanwhile, the expression profile of P14 Notch1+ cells were more similar to the expression profiles of P14, P21, and P28 Glast+ cells (Figure 3A, Supplementary Data S1). It should be noted that P14, P21, and P28 Glast+ cell profiles share the most similarity with each other. The identified subgroups reflect the steps in RPC differentiation into Müller glia. Our data suggest that differentiation of RPCs into Müller glia starts at P7, but these RPCs still retain the ability to differentiate into retinal neurons (high similarity between P0, P3, and P7 Notch1+ cell and P7 Glast+ cell expression profiles), yet RPCs (Notch1+ cells) at P14 are predisposed to differentiate into Müller glia. It is important to note that P14 Müller glia can already be recognized as adult, since cells at this time point share a high similarity to adult Müller glia (P21, and P28 Glast+ cells; Figure 3A, Supplementary Data S1).
Figure 2.
Quantitative RT-PCR analysis has confirmed the microarray data for a group of selected genes. For each gene, the results are expressed as percentages ± SEM of the corresponding values in the Notch1+ cells isolated from P0 developing retinas (P0 Notch1+ cells).
Figure 3.
The results of the hierarchical and k-means clustering revealed the changes that are undergone in RPCs (Notch1+ cells) and Müller glia (Glast+ cells) during retinal development. A) Hierarchical clustering of differentially expressed genes showed that many P7 and P14 Notch1+ cells (late RPCs) are predisposed to become Müller glia. While Glast+ cells at P7 are still very close to a multipotent progenitor state, P14, P21, and P28 Glast+ cells are already grouped together indicating the adult Müller glia state. B) The 10 clusters were identified by the k-means clustering algorithm. C) The majority of differentially expressed genes were located in cluster 1, 3, and 4. D) Meanwhile, significant changes were undergone in genes located in small clusters 5, 7, and 8. A value F (F statistic) was calculated using the ANOVA test.
In our next step, we performed k-means clustering of differentially expressed genes, which allowed us to identify 10 different clusters (Figure 3B, Supplementary Data S1). To characterize these clusters, we used the DAVID functional annotation bioinformatic analysis tool to pinpoint enriched pathways in the KEGG and PANTHER databases and The Gene Ontology (GO) Biological Processes. We noted that three clusters (1, 3, and 4) contain more than 70% of the studied genes (Figure 3C, Supplementary Data S1). The gene expression in these clusters was not significantly changed during development and the majority are housekeeping genes (Figure 3D, Supplementary Data S1). However, we observed significant changes in the expression of genes that belong to clusters 5, 7, and 8, containing a total of less than 10% of the studied genes (Figure 3D). The expression of genes in clusters 5 and 7 is increased during differentiation of RPCs into Müller glia, while the expression of genes from cluster 8 is reduced (Figure 3B). The genes that belong, in part, to cluster 5 and mostly to cluster 7 are involved in immune response (Supplementary Data S1 and S2), while the genes from cluster 8 are involved in the cell cycle. Cluster 8 also contains transcription factors, such as Foxn4, Ptf1a, Ascl1, Olig2, Sox11, Sox4, etc., that are critical for differentiation of RPCs into retinal neurons. It should also be noted that cluster 6 contains RPC markers, such as Sox2, Vsx2, Lhx2, Rax, Six6, Pax6, Meis1, Meis2, Otx2, etc. The expression of these genes was not significantly changed and was present in P14, P21, and P28 Glast+ Müller glia. Thus, analysis of our microarray data suggests that the Müller glia transcriptome is very similar to the RPC transcriptome. Meanwhile, significant changes affected a small group of genes: 1) expression of pro-neuronal markers and cell cycle-related genes was reduced reflecting the differentiation of RPCs into Müller glia; 2) expression of immune response-related genes was increased in differentiating Müller glia. It should be noted that we observed increased expression of pro-inflammatory and anti-inflammatory genes (like receptors Il10ra, Il10rb, Cx3cr1), which may regulate the Müller glia immune response (Supplementary Data S1 and S2).
Global epigenetic profile of Müller glia
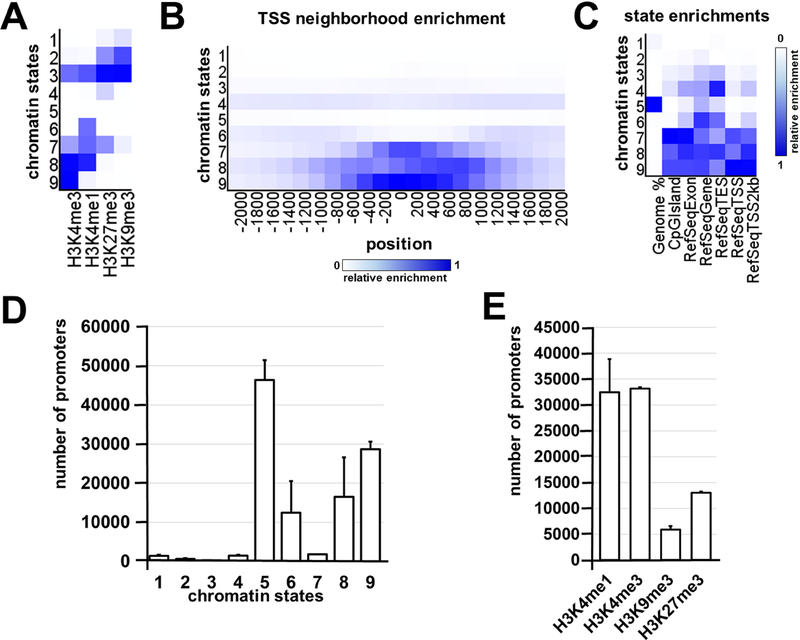
Since our microarray data suggested a high similarity between Müller glia and RPCs, we decided to test the epigenome of adult murine Müller glia. To characterize the epigenetic states of the adult Müller glia, we isolated Glast+ cells from P28 retinas. We analyzed these cells on the genome-wide histone modifications level using ChIP-seq technology and the DNA methylation level using the whole-genome bisulfite sequencing (WGBS) approach. To investigate genome-wide histone modifications in adult Müller glia, we selected H3K4me3, H3K4me1, H3K27me3, and H3K9me3 histone marks. These histone modifications, alone or in specific combinations, characterize the chromatin epigenetic state: active/open (permissive) chromatin (H3K4me3 alone or in combination with H3K4me1), bivalent/poised state (H3K27me3 and H3K4me3), temporarily inactive (repressive) Polycomb state (H3K27me3), and permanently inactive (repressive) state (H3K9me3 alone or in combination with H3K27me3). Two pellets (independent biological replicates) of fixed samples containing 1.5 – 2.0 × 105 Glast+ cells isolated from the retinas of P28 mice were used. The chromatin was extracted from the cells of each sample, sheared, and then H3K4me1, H3K4me3, H3K9me3, and H3K27me3 ChIP assays were performed on each chromatin preparation. Sequencing was performed, followed by the alignment, peak calling procedures, and annotation for studied histone modifications (Supplementary Data S3). We used computational, multivariate Hidden Markov Models (chromHMM) with all of our ChIP-seq data to identify the chromatin states [27]. Using this computational approach, we chose 9 chromHMM chromatin states for analysis (Figure 4A, 4B, 4C, Supplementary Data S4). Chromatin states 1 and 2 represent the H3K9me3 and H3K27me3 markers – permanently repressed (inactive) chromatin. Since state 3 was marked by all histone modifications tested in the study, we analyzed gene promoters linked to this state and found that it may label active genes located in inactive regions. Chromatin state 4 was marked by H3K27me3, the temporarily inactive polycomb-repressed chromatin marker. State 5 was empty chromatin (no tested histone marks). State 6 may be predominantly enhancers, since H3K4me1 is known as an enhancer mark [32]. Chromatin state 7 may determine bivalent/poised promoter regions, while states 8 and 9 had the epigenetic marks of active genes. The numbers of annotated gene promoters located in regions with matching chromatin states and individual histone marks are shown in Figures 4D and 4E. Our findings indicate that a significant number of Müller glia promoters are in open (no tested histone marks detected) or active (permissive) chromatin, which is specific for epigenetically mobile stem cells and progenitors [33–35].
Figure 4.
The 9 chromatin states were identified with the chromHMM software package using Müller glia ChIP-seq data. A) Heat map of the chromatin states. The darker blue color labels more abundant ChIP-seq marks within the chromatin state. B) The fold enrichment for each chromatin state at fixed positions (from −2000 bp in the promoter area up to 2000 bp in the first exon and intron) in regards to the transcription start site (TSS) suggests a high level of modified histone accumulation (mostly active H3K4me3 marks) around the TSS of genomic regions in chromatin states 9, 8, and 7. C) Heat map for the chromatin state functional enrichment displays on the genome, CpG islands, exons, genes, transcript end sites (TES), TSS, and 2000 base pair intervals around the TSS represented by each state. D) The columns correspond to the number of promoters located in genomic regions marked by identified chromatin states. E) The ChIP-seq data was annotated and presented as the number of the promoters that contain H3K4me1, H3K4me3, H3K9me3, and H3K27me3 histone marks.
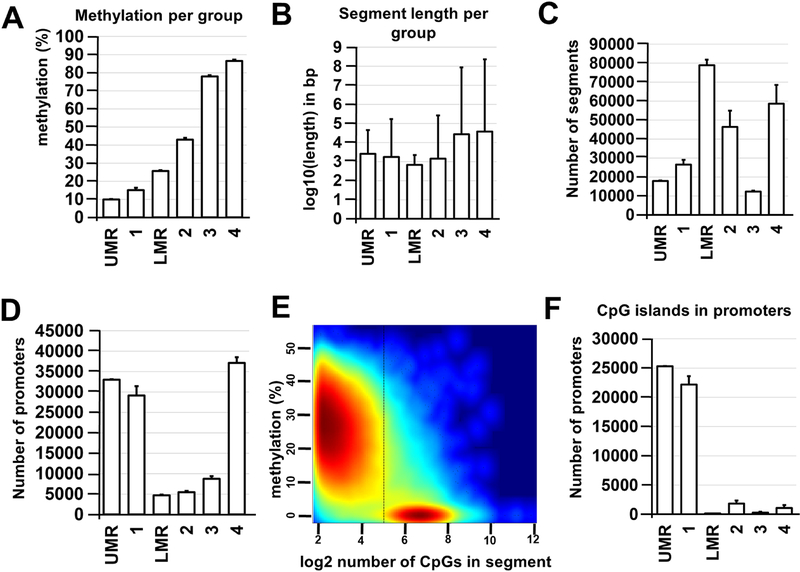
To study DNA methylation, we prepared two WGBS libraries using genomic DNA isolated from two independent samples containing 1.5 – 2.0 × 105 P28 Glast+ cells. These libraries were sequenced, bisulfite sequence reads were aligned using the Bismark software package, and then annotated. We performed an analysis of the global methylation level in Müller glia and found that the percentages of methylated CpG, CHG, and CHH contexts (where H is A, C, or T) were 80.2 % for CpG, 0.4% for CHG, and 0.6 % for CHH. We further focused our investigation on CpG methylation, because Müller glia DNA methylation was mostly restricted to CpG sequences. To analyze Müller glia DNA methylome segmentation and dynamics, we used the following computational approaches: 1) the Bioconductor R package “MethylSeekR” – a computational tool that accurately identifies unmethylated regions (UMRs) and low-methylated regions (LMRs) from bisulfite-sequencing data; 2) the Bioconductor R package “methylKit” – which segments the DNA methylome into four distinct features (segmentation classes) based only on the average DNA methylation level (Supplementary Data S5) [30, 31]. In agreement with previously published data, we found a high-concordance between the MethylSeekR and methylKit data: segmentation class 1 and UMRs, as well as segmentation class 2 and LMRs largely share similar segment lengths and methylation levels (Figure 5A–C, Supplementary Data S5) [36]. At the same time, segmentation classes 3 and 4 correspond to highly methylated regions [36]. The analysis of the annotated gene promoters revealed that Müller glia gene promoters were mostly located in either highly methylated or unmethylated regions (Figure 5D, Supplementary Data S5). Meanwhile, CpG islands were mostly located in the unmethylated promoters of the studied genes (87% of CpG islands had an average methylation level of 10%) (Figure 5E, F, Supplementary Data S5).
Figure 5.
MethylSeekR and methylKit R Bioconductor packages are robust and comparable methods for identification of Müller glia methylome states. A) Average methylation levels were calculated for each methylation region and segmentation class. B) Average lengths (log10-transformed base pairs; bp) were determined for identified segments and regions. C) Average number of segments vary between segmentation classes and methylation regions. D) The majority of promoters are located in unmethylated (segmentation class 1 and UMR) or highly methylated (segmentation class 4) regions of the Müller glia genome (UMR – unmethylated region; LMR – low-methylated region). E) HMM-based MethylSeekR allowed us to separate CpG-rich UMRs and CpG-poor LMRs. The figure generated by MethylSeekR demonstrates the log2-transformed number of CpGs (hypomethylated regions) versus its average methylation level. F) The CpG islands identified in promoters of Müller glia genomic DNA are mostly unmethylated (segmentation class 1 and UMR).
Müller glia epigenome contains epigenetic restrictions (barriers), which may prevent Müller glia dedifferentiation into early-born retinal neurons
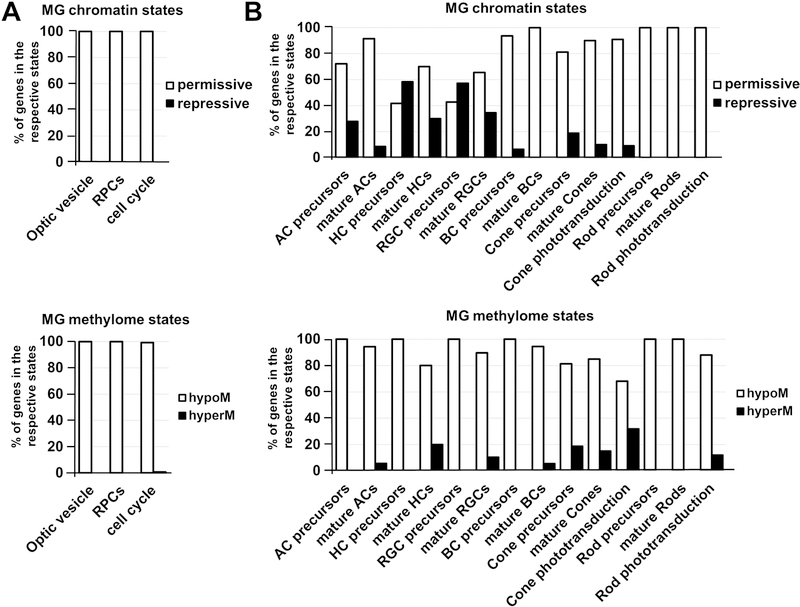
Using our global epigenetic profile of Müller glia, we evaluated the epigenetic states of promoters of key genes required for the development and function of optic vesicle progenitors (OVPs) and RPCs in the Müller glia genome. The key genes required for OVPs were collected from peer-reviewed articles (Supplementary Data S6 and S7). The lists of genes that regulate RPC development and function were obtained from LifeMap Discovery and The Stem Cell Research Database. We also studied cell cycle-related genes obtained from the KEGG database. All these genes were included in Supplementary Data S6 and S7. We recognized gene promoters that only contain H3K9me3 and/or H3K27me3 markers and have chromatin states 1, 2, 4, or 7 as promoters in an inactive (repressive) state (Supplementary Data S6). A promoter was considered highly methylated (hypermethylated) if it was located in segmentation class 3 or 4 genomic regions and UMRs or LMRs (size > 500 bp) were not present in the promoter area (Supplementary Data S7). Next, we calculated the % of genes found in a repressive chromatin state compared to the total number of genes required for either OVP or RPC phenotypes or for the cell cycle process (Figure 6A, Supplementary Data S6). We also calculated the percentage of hypermethylated genes (genes with hypermethylated promoters) relative to the total gene number required for OVPs, RPCs, and the cell cycle (Figure 6A, Supplementary Data S7). Our data revealed that promoters of all tested genes required for OVP and RPC development and function, as well as cell cycle-related genes, were in the permissive chromatin state in the genome of adult murine Müller glia (Figure 6A, Supplementary Data S6). In the same way, our analysis of the Müller glia DNA methylation showed that 100% of promoters of OVP- and RPC-related genes, and 99% of promoters of cell cycle-related genes, were in unmethylated or low-methylated regions of the Müller glia genome (Figure 6A, Supplementary Data S7). Thus, Müller glia are epigenetically very close to progenitor-like cell phenotypes.
Figure 6.
The chromatin and methylome states of key genes required for retinal development in the Müller glia epigenome. A) Promoters of the optic vesicle and RPC phenotypes, and cell cycle-related genes are in a permissive (active/open) chromatin state and unmethylated or low-methylated (% - percent of genes in the respective states). B) Many promoters of genes required for the development and function of early-born retinal neurons are in a repressive (inactive) chromatin state, but they are mostly unmethylated or low-methylated. Promoters of late-born neurons are mostly in a permissive (active/open) chromatin state and unmethylated or low-methylated. However, promoters of some genes required for cone and rod phototransduction are highly methylated, which may affect the function of photoreceptors dedifferentiated from Müller glia. (hypoM – hypomethylated genes; hyperM – hypermethylated genes)
Since RPCs can be divided in two subpopulations depending on their ability to differentiate into early-born retinal neurons (RGCs, cone photoreceptors, horizontal cells, and a subpopulation of early-born amacrine cells) and late-born retinal neurons (bipolar cells, rod photoreceptors, and a subpopulation of late-born amacrine cells), we began to wonder if Müller glia are epigenetically close to either early RPCs, late RPCs, or both. To this end, in the Müller glia epigenome we evaluated the promoters of genes required for retinal neuron development and function, as well as genes required for rod and cone phototransduction. The lists of genes for retinal neuronal cell types were obtained from LifeMap Discovery and The Stem Cell Research Database. The list of genes required for cone and rod phototransduction was collected from the RGD database. All of these genes can be found in Supplementary Data S6 and S7. To calculate the % of genes required for each retinal phenotype/process (phototransduction) found in a repressive chromatin state (% of hypermethylated genes) in the Müller glia genome, we used the approach described above. We found that many promoters of genes required for the development of precursors of early-born retinal neurons were in a repressive chromatin state in the Müller glia epigenome (57% of RGC precursors, 58% of horizontal cell precursors) (Figure 6B, Supplementary Data S6). Meanwhile, we found that only 28% of promoters of amacrine precursor-related genes and 19% of promoters of genes required for cone photoreceptor precursor development were in a repressive chromatin state. We also observed that promoters of genes required for mature RGCs and horizontal cells were mostly in a permissive chromatin state (66% and 70% respectively). The numbers were even higher for promoters of genes necessary for mature amacrine cell and cone photoreceptor phenotypes (91% and 90% respectively). We also found that 91% of cone phototransduction-related gene promoters were in a permissive chromatin state. Analysis of Müller glia genomic DNA methylation in promoters of genes required for early-born retinal neurons revealed that these regulatory elements were mostly present in unmethylated or low-methylated regions (Figure 6B, Supplementary Data S7). However, we found that promoters of an important group of genes required for cone photoreceptor function (cone phototransduction), including Opn1mw, Opn1sw, Arr3, Pde6c, Gucy2f, and Cnga3, were located in highly methylated regions (Supplementary Data S7). We also analyzed the chromatin and methylome states of promoters of genes required for the development and function of late-born retinal neurons (bipolar cells and rod photoreceptors) in the Müller glia epigenome. We found that the majority of these genes were in a permissive chromatin state and in unmethylated or low-methylated regions (Figure 6B, Supplementary Data S6 and S7). Our findings also indicate that promoters of rod phototransduction genes, Gngt1 and Gucy2f, were highly methylated (Supplementary Data S7). Thus, our data suggest that Müller glial cells are epigenetically closer to late-born retinal neurons, rather than early-born retinal neurons.
Discussion
In our study we investigated changes in the global transcriptome during differentiation of RPCs into Müller glia in the developing retina. We also analyzed the global epigenetic profile of adult Müller glia to evaluate the level of epigenetic plasticity of this cell type to be reprogrammed and differentiated into retinal neurons. Analysis of our global transcriptome data revealed that significant changes during the transition of RPCs into Müller glia affect a small group of genes. We found reduced expression of proneuronal markers and cell cycle-related genes, reflecting differentiation of RPCs into Müller glia. Meanwhile, the expression of immune response-related genes was increased in differentiating Müller glia. Concerning the remaining genes, however, our data suggest that Müller glia are very similar to late RPCs. Our epigenetic data strongly supports this conclusion. We observed a high similarity between Müller glia chromatin and methylome states with OVPs and RPCs. Our findings also indicate that Müller glia are epigenetically very close to late-born retinal neurons, but not early-born retinal neurons, pointing to the aforementioned similarity with late RPCs. Our data suggests that the obstacles prohibiting regeneration of the entire mammalian retina from Müller glia are repressed chromatin and highly methylated promoters of many genes required for the development of early-born retinal neurons within the Müller glia epigenome. However, since promoters of these genes are mostly marked by the H3K27me3 histone modification, which is related to the temporarily inactive polycomb-repressed state, they may be activated in the presence of some not-yet-identified pioneer transcription factors (PTFs) [37]. In addition, DNA demethylation may be required for proper activity of cone photoreceptor-related genes [38]. Our data also suggest that DNA demethylation in developing rod photoreceptors may also be required to promote the phototransduction process in these neurons.
Retinal development begins with a population of equivalent, proliferating early RPCs, which can only differentiate into early-born retinal neurons or late RPCs [15–17]. In turn, late RPCs produce only late-born retinal neurons and Müller glia [15–17]. The Notch signaling cascade is critical for maintaining a population of undifferentiated RPCs, preventing differentiation of RPCs into retinal neurons in the developing retina [15, 16]. To this end, when the Notch receptor is activated by its ligands, expression of Hes1 and Hes5 transcription factors is induced, which represses the expression of genes required for retinal neuronal phenotypes [15, 16]. Meanwhile, reduced Hes1 and Hes5 expression facilitates RPC differentiation into retinal neurons [15, 16]. After retinal neurogenesis, when all retinal neuronal phenotypes appear, a high and sustained expression of Hes1 and Hes5 in the remaining late RPCs prevents any possible neuronal phenotypes [15, 16]. These late RPCs become Müller glia [15, 16]. Since high and sustained Hes1 expression inhibits cell proliferation, the Müller glia phenotype may resemble “frozen” late RPCs [39–43]. Our findings indicate that Hes1 expression is increased during the transition from RPCs to Müller glia (Hes5 expression doesn’t change much between RPCs and Müller glia), while the expression of cell cycle-related genes and genes defining early-born retinal neuronal phenotypes were reduced (Supplementary Data S1 and S2). At the same time, many key progenitor-, RPC-, and late-born retinal neuron-related genes are still active in Müller glia, or their respective promoters are found in a permissive chromatin state. Thus, a correct “defrosting” protocol should be successful in Müller glia reprogramming and differentiation into late-born neurons. Some of these protocols were already developed; Reh’s lab is already successfully reprogramming Müller glia in vivo and in vitro into progenitor-like cells and have differentiated them into bipolar cells [11–13, 44]. Meanwhile, Chen’s lab recently demonstrated that increased WNT signaling activity and increased expression of rod photoreceptor transcription factors in mammalian Müller glia promote proliferation, reprogramming, and differentiation of the cells into new rod photoreceptors [14]. Though, our data suggest that de novo genesis of early-born retinal neurons from mammalian Müller glia may be prevented due to the repressive chromatin state in which many genes required for the development of these retinal neuronal phenotypes are found. However, they are mostly marked by H3K27me3 and may be activated in the presence of some unidentified PTFs [37].
Much like Müller glia chromatin states, promoters of progenitor- and cell cycle-related genes were found in unmethylated or low-methylated regions of the Müller glia genome, supporting the notion of the Müller glia progenitor state on the epigenetic level. In addition, while we observed a significant number of genes required for early-born retinal neurons in a repressive chromatin state in the Müller glia genome, the majority of promoters of these genes were in unmethylated or low-methylated regions. The regulatory elements of late-born neurons were also unmethylated or low-methylated. However, we found that promotors of some key genes required for phototransduction were in highly methylated regions of the Müller glia genome. The methylation affected the rod phototransduction pathway, but the effects were more profound for the cone phototransduction pathway. Thus, in addition to PTF activity, DNA demethylation is required for reprogramming Müller glia to restore the normal function of photoreceptors in the self-healing retina. The DNA demethylation signaling cascade includes the TET demethylase family (Tet1, Tet2, and Tet3), which catalyzes the sequential conversion of 5-methylcytosine into 5-hydroxymethylcytosine, then 5-formylcytosine and 5-carboxylcytosine are both converted back to unmethylated cytosine [45, 46]. TET proteins play an important role during neurogenesis [47]. Tet1 and Tet2 regulate neurogenesis by increasing the expression of target genes [48–52]. The importance of Tet2 and Tet3 was revealed during zebrafish retinal neurogenesis when Seritrakul et al. observed impaired differentiation of RGCs and photoreceptors in zebrafish Tet2/Tet3 double knockouts [38]. Xenopus Tet3, by itself, regulates early eye and neural development by directly activating a set of key developmental genes [53]. Meanwhile, Tet3 is required to facilitate photoreceptor differentiation in mice [54]. Thus, TET demethylase activities are critical in adult neurogenesis and retinogenesis (especially in photoreceptor differentiation). Therefore, increased expression of TET genes during reprogramming and differentiation of Müller glia into new photoreceptors may be required to promote phototransduction activity. Since Tet2 and Tet3 regulate retinal development in zebrafish, these animals may use TET demethylases to initiate and promote retinal regeneration after injury.
While we found a high similarity between late RPCs and Müller glia on the transcriptomic and epigenomic levels, a significant difference between these cell types was observed in increased expression of immune response-related genes. We observed increased expressions of pro-inflammatory and anti-inflammatory genes (such as receptors Il10ra, Il10rb, and Cx3cr1), which may help to regulate the Müller glia immune response. A balance between anti-inflammatory and pro-inflammatory gene activity in Müller glia may be required to keep pro-inflammatory genes under control, which aids in avoiding neuronal toxicity. Meanwhile, the balance may be switched in injured retina to increase pro-inflammatory responses, promoting retinal damage. However, the reason why the expression of immune response genes, including the Tnf and TLR signaling cascades, is increased in developing Müller glia remains unclear (Supplementary Data S2). Immune response is part of the renewal/regeneration mechanism in many tissues [55–57]. In particular, Tnf signaling and innate immune response play important roles in this process [55–57]. Thus, increased expression of immune response genes during retinal development in mammalian Müller glia might be part of an evolutionarily ancient mechanism of retinal regeneration. This mechanism, which is still present in certain species like zebrafish, may have been damaged during evolution and inactivated in mammals [6, 58, 59]. We can suggest that mammalian glial cells, while trying to activate the regeneration mechanism unsuccessfully after injury, actually produce more and more pro-inflammatory factors like Tnf, which is fatally toxic for CNS (including retinal) neurons in high concentrations [19, 60–62]. As a result, brain and retinal damage may be reinforced and multiplied, instead of leading to a regenerative effect. Meanwhile, many non-CNS tissues still successfully utilize this renewal/regeneration mechanism [55–57]. Thus, if we can restore the mechanism in mammalian glia (including Müller glia), we can solve two problems contemporaneously: 1) reduce the neurotoxic, pro-inflammatory response in injured tissue; 2) regenerate injured tissue. We suggest that such an approach may be similar for many other CNS tissues, and the retina could be a great model for further investigation in order to achieve this goal.
Conclusions
Our data suggest that Müller glia are epigenetically very close to progenitor-like cell phenotypes and late-born retinal neurons (bipolar cells and rod photoreceptors), pointing to a similarity with late RPCs. Since the epigenetic plasticity of Müller glia in adult teleost fish allows these animals to regenerate the entire retina and restore lost sight after injury, our data suggest that mammalian Müller glia should have no epigenetic barriers preventing reprogramming and differentiation into late-born retinal neurons (bipolar cells and rod photoreceptors). However, DNA demethylation may be required to allow, in part, the phototransduction process in rod photoreceptors. Our ChIP-seq data also suggests that obstacles for regeneration of the entire mammalian retina from Müller glia after injury is due to the repressive chromatin state of many genes required for the development of early-born retinal neurons. However, these repressive chromatin states are mostly marked by H3K27me3 and may be activated in the presence of specific pioneer transcription factors (PTFs) that remain unidentified. DNA demethylation in the Müller glia genome may also be required for proper activity of cone photoreceptor- and cone phototransduction-related genes. Thus, our findings suggest that the epigenetic plasticity of adult mammalian Müller glia can be restored in the presence of some PTF and DNA demethylase activities, allowing Müller glia to regenerate the entire retina after injury. Identification of the PTFs and DNA demethylases might be the key to restoring the self-healing ability of the mammalian retina.
Supplementary Material
Highlights.
RPCs and Müller glia have high similarity on the transcriptomic and epigenomic levels
Müller glia are epigenetically close to late-born, not early-born, retinal neurons
Phototransduction-related genes are highly methylated in the Müller glia genome
Acknowledgments
Supported in part by the National Institutes of Health/National Eye Institutes Grant R01 EY027311 (DI), National Institutes of Health Center Core Grant P30EY014801, Research to Prevent Blindness Unrestricted Grant, Inc. (New York, NY, USA). The authors thank Gabriel Gaidosh and Xenia Dvoriantchikova for their expert assistance.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Galina Dvoriantchikova, Bascom Palmer Eye Institute, Department of Ophthalmology, University of Miami Miller School of Medicine, Miami, FL, 33136, USA.
Rajeev J. Seemungal, Bascom Palmer Eye Institute, Department of Ophthalmology, University of Miami Miller School of Medicine, Miami, FL, 33136, USA
Dmitry Ivanov, Bascom Palmer Eye Institute, Department of Ophthalmology, University of Miami Miller School of Medicine, Miami, FL, 33136, USA; Department of Microbiology and Immunology, University of Miami Miller School of Medicine, Miami, FL, 33136, USA.
References
- [1].Frick KD, Gower EW, Kempen JH, Wolff JL, Economic impact of visual impairment and blindness in the United States, Arch Ophthalmol, 125 (2007) 544–550. [DOI] [PubMed] [Google Scholar]
- [2].Johnson T, Rovner B, Haller J, Suicide and visual loss: a case report reflecting the need for recognition and management in ophthalmological settings, Semin Ophthalmol, 29 (2014) 202–204. [DOI] [PubMed] [Google Scholar]
- [3].Hine TJ, Pitchford NJ, Kingdom FA, Koenekoop R, Blindness and high suicide risk?, Psychosomatics, 41 (2000) 370–371. [DOI] [PubMed] [Google Scholar]
- [4].Bernardos RL, Barthel LK, Meyers JR, Raymond PA, Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells, J Neurosci, 27 (2007) 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lenkowski JR, Raymond PA, Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish, Prog Retin Eye Res, 40 (2014) 94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goldman D, Muller glial cell reprogramming and retina regeneration, Nat Rev Neurosci, 15 (2014) 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ramachandran R, Fausett BV, Goldman D, Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway, Nat Cell Biol, 12 (2010) 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kassen SC, Ramanan V, Montgomery JE, C TB, Liu CG, Vihtelic TS, Hyde DR, Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish, Dev Neurobiol, 67 (2007) 1009–1031. [DOI] [PubMed] [Google Scholar]
- [9].Powell C, Cornblath E, Elsaeidi F, Wan J, Goldman D, Zebrafish Muller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons, Sci Rep, 6 (2016) 24851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Powell C, Grant AR, Cornblath E, Goldman D, Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration, Proc Natl Acad Sci U S A, 110 (2013) 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA, Stimulation of functional neuronal regeneration from Muller glia in adult mice, Nature, 548 (2017) 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ueki Y, Wilken MS, Cox KE, Chipman L, Jorstad N, Sternhagen K, Simic M, Ullom K, Nakafuku M, Reh TA, Transgenic expression of the proneural transcription factor Ascl1 in Muller glia stimulates retinal regeneration in young mice, Proc Natl Acad Sci U S A, 112 (2015) 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wohl SG, Reh TA, miR-124–9-9* potentiates Ascl1-induced reprogramming of cultured Muller glia, Glia, 64 (2016) 743–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, Demb JB, Chen B, Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas, Nature, 560 (2018) 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiang M, Intrinsic control of mammalian retinogenesis, Cell Mol Life Sci, 70 (2013) 2519–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ohsawa R, Kageyama R, Regulation of retinal cell fate specification by multiple transcription factors, Brain Res, 1192 (2008) 90–98. [DOI] [PubMed] [Google Scholar]
- [17].Kohwi M, Doe CQ, Temporal fate specification and neural progenitor competence during development, Nat Rev Neurosci, 14 (2013) 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dvoriantchikova G, Perea-Martinez I, Pappas S, Barry AF, Danek D, Dvoriantchikova X, Pelaez D, Ivanov D, Molecular Characterization of Notch1 Positive Progenitor Cells in the Developing Retina, PLoS One, 10 (2015) e0131054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dvoriantchikova G, Santos AR, Danek D, Dvoriantchikova X, Ivanov D, The TIR-domaincontaining adapter inducing interferon-beta-dependent signaling cascade plays a crucial role in ischemia-reperfusion-induced retinal injury, whereas the contribution of the myeloid differentiation primary response 88-dependent signaling cascade is not as pivotal, Eur J Neurosci, 40 (2014) 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dvoriantchikova G, Degterev A, Ivanov D, Retinal ganglion cell (RGC) programmed necrosis contributes to ischemia-reperfusion-induced retinal damage, Exp Eye Res, 123 (2014) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH, Amplified RNA synthesized from limited quantities of heterogeneous cDNA, Proc Natl Acad Sci U S A, 87 (1990) 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].de Hoon MJ, Imoto S, Nolan J, Miyano S, Open source clustering software, Bioinformatics, 20 (2004) 1453–1454. [DOI] [PubMed] [Google Scholar]
- [23].Saldanha AJ, Java Treeview--extensible visualization of microarray data, Bioinformatics, 20 (2004) 3246–3248. [DOI] [PubMed] [Google Scholar]
- [24].Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25 (2009) 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN, RNA-Seq gene expression estimation with read mapping uncertainty, Bioinformatics, 26 (2010) 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, 26 (2010) 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ernst J, Kellis M, ChromHMM: automating chromatin-state discovery and characterization, Nat Methods, 9 (2012) 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cavalcante RG, Sartor MA, annotatr: genomic regions in context, Bioinformatics, 33 (2017) 23812383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Krueger F, Andrews SR, Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications, Bioinformatics, 27 (2011) 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Burger L, Gaidatzis D, Schubeler D, Stadler MB, Identification of active regulatory regions from DNA methylation data, Nucleic Acids Res, 41 (2013) e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE, methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles, Genome Biol, 13 (2012) R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou VW, Goren A, Bernstein BE, Charting histone modifications and the functional organization of mammalian genomes, Nat Rev Genet, 12 (2011) 7–18. [DOI] [PubMed] [Google Scholar]
- [33].Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T, Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells, Dev Cell, 10 (2006) 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Keenen B, de la Serna IL, Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation, J Cell Physiol, 219 (2009) 1–7. [DOI] [PubMed] [Google Scholar]
- [35].Kraushaar DC, Zhao K, The epigenomics of embryonic stem cell differentiation, Int J Biol Sci, 9 (2013) 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wreczycka K, Gosdschan A, Yusuf D, Gruning B, Assenov Y, Akalin A, Strategies for analyzing bisulfite sequencing data, J Biotechnol, 261 (2017) 105–115. [DOI] [PubMed] [Google Scholar]
- [37].Iwafuchi-Doi M, Zaret KS, Cell fate control by pioneer transcription factors, Development, 143 (2016) 1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Seritrakul P, Gross JM, Tet-mediated DNA hydroxymethylation regulates retinal neurogenesis by modulating cell-extrinsic signaling pathways, PLoS Genet, 13 (2017) e1006987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R, Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system, Development, 133 (2006) 2467–2476. [DOI] [PubMed] [Google Scholar]
- [40].Noda N, Honma S, Ohmiya Y, Hes1 is required for contact inhibition of cell proliferation in 3T3-L1 preadipocytes, Genes Cells, 16 (2011) 704–713. [DOI] [PubMed] [Google Scholar]
- [41].Kageyama R, Ohtsuka T, Kobayashi T, Roles of Hes genes in neural development, Dev Growth Differ, 50 Suppl 1 (2008) S97–103. [DOI] [PubMed] [Google Scholar]
- [42].Kobayashi T, Kageyama R, Expression dynamics and functions of Hes factors in development and diseases, Curr Top Dev Biol, 110 (2014) 263–283. [DOI] [PubMed] [Google Scholar]
- [43].Castella P, Sawai S, Nakao K, Wagner JA, Caudy M, HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression, Mol Cell Biol, 20 (2000) 6170–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pollak J, Wilken MS, Ueki Y, Cox KE, Sullivan JM, Taylor RJ, Levine EM, Reh TA, ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors, Development, 140 (2013) 2619–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tan L, Shi YG, Tet family proteins and 5-hydroxymethylcytosine in development and disease, Development, 139 (2012) 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li D, Guo B, Wu H, Tan L, Lu Q, TET Family of Dioxygenases: Crucial Roles and Underlying Mechanisms, Cytogenet Genome Res, 146 (2015) 171–180. [DOI] [PubMed] [Google Scholar]
- [47].Yao B, Jin P, Unlocking epigenetic codes in neurogenesis, Genes Dev, 28 (2014) 1253–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD, TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation, Neuron, 79 (2013) 1086–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, Liu W, Xu ZM, Yang L, Ding YQ, Tang F, Meissner A, Ding C, Shi Y, Xu GL, Tet1 regulates adult hippocampal neurogenesis and cognition, Cell Stem Cell, 13 (2013) 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H, Neuronal activity modifies the DNA methylation landscape in the adult brain, Nat Neurosci, 14 (2011) 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH, Tet1 is critical for neuronal activity-regulated gene expression and memory extinction, Neuron, 79 (2013) 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li X, Yao B, Chen L, Kang Y, Li Y, Cheng Y, Li L, Lin L, Wang Z, Wang M, Pan F, Dai Q, Zhang W, Wu H, Shu Q, Qin Z, He C, Xu M, Jin P, Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis, Nat Commun, 8 (2017) 15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, Liu XS, Sugrue SP, He X, Min J, Kato Y, Shi YG, Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development, Cell, 151 (2012) 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Perera A, Eisen D, Wagner M, Laube SK, Kunzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, Muller M, Biel M, Carell T, Michalakis S, TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression, Cell Rep, 11 (2015) 283–294. [DOI] [PubMed] [Google Scholar]
- [55].Aurora AB, Olson EN, Immune modulation of stem cells and regeneration, Cell Stem Cell, 15 (2014) 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Eming SA, Wynn TA, Martin P, Inflammation and metabolism in tissue repair and regeneration, Science, 356 (2017) 1026–1030. [DOI] [PubMed] [Google Scholar]
- [57].Julier Z, Park AJ, Briquez PS, Martino MM, Promoting tissue regeneration by modulating the immune system, Acta Biomater, 53 (2017) 13–28. [DOI] [PubMed] [Google Scholar]
- [58].Nelson CM, Ackerman KM, O’Hayer P, Bailey TJ, Gorsuch RA, Hyde DR, Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration, J Neurosci, 33 (2013) 6524–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Conner C, Ackerman KM, Lahne M, Hobgood JS, Hyde DR, Repressing notch signaling and expressing TNFalpha are sufficient to mimic retinal regeneration by inducing Muller glial proliferation to generate committed progenitor cells, J Neurosci, 34 (2014) 14403–14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tse BC, Dvoriantchikova G, Tao W, Gallo RA, Lee JY, Pappas S, Brambilla R, Ivanov D, Tse DT, Pelaez D, Tumor Necrosis Factor Inhibition in the Acute Management of Traumatic Optic Neuropathy, Invest Ophthalmol Vis Sci, 59 (2018) 2905–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McCoy MK, Tansey MG, TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease, J Neuroinflammation, 5 (2008) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dvoriantchikova G, Ivanov D, Tumor necrosis factor-alpha mediates activation of NF-kappaB and JNK signaling cascades in retinal ganglion cells and astrocytes in opposite ways, Eur J Neurosci, 40 (2014) 3171–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw files from the microarray study, ChIP-Seq, and WGBS have been deposited in the NCBI Gene Expression Omnibus (GEO) database. They are accessible through GEO accession number GSE122337 (GSE122301, GSE122302, GSE122356).