Abstract
BACKGROUND:
Colorectal Adenocarcinoma (ADCCR) is the third most cancer not only in the world but also in Indonesia. There were 623 cases of ADCCR at Dr Hasan Sadikin hospital within 2015-2017. Both KRAS and TP53 mutation are known as genes which involve in carcinogenesis through the same pathway, namely the chromosomal instability pathway. In West Java, researches focusing on mutation KRAS and p53 also a correlation between both biomarkers among ADCCR patients are still limited.
AIM:
Therefore, this research aimed to perceive a correlation between KRAS gene expression with p53 immunoexpression in ADCCR.
METHODS:
Cross section research design was performed to 62 cases of ADCCR as paraffin block taken from 4 hospitals in West Java, including Dr Hasan Sadikin hospital Bandung, Santosa hospital Bandung, Borromeus hospital Bandung and Syamsudin hospital Sukabumi from January 1st 2014 to 31s November 2018. KRAS mutation gene data taken from secondary data at molecular laboratory in Ciptomangunkusumo Hospital Jakarta and Dr Sardjito Hospital Jogjakarta, while the detection of p53 immunoexpression data using immunohistochemical staining was carried out in the Laboratorium of Anatomical Pathology of Padjadjaran University (Dr Hasan Sadikin Hospital). All data were analysed using Chi-Square test with p-value < 0,05 of significant level then proceeded with Stata ver.11 for windows.
RESULTS:
The results of this study showed that KRAS gene expressions from 62 sample consist of 39 wild type KRAS (62.39%) and 23 mutant KRAS (37.1%). The p53 immunoexpression consists of 27 negative cases (non-mutant p53) and 35 mutant p53, which includes 10 cases as focal expression (16.33%) and 25 cases as diffuse expressions (40.33%). There is a significant association between KRAS gene expression and p53 immunoexpressions in ADCCR (p = 0.04), with mild positive correlation (Rho 0.28).
CONCLUSION:
This study concluded that KRAS and p53 mutations are involved in carcinogenesis, and the p53 mutation is a more dominant risk factor than KRAS mutation among West Java people. P53 mutations with diffuse pattern tend to express mutant KRAS while p53 negative and having a focal pattern tend to express wt KRAS.
Keywords: KRAS, p53, Adenocarcinoma Colorectal (ADCCR)
Introduction
Colorectal carcinoma (CRC) is a malignant epithelial tumour originating in the large bowel. Colorectal carcinoma ranks as the third most frequent cancer not only in the world but also in Indonesia [1], [2]. The worldwide mortality rate is about 608.000 deaths [2]. In Indonesia, the mortality rate is about 9.5% of all cancer deaths [3], [4]. According to data from the Department of Anatomy Pathology Dr Hasan Sadikin Hospital Bandung, the frequency of CRC is about 224 cases in 2015, 187 cases in 2016 and 212 cases in 2017.
Colorectal Adenocarcinoma (ADCCR) is the most frequent type 0f CRC in the world [1]. There are many mutation genes occur in ADCCR, such as Adenomatous Polyposis Coli (APC), TP53, Kirsten rat sarcoma virus (KRAS), PIK3CA, etc. [5]. These genes are involved in carcinogenesis through three major pathways such as chromosomal instability, mismatch repair and CpG island methylator phenotype (CIMP). The most frequent pathway is chromosomal instability, about 65% [6], [7]. Three genes are involved in this pathway; they are: 1) APC gene mutation as an initiator in the early phase of carcinogenesis. 2) Kirsten rat sarcoma virus gene mutation, which occurs in 50% of ADCCR from intermediate phase to advance phase. 3) TP53 gene mutation in the late phase of carcinogenesis is about 40-60% (Figure 1) [6].
Figure 1.
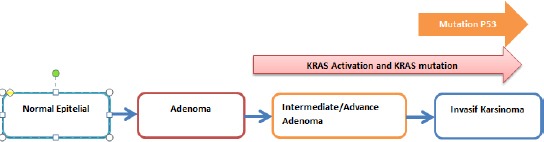
KRAS mutation can occur together with p53 mutation in the late phase of carcinogenesis. Both of them involved in the same pathway of carcinogenesis (instability chromosome pathway)
KRAS gene expression consists of wild type (wt) KRAS and mutant KRAS. Wild type KRAS gene is located in short arm of chromosome number 12, in 12 p12.1. This product is wt KRAS protein, which can flip back and forth between an excited signal transmitting state and quiescent state. RAS is inactive when bound to GDP, but stimulation of cells by growth factors such as EGF and PDGF leads to an exchange of GDP for GTP and subsequent conformational changes that generate active RAS and stimulate downstream regulators of cells proliferation [6], [8].
This excited signal emitting state, in short, lived, however, because the intrinsic guanosine triphosphatase (GTPase) activity of wt RAS hydrolyses GTP to GDP return the active wt KRAS to its inactive state [8]. In ADCCR wt KRAS will always be active because EGF stimulates continuously. ADCCR patients with wt KRAS have a good response to anti-epidermal Growth Factor Receptor (EGFR) drugs such as cetuximab and panitumumab [1], [9], [10], [11]. In contrast, mutant KRAS was not responsive to anti-EGFR inhibition [1], [3], [12]. Mutant KRAS can be active continuously although having limited EGFR stimulation. This gene has GTP-ase activity lower than wt KRAS, resulting in a decrease of hydrolysing activity 3-9 times lower than wt KRAS. This situation makes the irreversible signal to downstream regulation to provide uncontrolled proliferation and differentiation cells [8].
Mutant KRAS gene occurs in exon 2, 3 and 4. Exon 2 is the frequent site of mutation [12] — mutant KRAS gene as a predictive negative response from anti-EGFR therapy. Patient with mutant KRAS has a worse prognosis than those with wt KRAS. Product of mutant KRAS gene is mutant KRAS protein [1], [9], [10], [12].
In ADCCR another abnormality is Tumor Protein (TP) 53 pathway consists of loss of function wt TP53 and the increase of TP53 mutant concentration. Wild type TP53 is normally in the latent phase and will be stabilised and activated by genotoxic agent and another stress signal. Regulatory of TP53 is Murine Double minute 2 (Mdm2), a protein which can bind TP53 in unstable or inactivated conditions. TP53 gene can be activated by the detachment of TP53 from Mdm2. This condition makes this gen stop the cell cycle and repairing DNA breakdown and involving in apoptosis and ageing [6].
In the state of activated p53 due to release from Mdm2, p53 will have the ability to trigger cell cycle discontinuation, repair DNA damage and be involved in the process of apoptosis and cell ageing [6]. One of the p53 gene mutations is caused by the loss of alleles on the chromosome on the short arm of chromosome 17, resulting in genomic disorders that occur in the later phase of tumour progression [8]. Most TP53 mutations occur in exons 5 to 8, which encode residues 130-286, which are important and responsible for folding and stabilising tertiary structures of proteins [8]. Mutations in the P53 gene can result in loss of wild type TP53 ability for DNA repair, apoptosis, checkpoints, and cell ageing processes [8], [9], [10]. Also, the TP53 mutation produces a mutant P53 gene which acts as an oncogene, thereby increasing the aggressiveness, survival and metastasis ability of the tumour cell [13], [14]. The product of the P53 gene mutation is a mutant p53 protein that can be examined by several methods, including the Immunohistochemical (CPI) method [8], [9], [10].
The expression of the KRAS and TP53 genes are mostly associated with pathological features, lymph node metastasis, distant metastasis and ADCCR therapy response [10, [15], [16], [17]. The role of these two molecular markers is widely used as a predictor of prognosis of ADCCR therapy. The Assesment of the anti p53 antibody with immunohistochemistry methods is lower in cost but more effective and efficient. Thus it can be considered as one alternative to predict KRAS gen expressions in ADCCR patients. However, studies involving the relationship of KRAS mutations as one of the stress signals in tumour cells with mutant Immunoexpression p53 are still not widely performed and documented [6], [10], [13], [14], [15], [16], [17].
The objective of this study is to understand the relationship and correlation between KRAS gene expression and p53 immunoexpression in ADCCR.
Methods
Samples were obtained from tissue biopsy and or colectomy of surgery patients registered at Dr Hasan Sadikin Hospital Bandung, Santosa hospital Bandung, Borromeus hospital Bandung and Syamsudin hospital Sukabumi. Histopathologically diagnosed with ADCCR from January 1st, 2014 until November 31st, 2018. Samples were attained after approved by the Ethical Committee with assessment number 105/UN6.KEP/EC/2019, and then drawn according to inclusion criteria.
Sixty-two samples of KRAS gene expression had been checked in the molecular laboratory, Ciptomangunkusumo Hospital Jakarta and Sardjito Hospital Jogjakarta. To see if mutation in exon 2 occur, the KRAS mutation using PCR-HRM (High-Resolution Melting-Polymerase Chain Reaction) methods was combined with RFLP (Restriction fragment length polymorphism) and to see if mutation in exon 3 and 4 occur, the KRAS mutation using PCR-HRM (High-Resolution Melting-Polymerase Chain Reaction) methods was combined with direct DNA sequencing. KRAS mutation is positive if there is ‘split peaks’ in the melting curve that shows two populations (wild type and mutant allele) and occurs in at least 1 out of 3 exons. KRAS mutation is negative (wild type) if there are no ‘split peaks. These gene expressions data were recorded and collected as secondary data in this research
P53 immunoexpression had been checked by immunohistochemistry staining procedure at the Laboratorium of Anatomical Pathology of Padjadjaran University (Dr. Hasan Sadikin Hospital. Immunohistochemistry staining using mouse monoclonal antibody p53 (Santa Cruz, Pab-1801, SC-98) with dilution 1:150 was used in standard immunohistochemistry (IHC) staining procedure. Immunoexpression of p53 was categorised by calculating the distribution of the cells that showed immunoreactivity, which was the nucleus-stained cells. Distribution score was explained as 0 = negative; Focal = 1-50%; Diffuse = ≥ 50%. Intensity compares with breast cancer cells as an internal positive control. IHC staining result was examined by two experts in the IHC technique using light microscope Olympus CX31.
Statistical analysis
The data obtained from this research was analysed using Chi-Square test and Spearman Rho correlation. A significant association was interpreted from p-value where p ≤ 0.05 showed a statistically significant association, while p ≥ 0.05 showed otherwise. If the Spearman rho was not equal to 0, the significant correlation showed. If Spearman rho = 0, it showed otherwise. The data attained from laboratory procedure was recorded in a distinct form, and Stata ver 11 for Windows was used to be analysed statistically.
Results
The characteristics of the research subjects are based on age, gender and the degree of ADCCR differentiation as mention in table 1. Age < 40 years as many as 8 cases (12.90%), age 40-60 years as many as 35 cases (56.45%) and age > 60 years as many as 19 cases (30.65%). The youngest is 26 years old, and the oldest is 81 years old, and the middle value is 54 years. The males are 33 people (53.23%), and females are 29 people (46.77%). Histological degree in ADCCR consists of 42 well-differentiated cases (67.74%), 10 cases for each moderately and poorly differentiated (16.13%).
Table 1.
Research patient’s characteristic
| Characteristics | ADCCR (n = 62) |
|---|---|
| Age (years) | |
| Mean ± Std | 54 ± 12 |
| Median (min, maks) | 54 (26.81) |
| ≤40years | 8 (12.9%) |
| 40-60 years | 35 (56.45%) |
| ≥60years | 19 (30.65%) |
| Sex | |
| Male | 33 (53.23%) |
| Female | 29 (46.77%) |
| Histological degree | |
| Well | 42(67.74%) |
| Moderately | 10 (16.13%) |
| Poorly | 10 (16.13%) |
Based on secondary data from PCR examination, KRAS gene expression includes wt KRAS (negative mutation) and KRAS mutants. P53 immunoexpression in this study was assessed from the brown stained of the tumour cell nucleus. The result of p53 immunohistochemical staining was assessed based on tumour cell distribution consisting of negative, focal and diffuse. The results of the data and calculations are shown in Table 2.
Table 2.
KRAS Gene Expression and p53 Immunoexpression
| Characteristics | ADCCR (n = 62) |
|---|---|
| KRAS gene expression | |
| Wt KRAS (negative mutation) | 39 (62.39%) |
| Mutant KRAS | 23 (37.1%) |
| P53 immunoexpression | |
| Negative | 27 (43.55%) |
| Focal | 10 (16.13%) |
| Diffuse | 25 (40.33%) |
Table 2 shows the descriptive data distribution regarding the frequency of the KRAS gene expression and p53 immunoexpression. It is shown that the most visible KRAS gene expression is wt KRAS as many as 39 cases (62.39%), KRAS mutations occur in only 23 cases (37.1%). The highest p53 Immunoexpression is in the negative distribution of 27 cases (43.55%), diffuse distribution of 25 cases (40.32%), and the focal distribution of 10 cases (16.13%).
In this study, the statistical tests were using Chi-square to analyse the relationship between expression of the KRAS gene and immunoexpression of TP53 in ADCCR and using Spearman Rho to analyse the correlation between expression of the KRAS gene and immunoexpression of TP53 in ADCCR. The result is a significant relationship with a value of p = 0.04 (p < 0.05)), with positive mild correlation (Rho 0.28) as shown in Table 3.
Table 3.
Correlation of KRAS Gene Expression with p53 Immunoexpression
| KRAS | P53 Immunoexpression | P value (Chi Square) | Spearman Rho (Correlation) | P(Spearman) | ||
|---|---|---|---|---|---|---|
| Negative | Focal | Diffuse | ||||
| Wt | 20 (74.07%) | 8 (80%) | 11 (44%) | 0.04 | 0.28 | 0.03 |
| Mutant | 7 (25.93%) | 2 (20%) | 14 (56%) | |||
Immunoexpression of p53 was categorised by calculating the distribution of the cells that showed immunoreactivity, which was the nucleus-stained cells. Distribution score was explained as 0 = negative; Focal = 1-50%; Diffuse = ≥ 50%. The figure below showed the distribution of p53 in tumour cells more than 50% categorised as Diffuse distribution.
Figure 2.
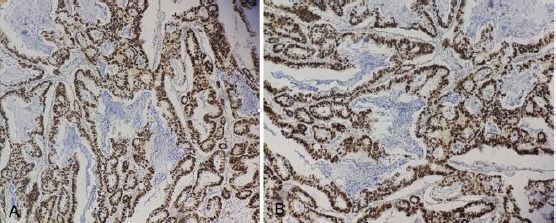
A) Immunoexpression p53 diffuse distribution (magnification 40 x); B) Immunoexpression p53 diffuse distribution (magnification 100 x)
Discussion
The collection of KRAS gene expression examination data in all ADCCR cases was carried out. The result is that the KRAS mutation only occurs in 23 cases (37.1%). This figure is lower than the data obtained from the literature (50%), but it is higher than the research conducted by Sameer et al. in Kashmir ethnicity which was 22.6% [4], [6]. The results of the KRAS secondary data collection showed that 62.9% of ADCCR in this study has a wild type gene expression profile. In ADCCR through the wt KRAS pathway, continuous KRAS activation will occur because it is stimulated continuously by growth factors, one of which is EGF which will bind to EGFR to subsequently induce various proteins that help KRAS phosphorylation process. Phosphorylated KRAS is KRAS, which is bound to GTP, which will give hyperproliferative signals to cancer cells [1], [6], [18].
ADCCR patients with wt KRAS status have a good response to anti-EGFR therapy [8], [18]. This study shows that 62.9% of ADCCR cases in this study had a good response to anti-EGFR, such as cetuximab and panitumumab [1], [3], [11], [18].
Patients with wt KRAS will have a better prognosis than patients with mutant KRAS [1], [8], [18]. The ability of tumour cells to proliferate, differentiate, invade, and metastasis is not as bad as in ADCCR patients with mutant KRAS status. This is by the most differentiation degrees in this study, namely low degrees with better prognosis (67.74%). KRAS mutations only occurred in 23 cases (37.1%). These results indicate that it is necessary to consider other alternative carcinogenesis pathways in ADCCR in West Java in addition to the CIN pathway.
In ADCCR, the TP53 pathway can experience abnormalities, including loss of function wt TP53 and increased mutant TP53 levels [10]. The products of the two genes are wt p53 and p53 mutant proteins. In this study, the measured protein is the p53 mutant using the IHK method, and the results of the assessment are thus referred to as p53 immunoexpression. The results of this study showed that the highest p53 immunoexpression was 27 cases of negative distribution (43.55%), 25 cases of diffuse distribution (40.32%) and 10 cases of focal distribution (16.13%). These results showed mutant p53 occurred in 35 cases (56, 45%). These results are in line with the literature, which states that p53 mutations can occur in as many as 40-60% of ADCCR cases [10]. It was higher than the incidence of ADCCR which was identified as having a TP53 gene mutation in Saudi Arabia of 33.7% but data from WHO showed a higher mutation of 80% [1], [16].
In this study, the statistical tests were using Chi-square to analyse the relationship between expression of the KRAS gene and immunoexpression of TP53 in ADCCR and using Spearman Rho to analyse the correlation between expression of the KRAS gene and immunoexpression of TP53 in ADCCR. The result is a significant relationship with a value of p = 0.04 (p < 0.05)), with positive mild correlation (Rho 0.28). This may be due to the activation of the wt KRAS by excessive growth factors signal that will cause cell stress due to hyperproliferative signals caused by stimulation of proteins involved in the KRAS pathway which activates the cell nucleus for the process of transcription and proliferation [13], [14]. This causes various activation of E2F protein, a protein involved in triggering the role of tumour suppressor genes such as TP53, RB, and P21 [13], [14].
The signalling pathway that connects the KRAS mutation with wildtype TP53 has been widely documented, one of which is research on mice that have become experimental animal models for lung cancer. The results of the study stated that the KRAS mutations that occur could lead to excessive cell proliferation. This Hyperproliferative signal is considered a stress signal which triggers the release of wt TP53 from Murine Double Minute 2 (Mdm2). This results in wt TP53 being able to stop the cell cycle and trigger the apoptosis program and protect cells from abnormal growth factors [13].
One study reported that the correlation of KRAS mutations in ADCCR patients had a low prevalence of TP53 mutations. This result contrary to this study, which wt KRAS tend to show high frequent of negative (20/27; 74.07%) and focal (8/10; 80%) TP53 mutant. This is possible because the mutations occurred in ADCCR in the study were indeed low so that the wt KRAS predominant as hyperproliferative stress compare than mutant KRAS and will trigger the wt TP53 pathway, especially for well-differentiated and less aggressive tumours, characterised by slow growth and low metastatic ability. This was caused by the fact that TP53 still functions as a tumour suppressor and have a little oncogenic ability [13], [14], [20]. In this study, the proportion of KRAS mutations was low in the wt KRAS, whereas positive p53 immunoexpression (mutant p53) was higher than the negative (non-mutant p53). This is possible because the KRAS mutation in ADCCR in this study had little role in carcinogenesis in the CIN pathway compared to TP53 mutations. This result shows that ADCCR among west java people mostly occurred sporadically.
In conclusion, a mild correlation occurs in the relationship between the expression of the KRAS gene and p53 immunoexpression in ADCCR In the case of ADCCR with diffuse p53 immunoexpression, it expresses or predicts the expression of mutant KRAS genes. On the contrary, in the case of ADCCR with negative and focal p53 immunoexpression, it tends to express or predict the expression of the wt KRAS gene.
Acknowledgement
This research was funded by an internal grant for Padjadjaran University batch 2 no: 4851/UN6.C/LT/2018. We thanked Mr Wahyudin, Mrs Nur, Mr Herman, for excellent technical assistance. We also thanked Mrs Arief Budi Yulianti and Mr Fajar Awaliya and for excellent manuscript writing and statistician assistance.
Footnotes
Funding: This research was funded by an internal grant for Padjadjaran University, Indonesia batch 2 no: 4851/UN6.C/LT/2018
Competing Interests: The authors have declared that no competing interests exist
References
- 1.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. World Health Organization; 2010. pp. 131–181. [Google Scholar]
- 2.Mahasneh A, Al-Shaheri F, Jamal E. Molecular biomarkers for early diagnosis, effective treatment and prognosis of colorectal cancer:Current updates. Experimental and molecular pathology. 2017;102(3):475–83. doi: 10.1016/j.yexmp.2017.05.005. https://doi.org/10.1016/j.yexmp.2017.05.005 PMid:28506769. [DOI] [PubMed] [Google Scholar]
- 3.Kementrian Kesehatan RI. Panduan Penatalaksanaan Kanker kolorektal. 2015:1–67. [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. https://doi.org/10.3322/caac.21492 PMid:30207593. [DOI] [PubMed] [Google Scholar]
- 5.Testa U, Pelosi E, Castelli G. Colorectal cancer:genetic abnormalities, tumour progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Medical Sciences. 2018;6(2):31. doi: 10.3390/medsci6020031. https://doi.org/10.3390/medsci6020031 PMid:29652830 PMCid:PMC6024750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar V, Abbas AK, Aster JC. 10. Elsevier Health Sciences; 2017. Robbins basic pathology; pp. 261–71. [Google Scholar]
- 7.Abdullah M, Sudoyo AW, Utomo AR, Fauzi A, Rani AA. Molecular profile of colorectal cancer in Indonesia:is there another pathway? Gastroenterology and Hepatology from bed to bench. 2012;5(2):71–5. [PMC free article] [PubMed] [Google Scholar]
- 8.Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World journal of gastroenterology: WJG. 2012;18(37):5171. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui X, Zhu J, Tang H, Wang C, Zhou J, Han W, Wang X, Fang Y, Xu Y, Li D, Chen R. p53 controls colorectal cancer cell invasion by inhibiting the NF-κB-mediated activation of Fascin. Oncotarget. 2015;6(26):22869. doi: 10.18632/oncotarget.5137. https://doi.org/10.18632/oncotarget.5137 PMid:26362504 PMCid:PMC4673205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaar I, Amara S, Elamine OE, Khiari M, Ounissi D, Khalfallah T, Mzabi S, Bouraoui S. Biological significance of promoter hypermethylation of p14/ARF gene:relationships to p53 mutational status in Tunisian population with colorectal carcinoma. Tumor Biology. 2014;35(2):1439–49. doi: 10.1007/s13277-013-1198-9. https://doi.org/10.1007/s13277-013-1198-9 PMid:24065196 PMCid:PMC3932170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levi M, Prayogi G, Sastranagara F, Sudianto E, Widjajahakim G, Gani W, Mahanadi A, Agnes J, Khairunisa BH, Utomo AR. Clinicopathological associations of K-RAS and N-RAS mutations in Indonesian colorectal cancer cohort. Journal of gastrointestinal cancer. 2018;49(2):124–31. doi: 10.1007/s12029-016-9901-x. https://doi.org/10.1007/s12029-016-9901-x PMid:28044264. [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Kohne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer:updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–9. doi: 10.1200/JCO.2010.33.5091. https://doi.org/10.1200/JCO.2010.33.5091 PMid:21502544. [DOI] [PubMed] [Google Scholar]
- 13.Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, Van Pelt C, Terzian T, Lozano G. Multiple stress signals activate mutant p53 in vivo. Cancer research. 2011;71(23):7168–75. doi: 10.1158/0008-5472.CAN-11-0459. https://doi.org/10.1158/0008-5472.CAN-11-0459 PMid:21983037 PMCid:PMC3320147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. The Journal of pathology. 2010;222(2):129–37. doi: 10.1002/path.2748. https://doi.org/10.1002/path.2748 PMid:20662002. [DOI] [PubMed] [Google Scholar]
- 15.Ross JS. Clinical implementation of KRAS testing in metastatic colorectal carcinoma:the pathologist's perspective. Archives of pathology &laboratory medicine. 2012;136(10):1298–307. doi: 10.5858/arpa.2011-0478-RA. https://doi.org/10.5858/arpa.2011-0478-RA PMid:22272560. [DOI] [PubMed] [Google Scholar]
- 16.Al-Kuraya KS. KRAS and TP53 mutations in colorectal carcinoma. Saudi journal of gastroenterology. 2009;15(4):21–7. doi: 10.4103/1319-3767.56087. https://doi.org/10.4103/1319-3767.56087 PMid:19794264 PMCid:PMC2981835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi M. Colorectal carcinoma:a general overview and future perspectives in colorectal cancer. International journal of molecular sciences. 2017;18(1):197. doi: 10.3390/ijms18010197. https://doi.org/10.3390/ijms18010197 PMid:28106826 PMCid:PMC5297828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, Garcia-Aguilar J, Kim J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. International journal of molecular sciences. 2012;13(10):12153–68. doi: 10.3390/ijms131012153. https://doi.org/10.3390/ijms131012153 PMid:23202889 PMCid:PMC3497263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyiraneza C, Jouret-Mourin A, Kartheuser A, Camby P, Plomteux O, Detry R, Dahan K, Sempoux C. Distinctive patterns of p53 protein expression and microsatellite instability in human colorectal cancer. Human pathology. 2011;42(12):1897–910. doi: 10.1016/j.humpath.2010.06.021. https://doi.org/10.1016/j.humpath.2010.06.021 PMid:21665242. [DOI] [PubMed] [Google Scholar]
- 20.Knickelbein K, Zhang L. Mutant KRAS as a critical determinant of the therapeutic response of colorectal cancer. Genes & diseases. 2015;2(1):4–12. doi: 10.1016/j.gendis.2014.10.002. https://doi.org/10.1016/j.gendis.2014.10.002 PMid:25815366 PMCid:PMC4372129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duldulao MP, Lee W, Nelson RA, Li W, Chen Z, Kim J, et al. Mutations in specific codons of the KRAS oncogene are associated with variable resistance to neoadjuvant chemoradiation therapy in patients with rectal adenocarcinoma. Annals of surgical oncology. 2013;20(7):2166–71. doi: 10.1245/s10434-013-2910-0. https://doi.org/10.1245/s10434-013-2910-0 PMid:23456389 PMCid:PMC5584556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller PAJ, Vousden KH. Mutant p53 in cancer:new functions and therapeutic opportunities. Cancer cell. 2014;25(3):304–17. doi: 10.1016/j.ccr.2014.01.021. https://doi.org/10.1016/j.ccr.2014.01.021 PMid:24651012 PMCid:PMC3970583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Molecular and cellular biology. 2007;27(23):8284–95. doi: 10.1128/MCB.00050-07. https://doi.org/10.1128/MCB.00050-07 PMid:17908790 PMCid:PMC2169174. [DOI] [PMC free article] [PubMed] [Google Scholar]


