Abstract
In eukaryotes, a nascent peptide entering the endoplasmic reticulum (ER) is scanned by two Sec61-translocon-associated large membrane machines for protein N-glycosylation and protein O-mannosylation, respectively. While the structure of the eight-protein oligosaccharyltransferase complex has been determined recently, the structures of mannosyltransferases of the PMT family, which are an integral part of ER protein homeostasis, are still unknown. Here we report cryo-EM structures of the S. cerevisiae Pmt1–Pmt2 complex bound to a donor and an acceptor peptide at 3.2-Å resolution, showing that each subunit contains 11 transmembrane helices and a lumenal β-trefoil fold termed the MIR domain. The structures reveal the substrate recognition model and confirm an inverting mannosyl-transferring reaction mechanism by the enzyme complex. Furthermore, we found that the transmembrane domains of Pmt1 and Pmt2 share a structural fold with the catalytic subunits of oligosaccharyltransferases, confirming a previously proposed evolutionary relationship between protein O-mannosylation and protein N-glycosylation.
Protein O-mannosylation is an essential posttranslational protein modification in eukaryotes and certain bacteria1,2. It is evolutionarily conserved and plays crucial roles in numerous cellular processes, including unfolded protein O-mannosylation and protein quality control3. The protein O-mannosyltransferases (POMT in mammals, or PMT in yeast) catalyze the transfer of a mannose from dolichyl phosphate-activated mannose (Dol-P-Man) to a serine or threonine residue of proteins4. Dolichol is a linear, membrane-embedded molecule comprising a dozen or so isoprene repeats5. Dol-P-Man is synthesized at the cytosolic face of the ER membrane by the dolichol phosphate mannose synthase (DPMS) which transfers a mannose from GDP-Man to the dolichol monophosphate carrier Dol-P6. Dol-P-Man is then flipped to the lumenal face of the ER to serve as the mannose donor of the PMTs7. In contrast, protein N-glycosylation is catalyzed by the oligosaccharyltransferase (OST), an eight-protein complex embedded in the ER membrane in eukaryotes8,9. The OST is an inverting glycosyltransferase that transfers a preassembled 14-sugar oligosaccharide (OS) carried by a dolichol diphosphate (Dol-PP-OS) to the NxS/T peptide motif in acceptor proteins10. The Dol-PP is synthesized from Dol-P by adding a phosphate at the cytosolic face of the ER. After attaching seven sugars (two GlcNAc and five mannoses), Dol-PP is flipped to the ER lumenal face where seven additional sugars (four mannoses and three glucoses) are added to become Dol-PP-OS, the ultimate donor substrate of the OSTs11. Therefore, the initial synthesis of the Dol-P carrier is shared between the protein N-glycosylation and protein O-mannosylation pathways, and this process is well conserved in eukaryotes. Furthermore, because OST and PMT are both physically associated with the Sec61 translocon (Fig. 1a)12 and their target sites may overlap (i.e., NxS/T versus S/T), protein O-mannosylation and N-glycosylation are inherently interconnected and affect one another13,14. In fact, PMTs are suggested to be evolutionarily related to OSTs, although structural evidence has been lacking (https://pfam.xfam.org/family/PMT)15,16
Fig. 1. Structure of the yeast Pmt1–Pmt2 complex.
(a) Both protein O-mannosylation and protein N-glycosylation can be carried out co-translationally and co-translocationally, because both Pmt1–Pmt2 and OST physically associate with the Sec61 translocon. (b) The overall structure of Pmt1–Pmt2 in cartoons is shown in front and back views (green for Pmt1 and purple-blue for Pmt2). The Dol-P and N-glycans are shown as orange and red sticks, respectively. The acceptor peptide is shown as red spheres. (c) A ribbon diagram and topological sketch of Pmt1. The 11 TMHs are numbered and colored in a rainbow scheme. The MIR domain is drawn as a green circle. The number and identity of the first residue on each TMH is labeled. The red asterisk marks the DE motif at the beginning of the first horizontal helix (HH1).
Protein O-mannosylation is implicated in several human diseases, including congenital muscular dystrophy, cancer metastasis, and viral entry1,17. The mammalian mannosyltransferase is a heterodimeric POMT1–POMT2 complex. S. cerevisiae contains six PMT proteins that can be classified into three subfamilies based on sequence similarity: PMT1 (Pmt1 and Pmt5), PMT2 (Pmt2, Pmt3, and Pmt6), and PMT4 (Pmt4)13. Among the six, Pmt1 primarily forms a heterodimer with Pmt2, and Pmt4 homo-dimerizes, and they together account for the majority of the transferase activities. Simultaneous knockout of Pmt1, 2, and 4 is lethal in yeast18,19. Because mannose is β-linked to the phosphate in Dol-P-Man, but is α-linked to the S/T in the acceptor proteins20, PMTs are inverting transferases4,10. Pmt1 and Pmt2 are large, multi-pass transmembrane proteins that have 817 and 759 amino acids, respectively, with a calculated mass of 173 kDa for the heterodimer. Pmt1 and Pmt2 belong to the glycosyltransferase 39 (GT39) family (http://www.cazy.org), for which no structure has been reported4.
Results
Purification and structural determination of the Pmt1–Pmt2 complex
We produced the Pmt1–Pmt2 complex endogenously from baker’s yeast by inserting a 3× FLAG tag to the carboxyl terminus of Pmt1. We purified about 0.5 mg of the complex from 18 L of yeast culture by anti-FLAG affinity resin followed by size-exclusion chromatography. The quality and identity of the protein complex were verified by SDS-PAGE and mass spectrometry, respectively (Supplementary Note 1). To determine the structure, we first performed single-particle cryo-EM of the purified Pmt1–Pmt2 alone using a 300-kV Titan Krios microscope equipped with a K2 detector. A 3.2-Å-resolution cryo-EM 3D map was derived from 581,966 particles that were selected from about 2800 raw electron micrographs (Supplementary Note 2, Table 1). To understand the substrate binding mechanism, we also performed single-particle cryo-EM of the purified Pmt1–Pmt2 in complex with a tetrapeptide acceptor (PYTV), which was shown to be an effective acceptor in an in vitro assay21. A 3.2 Å resolution cryo-EM 3D map was derived from 521,848 particles that were selected from about 3000 raw electron micrographs (Supplementary Note 3, Supplementary Figure 1a, Supplementary Video 1, Table 1). Superposition of these two maps revealed they were highly similar except for an extra density that corresponded to the tetrapeptide in the later map (Supplementary Figure 1b–e). Therefore, we will mainly discuss the map of Pmt1–Pmt2 in complex with the peptide in this paper. Most regions of the map had excellent main-chain connectivity and side-chain densities (Supplementary Figures 1–2), except for the lumenal domain of Pmt2. This domain is termed the MIR domain because of its presence in the mannosyltransferase, inositol triphosphate, and ryanodine receptor22. We subsequently determined the crystal structure of the Pmt2 MIR domain at 1.35-Å resolution (Supplementary Figure 3, Table 2), which was fitted into the cryo-EM 3D map as a rigid body. Finally, an atomic model of the full Pmt1–Pmt2 complex was built and refined. The final model contained residues 15 –749 in Pmt1 and residues 57 – 754 in Pmt2 — missing N-terminal 14 residues and C-terminal 67 residues in Pmt1, and missing N-terminal 56 residues and C-terminal 5 residues in Pmt2. Pmt1 is unique among PMTs in having a longer C-terminal tail. However, deletion of the C-terminal 86 residues (732–817) in Pmt1 did not affect its dimerization with Pmt2, nor the enzyme activity of the Pmt1–Pmt2 complex23, suggesting that this region is disordered and is not important for the enzyme function.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Pmt1–Pmt2–donor (EMD-20240, PDB 6P2R) | Pmt1–Pmt2–donor–acceptor (EMD-20236, PDB 6P25) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 60 | 60 |
| Defocus range (μm) | −1.0 – −2.0 | −1.0 – −2.0 |
| Pixel size (Å) | 1.029 | 1.029 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 913,730 | 953,292 |
| Final particle images (no.) | 581,966 | 521,848 |
| Map resolution (Å) | 3.2 | 3.2 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 278.5–3.2 | 278.5–3.2 |
| Refinement | ||
| Initial model used (PDB code) | N/A | N/A |
| Model resolution (Å) | 3.6 | 3.4 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | ||
| Map sharpening B factor (Å2) | 177 | 169 |
| Model composition | ||
| Nonhydrogen atoms | 11,748 | 11,774 |
| Protein residues | 1,413 | 1,416 |
| Ligands | 6 | 6 |
| B factors (Å2) | ||
| Protein | 89.9 | 82.8 |
| Ligand | 26.1 | 17.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | 0.01 |
| Bond angles (°) | 1.01 | 1.12 |
| Validation | ||
| MolProbity score | 1.88 | 1.89 |
| Clashscore | 8.05 | 8.42 |
| Poor rotamers (%) | 0.4 | 0.4 |
| Ramachandran plot | ||
| Favored (%) | 93.2 | 93.3 |
| Allowed (%) | 7.8 | 7.7 |
| Disallowed (%) | 0 | 0 |
Table 2.
X-ray diffraction Data collection and refinement statistics (molecular replacement)
| Pmt2 MIR domain (PDB code 6P28) | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 34.28, 71.40, 37.88 |
| α, β, γ (°) | 90, 96.37, 90 |
| Resolution (Å) | 1.35 (1.38–1.35)a |
| Rmerge | 6.8 (58.5) |
| I/σ(I) | 13.4 (2.3) |
| CC1/2 | N/A |
| Completeness (%) | 96.7 (85.4) |
| Redundancy | 3.5 (3.4) |
| Refinement | |
| Resolution (Å) | 26.79–1.35 |
| No. reflections | 38,941 (2,627) |
| Rwork / Rfree | 0.184 / 0.208 |
| No. atoms | |
| Protein | 1555 |
| Ligand/ion | 0 |
| Water | 199 |
| B factors | |
| Protein | 68.6 |
| Ligand/ion | N/A |
| Water | 68.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.875 |
Diffraction data from one crystal was used for structure solution.
Values in parentheses are for highest-resolution shell.
Overall structure of the Pmt1–Pmt2 complex
The overall structure is approximately 110 × 100 × 110 Å and reveals a 1:1 heterodimer of Pmt1 and Pmt2 with a pseudo twofold symmetry (Fig. 1b). Both Pmt1 and Pmt2 contain a transmembrane region and a lumenal MIR domain. The MIR domain contains three MIR motifs forming a β-trefoil, and each MIR motif contains four β-strands and a short α-helix (Supplementary Figure 3). Remarkably, the transmembrane regions of Pmt1 and Pmt2 don’t interact directly; they are held together by contacts located in a cytosolic region and a lumenal region. Such arrangement generates a sizable rhombic cavity in the middle of the structure that is about 20 Å long on each side. The central cavity likely allows the membrane-embedded donor substrate, Dol-P-Man, to diffuse into the catalytic site of the complex.
There are five potential N-glycosylation sites in the complex, three (N390, N513, and N743) in Pmt1 and two (N131 and N403) in Pmt2; the presence of N-glycans on PMT enzymes can affect their activity in vitro24. We observed two N-glycans in our EM map, one on N390 of Pmt1 and the other on N131 of Pmt2 (Fig. 1b, Supplementary Figure 2). The N513 and N743 of Pmt1 was clearly unmodified, but the N-glycosylation state of the N403 in Pmt2 was undetermined because they are located in flexible regions of the structure.
Asymmetric interactions between Pmt1 and Pmt2
Pmt1 and Pmt2 each comprises 11 TMHs plus a cytosolic N-terminus and a lumenal C-terminus (Fig. 1c). The N-terminal region of Pmt1 contains an extended sequence that folds into a two-stranded β-sheet at the bottom cytosolic face of the transmembrane region, apparently stabilizing the Pmt1 structure, but the corresponding region in Pmt2 is disordered. The 11 PMT TMHs are mostly connected by short loops except for the lumenal loops LL1 and LL4. LL1, connecting TMH1 and TMH2, forms two horizontal helices (HH1 and HH2). LL4, connecting TMH7 and TMH8, contains an MIR domain followed by another horizontal helix (HH3) and another two-stranded β-sheet. Consistent with their high sequence identity of 35%, Pmt1 and Pmt2 are superimposable, with a root-mean-square-deviation (rmsd) of 1.8 Å in the membrane regions and an rmsd of 1.6 Å in the MIR domains (Supplementary Figure 4). However, the MIR domain of Pmt2 rotates about 17° away from the membrane region relative to that of Pmt1. This rotation disengages the MIR domain of Pmt2 from Pmt1. Therefore, only the MIR domain of Pmt1 interacts with Pmt2 in our structure (Fig. 2a). This interaction is mediated by stacking of W423, H482, S501 and R503 of the Pmt1 MIR domain with D571, K572 and F573 of the Pmt2 LL4. As a result, the lumenal access of Dol-P-Man to the central substrate cavity is closed from the Pmt1-MIR side and is open only from the Pmt2-MIR side. On the cytosolic side, the interface between Pmt1 and Pmt2 is mainly formed by interactions between the ends of TMH6 and TMH8, as well as between cytosolic loop 3 (CL3) of one subunit with CL4 of the other subunit. Specifically, Q612 and L613 of Pmt1 CL3 interact with W269 and L272 of Pmt2 CL4, and Q633, R634 and Q635 of the Pmt2 CL3 interact with W253 and I256 of Pmt1 CL4 (Fig. 2b). In addition, The N-terminal loop (NTL) of Pmt1 contributes to this cytosolic interface via its V26 and R27 interacting with W632 and Q635 of the Pmt2 CL3. The N-terminal loop of Pmt2 is flexible and thus does not contribute to the interface. Most residues at the cytosolic interfaces between Pmt1 and Pmt2 are conserved (Supplementary Note 4). In fact, there is an ordered lipid molecule that binds hydrophobically to TMH6 and TMH7 of Pmt1 and TMH8 of Pmt2, thereby contributing to the dimerization between Pmt1 and Pmt2 (Fig. 2b).
Fig. 2. The interfaces between Pmt1 and Pmt2.
A lumenal view (a) and a cytosolic view (b) of the Pmt1–Pmt2 structure. The interface areas between the two subunits are highlighted by a dashed black rectangle in (a) and by a dashed orange rectangle in (b). The areas in the rectangles are enlarged at the right. The lipid molecule mediating the Pmt1–Pmt2 interaction at the cytosolic side is shown as sticks.
Peptide acceptor and dolichol donor binding sites in Pmt1 and Pmt2
By comparing the maps of Pmt1–Pmt2 alone with Pmt1–Pmt2–peptide, we observed clear extra density for the bound peptide in a lumenal pocket of Pmt2 (Supplementary Figure 1b-e, bottom panel of Supplementary Figure 2, Figs. 1b, 2a). Furthermore, we observed an elongated density at the left side of the central cavity stacking against the TMH6 of Pmt1, and this density resembled the native donor substrate in a zig-zag shape (bottom panel of Supplementary Figure 2, Figs. 1b, 3a). The donor was likely co-purified endogenously with the Pmt1–Pmt2 complex. Interestingly, the peptide acceptor and the donor were observed in different subunits; the peptide was found in Pmt2 and the donor in the Pmt1. However, both the donor and acceptor substrate were located on the same (lumenal) side of the Pmt1 and Pmt2 (Fig. 2a). On the lumenal side, the above-mentioned asymmetric interaction between Pmt1 and Pmt2 has two consequences (Fig. 2a): 1) Pmt2 has a larger and more open acceptor-binding site than Pmt1, and 2) the donor-entry or product-release pathway in Pmt1 is locked closed relative to the open pathway in Pmt2. These structural features likely explain why the peptide acceptor was accommodated only in Pmt2, but not in Pmt1, and why the donor-like density was trapped in Pmt1 but not in Pmt2 during protein purification.
Because the elongated donor-like density lacked the corresponding mannose moiety, we modeled the density as the enzyme product Dol-P rather than the donor substrate Dol-P-Man. The phosphate group of the reaction product had well-defined density. The yeast dolichol has 14–18 isoprene units25, but in our cryo-EM maps, the product density was only long enough to accommodate the first 7 isoprene repeats. Therefore, the second half of the yeast dolichol is likely disordered in the enzyme complex. We modeled the tetrapeptide PYTV into the identified acceptor density with the threonine side chain facing the donor binding site. By superimposing the structure of Pmt1 in complex with Dol-P on the structure of Pmt2 complexed with the acceptor peptide, we were able to examine the acceptor and donor binding sites in both Pmt1 and Pmt2 (Fig. 3a–e).
Fig. 3. The active site of the Pmt1–Pmt2 complex.
(a) Pmt1 is shown as a cartoon and a transparent surface view. The transmembrane region of Pmt2 (purple-blue) is superposed with Pmt1. Dol-P is shown as orange sticks, and the acceptor peptide is shown as a red cartoon. (b) Close-up view of the highly conserved DE motif of Pmt1 and Pmt2. (c) Close-up view of the active site of Pmt1. (d) Close-up view of the active site of Pmt2. (e) LIGPLOT scheme for Dol-P and acceptor peptide binding residues of Pmt1. The pink circle represents the active site of the enzyme.
In the Pmt1 structure, the phosphate of Dol-P is stabilized by H98, K234, R649, H654, and H655, and the acceptor threonine-binding site is formed by F76, D77, H80 and F652 (Fig. 3c, 3e). All these residues in the active site are conserved among the yeast and human mannosyltransferases (Supplementary Note 4). A K234A mutant Pmt1 retained about 80% of the wild type enzyme activity23. However, the previously identified invariant DE motif, D77E78 in Pmt1 and D92E93 in Pmt215, plays a critical role in the active site. Specifically, Pmt1 D77 (Pmt2 D92) is ideally positioned to activate the hydroxyl of the acceptor threonine, and Pmt1 E78 (Pmt2 E93) forms a salt bridge with the invariant lysine, R138 in Pmt1 and R152 in Pmt2 (Fig. 3b). This salt bridge is located at the beginning of HH1, apparently playing a structural role to force the preceding aspartate residue (D77 in Pmt1 and D92 in Pmt2) to point towards the substrate binding pocket and the acceptor threonine. Replacement of any residue in the DE motif in the homodimeric Pmt4 or replacing the DE motif with two alanine residues in Pmt1–Pmt2 has been shown to completely abolish the enzyme activities15. In contrast to the conserved threonine binding site, the binding pocket of other part of acceptor peptide is not conserved among PMTs, in agreement with the fact that these enzymes do not have a preferred amino acid sequence except for the single mannose-receiving serine or threonine.
The dolichol was bound to an elongated hydrophobic groove formed by TMH6, TMH7, TMH9 and HH3, and was stabilized by W235, L238, F239, V241, T242, G245, and L252 of TMH6; A270, L274, L277, and L278 of TMH7; W545 and N548 of HH3; and M647 of TMH9 (Fig. 3e). Given the extensive interface, individual point mutations are not expected to abolish donor binding or the enzyme activity. For example, the W253A mu2tant Pmt1 retained about 70% of the wild type enzyme activity23. Because Dol-P-Man — the donor substrate of protein mannosyltransferases — is the product of the membrane-embedded DPMS, the Dol-P-Man synthase, we compared our Pmt1 structure with the structure of DPMS (PDB 5MM1)26, and we found that the pockets for the dolichol-linked ligands were lined by conserved residues in both enzymes (Supplementary Figure 5a-b). This observation suggests that Dol-P-Man is similarly coordinated in these different enzymes.
Conserved fold and similar catalytic mechanism between protein O-mannosylation and protein N-glycosylation
To better understand the mechanism of Pmt1–Pmt2, we performed a structure-based homology search using the online Dali server27. We found that the transmembrane region of Pmt1 and Pmt2 share a structural fold with the bacterial oligosaccharyltransferase PglB, the archaeal oligosaccharyltransferase AglB, and the catalytic subunit Stt3 of eukaryotic OST (Fig. 4)28–33. Structures of Pmt1 and PglB (PDB: 5OGL) are superimposable with an rmsd of 3.2 Å (Fig. 4). This observation confirms the previous suggestions that PMTs are evolutionarily linked to the oligosaccharyltransferases15,16. Both PMTs and oligosaccharyltransferases belong to the inverting glycosyltransfereases in which the covalent bond with the anomeric carbon is on the opposite side of the sugar in the donor and the acceptor, respectively4,10. Remarkably, when the structures of the two enzymes are superimposed, the acceptor and donor binding pocket in Pmt1–Pmt2 is almost in the same position as in PglB (Fig. 4). Because the mannose moiety of the donor is missing in our structure, we didn’t observe a strong enough density corresponding to the Mn2+, although conserved residues for coordinating the divalent metal ion were present in both Pmt1 and Pmt2. The phosphate group of the modeled Dol-P in Pmt1 is near the phosphate of the Dol-PP-OS in PglB, and the acceptor threonine in Pmt2 is also close to the acceptor asparagine in PglB. Therefore, both the acceptor threonine in Pmt2 and the acceptor asparagine in PglB are located on the opposite side of their respective donors, in a similar configuration that is consistent with the inverting transfer reaction mechanism34–37.
Fig. 4. Superimposition of the transmembrane domain of Pmt1 (green) with that of the PglB structure (grey).
A side view (a) and a top view (b) of the superimposition of the transmembrane domain of Pmt1 (green) with that of the PglB structure (PDB ID 5OGL, grey), which is in complex with an acceptor peptide, Mn2+, and a nonhydrolyzable lipid-linked oligosaccharide analog, (ωZZZ)-PPC-GlcNAc (blue). The red asterisk denotes the overlapping phosphate groups and the acceptor residues in the two structures. The acceptor residues, a Thr in Pmt1–Pmt2 and an Asn in PglB, are located on the opposite side of the donor group, in consistent with the fact that both enzymes are inverting glycosyltransferases.
Another similarity between PMTs and oligosaccharyltransferases is a horizontal helix positioned right above the donor binding site. The horizontal helix in PglB is a flexible loop in the apo enzyme, perhaps allowing the large dolichol-linked oligosaccharide to diffuse into the substrate pocket, because the flexible peptide becomes an ordered helix once the dolichol-linked oligosaccharide is present in the substrate pocket28. The corresponding horizontal helix in Pmt1 and Pmt2 is HH3, which contributes to the binding of Dol-P. Because HH3 is ordered even in the absence of a substrate, this helix may participate in the selection of the donor substrate, perhaps by preventing the dolichol-linked ligands with large sugar moieties (such as an OS) from accessing the substrate pocket.
Discussion
Glycosyltransfereases are structurally classified into three different folds: GT-A, GT-B, and GT-C. The GT-A and GT-B folds are better represented in the structural databases10,35,38. There have been very few structures determined that have the GT-C fold, due to the fact that enzymes with this fold are mostly membrane-embedded, and in many cases they form multi-subunit complexes and contain post-translational modifications, making structural determination by traditional X-ray crystallography very difficult29,32. These problems have recently been alleviated due to the advance of cryo-EM methodology39, as exemplified by the three cryo-EM structures of the yeast and human oligosaccharyltransferases30,31,33. There are currently nine GT families in the CAZy database that have the GT-C fold (GT22, GT39, GT50, GT57, GT58, GT59, GT66, GT83, and GT85) (http://www.cazy.org). However, all of the GT-C fold structures published so far are from the GT66 family. Therefore, the Pmt1–Pmt2 complex structure represents the first structural solution of the second glycosyltransferease family (GT39).
The Pmt1–Pmt2 structure explains a large body of biochemical and mutational work done over the past decades13, and in particular, the role of the invariant residues in setting up the catalytic pocket of the enzyme4,40. Our structure also provides direct evidence for the evolutionary relationship of PMT proteins with OST, which was initially proposed based on sequence analysis15,16. The structure of the Pmt1–Pmt2 in complex with Dol-P and the tetrapeptide further confirms the inverting transfer reaction mechanism of protein O-mannosyltransferases.
Mutations in the human mannosyltransferase POMT1–POMT2 cause congenital muscular dystrophies (CMDs), characterized by abnormal glycosylation of α-dystroglycan with neuronal migration defects17,41. Based on the clinical phenotypes, these diseases include Walker-Warburg syndrome, muscle-eye-brain disease, and less severe forms of muscular dystrophy. We surveyed the reported POMT1–POMT2 mutations of hundreds of CMD patients and identified dozens of missense mutations in POMT1 and POMT242–65 (Supplementary Table 1). Many of these mutations either reduced or abolished the enzyme activity. Yeast Pmt1 and Pmt2 share 29% and 36% identity with human POMT1 and POMT2, respectively, suggesting a similar overall structure for the human POMT1–POMT2 complex. Furthermore, most pathological mutations in human POMT1–POMT2 are conserved in yeast Pmt1–Pmt2 (Supplementary Note 4). Therefore, we mapped the disease mutations onto the Pmt1–Pmt2 structure and found that most mutations were clustered to the ER lumenal half of the structure (Fig. 5). Because the active site and the donor and acceptor binding pockets are on the lumenal side, their distribution pattern suggests that these mutations likely affect PMT enzymatic activity.
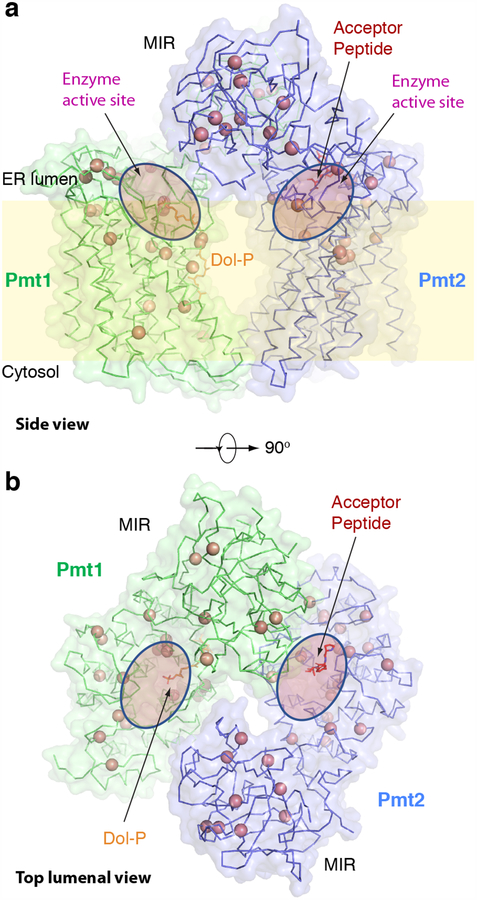
Fig. 5. Mapping of the congenital muscular dystrophy mutations found in human POMT1–POMT2 on to the structure of the yeast Pmt1–Pmt2.
(a, b) A side and a top lumenal views of the Pmt1-Pmt2 in ribbon presentation. The Cα atoms of disease-related residues are shown as salmon spheres. The active sites are shown by transparent pink ovals. The acceptor peptide and Dol-P are shown as sticks. Individual mutations are summarized in Supplementary Table 1.
Unexpectedly, many disease mutations were located in the MIR domains that are away from the catalytic site, suggesting that the MIR domains may play an important role. Indeed, previous work has shown that point mutations in the Pmt1 MIR domain reduced the enzymatic activity, and deletion of the entire MIR domain of Pmt1 (304–531) nearly abolished the activity of the Pmt1–Pmt2 complex23. Although the exact function of the MIR domain is unknown, we performed a structure-based homologue search and found that the MIR domain was similar to a sugar-binding mushroom lectin (PDB: 2IHO), with an rmsd of 2.5 Å (Supplementary Figure 5c)66. Based on the structural homology and the fact that the MIR domain is located just above the Dol-P-Man entry or product release path, we suggest that it may play a role in recognition of the mannose moiety of the donor substrate. But more studies are needed to determine the function of this domain. Our structure provides a platform for further structure–function investigations of Pmt1–Pmt2 and facilitates the future development of small molecules to modulate the enzyme activity.
METHODS
Purification of the endogenous Pmt1–Pmt2 complex.
We generated a C-terminal triple-FLAG-tagged Pmt1 construct using a PCR-based genomic epitope-tagging method on the yeast W303–1a (MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11). The strain was grown in YPD medium and then was harvested and resuspended in lysis buffer containing 20 mM Tris-HCl (pH 7.4), 0.2 M sorbitol, 50 mM potassium acetate, 2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed using a French press at 15,000 psi and then were centrifuged at 10,000 × g for 30 min at 4 °C. The supernatant was then centrifuged at 100,000 × g for 60 min at 4 °C. The membrane pellet was collected and then resuspended in buffer A containing 10% glycerol, 20 mM Tris-HCl (pH 7.4), 1.5% DDM, 0.5 M NaCl, 1 mM MgCl2, 1 mM MnCl2, 1 mM EDTA, and 1 mM PMSF. After incubation for 30 min at 4 °C, the mixture was centrifuged for 30 min at 120,000 × g, and the clarified supernatant was mixed with pre-washed anti-FLAG (M2) affinity gel at 4 °C overnight with shaking. The affinity gel was collected by centrifugation and washed three times in buffer B containing 0.025% n-dodecyl β-D-maltoside (DDM), 0.0025% cholesteryl hemisuccinate tris salt (CHS), 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1 mM MgCl2, and 1 mM MnCl2. We found that CHS did not affect Pmt1 and Pmt2 dimerization. Also, including CHS in the detergent mix improved the sample quality, as judged by the improved structural details present in 2D class averages of the cryo-EM particles. Finally, the Pmt1–Pmt2 complex was eluted with buffer B containing 0.15 mg/ml 3×FLAG peptide and was further purified in a Superose 6 10/300 gel filtration column in buffer C containing 0.01% lauryl maltose-neopentyl glycol (LMNG), 0.001% CHS, 150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 1 mM MgCl2, and 1 mM MnCl2. The final sample was assessed by SDS-PAGE gel and was identified by tryptic digestion and mass spectrometry. From 18 L yeast culture, about 0.5 mg of purified complex can be routinely obtained with this procedure. To make the Pmt1–Pmt2–peptide complex, the peptide (PYTV) was synthesized by Genscript (Piscataway, NJ, USA), and incubated with purified Pmt1–Pmt2 sample at a 5:1 molar ratio for 1h before preparing the cryo-EM grids.
Purification of the MIR domain of Pmt2.
The polymerase chain reaction (PCR) product of the predicted MIR domain (amino acids 337–532) of Pmt2 was cloned into a pSUMO expression vector. It was expressed in E. coli BL21 (DE3) and purified as an N-terminal 6×His–Sumo fusion protein. Cells were grown in Luria-Bertani medium at 24 °C to 0.8 at OD600 and then were induced by 0.05 mM isopropyl β-D-1-thiogalactopyranoside at 16 °C for overnight. Proteins were first purified by Ni-affinity chromatography in buffer containing 20 mM Tris, pH 8.0, 200 mM NaCl, and 10% glycerol, and they then were eluted in same buffer with 300 mM imidazole. The 6×His–Sumo tag was then removed by UIP proteolytic cleavage and rebound to a Nickel HP column. Finally, the protein was purified by gel filtration chromatography (Superose 6 10/300) in 20 mM Tris, pH 8.0, and 200 mM NaCl.
Crystallization, data collection, and structure determination of the MIR domain of Pmt2.
The crystallization screen was carried out using the sitting-drop vapor diffusion method at 20 °C. Crystals appeared after 3 d from drops consisting of 1 μl of protein solution at 10 mg/ml and 1 μl of reservoir solution containing 25% PEG 2K MME. Crystal diffraction data sets were collected at 100 K at the LS-CAT beamlines at the Argonne National Laboratory. The X-ray wavelength was 1.078 Å. Diffraction data were processed with HKL20001. The initial phases were obtained using the molecular replacement method with our EM structure of MIR domain of Pmt1 as the search model in the Phenix program2. Coot was used to manually adjust and rebuild the model3. Further model refinement was carried out using Refmac54 in the CCP4 suite and Phenix. The refinement and manual rebuilding were iterated multiple times until acceptable Rwork (18.4%) and Rfree (20.8%) values were obtained. Ramachandran statistics were 95.79% in the favored region, 4.21% in the allowed region, and 0% outlier. MolProbity score was 1.53. The Clashscore was 4.63 with 0% residues having poor rotamers.
Cryo-electron microscopy.
Aliquots of 3 μl of purified Pmt1–Pmt2 or Pmt1–Pmt2–peptide complex at a concentration of 5 mg/ml were placed on glow-discharged holey carbon grids (Quantifoil Au R2/1, 300 mesh) and were flash-frozen in liquid ethane using a Thermo Fisher Scientific (TFS) Vitrobot Mark IV. We initially determined a 3.8 Å resolution cryo-EM map based on a data set of approximately 470,000 particles selected from 900 movie micrographs collected on a TFS Arctica microscope operated at 200 kV. The micrographs were collected automatically with EPU at a nominal magnification of 130,000× and a pixel size of 1.21 Å per pixel with defocus values from −1.5 to −2.5 μm. A Falcon III direct detector was used for image recording under counting mode. The dose rate was 0.6 electron per Å2 per second, and the total exposure time was 80 s. The total dose was divided into 180 frames per movie, so each frame was exposed for 0.44 s. However, the two final datasets that resulted in the two reported 3D maps were collected in the TFS Titan Krios. The Titan Krios electron microscope operated at a high tension of 300 kV. Micrographs were collected automatically with SerialEM at a nominal magnification of 130,000× and a pixel size of 1.029 Å per pixel with defocus values from −1.0 to −2.0 μm. Gatan K2 summit detector was used for image recording under counting mode. The dose rate was 10 electrons per Å2 per second, and the total exposure time was 6 s. The total dose was divided into 30 frames per movie so each frame was exposed for 0.2 s.
Image processing.
For the sample of Pmt1–Pmt2 only, about 2,787 raw movie micrographs were collected and motion-corrected using the program MotionCorr 2.05. Contrast transfer function parameters of each aligned micrograph were calculated using CTFFIND 4.16. All the remaining steps were performed using RELION 37. Templates for automatic picking were generated from a 2D average of about 1,000 manually picked particles. A total of 913,730 particles were picked automatically. 2D classification was then performed and particles in the classes with features unrecognizable by visual inspection were removed. A total of 869,556 particles was used for further 3D classification, and 581,966 particles were selected for further 3D refinement and postprocessing, resulting in the 3.2-Å 3D density map. The resolution of the map was estimated by the gold-standard Fourier shell correlation at a correlation cutoff value of 0.143.
For cryo-EM structural analysis of Pmt1–Pmt2 in complex with the synthetic tetrapeptide, about 2,988 raw movie micrographs were collected and motion-corrected using the program MotionCorr 2.05. Contrast transfer function parameters of each aligned micrograph were calculated using CTFFIND 4.16. All the remaining steps were performed using RELION 37. Templates for automatic picking were generated from a 2D average of about 1,000 manually picked particles. A total of 953,292 particles were picked automatically. 2D classification was then performed and particles in the classes with features unrecognizable by visual inspection were removed. A total of 949,765 particles was used for further 3D classification, and 521,848 particles were selected for further 3D refinement and postprocessing, resulting in the 3.2-Å 3D density map. The resolution of the map was estimated by the gold-standard Fourier shell correlation at a correlation cutoff value of 0.143.
Structural modeling, refinement, and validation.
The initial models of Pmt1 and Pmt2 were first automatically built into the 3.2-Å EM map using the model_to_map in the PHENIX program2. Models in the remaining density was then manually built in the program COOT3. Sequence assignment was guided by bulky residues such as Phe, Tyr, Trp, and Arg. In the 3D map of Pmt1–Pmt2–peptide, only the first three residues (PYT) of the acceptor tetrapeptide (PYTV) were modeled, because the map lacked solid density for the last valine residue. The complete Pmt1–Pmt2 model was refined by real-space refinement in the PHENIX program and subsequently adjusted manually in COOT. Finally, the atomic model was validated using MolProbity8. We then correlated the refined model with the 3D maps of the final map and the two half-maps (Half1 and Half2) to produce three FSC curves: model vs. final map, FSCwork (model vs. Half1 map) and FSCfree (model vs. Half2 map), the general agreement of which was taken as an indication that the model was not over-fitted. Structural figures were prepared in Chimera9 and PyMOL (https://pymol.org/2/).
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Li for advice on constructing yeast strains and D. Nadziejka for critical reading of the manuscript. Cryo-EM data were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite at Van Andel Research Institute. We thank G. Zhao and X. Meng for assistance in data collection. This work was supported by the U.S. National Institutes of Health grant R01 CA231466 (to H.L.). X-ray diffraction data was collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline at the Advanced Photon Source, which was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Footnotes
Competing Interests
The authors declare no competing financial interests.
REPORTING SUMMARY
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
DATA AVAILABILITY
The cryo-EM 3D maps of the S. cerevisiae Pmt1–Pmt2 alone or complexed with the acceptor peptide have been deposited at the EMDB database with accession codes EMD-20240 and EMD-20236, respectively. Their corresponding atomic models were deposited at the RCSB PDB with accession codes 6P2R and 6P25, respectively. The crystal structure of the Pmt2 MIR domain was deposited at the RCSB PDB with accession code 6P28.
REFERENCES
- 1.Panin VM & Wells L Protein O-mannosylation in metazoan organisms. Curr Protoc Protein Sci 75, 12–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VanderVen BC, Harder JD, Crick DC & Belisle JT Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science 309, 941–3 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Xu C, Wang S, Thibault G & Ng DT Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science 340, 978–81 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Loibl M & Strahl S Protein O-mannosylation: what we have learned from baker’s yeast. Biochim Biophys Acta 1833, 2438–46 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Burda P & Aebi M The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta 1426, 239–57 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Maeda Y & Kinoshita T Dolichol-phosphate mannose synthase: structure, function and regulation. Biochim Biophys Acta 1780, 861–8 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Hirschberg CB & Snider MD Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem 56, 63–87 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Schwarz F & Aebi M Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol 21, 576–82 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Bai L & Li H Cryo-EM is uncovering the mechanism of eukaryotic protein N-glycosylation. FEBS J (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gloster TM Advances in understanding glycosyltransferases from a structural perspective. Curr Opin Struct Biol 28, 131–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebi M N-linked protein glycosylation in the ER. Biochim Biophys Acta 1833, 2430–7 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Loibl M et al. Protein O-mannosyltransferases associate with the translocon to modify translocating polypeptide chains. J Biol Chem 289, 8599–611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neubert P & Strahl S Protein O-mannosylation in the early secretory pathway. Curr Opin Cell Biol 41, 100–8 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Ecker M et al. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep 4, 628–32 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lommel M, Schott A, Jank T, Hofmann V & Strahl S A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J Biol Chem 286, 39768–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard J The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biol Direct 11, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobson CM, Hempel SJ, Stalnaker SH, Stuart R & Wells L O-Mannosylation and human disease. Cell Mol Life Sci 70, 2849–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Immervoll T, Gentzsch M & Tanner W PMT3 and PMT4, two new members of the protein-O-mannosyltransferase gene family of Saccharomyces cerevisiae. Yeast 11, 1345–51 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Gentzsch M & Tanner W The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J 15, 5752–9 (1996). [PMC free article] [PubMed] [Google Scholar]
- 20.Lehle L & Tanner W Glycosyl transfer from dolichyl phosphate sugars to endogenous and exogenous glycoprotein acceptors in yeast. Eur J Biochem 83, 563–70 (1978). [DOI] [PubMed] [Google Scholar]
- 21.Strahl-Bolsinger S & Tanner W Protein O-glycosylation in Saccharomyces cerevisiae. Purification and characterization of the dolichyl-phosphate-D-mannose-protein O-D-mannosyltransferase. Eur J Biochem 196, 185–90 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Ponting CP Novel repeats in ryanodine and IP3 receptors and protein O-mannosyltransferases. Trends Biochem Sci 25, 48–50 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Girrbach V, Zeller T, Priesmeier M & Strahl-Bolsinger S Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J Biol Chem 275, 19288–96 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Manya H, Akasaka-Manya K, Nakajima A, Kawakita M & Endo T Role of N-glycans in maintaining the activity of protein O-mannosyltransferases POMT1 and POMT2. J Biochem 147, 337–44 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Jung P & Tanner W Identification of the lipid intermediate in yeast mannan biosynthesis. Eur J Biochem 37, 1–6 (1973). [DOI] [PubMed] [Google Scholar]
- 26.Gandini R, Reichenbach T, Tan TC & Divne C Structural basis for dolichylphosphate mannose biosynthesis. Nat Commun 8, 120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L & Laakso LM Dali server update. Nucleic Acids Res 44, W351–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Napiorkowska M et al. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat Struct Mol Biol 24, 1100–1106 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto S et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci U S A 110, 17868–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai L, Wang T, Zhao G, Kovach A & Li H The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 555, 328–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild R et al. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 359, 545–550 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Lizak C, Gerber S, Numao S, Aebi M & Locher KP X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–5 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Braunger K et al. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 360, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unligil UM & Rini JM Glycosyltransferase structure and mechanism. Curr Opin Struct Biol 10, 510–7 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Lairson LL, Henrissat B, Davies GJ & Withers SG Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77, 521–55 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Breton C, Fournel-Gigleux S & Palcic MM Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol 22, 540–9 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Sinnott ML Catalytic mechanism of enzymic glycosyl transfer. Chemical Reviews 90, 1171–1202 (1990). [Google Scholar]
- 38.Zhang L & Ten Hagen KG O-Linked glycosylation in Drosophila melanogaster. Curr Opin Struct Biol 56, 139–145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Y Membrane protein structural biology in the era of single particle cryo-EM. Curr Opin Struct Biol 52, 58–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lommel M & Strahl S Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19, 816–28 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Yoshida-Moriguchi T et al. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science 327, 88–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H et al. Analysis of phenotype, enzyme activity and genotype of Chinese patients with POMT1 mutation. J Hum Genet 61, 753–9 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Chong YK et al. Dystroglycanopathy with two novel POMT1 mutations in a Chinese boy with developmental delay and muscular dystrophy. Eur J Paediatr Neurol 18, 532–5 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Bello L et al. Cardiomyopathy in patients with POMT1-related congenital and limb-girdle muscular dystrophy. Eur J Hum Genet 20, 1234–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Reeuwijk J et al. The expanding phenotype of POMT1 mutations: from Walker-Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum Mutat 27, 453–9 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Johnson K et al. Detection of variants in dystroglycanopathy-associated genes through the application of targeted whole-exome sequencing analysis to a large cohort of patients with unexplained limb-girdle muscle weakness. Skelet Muscle 8, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Bernabe Beltran-Valero D. et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet 71, 1033–43 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu P, Yuan L & Deng H Molecular genetics of the POMT1-related muscular dystrophy-dystroglycanopathies. Mutat Res 778, 45–50 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Balci B et al. An autosomal recessive limb girdle muscular dystrophy (LGMD2) with mild mental retardation is allelic to Walker-Warburg syndrome (WWS) caused by a mutation in the POMT1 gene. Neuromuscul Disord 15, 271–5 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Snoei J et al. Genetic alterations of protein-o-mannosyltransferase-1 in glioneuronal and glial brain tumors with subarachnoid spread. Neuropathology 29, 116–24 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Currier SC et al. Mutations in POMT1 are found in a minority of patients with Walker-Warburg syndrome. Am J Med Genet A 133A, 53–7 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Cotarelo RP et al. A double homozygous mutation in the POMT1 gene involving exon skipping gives rise to Walker-Warburg syndrome in two Spanish Gypsy families. Clin Genet 76, 108–12 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Mercuri E et al. Congenital muscular dystrophies with defective glycosylation of dystroglycan: a population study. Neurology 72, 1802–9 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Messina S et al. POMT1 and POMT2 mutations in CMD patients: a multicentric Italian study. Neuromuscul Disord 18, 565–71 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Chen C, Mei S, Zhu C, Ren Y & Kong X [Analysis of POMT1 gene mutation in a pedigree affected with congenital muscular dystrophy]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 35, 78–80 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Manya H et al. Protein O-mannosyltransferase activities in lymphoblasts from patients with alpha-dystroglycanopathies. Neuromuscul Disord 18, 45–51 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Hafner P et al. Skeletal muscle MRI of the lower limbs in congenital muscular dystrophy patients with novel POMT1 and POMT2 mutations. Neuromuscul Disord 24, 321–4 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Yanagisawa A et al. POMT2 intragenic deletions and splicing abnormalities causing congenital muscular dystrophy with mental retardation. Eur J Med Genet 52, 201–6 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Biancheri R et al. POMT2 gene mutation in limb-girdle muscular dystrophy with inflammatory changes. Biochem Biophys Res Commun 363, 1033–7 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Murakami T et al. A novel POMT2 mutation causes mild congenital muscular dystrophy with normal brain MRI. Brain Dev 31, 465–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdullah S et al. Noncompaction cardiomyopathy in an infant with Walker-Warburg syndrome. Am J Med Genet A 173, 3082–3086 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Brun BN et al. Uniparental disomy unveils a novel recessive mutation in POMT2. Neuromuscul Disord 28, 592–596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nabhan MM et al. Cystic kidneys in fetal Walker-Warburg syndrome with POMT2 mutation: Intrafamilial phenotypic variability in four siblings and review of literature. Am J Med Genet A 173, 2697–2702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagisawa A et al. New POMT2 mutations causing congenital muscular dystrophy: identification of a founder mutation. Neurology 69, 1254–60 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Martinez HR et al. Novel cardiovascular findings in association with a POMT2 mutation: three siblings with alpha-dystroglycanopathy. Eur J Hum Genet 22, 486–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grahn E et al. Crystal structure of the Marasmius oreades mushroom lectin in complex with a xenotransplantation epitope. J Mol Biol 369, 710–21 (2007). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 1.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murshudov GN, Vagin AA & Dodson EJ Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–255, doi: 10.1107/S0907444996012255 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mindell JA & Grigorieff N Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.