SUMMARY
Many stem cells undergo asymmetric division to produce a self-renewing stem cell and a differentiating daughter cell. Here we show that, similarly to H3, histone H4 is inherited asymmetrically in Drosophila melanogaster male germline stem cells undergoing asymmetric division. In contrast, both H2A and H2B are inherited symmetrically. By combining superresolution microscopy and chromatin fiber analyses with proximity ligation assays on intact nuclei, we find that old H3 is preferentially incorporated by the leading strand whereas newly synthesized H3 is enriched on the lagging strand. Using a sequential nucleoside analog incorporation assay, we detect a high incidence of unidirectional replication fork movement in testes-derived chromatin and DNA fibers. Biased fork movement coupled with a strand preference in histone incorporation would explain how asymmetric old and new H3 and H4 are established during replication. These results suggest a role for DNA replication in patterning epigenetic information in asymmetrically dividing cells in multicellular organisms.
INTRODUCTION
Epigenetic mechanisms play important roles in cell fate specification by altering chromatin structure and gene expression patterns while preserving DNA sequences. Asymmetric cell division (ACD) is essential to generate cells with distinct fates in development, homeostasis, and tissue regeneration1–4. Stem cells often use ACD to give rise to one daughter cell capable of self-renewal and another daughter cell in preparation for terminal differentiation. Despite the crucial role of epigenetic mechanisms in regulating cell fate decisions during development, it remains unclear how stem cells and differentiating daughter cells establish different epigenomes following ACD5.
The Drosophila male germline stem cell (GSC) system provides a great model to investigate the molecular and cellular mechanisms underlying ACD6. Previous work has demonstrated that during the process of GSC ACD, old histone H3 are selectively segregated to the GSC, whereas new H3 are enriched in the gonialblast (GB) committed for differentiation7. Furthermore, subsequent studies have revealed that asymmetric histone inheritance is required for germline function, as disruption of histone inheritance can lead to phenotypes ranging from cell death to tumorigenesis8.
In eukaryotic cells, chromatin must be reestablished on both DNA strands during and after replication9,10. Accordingly, the bulk of canonical histones (i.e. H3, H4, H2A, and H2B) are synthesized and incorporated during DNA replication11. Old nucleosomes on parental DNA must be disassembled ahead of the replication fork and reassembled onto one of the two new double-stranded DNA (dsDNA) daughter strands following passage of the replication fork12,13. Although the process of new histone incorporation onto DNA has been well studied, how old histones are recycled during DNA replication is less clear, particularly in the context of asymmetric cell divisions9,14,15 during animal development. Previous studies have shown that old histones can display a strand preference towards either the leading strand16–18, or the lagging strand19,20 during replication-coupled nucleosome assembly in different systems. Notably, the mode of histone incorporation has not been systematically studied in the context of cellular differentiation and asymmetric cell division in multicellular organisms. Furthermore, previous studies using biochemistry or high-throughput sequencing methods have not allowed for visualization of histone incorporation patterns at the single-molecule level. Characterizing patterns of histone incorporation during DNA replication in cells under physiological condition is critical to our understanding of epigenetic regulation in animal development and diseases such as cancer and tissue dystrophy21.
RESULTS
Asymmetric inheritance of H4 in Drosophila male GSC asymmetric division
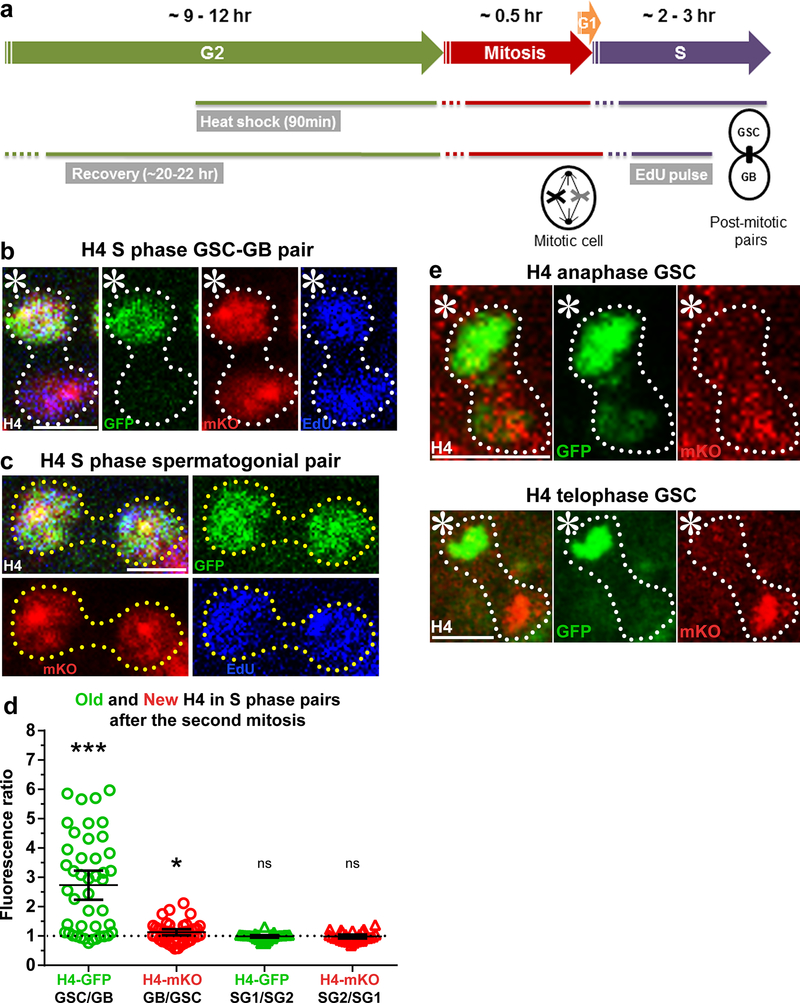
Using a heat shock-controlled switching system to label old histone with GFP (green fluorescent protein) and new histone with mKO (monomeric Kusabira Orange fluorescent protein) (Fig. 1a and Supplementary Fig. 1a), we explored the inheritance pattern for all canonical histones following the asymmetric division of male Drosophila GSCs. The distributions of old histone (GFP) and new histone (mKO) were measured following the second mitosis after heat shock-induced genetic switch7. Since mitotic GSCs account for less than 2% of the total population of GSCs22, post-mitotic GSC-GB pairs derived from the asymmetric GSC divisions were used to visualize and quantify histone inheritance patterns in fixed images7,8. For H4, we found that old H4-GFP was enriched in the GSCs (Fig. 1b), similar to what was previously reported for old H37,8. By contrast, an asymmetric old H4 inheritance pattern was not observed in spermatogonial (SG) pairs after symmetrical cell divisions (Fig.1c). Quantification of post-mitotic pairs revealed an average 2.7-fold enrichment for old H4 in the GSC relative to the GB, whereas spermatogonial pairs showed no significant enrichment of old H4 relative to one another (Fig. 1d). New H4-mKO displayed a more symmetric pattern between GSCs and GBs (Fig. 1b,d). The presence of newly synthesized H4-mKO in both nuclei of the GSC/GB pairs was consistent with the fact that both cells underwent S phase after the second mitosis following heat shock, as indicated by ~ 30-minute nucleoside analog EdU (5-ethynyl-2’- deoxyuridine) incorporation (Fig. 1a,b). As the incorporation of new H4-mKO during the subsequent S phase may change the H4-mKO pattern in post-mitotic GSC-GB pairs, we also examined the H4 segregation pattern in mitotic GSCs. In mitotic GSCs, both old H4-GFP and new H4-mKO showed asymmetric segregation patterns (Fig. 1e). Together, these results establish that histone H4 segregates asymmetrically during ACD, similar to H3.
Figure 1: Histone H4 shows asymmetric inheritance pattern during Drosophila GSC asymmetric divisions.
(a) A cartoon depicting the experimental design. (b) H4 distribution in a post-mitotic GSC-GB pair labeled with EdU, showing H4-GFP is distributed asymmetrically towards the GSC, whereas H4-mKO is distributed more evenly between the GSC and the GB. (c) H4 distribution patterns in a post-mitotic SG pair, showing both H4-GFP and H4-mKO are symmetrically distributed between the two SG nuclei. (d) Quantification of H4- GFP and H4-mKO distributions in GSC-GB pairs (n=44) and SG1-SG2 pairs (n=27). Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. *** P< 0.0001, * P < 0.05; two-tailed Wilcoxon signed rank test. See Supplementary Tables 1 and 2 for data values and online Methods for additional statistical information. (e) An anaphase and telophase GSC showing asymmetric segregation of H4-GFP towards the GSC and H4-mKO towards the GB. Scale bars for panels b, c and e, 5μm; asterisk: hub.
Histones H2A, H2B and H1 display symmetric inheritance patterns
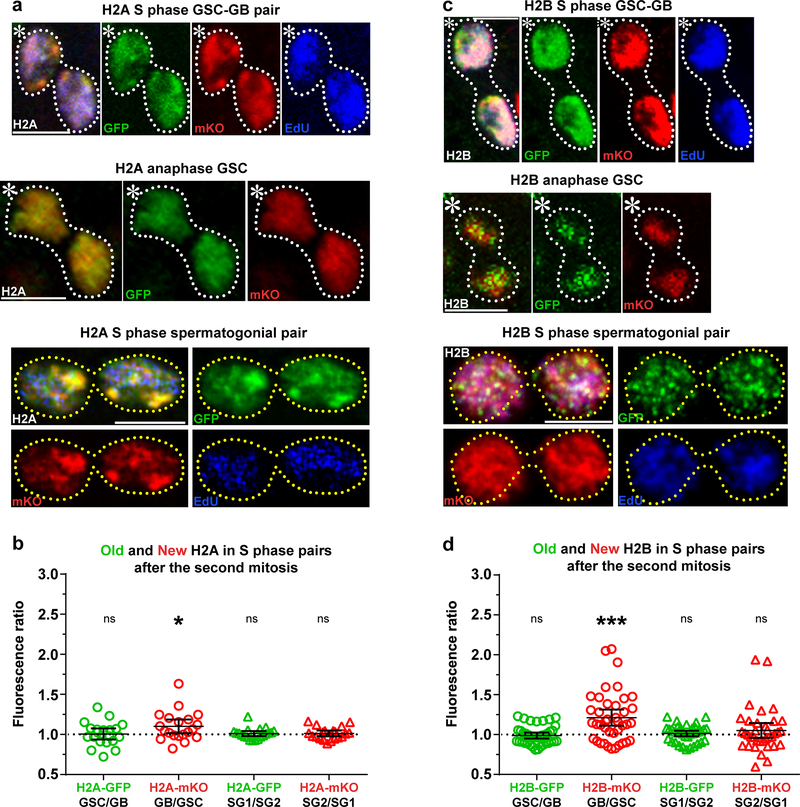
Next, we characterized the inheritance patterns of the rest of the canonical histones: H2A and H2B, as well as the linker histone H1 (Supplementary Fig. 1a). Using a similar heat shock-induced switching scheme (Fig. 1a), we found that old and new H2A (Fig. 2a,b) as well as old and new H2B (Fig. 2c,d) showed more symmetric inheritance patterns in mitotic cells as well as post-mitotic GSC-GB pairs. Additionally, both H2A (Fig. 2a,b) and H2B (Fig. 2c,d) displayed symmetric old and new histone inheritance patterns in post-mitotic SG pairs. Finally, the linker histone H1 also showed globally symmetric inheritance pattern in post-mitotic GSC-GB pairs (Supplementary Fig. 1b).
Figure 2: Histones H2A and H2B show symmetric distribution during Drosophila GSC asymmetric division.
(a) Symmetric H2A inheritance pattern in a post-mitotic GSC-GB (top), mitotic GSC (middle) and post-mitotic spermatogonial (bottom) pairs. (b) Quantification of H2A-GFP and H2A-mKO distribution in GSC-GB pairs (n=20) and SG1-SG2 pairs (n=20). Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. * P< 0.05, two-tailed Student’s t-test if average significantly different than 1. (c) Symmetric H2B inheritance pattern in a post-mitotic GSC-GB (top), mitotic GSC (middle) and post-mitotic spermatogonial (bottom) pairs. (d) Quantification of H2B-GFP and H2B-mKO distribution in GSC-GB pairs (n=40) and SG1-SG2 pairs (n=36). Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. *** P< 0.0001, two-tailed Wilcoxon signed rank test. See Supplementary Tables 1 and 2 for data values and online Methods for additional statistical information. Scale bar for panels a and c, 5μm. Asterisk: hub.
Overall, histones H3 and H4 showed significantly greater asymmetric distributions in asymmetrically dividing GSCs when compared to H2A and H2B. These findings are important for our understanding of epigenetic inheritance, as previous studies have demonstrated that H3 and H4 have the majority of known post-translational modifications and may act as the main epigenetic information carriers23. Furthermore, these findings indicate that even though canonical histones are incorporated in a replication-dependent manner, different histones display distinct inheritance patterns in Drosophila male GSCs. Previous studies have established that during DNA replication, H3 and H4 are incorporated as a tetramer (H3-H4)2, while H2A and H2B are incorporated as dimers24–28. Moreover, H2A and H2B exhibit much more dynamic behavior throughout the cell cycle when compared to (H3-H4)2 tetramers29. Taken together, the distinct biochemical properties of H2A-H2B dimers and (H3-H4)2 tetramers could account for differences observed in histone inheritance patterns in GSCs. Indeed, the similar asymmetric inheritance pattern of old H3 and old H4 suggests that preexisting (H3-H4)2 tetramers are inherited as a whole unit, consistent with previous reports24,30. In order to better understand when differences between histone inheritance patterns first become apparent, we sought to develop a methodology to directly visualize histone inheritance patterns at the replication fork.
Chromatin fiber technique to directly visualize sister chromatids
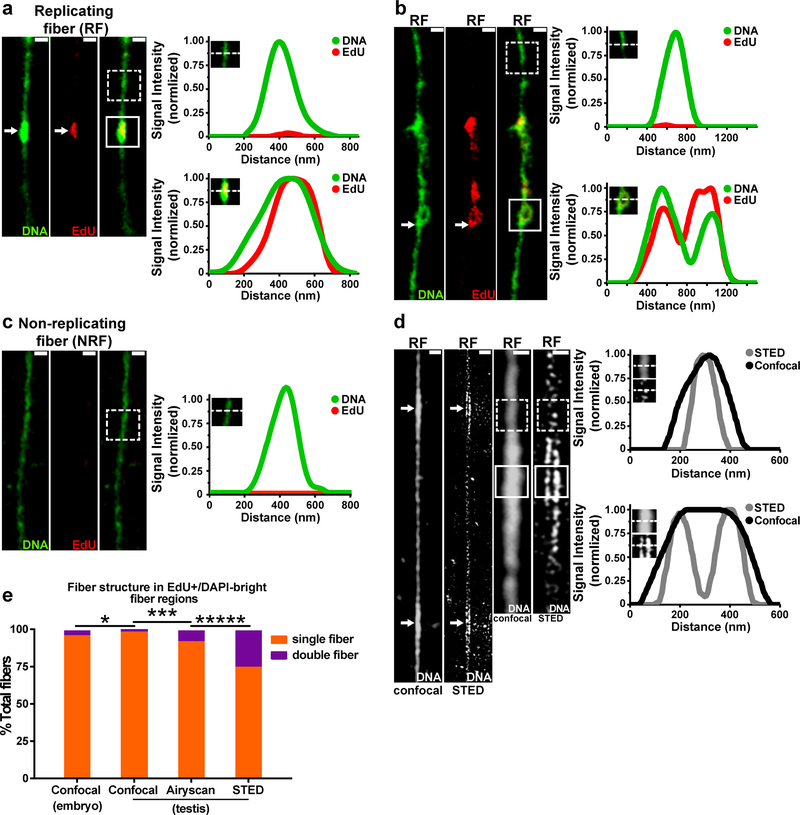
In order to directly examine histone incorporation patterns on newly replicated DNA, we adapted the chromatin fiber technique31–33 to visualize EdU pulse-labeled DNA with associated proteins outside the confines of the nucleus (see online Methods). To validate this technology, chromatin fibers were isolated from Drosophila embryos at the syncytial blastoderm stage and compared with previous electron microscopy images34. Replicating regions (EdU-positive) of chromatin fibers ranged from 250nm to 8μm long, similar in size to replicating regions identified with electron microscopy34. EdU incorporation clearly distinguished unreplicated and newly replicated regions of chromatin fibers (Fig. 3a,b). Consistent with previous findings31, EdU-positive regions showed wider fiber structure and brighter DNA staining with the DNA dye DAPI (Fig. 3a,b, Supplementary Fig. 2a). In a small subset of EdU-labeled fibers, sister chromatids could be resolved at EdU-positive regions (Fig. 3b).
Figure 3: Superresolution microscopy helps visualize sister chromatids on isolated chromatin fibers.
(a,b) Confocal images of chromatin fibers isolated from replicating cells in the Drosophila embryo. Images show replication “bubble” structure with EdU and brighter DNA label (DAPI) signal (white arrow). Line-plots show DNA label (DAPI) and EdU distributions across unreplicated regions without EdU (top) or on replicated regions with EdU (bottom). In a subset of fibers, sister chromatids could be resolved at EdU-positive regions, as shown in (b). (c) Confocal image of chromatin fiber isolated from non-replicating cells in the Drosophila adult eye. Line-plot showing DNA label (DAPI) and EdU distribution on randomly selected fiber region (dashed line box). (d) Confocal and STED images of chromatin fiber isolated from Drosophila male germline stained with DNA label (Yoyo3). Line-plot of Yoyo3 signal in Yoyo3 dim region shows a single fiber structure with both confocal and STED (dashed line box). Line-plot of Yoyo3 signal in Yoyo3-bright region shows double fiber structure with STED but not with confocal (solid line box). (e) Frequency of single and double fiber structures at EdU-positive, DAPI-bright regions. n = 250 for confocal using embryos; for testes analyses, n = 192 for confocal; n = 232 for Airyscan; n = 256 for STED. ***** P< 0.00001, *** P< 0.001, * P< 0.05, Chi-squared test. See Supplementary table 2 and online Methods for additional statistical information. Scale bars for panels a, b, c, d, 500nm.
To confirm that DAPI-bright, EdU-positive fiber structures represent replicating regions, fibers were isolated from non-replicating cells (i.e. Drosophila adult eye) incubated with EdU. Fibers isolated from non-replicating cells showed uniform DNA staining with no identifiable regions of EdU incorporation (Fig. 3c, Supplementary Fig. 2b) when compared to chromatin fibers derived from replicating cells (Fig. 3a,b,d and Supplementary Fig. 2a). These data demonstrate that DAPI-bright, EdU-positive chromatin fibers represent regions of DNA synthesis.
Using confocal microscopy, only 3.2% of DAPI-bright, EdU-positive regions on embryo-derived chromatin fibers could be clearly resolved into two sister chromatids (Fig. 3e). To overcome resolution limits, we used two high resolution microscopy methods: Stimulated Emission-Depletion (STED) microscopy35 and Airyscan imaging36. Both STED (Fig. 3d,e and Supplementary Fig. 2c) and Airyscan (Fig. 3e, Supplementary Fig. 2d) greatly improved the frequency of resolving sister chromatids at actively replicating regions of chromatin fibers. Overall, the percentage of spatially resolvable sister chromatids from EdU-positive chromatin fibers ranged from 8.6% using Airyscan to 25.0% using STED (Fig. 3e). Differences in the relative frequency of resolvable sisters between these two methods likely reflects the lower resolution of Airyscan (~150 nm)37 compared to STED (~35 nm)35. The application of superresolution microscopy to imaging replicating chromatin fibers provides a new methodology to study nucleosome assembly during DNA replication.
Distinct patterns for old versus new H3 and H2A on sister chromatids
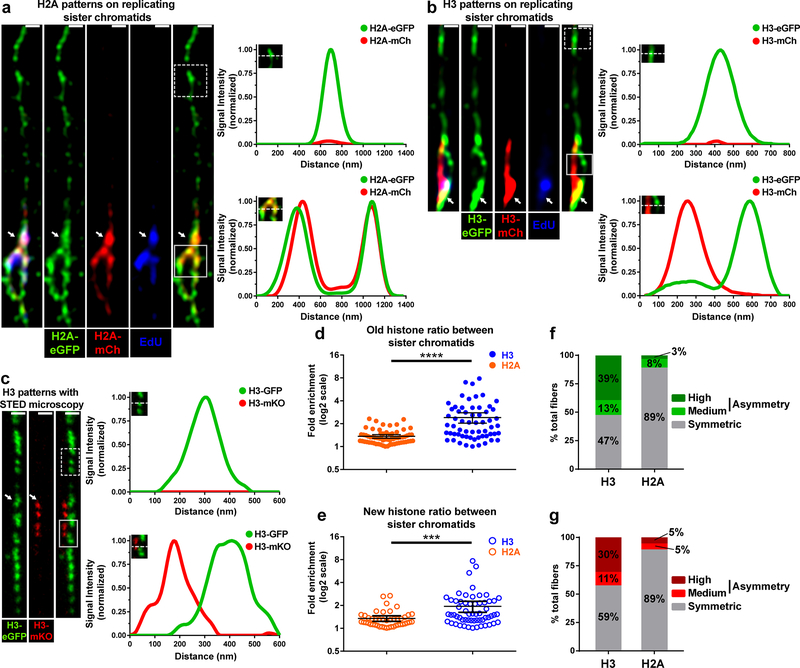
We next explored old and new histone distribution on chromatin fibers derived from the early-stage Drosophila male germ cells, which were labeled with histones driven by an early-stage germline driver nanos-Gal438 (see online Methods). Using Airyscan imaging, unreplicated EdU-negative regions were detected as a single fiber structure enriched with predominantly old histones (Fig. 3). By contrast, replicating or newly replicated regions on chromatin fibers were EdU-positive and showed the double fiber structure indicative of sister chromatids (Fig. 3). To explore histone incorporation patterns on replicating or newly replicated chromatin fibers, we compared the distribution of old versus new H2A and old versus new H3 on sister chromatids. Old and new H2A showed a largely symmetric distribution on chromatin fibers (Fig. 4a). By contrast, old and new H3 showed a more asymmetric distribution pattern on newly replicated sister chromatids (Fig. 4b). These results with H3 were further confirmed using two-color STED imaging (Fig. 4c).
Figure 4: Asymmetric H3 and symmetric H2A distribution on replicating sister chromatids.
(a,b) Airyscan images of chromatin fibers labeled with EdU showing distribution of old H2A-eGFP and new H2A-mCherry (a) or old H3-eGFP and new H3-mCherry (b) on nonreplicated regions lacking EdU label (dashed line boxes) and replicating regions labeled with EdU (solid line boxes). Line-plots show old and new histone distribution across unreplicated (top) or replicated (bottom) regions. (c) Two-color STED image of chromatin fiber showing old H3-GFP and new H3-mKO distribution across unreplicated (dashed line box) and replicating (solid line box) chromatin regions. The transition from single to double-fiber occurs at the point where new histone incorporation begins (white arrow). Line-plots show old H3-GFP and new H3-mKO distribution across unreplicated region without new H3 (top) or on replicated region with new H3 (bottom). (d,e) Quantification of distribution of old H2A and H3 (d) or new H2A and H3 (e) between sister chromatids at replication regions on chromatin fibers. Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. *** P< 0.001; **** P< 0.0001, Mann-Whitney U test. In d, old H2A (Avg.= 1.36; n= 65 replicating regions from 33 chromatin fibers) and old H3 (Avg.= 2.41; n= 61 replicating regions from chromatin fibers) are shown. . In e, new H2A (Avg.= 1.24; n = 45 replicating regions from 25 chromatin fibers) and new H3 (Avg.= 1.94; n = 57 replicating regions from 32 chromatin fibers) are shown. (f) Classification of histone distribution patterns for old H3 and H2A. See online Methods for classification information. The frequency of highly asymmetric fibers was significantly different between old H3 and H2A (P< 0.00001). Moderately asymmetric fibers showed no statistically significant differences. Symmetric fibers were significantly different between H3 and H2A (P<0.00001). (g) Similar to f, showing classification of histone distribution patterns for new H3 and H2A. The frequency of highly asymmetric fibers was significantly different between H3 and H2A (P< 0.01). Moderately asymmetric fibers showed no statistically significant differences. (P = 0.37). Symmetric fibers were significantly different between H3 and H2A (59% H3 fibers vs. 89% of H2A fibers: P< 0.01). See Supplementary table 2 and online Methods for additional statistical information. Scale bar in panels a, b, c, 500nm.
To systematically compare histone distribution patterns of H2A and H3 along sister chromatids, we divided resolved sister chromatid fibers into 2μm units and measured the fluorescence levels for both old and new H2A and H3 on each unit (Supplementary Fig. 2e, online Methods). Overall, old H3 displayed a significantly higher frequency and magnitude of asymmetry than did H2A fibers (Fig. 4d). Old H3 showed on average a 2.41-fold ratio while old H2A showed a 1.36-fold ratio between sister chromatids. Similarly, new H3 also displayed a significantly higher frequency and magnitude of asymmetry when compared to new H2A (Fig. 4e). New H3 showed on average a 1.94-fold difference while new H2A showed a 1.24-fold difference between sister chromatids.
To further compare the differences in old and new histone incorporation patterns between H3 and H2A, we classified fibers as symmetric, moderately asymmetric or highly asymmetric (see online Methods). Using these criteria, 39% of H3 fibers were found to be highly asymmetric compared to just 3% of H2A fibers for old histones (Fig. 4f, P< 10−4). Similarly, 30% of H3 fibers were found to be highly asymmetric compared to just 5% of H2A fibers for new histones (Fig. 4g, P< 10−2). For the moderately asymmetric fibers, H3 and H2A fibers showed comparable frequencies: 13% of H3 fibers were moderately asymmetric compared to 8% of H2A fibers for old histones (Fig. 4f, P = 0.32); and 11% of H3 fibers showed moderate asymmetry compared to 5% for H2A for new histones (Fig. 4g, P = 0.37). In summary, these results demonstrate that both old and new H3 are more asymmetrically incorporated during DNA replication compared to old and new H2A, consistent with their distinct segregation patterns during ACD of GSCs (Fig. 1 and 2).
Old H3 incorporation is anticorrelated with strand-enriched replication factors
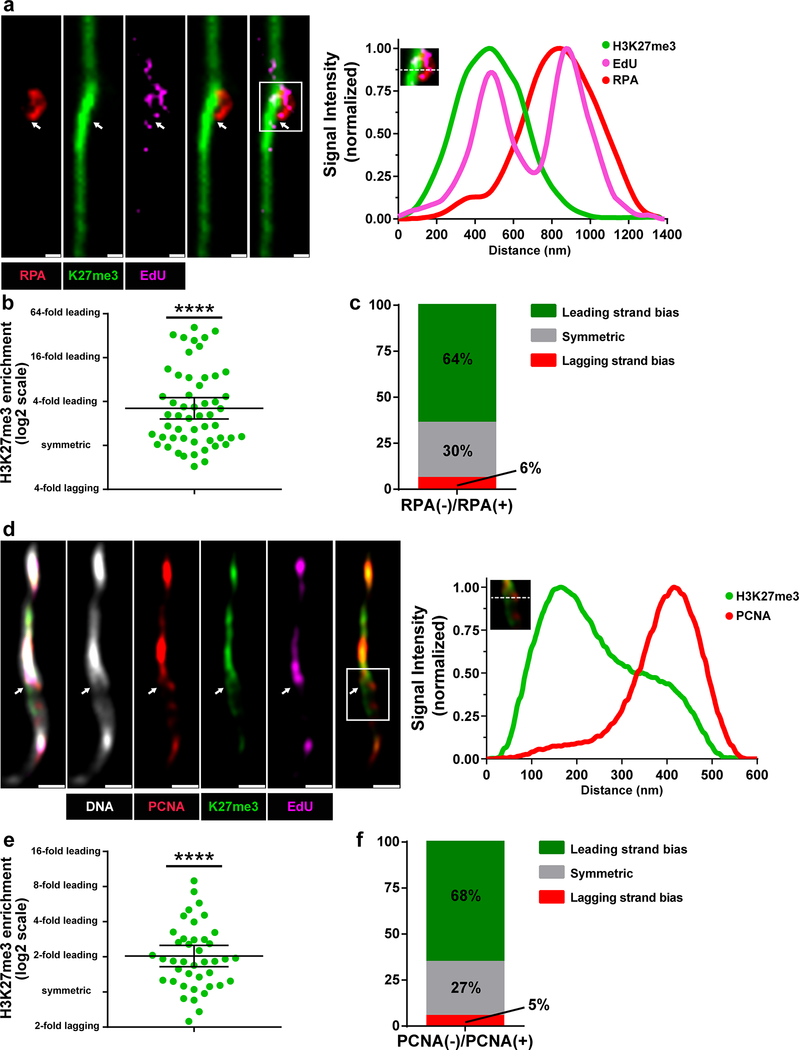
As old and new H3 show asymmetries during the process of replication-coupled nucleosome assembly, we next tested whether old versus new H3 asymmetry correlates in any way with strand-enriched DNA replication machinery components. To determine strand specificity, chromatin fibers were isolated from flies expressing eGFP-RPA70 (replication protein-A) fusion protein under the control of the endogenous regulatory elements of the RPA70 gene (RPA70>RPA70-eGFP)39. RPA70 represents a highly conserved single-stranded DNA-binding protein significantly enriched on the lagging strand40. To visualize old histones, we utilized an antibody against the H3K27me3 histone modification, which has been shown to be enriched on old H341. At EdU-positive regions where the sister chromatids could be resolved, RPA70 and H3K27me3 occupied opposite strands of the bubble structure (Fig. 5a), suggesting that old H3 is recycled to the leading strand. Quantification showed an average of 3.2-fold more H3K27me3 at the RPA70-depleted leading strand compared to the RPA70-enriched lagging strand (Fig. 5b). Furthermore, using a set of criteria to classify fibers as leading strand enriched, symmetric, or lagging strand enriched (see online Methods), we found that 64% of fibers showed leading strand bias, 30% of fibers were symmetric and only 6% of fibers showed lagging strand enrichment (Fig. 5c).
Figure 5: Old H3 preferentially associate with the leading strand on chromatin fibers.
(a) Confocal image of chromatin fiber labeled with EdU showing anti-correlated H3K27me3 and RPA70 distributions. The transition from single to double fibers is correlated with the EdU incorporation site (white arrow). Line-plot shows EdU, H3K27me3 and RPA70 distribution across the replicating region (box with solid white lines). (b) Quantification of the log2 average of H3K27me3 fluorescence intensity on RPA70-depleted sister chromatid/ average H3K27me3 fluorescence intensity on RPA70-enriched sister chromatid (Avg. fold enrichment = 3.20; n= 53 replicating regions from 28 chromatin fibers). Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. Data are significantly different from log2=0, **** P< 0.0001, two-tailed Wilcoxon signed rank test. (c) Classification of RPA70-labeled sister chromatids into leading-strand enriched (inter-sister ratio >1.4), lagging-strand enriched (inter-sister ratio <1.4) and symmetric (−1.4< inter-sister ratio< 1.4). (d) Airyscan image of chromatin fiber labeled with EdU showing anti-correlated H3K27me3 and PCNA distribution. The white arrow indicates the replication bubble. Line-plot shows EdU, H3K27me3 and PCNA distribution across replicating region (box with solid white lines). (e) Quantification of the log2 average of H3K27me3 fluorescence intensity on PCNA-depleted sister chromatid/ average H3K27me3 fluorescence intensity on PCNA-enriched sister chromatid (Avg. fold enrichment = 2.04; n=41 replicating regions from 26 chromatin fibers). Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. Data is significantly different from log2=0, **** P< 0.0001, two-tailed Wilcoxon signed rank test. (f) Classification of PCNA-labeled fibers into leading-strand enriched (inter-sister ratio >1.4), lagging-strand enriched (inter-sister ratio <1.4) and symmetric (−1.4<inter-sister ratio<1.4). Y-axis is with log2 scale). See Supplementary table 2 and online Methods for additional statistical information. Scale bar in panels a and d, 500nm.
We also investigated H4 incorporation patterns at replicating regions using an old H4-enriched H4K20me2/3 modification16,41. At EdU-positive regions of germline-derived chromatin fibers, H4K20me2/3 levels were more abundant on the RPA70-negative leading strand when compared to the RPA70-positive lagging strand (Supplementary Fig. 3a). Quantification showed a 1.8-fold difference on average in H4K20me2/3 levels of leading strand compared to the lagging strand (Supplementary Fig. 3b). Further analysis demonstrated that 54% of fibers showed old histone bias towards the leading strand, 31% showed symmetry, while only 15% of fibers showed enrichment towards the lagging-strand (Supplementary Fig. 3c). Taken together, these results suggest that old H4, similar to old H3, is preferentially recycled to the leading strand.
To further validate histone inheritance patterns at the replication fork, similar experiments were performed using another lagging-strand-enriched component, Proliferating Cell Nuclear Antigen (PCNA), which was expressed in its endogenous genomic context (pcna>PCNA-eGFP)39. At EdU-positive sister chromatid regions, PCNA and H3K27me3 were enriched at the opposite sides of sister chromatids (Fig. 5d), further demonstrating that old H3 is preferentially recycled to the leading strand. Quantification showed an average of 2.0-fold more H3K27me3 at the PCNA-depleted leading strand compared to the PCNA-enriched lagging strand (Fig. 5e). Further analysis revealed that 68% of fibers showed old histone bias towards the leading strand, 27% showed symmetry, while only 5% of fibers showed enrichment towards the lagging-strand (Fig. 5f). Taken together, these results demonstrate that during DNA replication, old (H3-H4)2 tetramers are preferentially recycled by the leading strand.
Strand preferences of old and new H3 during replication-coupled nucleosome assembly
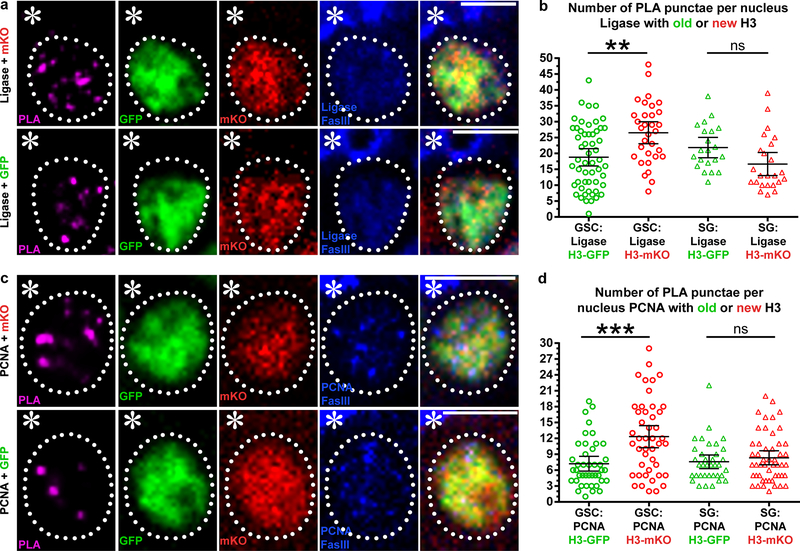
As a complementary method to explore histone inheritance patterns at the replication fork, we used an imaging-based proximity ligation assay (PLA) to probe the spatial proximity between histones (old versus new) and different strand-enriched DNA replication components in intact nuclei. We used CRISPR/Cas9-mediated genome editing to tag the lagging strand-enriched DNA Ligase at its endogenous genomic locus using a 3xHA epitope. We then applied anti-HA for the PLA assay to probe the spatial proximity between DNA Ligase and old versus new histones. We observed more PLA fluorescent puncta between ligase and new H3-mKO than those between ligase and old H3-GFP (Fig. 6a). Quantification of the overall PLA signals in GSCs showed significantly more PLA fluorescent puncta between ligase and new H3 than those between ligase and old H3 (Fig. 6b).
Figure 6: Proximity ligation assay shows distinct proximity between histones (old versus new) and lagging strand-enriched DNA replication machinery components in GSCs.
(a) A representative GSC showing PLA signals between lagging-strand-specific ligase-HA and new H3-mKO (top), and a representative GSC showing PLA signals between ligase-HA and old H3-GFP (bottom). (b) Quantification of the number of PLA puncta per nucleus between ligase and histones (old versus new) in GSCs and SGs. Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. In GSCs, PLA puncta between ligase and new H3-mKO: 26.5; (n=35); between ligase and old H3-GFP: 18.5; (n=53). In SGs, PLA puncta between ligase and new H3-mKO: 16.7; (n=24); between ligase and old H3-GFP: 21.9; (n=21) **: P< 0.01, Kruskal-Wallis multiple comparisons of non-parametric data with Dunn’s multiple comparisons corrections test. (c) A representative GSC showing PLA signals between lagging-strand enriched PCNA and new H3-mKO (top), and a representative GSC showing PLA signals between PCNA and old H3-GFP (bottom). (d) Quantification of the number of PLA puncta per nucleus between PCNA and histones (old versus new) in GSCs and SGs. Individual data points (circles) and mean values are shown. Error bars represent 95% confidence interval. In GSCs, PLA puncta between PCNA and new H3-mKO: 12.3; between PCNA and old H3-GFP: 7.2; (n=42). In SGs, PLA puncta between PCNA and new H3-mKO: 8.3; (n=50); between PCNA and old H3-GFP: 7.6; (n=36), ***: P< 0.001, Kruskal-Wallis multiple comparisons of non-parametric data with Dunn’s multiple comparisons corrections test. See Supplementary table 2 and online Methods for additional statistical information. Scale bar for panels a and c, 5μm.
Using another lagging strand-enriched component, PCNA, as a marker for the PLA experiments, we also observed more PLA fluorescent puncta between PCNA and new H3-mKO than those between PCNA and old H3-GFP (Fig. 6c). Again, quantification of the overall PLA signals showed significantly more PLA fluorescent puncta between PCNA and new H3-mKO than those between PCNA and old H3-GFP in GSCs (Fig. 6d).
As a control, we also performed PLA experiments using a strain where the tags for old H3 and new H3 were swapped, resulting in old H3-mKO and new H3-GFP. Consistent with the previous results, more PLA fluorescent puncta were obtained between PCNA and new H3-GFP than the signals between PCNA and old H3-mKO (Supplementary Fig. 4a,b).
To confirm the specificity of our PLA signal, we also performed PLA in non-replicating somatic hub cells as well as between histones and a cytoplasmic protein Vasa (Supplementary Fig. 4c). In these experiments, we observed negligible PLA signal, confirming that PLA signals were specific to replicating nuclei and false positive signals were minimal in our experimental conditions. These results are consistent with the chromatin fiber results shown above (Fig. 5), and further suggest that new H3 preferentially associates with the lagging strand.
Compared to GSCs, SGs showed no significant strand preference between old and new histones for either ligase (Fig. 6b) or PCNA (Fig. 6d). Together, these results demonstrate that histone distribution patterns show a cellular specificity not only during mitosis7 (Fig. 1), but also during DNA replication. We therefore conclude that differences in epigenetic inheritance at the replication fork likely underlie differences in global epigenetic inheritance patterns observed between GSCs and SGs.
High incidence of unidirectional fork progression in Drosophila testis
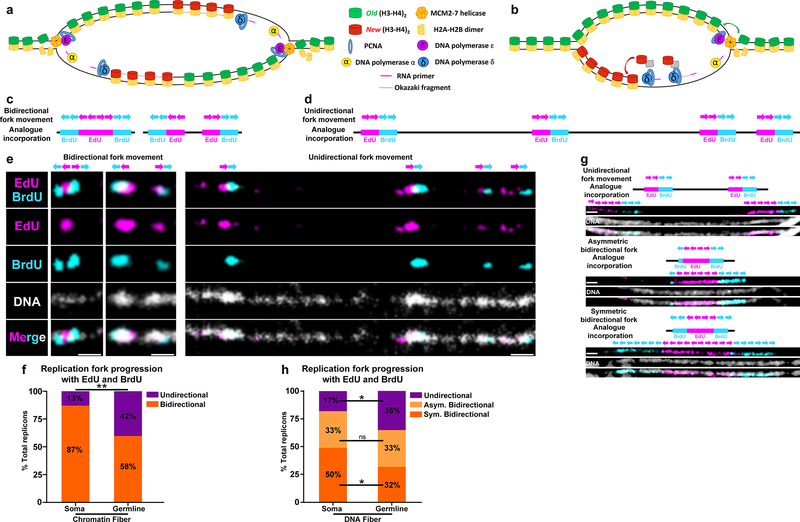
We have demonstrated that old H3 are incorporated on the leading strand whereas new H3 are preferentially incorporated on the lagging strand during the process of replication-coupled nucleosome assembly. However, if replication forks are proceeding outward from replication origins in a bidirectional manner, asymmetries in histone inheritance at the replication fork alone would lead to alternating stretches of leading strand-incorporated old histones and lagging strand-incorporated new histones on each of the two duplicating sister chromatids (Fig. 7a), which would not be sufficient to explain the globally asymmetric histone inheritance pattern we have observed. Therefore, we hypothesize that replication forks are coordinated to achieve long-range asymmetric histone patterns (Fig. 7b).
Figure 7: Germline-derived chromatin and DNA fibers show more unidirectional fork progression compared to soma-derived chromatin and DNA fibers.
(a,b) A cartoon showing strand biased histone incorporation at a bidirectional replication fork (a) and a unidirectional replication fork (b). (c,d) Predicted bidirectional fork progression result (c) and unidirectional fork progression result (d). (e) Bidirectional fork progression pattern from somatic cell-derived chromatin fiber. Replicons show early label (EdU) flanked by late label (BrdU) on both sides. Unidirectional fork progression pattern from germline-derived chromatin fiber. Multiple replicons show alternation between early label (EdU) and late label (BrdU) along the chromatin fiber toward the same direction. (f) Quantification of fork progression patterns in somatic cell-derived versus germline-derived chromatin fibers. Testis-derived fibers show a significantly higher incidence of unidirectional fork progression: 42% in testis-derived chromatin fiber (n=54) versus 13% in eye imaginal disc-derived chromatin fiber (n=31) **: P< 0.01, Chi-squared test. (g) Fork progression patterns in DNA fibers: unidirectional fork progression, asymmetric bidirectional fork progression and bidirectional fork progression. (h) Quantification of fork progression patterns in somatic cell-derived versus germline-derived DNA fibers. Germline-derived fibers show a significantly higher incidence of unidirectional fork progression: 35% in germline DNA fiber (n=109), 17% soma DNA fiber (n=48). *: P< 0.05, Chi-squared test. See online Methods for additional statistical information. Scale bar for panels e and g, 1μm
To explore the fork movement, we applied sequential nucleoside analog incorporation to the chromatin fibers (see online Methods). In these experiments, active DNA replication regions were labeled first by EdU and subsequently by BrdU (Supplementary Fig. 5a). A DNA dye was added to verify fiber continuity. Only continuous fibers containing multiple replicons in tandem were included for fork movement analysis (Supplementary Fig. 5b,c). The progression of replication forks in a bidirectional (Fig. 7c) or a unidirectional manner (Fig. 7d) would produce distinct patterns. Chromatin fibers derived from somatic cells, such as larval eye imaginal disc cells, displayed largely bidirectional fork movement (Fig. 7e), as 87% of replicons on chromatin fibers showed typical bidirectional fork movement while only 13% of replicons showed unidirectional fork movement. In contrast, a substantial fraction (42%) of germ cell-enriched, testis-derived chromatin fibers contained replicons with unidirectional replication progression (Fig. 7e and 7f). Furthermore, fork movement in unidirectional replicons appeared to be coordinated, as multiple unidirectional forks appeared to move in the same direction (Fig. 7e).
To further explore replication fork movement pattern in the Drosophila testes, we utilized a similar sequential nucleoside analog incorporation method to track fork movement using DNA fibers (Supplementary Fig. 5a–c). As DNA fibers have been stripped of DNA-associated proteins, these fibers lack protein-mediated compaction and show more details of replication fork movement. Therefore, we were able to classify replication fork movement based on DNA fibers into three categories: unidirectional, asymmetric bidirectional and symmetric bidirectional (Fig. 7g). We classified bidirectional forks as asymmetric in cases where the BrdU signal on one side was more than 2-fold longer than the BrdU signal on the other side flanking the EdU label (Fig. 7g: Asymmetric bidirectional fork pattern). Consistent with the chromatin fiber data, DNA fibers derived from testes showed a significantly higher incidence of unidirectional fork movement (35%) compared to those derived from somatic tissue control (17%, Fig. 7h, P< 0.05). DNA fibers derived from testes also show a significantly lower incidence of symmetric bidirectional fork movement (32%) compared to fibers derived from somatic tissue control (50%, Fig. 7h, P< 0.05). Incidences of asymmetric bidirectional fork movement were not significantly different between these two samples.
In order to investigate replication fork movement in distinct stages of germline development, we derived DNA fibers from bag-of-marbles (bam) mutant testes42, which lack meiotic germ cells and are instead filled with mitotic germ cells (Supplementary Fig. 5d). The bam testes-derived DNA fibers showed replication patterns similar to wild-type testes, suggesting that biased replication fork movement might be more prevalent in the germline when compared to somatic cell types (Supplementary Fig. 5e). However, these data cannot pinpoint whether GSCs show an even more pronounced bias in fork movement when compared to more differentiated germ cells.
DISCUSSION
Our results using both chromatin fiber and DNA fiber methods suggest that replication fork direction is coordinated in the Drosophila germline. Together with the strand bias found between old and new H3 and H4 histones, these mechanisms could expand asymmetric histone incorporation at individual forks to global asymmetries between sister chromatids.
While asymmetries in the deposition of histone proteins have been observed experimentally17,18,43–46, a majority of studies have demonstrated that, on a global scale, old and new histones are equally associated with leading and lagging strands following replication9,25,47. However, this question had not been addressed in a multicellular organism in a developmental context. Interestingly, studies on histone segregation in symmetrically dividing mouse embryonic stem cells16 and yeast20 have revealed that histones can show different biases in inheritance patterns: in mouse embryonic stem cells, old histones are inherited with a bias towards the leading strand, whereas in yeast old histones have a bias towards the lagging strand. Importantly, both studies demonstrated that molecular mechanisms exist that act at the replication fork to counteract asymmetric histone incorporation in order to achieve a more symmetric outcome. Here, our studies in Drosophila melanogaster suggest that in a developmental context of a multicellular organism, asymmetries at the replication fork can be utilized as a tool to generate epigenetically distinct sister chromatids to regulate cell fate decisions in vivo8.
A multitude of factors could be responsible for the differences in histone inheritance patterns observed in yeast, mammalian cell culture or Drosophila male germline. One possible difference could be the identity of the H3 species present in the nucleosome. The mouse and fly genomes encode distinct H3 variants, whereas the yeast genome contains a single H3 gene, an ancestral variant that more closely resembles the histone variant H3.3. By contrast, the dominant H3 species found in mice or flies is the canonical replication-dependent histone H348. These histones differ considerably in multitude of aspects, ranging from mode of incorporation, to post-translational modifications, to stability of the (H3-H4)2 tetramer49,50. Based on the reported differences between H3 and H3.3, it is conceivable that these two histones may have distinct incorporation behavior at the replication fork, with H3.3 associating more frequently with the lagging strand, and H3 with the leading strand. Other factors, such as the histone modification studied or the genomic context, could also play important roles in biasing histone inheritance. As our chromatin fiber experiments were conducted using a mixed population of cells containing both asymmetrically dividing GSCs and symmetrically dividing SGs, we speculate that the heterogeneity observed in histone inheritance patterns in our dataset could be due to stage-specific histone incorporation modes in GSCs versus SGs. Results from our PLA assays support this hypothesis, as differentiated (SGs) germ cells show significantly less asymmetry at the fork when compared to GSCs. Further methodological improvements to generate pure population of cells would be required to better understand histone incorporation patterns in a cell-type- and stage-specific manner using the chromatin fiber method. More technology development will also be needed to address histone incorporation patterns at particular genomic regions of interest.
Recent studies have indicated that replication fork rate could also play an important role in regulating differential histone incorporation patterns. Replication fork rate varies depending on interactions with transcriptional machinery. Head-on collisions, where the replication fork and transcription machinery directly collide when progressing in opposite directions, are more likely to slow fork progression when compared to codirectional interactions51. Importantly, the type of interaction between replication fork and transcription also affects the relative rates of chromatin maturation on the daughter strands: the leading strand matures faster in cases of codirectional replication-transcription interactions, whereas the lagging strand matures faster in cases of head-on collisions13. As histone recycling is the first step in chromatin maturation, these studies suggest that transcription-induced changes in fork rate may play an important role in regulating histone recycling patterns behind the replication fork. In this model, slow fork progression rates in cases of head-on collisions would give Okazaki fragments more time to complete maturation to allow for old histone reincorporation by the lagging strand. Conversely, faster fork rates in the case of codirectional replication-transcription interactions would produce a more pronounced leading strand bias for recycling old histones52. Because head-on collisions are generally minimized during DNA replication53, it is possible that the few cases of lagging strand bias in old histone incorporation observed in our dataset (Fig. 5d,h) reflect instances of head-on collisions between replication and transcription. Conversely, the more common pattern of leading strand bias in old histone inheritance (Fig. 5d,h) likely represents no collision or co-directional collisions. Lastly, the possibility that transcription could influence fork movement in vivo raises an intriguing possibility that transcription may function to regulate both histone incorporation pattern and fork movement during DNA replication.
Unidirectional replication and biased fork movement are by no means unprecedented observations54–60. Fork block systems in the replicating S. pombe are utilized during mating-type switching to help coordinate fork movement across the mating type locus to create a DNA lesion necessary for initiating the DNA repair mechanisms involved in the process of mating-type switching61. In multiple organisms, it has been shown that fork movement at the rDNA region is unidirectional56,62,63. Fork block systems have also been found in metazoan systems to ensure that replication/transcription collisions do not occur in the context of heavily transcribed loci64. A majority of studies on mammalian replicons have identified that approximately 5–14% of origins are replicated in a unidirectional manner whereas 86–95% are bidirectional65,66. Some studies have observed that higher incidences of unidirectional fork movement can be detected in late-replicating regions of the genome when compared to early replicating regions67. However, fork coordination across broad stretches of the genome as a means to regulate epigenetic inheritance represents a previously uncharacterized aspect of cell-type specific regulation of DNA replication. Currently, the molecular mechanisms responsible for biasing fork movement in the Drosophila germline remain unclear. However, studies have demonstrated that across multiple species, levels of transcription are significantly higher in the testes when compared to other tissues such as the brain or the liver68,69. If transcription is indeed involved in biasing fork movement, then it possible that the high levels of transcription present in the Drosophila testes could play an important role in influencing fork movement genome wide. Further studies will be needed to fully understand the molecular players and sequence features responsible for coordinating fork movement in the Drosophila germline.
Together, our findings suggest that DNA replication may play a novel, unappreciated role in directing histone incorporation to differentially establish epigenetic information on two genetically identical sister chromatids. Furthermore, these results identify that DNA replication can be exploited in a cell-type specific manner. While the molecular players responsible for this cell-type-specificity remain unclear, this demonstration of a potential regulatory role for DNA replication represents an important step forward in understanding how DNA replication and replication-coupled nucleosome assembly could act to regulate asymmetric cell division and cell fate specification.
Online Methods
Fly strains and husbandry
Fly stocks were raised using standard Bloomington medium at 18°C, 25°C, or 29°C as noted. The following fly stocks were used: hs-flp on the X chromosome (Bloomington Stock Center BL-26902), nos-Gal4 on the 2nd chromosome38, UASp-FRT-H3-GFP-PolyA-H3-mKO on the 3rd chromosome and UASp-FRT-H2B-GFP-PolyA-H2B-mKO, as reported previously7. Other new histone transgenic strains generated for this work are described as follows and are all on either the 2nd or the 3rd chromosome as a single-copy transgene. To obtain bam mutant flies, the bamΔ86 (Bloomington Stock Center BL5427) and bam1 alleles70 were used to generate bamΔ86/bam1 flies.
Generation of fly strains with different switchable dual-color transgenes
Standard procedures were used for all molecular cloning experiments. Enzymes used for plasmid construction were obtained from New England Biolabs (Beverly, MA). The new histone sequences, including histone-mKO, histone-mCherry or histone-GFP, were recovered as an XbaI flanked fragment and were subsequently inserted into the XbaI site of the UASp plasmid to construct the UASp-new histone plasmid. The old histone sequences, including histone-GFP, histone-EGFP, or histone-mKO, were inserted to pBluescript-FRT-NheI-SV40 PolyA-FRT plasmid at the unique NheI site. The entire NotI-FRT-old histone-SV40 PolyA-FRT-EcoRI sequences were then subcloned into the UASp-new histone plasmid digested by NotI and EcoRI. The final UASp-FRT-old histone-PolyA-FRT-new histone plasmids were introduced to w1118 flies by P-element-mediated germline transformation (Bestgene Inc.). Transgenic flies with the following transgenes were newly generated in studies reported here:
UASp-FRT-H4-GFP-PolyA-FRT-H4-mKO, UASp-FRT-H2A-GFP-PolyA-FRT-H2A-mKO, UASp-FRT-H2A-EGFP-PolyA-FRT-H2A-mCherry, UASp-FRT-H1-GFP-PolyA-FRT-H1-mKO, UASp-FRT-H3-mKO-PolyA-FRT-H3-GFP, and UASp-FRT-H3-EGFP-PolyA-FRT-H3-mCherry.
To assess the impact of transgene expression on cell cycle dynamics, live cell imaging was performed on fly strains expression transgenic histones (nos-Gal4; UASp-histone transgene) versus a control fly strain (nos-Gal4; UASp-tubulin-GFP). The average GSC cell cycle lengths between these two groups were not significantly different (P = 0.88; students t-test): Average time= 850.0 minutes per GSC cell cycle for the fly lines with histone transgenes (from M-phase of one cell cycle to the subsequent M-phase; n = 10) versus Average time= 842.5minutes per GSC cell cycle for the control (from M-phase of one cell cycle to the subsequent M-phase; n = 12).
Generating knock-in fly strains to tag genes encoding key DNA replication components
In collaboration with Fungene Inc. (Beijing, China), the following fly line was generated using the CRISPR-Cas9 technology: CG5602 (DNA ligase I, major replicative ligase) with 3xHA tag at the 3’ immediately upstream of the STOP codon, generating the fusion protein: DNA ligase-3HA.
Heat shock scheme
Flies with UASp-dual color histone transgenes were paired with nos-Gal4 drivers. Flies were raised mostly at 18°C throughout development until adulthood to avoid pre-flipping. In all experiments, flies without heat shock were always checked to evaluate pre-flip events. Samples showing pre-flipping activity were excluded from all data acquisition.
For adult males: Before heat shock, 0–3 day old males were transferred to vials that had been air dried for 24 hours. Vials were submerged underneath water up to the plug in a circulating 37°C water bath for 90 minutes and recovered in a 29°C incubator for indicated time before dissection, followed by immunostaining or live cell imaging experiments.
For wandering third-instar larvae: bottles containing third instar larvae (pre-wandering stage) were submerged underneath water up to the plug in a circulating 37°C water bath for 90 minutes and recovered in a 29°C incubator for indicated time before dissection, followed by fiber preparation and immunostaining experiments.
Immunostaining experiments
Immunofluorescence staining was performed using standard procedures7,71. Primary antibodies used were mouse anti-Fas III (1:200, DSHB, 7G10), anti-HA (1:200; Sigma-Aldrich H3663), anti-PCNA (1:200; Santa Cruz sc-56), anti-GFP (1:1,000; Abcam ab 13970), anti-mKO (1:200; MBL PM051M), anti-mCherry (1:1000; Invitrogen M11217), anti-H3K27me3 (1:200; Millipore 07–449), anti-H4K20me2/3 (1:400; Abcam ab7817), anti-ssDNA (1:100, DSHB) and anti-BrdU (1:200; Abcam ab6326). BrdU analogue was Invitrogen B23151 5-bromo-2´-deoxyuridine (BrdU). Secondary antibodies were the Alexa Fluor-conjugated series (1:1000; Molecular Probes). Confocal images for immunostained fixed sample were taken using Zeiss LSM 700 Multiphoton confocal microscope with 63x or 100x oil immersion objectives and processed using Adobe Photoshop software.
Quantification of GFP and mKO intensity in whole testis
No antibody was added to enhance either GFP or mKO signal. Values of GFP and mKO intensity were calculated using Image J software: DAPI signal was used to determine the area of nucleus for measuring both GFP and mKO fluorescent signals, the raw reading was subsequently adjusted by subtracting fluorescence signals in the hub region used as background in both GSC and GB nuclei and compared between each other.
Chromatin fiber preparation with nucleoside analogue incorporation and immunostaining
Testes were dissected in Schneider’s Drosophila medium (Gibco™, Catalog# 21720001) at room temperature (RT) and incubated in Schneider’s medium containing 10μM EdU analogue (Invitrogen™ Click-iT™ EdU Imaging Kit, Catalog# C10340). Testes were incubated for 30 minutes, rotating, at RT unless otherwise specified in the protocol. At the end of the 30 minutes, testes were washed for three times with Schneider’s medium at RT. Following wash, testes were incubated in the dissociation buffer [Dulbecco’s PBS with Mg2+ and Ca2+ with collagenase/dispase (MilliporeSigma®) added to a final concentration of 2mg/ml] in a 37°C water bath for five minutes. Cells were pelleted at 1000G for five minutes, after which the dissociation buffer was drained. Cells were suspended in 60μl of lysis buffer (80mM NaCl, 150mM Tris-base, 0.2% Joy detergent, pH 10). Following resuspension, 20μl of lysis buffer/cell mixture was transferred to a clean glass slide (Fisherbrand™ Superfrost™ Plus Microscope Slides) and allowed to sit in lysis buffer until cells were fully lysed (~5 minutes). 10μl of sucrose/formalin solution (1M sucrose; 10% formaldehyde) was then added and incubated for two minutes. A coverslip (Fisherbrand™ Microscope Cover Glass, 12–545-J 24×60mm) was placed on top of the lysed chromatin solution, after which, the slide was transferred immediately to liquid nitrogen and allowed to sit for two minutes. Cover slip was then removed with a razor blade, after which the slide was transferred to cold (−20°C) 95% ethanol for ten minutes. Next, slide was incubated with fixative solution [0.5% formaldehyde in 1xPBST (1xPBS with 0.1% Triton)] for one minute. The fixative solution was drained and the slides were placed into a coplin jar containing 50ml 1xPBS. Slides were washed twice with 50ml 1xPBS each time and placed in a humid chamber with 1ml of blocking solution (2% BSA in 1xPBST) for 30-minute pre-blocking. Blocking buffer was then drained and primary antibodies were added for incubation overnight at 4°C. Slides were then washed twice with 50ml 1xPBS each time and incubated with secondary antibodies for two hours at RT. Slides were then washed twice with 50ml 1xPBS each time and mounted with ProLong® Diamond mounting media.
For BrdU labeling, fibers were treated with 1M HCL for two hours at 37°C to expose BrdU epitope prior to addition of anti-BrdU primary antibody. For EdU visualization: EdU analogue was conjugated to Alexa-647 dye using CLICK chemistry [reviewed by 72,73].
Identification of replicating regions at chromatin fibers
Replicating regions were identified by EdU incorporation into the chromatin fiber. Replication bubbles were identified as EdU-positive regions showing a transition from a single fiber structure to a double fiber structure, which was co-localized with EdU incorporation. EdU-positive regions along the chromatin fiber were also associated with a significant increase in DNA dye intensity, referred to as DAPI-bright regions. DAPI-bright regions were identified as regions showing greater than a 2-fold increase in DAPI-intensity relative to surrounding DAPI-dim regions from the same chromatin fiber. This greater than 2-fold difference in DAPI intensity likely reflects both the two-fold increase in DNA content associated with replication and the differences in super-helical torsion associated with the advancing replication fork. Previous studies have reported that negative-supercoiling is enriched behind the replication fork, whereas positive-supercoiling is enriched ahead of the replication fork74–76. The unwound nature of the negatively supercoiled DNA favors the binding of intercalating DNA dyes, such as DAPI and Yoyo-III77,78. Conversely, positive supercoiling structure limits the accessibility of DNA to intercalating small molecules.
Superresolution STED imaging
Superresolution images were acquired using a Leica TCS SP8 STED microscope with a 1.4 NA 100X STED white objective. Immunostaining experiments were performed to enhance specimen brightness and photo-stability for STED microscopy. Secondary antibody fluorophore conjugates were empirically selected for STED performance. The optimal 3-colour separation was performed with the 592 nm continuous wave (CW) and 775 nm pulsed depletion lasers (Alexa 488 with STED 592nm, Alexa 568 with STED 775nm, and Alexa 647 with STED 775nm). Images were acquired as single z-planes for all tissue types, including whole mount and squash tissues, as well as isolated fibers, in order to minimize drift between channel acquisitions. Specimens included 100 nm TetraSpeck microsphere beads as fiducial markers (Thermo Fisher Catalog# T7279). Instrument aberration and blurring was corrected with post-acquisition deconvolution using the Scientific Volume Imagine (SVI) Huygens Professional software package, which achieved improved calculated/theoretical PSFs via complete integration with the Leica LAS-X software and hardware. Detailed instrument acquisition and post-processing settings are available upon request.
Quantification of proteins on sister chromatids without strandness information
All fiber analyses were performed using Java image processing program FIJI. To capture localized distribution of histones and other proteins on chromatin fibers, images were imported into FIJI and line plots were drawn across sister chromatids to measure average fluorescence intensity at the specified region. To measure histone distribution differences between sister chromatids, replication regions longer than 2μm in length were subdivided into 2μm-long segments along the length of the chromatin fiber. Two microns were chosen as this was the average size of individual replicons with 30 minute EdU pulses (Figure S2I). Given the estimated average rate of DNA polymerase to synthesize ~0.5– 2.0 kb DNA per minute79, this 2μm chromatin fiber reflects approximately 15–60 kb of genomic DNA. For replication region shorter than 2μm, the entire length of the region containing resolvable sister chromatids was used to assess differences in histone distribution. To effectively compare histone distribution patterns across multiple data sets, we normalized them using the following strategy: First, we quantified fluorescence levels for histone signals [e.g. old histones (GFP or EGFP), new histones (mKO or mCherry)] for each sister chromatid fiber segment. We then divided fluorescence intensity from the brighter sister chromatid fiber segment by the fluorescence intensity from the less bright sister chromatid fiber segment, to generate a ratio of the relative difference between sister chromatids. This quantification scheme was used for Figure 4.
Classification of different categories of chromatin fibers without strandness information
For old histone on sister chromatids, we classified fibers as symmetric (ratio <1.80), moderately asymmetric (1.8< ratio<2.44), or highly asymmetric (ratio >2.44). We used 2.44 as a standard for calling highly asymmetric, as it is two standard deviations above the average ratio observed for old H2A between sister chromatids. For new histones on sister chromatids, we classified fibers as symmetric (ratio <1.70), moderate asymmetric class (1.70< ratio<2.16), or highly asymmetric (ratio >2.16). We used 2.16 as a standard for calling highly asymmetric, as it is two standard deviations above the average ratio observed for new H2A between sister chromatids.
Quantification of proteins on sister chromatids with strandness information
To capture localized distribution of post-translational histone modifications and other proteins on replicating or newly replicated chromatin fibers, replication region longer than 2μm in length were divided into 2μm-segments, as described above. For replication region shorter than 2μm, the entire resolvable sister chromatids region was used to assess differences in the distribution of post-translational histone modifications. Lagging strand-enriched proteins, such as RPA70 and PCNA, were used as a proxy for lagging strands. To compare distribution of post-translational histone modifications on replicating or newly replicated chromatin fibers, the leading strand (RPA70-depleted or PCNA-depleted strand) was divided by the lagging strand (RPA70-enriched or PCNA-enriched) to generate: Ratio = leading strand protein levels/lagging strand protein levels. Ratios were then log2-transformed such that leading strand enrichment would appear as a positive value and lagging strand enrichment would appear as a negative value.
To retrieve the information of average fold-enrichment: Average fold enrichment = 2^ (Average of log2 values).
This quantification scheme was used to analyze Figure 5 and Supplemental figure 3.
Classification of chromatin fibers with strandness information
To allow for comparison between different data sets of chromatin fibers with leading/lagging strand information, the following criteria were used: Fibers with a log2 (leading strand/lagging strand ratio) ≥ 0.5 were classified as leading strand enriched. Fibers with a −0.5< log2 (leading strand/lagging strand ratio) < 0.5 were classified as symmetric. Fibers with log2 (leading strand/lagging strand ratio) ≤ −0.5 were classified as lagging strand enriched.
Sequential incorporation of EdU and BrdU
EdU labeling was performed using Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Life Science C10640) according to manufacturer’s instructions. Dissected testes were immediately incubated in in Schneider’s Drosophila medium (Gibco™, Catalog# 21720001) with 10 μM EdU for 30 minutes at RT. The testes were subsequently fixed and incubated with primary antibody, as described above. Fluorophore conjugation to EdU was performed along manufacturer’s instructions and followed by secondary antibody incubation.
For double-labelling (EdU followed by BrdU), testes were first incubated for ten minutes in Schneider’s medium containing 10μM of EdU. Samples were then washed three times quickly: Washes entailed re-suspending testes in fresh Schneider’s medium, allowing testes to settle to the bottom of the eppendorf tube, and then quickly pipetting away extra Schneider’s medium. All three washes were completed within two minutes. Testes were then transferred to Schneider’s medium containing 25μM of BrdU analog and incubated for ten minutes at RT, after which, testes were rigorously washed three times as described above. Chromatin fibers were then generated as described above and DNA fibers were then generated as following.
DNA fiber preparation
Testes (or eye imaginal disks) were dissected in fresh Schneider’s Drosophila medium (Gibco™, Catalog# 21720001) at RT and incubated in Schneider’s medium containing 10 μM EdU analogue (Invitrogen™ Click-iT™ EdU Imaging Kit, Catalog# C10340). Testes were incubated for 10 minutes. At the end of the 10 minutes, testes were washed once with Schneider’s medium at RT. Following wash, testes were incubated in the Schneider’s medium containing 25 μM BrdU. Testis were then wash and transferred to dissociation buffer [Dulbecco’s PBS with Mg2+ and Ca2+ with collagenase/dispase (MilliporeSigma®) added to a final concentration of 2mg/ml] in a 37°C water bath for five minutes. Cells were pelleted onto glass slide using cytospin 3000 at 1200RPM for 3 minutes. Cells were then incubated in 50µl conical containing 40µls of DNA lysis buffer (200 mM Tris–HCl, pH 7.5, 50 mM EDTA, 0.5% SDS). This conical was modified with a small (1mm) hole in the bottom to allow lysis buffer to flow out. Slides were incubated for ten minutes in lysis buffer. (Note: Cap was tightly screwed on conical tube to prevent lysis buffer from flowing out). After 10-minute incubation, cap was unscrewed from conical tube, and lysis buffer was allowed to flow out at a slow, steady rate. Slides were dried for 10 minutes at RT, after which they were fixed in freshly prepared methanol:acetic acid (v/v 3:1) for five minutes. Slides were allowed to dry again (~ 10 minutes) after which slides were washed with 1xPBS. Slides were then stained for EdU, BrdU, or anti-ssDNA antibody that recognizes all DNA after HCl treatment80.
DNA fiber staining
Slides were washed with 1xPBS and then transferred to coplin jar containing 50ml of 1M HCL. Slides were incubated for 2 hrs at 37°C to expose BrdU epitope for immunostaining. To minimize cross-reactivity of BrdU antibody to EdU, visualization of EdU was done using Click-iT Plus EdU Alexa Fluor 568 Imaging Kit (Life Science Catalog# C10640) according to manufacturer’s instructions prior to addition of BrdU primary antibody. Following click-chemistry reaction, slides were washed twice in 1xPBS and then blocked for 30 minutes with 2.5% bovine serum albumin (BSA). Following blocking, 200 μls of anti-BrdU antibody was added to the slide. Slides were placed in a humid chamber and incubated overnight at 4°C. Slides were washed in 1xPBS and then blocked for 30 minutes in 5% normal goat serum (NGS). Secondary antibody that recognizes anti-BrdU was added and slides were incubated for 2hrs at RT in a humid chamber. Slides were then washed with 1xPBS and blocked for 30 minutes with 5% BSA. Following blocking, 200μls of anti-ssDNA antibody was added to the slide and incubated in a humid chamber for 1 hr at RT. Slides were washed in 1xPBS and blocked for 30 minutes with 5% NGS. Secondary antibody that recognizes anti-ssDNA was then added and slides were incubated at RT for 30 minutes in a humid chamber. Slides were washed in 1xPBS, dried and mounted with 20μls of Vectashield® mounting medium.
Determining fork movement in chromatin fibers and DNA fibers
Linearized fibers containing multiple replicons were analyzed to determine fork movement patterns in chromatin fibers and DNA fibers. Bidirectional replicons were identified by the presence of the early label (EdU) flanked by the late label (BrdU) at both sides (e.g. Figure 7e, 7h). Unidirectional replicons were identified by an alternating pattern of early (EdU) and late (BrdU) along the length of the fiber (e.g. Figure 7f, 7h). Only fibers with multiple identified replicons were included for data analysis. DNA labels (e.g. DAPI, Yoyo-III or anti-ssDNA) were included to ensure the continuity of the analyzed fibers.
PLA assay
Following incubation with primary antibodies, proximity ligation assay (PLA) was performed using 20 μL of reaction per step per slide according to Sigma-Aldrich Duolink In Situ PLA manufacturer’s instruction (Catalog# DUO92101). In brief, two PLA secondary probes, anti-mouse MINUS (e.g. targeting anti-HA mouse primary) and anti-rabbit PLUS (e.g. targeting either anti-GFP or anti-mKO rabbit primaries), were diluted 1:5 in Ab Diluent buffer provided by manufacturer and incubated overnight at 4 °C. Slides were washed in 1X wash buffer A for 10 minutes, followed by the ligation reaction, in which PLA ligation stock was diluted 1:5 in dH2O with ligase (added at 1:40), followed by incubation for one hour at 37 °C. Slides were then washed in wash buffer A for five minutes, followed by addition of the PLA amplification reaction (1:5 amplification stock and 1:80 polymerase diluted in dH2O) and incubated for two hours at 37 °C. Slides were then washed with 1X wash buffer B for ten minutes, 0.01X wash buffer B for one minute, and 1X PBS for one minute. Following washes, 100μls anti-FasIII (hub cell marker) was then added to slides and allowed to incubate for 30 minutes at RT. Slides were then washed with 1X PBS, after which anti-mouse secondary (Alexa Fluor 405; 1:1000 Molecular Probes) was added to recognize anti-FasIII (labeling hub cells) and anti-PCNA/anti-HA (labeling S phase cells). Slides were incubated for 2hrs at RT and then washed in 1xPBS and mounted. Images were taken using the Zeiss LSM 700 Multiphoton confocal microscope with a 63× oil immersion objectives and processed using Adobe Photoshop software.
Statistics and Reproducibility:
Figure 1d: H4 old GSC/GB data: Shapiro-Wilk normality test P = 0.0013, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of ranks = 883. P < 0.0001. H4 new GB/GSC data: Shapiro-Wilk normality test P = 0.036, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of ranks = 396. P = 0.0201. Old H4 SG1/SG2: Shapiro-Wilk normality test P = 0.003, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of ranks = 181. P = 0.8958. New H4 SG2/SG1: Shapiro-Wilk normality test P = 0.5785, data normally distributed. One sample t-test. Two-tailed test. t = 0.5393 df = 26. P = 0.5943.
Figure 2b: H2A old GSC/GB data: Shapiro-Wilk normality test P = 0.9201, data normally distributed. One sample t-test. Two-tailed test. t =0.1854 df = 19. P = 0.8549. H2A new GB/GSC data: Shapiro-Wilk normality test P = 0.0762, data normally distributed. One sample t-test. Two-tailed test. t=2.474 df = 19. P = 0.0230. Old H2A SG1/SG2: Shapiro-Wilk normality test P = 0.0215, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of signed ranks = 20. P = 0.7285. New H2A SG2/SG1: Shapiro-Wilk normality test P = 0.4955, data normally distributed. One sample t-test. Two-tailed test. t = 0.7504 df = 19. P = 0.4622. New H2A and new H2B show a subtle, but statistically significant enrichment in GB compared to GSC in post-mitotic pairs, likely due to asynchronous ongoing S phase in both GB and GSC nuclei.
Figure 2d: H2B old GSC/GB data: Shapiro-Wilk normality test P = 0.0185, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of signed ranks = −126. P = 0.4049. H2B new GB/GSC data: Shapiro-Wilk normality test P = 0.0020, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of signed ranks = 554. P < 0.0001 Old H2B SG1/SG2: Shapiro-Wilk normality test P = 0.3782, data normally distributed. One sample t-test. Two-tailed test. t = 0.7555 df = 35. P = 0.4550. New H2B SG2/SG1: Shapiro-Wilk normality test P < 0.0001, data not normally distributed Wilcoxon Signed Rank Test. Two-tailed test. Sum of signed ranks = 68. P = 0.6028. New H2B show a subtle, but statistically significant enrichment in GB compared to GSC in post-mitotic pairs, likely due to asynchronous ongoing S phase in both GB and GSC nuclei.
Figure 3e: Chi-square test: Confocal embryo vs. confocal testis: The chi-square statistic is 3.9078. The p-value is .048062. Confocal testis vs. airyscan testis: The chi-square statistic is 14.6415. The p-value is .00013. Airyscan testis vs. STED testis: The chi-square statistic is 22.9128. The p-value is < .00001
Figure 4d: Mann-Whitney U test: Old H3-eGFP asymmetry between sisters vs. Old H2A-eGFP asymmetry between sisters. Mann-Whitney U = 841.5; P < 0.0001; Two-tailed test.
Figure 4e: Mann-Whitney U test: New H3mCherry asymmetry between sisters vs. Old H2AmCherry asymmetry between sisters. Mann-Whitney U = 790.5; P = 0.0004; Two-tailed test.
Figure 4f: Chi-squared test: Highly asymmetric H3 old vs. H2A old. The chi-square statistic is 25.2739. The p-value is < .00001. Moderately symmetric H3 old vs. H2A old. The chi-square statistic is 1. The p-value is .31732. Symmetric H3 old vs. H2A old. The chi-square statistic is 25.5911. The p-value is < .00001.
Figure 4g: Chi-squared test: Highly asymmetric H3 new vs. H2A new. The chi-square statistic is 8.5965. The p-value is .003368. Moderately symmetric H3 new vs. H2A new. The chi-square statistic is 0.819. The p-value is .365483 Symmetric H3 new vs. H2A new. The chi-square statistic is 9.9691. The p-value is .001592.
Figure 5b: H3K27me3 Enrichment on RPA70-depleted sister chromatids: Shapiro-Wilk normality test P < 0.0028, data not normally distributed. Wilcoxon Signed Rank Test. Two-tailed test. Sum of signed ranks = 1235. P < 0.0001.
Figure 5e: H3K27me3 Enrichment on PCNA-depleted sister chromatids: Shapiro-Wilk normality test P = 0.8090, data normally distributed. One sample t-test. Two-tailed test t = 7.075 df = 40. P < 0.0001.
Figure 6b: Ligase + H3-GFP GSC Shapiro-Wilk normality test P = 0.0722, data normally distributed. Ligase + H3-mKO GSC Shapiro-Wilk normality test P = 0.9059; data normally distributed. Ligase + H3-GFP SG Shapiro-Wilk normality test P = 0.5180, data normally distributed. Ligase + H3-mKO SG Shapiro-Wilk normality test P = 0.0404; data not normally distributed. Kruskal-Wallis test: medians vary significantly; P = 0.0004. Kruskal-Wallis multiple comparisons of non-parametric data with Dunn’s multiple comparisons corrections test significant groups: Ligase + H3-GFP GSC vs. Ligase +H3-mKO GSC P < 0.01. Ligase-mKO GSC vs. Ligase + H3-mKO SG P < 0.001. Remaining comparisons were not statistically significantly different; ns = P > 0.05.
Figure 6d: PCNA + H3-GFP GSC Shapiro-Wilk normality test P = 0.0015, data not normally distributed. PCNA + H3-mKO GSC Shapiro-Wilk normality test P = 0.0842; data normally distributed. PCNA + H3-GFP SG Shapiro-Wilk normality test P = 0.0004, data not normally distributed. PCNA + H3-mKO SG Shapiro-Wilk normality test P < 0.0001; data not normally distributed. Kruskal-Wallis test: medians vary significantly; P = 0.0008. Kruskal-Wallis multiple comparisons of non-parametric data with Dunn’s multiple comparisons corrections test significant groups: PCNA + H3-GFP GSC vs. PCNA +H3-mKO GSC P < 0.001. PCNA-mKO GSC vs. PCNA + H3-mKO SG P < 0.05. PCNA-mKO GSC vs. PCNA + H3-GFP SG P < 0.05. Remaining comparisons were not statistically significantly different; ns = P > 0.05.
Figure 7g: Chi-squared test: Testis vs. Soma Chromatin fibers. The chi-square statistic is 8.0091. The p-value is .004654
Figure 7k: Chi-squared test: Testis vs. Soma DNA fibers. Unidirectional frequency: The chi-square statistic is 5.3263. The p-value is .021006. Asymmetric bidirectional frequency: The chi-square statistic is 0.0014. The p-value is .970079. Symmetric bidirectional frequency: The chi-square statistic is 4.5468. The p-value is .03298.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data behind graphs shown in Fig. 1d, Fig. 2b,d and Suppl. Fig. 1b are available in Suppl.Tables 1 and 2. Other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank B. Shelby and E. Wieschaus for the RPA70-GFP fly line. We thank E. Moudrianakis, A. Spradling, J. Berger, M. Van Doren, R. Johnston and X.C. lab members for suggestions. We thank Barbara Mellone and Sharon Pavanacherry and Lydia Sohn for help with chromatin fiber technique. We thank Johns Hopkins Integrated Imaging Center for confocal imaging and Carnegie Institute Imaging Center for STED microscopy work. Supported by NIH 5T32GM007231 and F31GM115149–01A1 (M.W.), NIH R01GM112008 (J.X.), NIH R01GM33397 (J.G.), NIH R01GM112008, R35GM127075, the Howard Hughes Medical Institute, the David and Lucile Packard Foundation, and Johns Hopkins University startup funds (X.C.)
Footnotes
Competing interests: The authors declare no competing interests.
REFERENCES
- 1.Betschinger J & Knoblich JA Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol 14, R674–85 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Clevers H Stem cells, asymmetric division and cancer. Nat Genet 37, 1027–8 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Inaba M & Yamashita YM Asymmetric stem cell division: precision for robustness. Cell Stem Cell 11, 461–9 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Morrison SJ & Kimble J Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–74 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Kahney EW, Ranjan R, Gleason RJ & Chen X Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harb Symp Quant Biol 82, 305–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spradling A, Fuller MT, Braun RE & Yoshida S Germline stem cells. Cold Spring Harb Perspect Biol 3, a002642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran V, Lim C, Xie J & Chen X Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338, 679–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J et al. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell 163, 920–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alabert C & Groth A Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13, 153–67 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Bellush JM & Whitehouse I DNA replication through a chromatin environment. Philos Trans R Soc Lond B Biol Sci 372(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzluff WF, Wagner EJ & Duronio RJ Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9, 843–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S & Henikoff S Replicating Nucleosomes. Sci Adv 1(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasseur P et al. Dynamics of Nucleosome Positioning Maturation following Genomic Replication. Cell Rep 16, 2651–2665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad K & Henikoff S No strand left behind. Science 361, 1311–1312 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Serra-Cardona A & Zhang Z Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity. Trends Biochem Sci 43, 136–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petryk N et al. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Seale RL Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell 9, 423–9 (1976). [DOI] [PubMed] [Google Scholar]
- 18.Seidman MM, Levine AJ & Weintraub H The asymmetric segregation of parental nucleosomes during chrosome replication. Cell 18, 439–49 (1979). [DOI] [PubMed] [Google Scholar]
- 19.Roufa DJ & Marchionni MA Nucleosome segregation at a defined mammalian chromosomal site. Proc Natl Acad Sci U S A 79, 1810–4 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C et al. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snedeker J, Wooten M & Chen X The Inherent Asymmetry of DNA Replication. Annu Rev Cell Dev Biol 33, 291–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita YM, Jones DL & Fuller MT Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–50 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Young NL, Dimaggio PA & Garcia BA The significance, development and progress of high-throughput combinatorial histone code analysis. Cell Mol Life Sci 67, 3983–4000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu M et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–8 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Jackson V & Chalkley R A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell 23, 121–34 (1981). [DOI] [PubMed] [Google Scholar]
- 26.Russev G & Hancock R Formation of hybrid nucleosomes cantaining new and old histones. Nucleic Acids Res 9, 4129–37 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katan-Khaykovich Y & Struhl K Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci U S A 108, 1296–301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson V Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27, 2109–20 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Kimura H Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst) 4, 939–50 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Annunziato AT Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280, 12065–8 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Cohen SM, Chastain PD 2nd, Cordeiro-Stone M & Kaufman DG. DNA replication and the GINS complex: localization on extended chromatin fibers. Epigenetics Chromatin 2, 6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad K & Henikoff S Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A 99 Suppl 4, 16477–84 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blower MD, Sullivan BA & Karpen GH Conserved Organization of Centromeric Chromatin in Flies and Humans. Developmental Cell 2, 319–330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKnight SL & Miller OL Jr. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12, 795–804 (1977). [DOI] [PubMed] [Google Scholar]
- 35.Hell SW & Wichmann J Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 19, 780–2 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Sivaguru M et al. Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc Res Tech 81, 101–114 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Ke MT et al. Super-Resolution Mapping of Neuronal Circuitry With an Index-Optimized Clearing Agent. Cell Rep 14, 2718–32 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Van Doren M, Williamson AL & Lehmann R Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8, 243–6 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Blythe SA & Wieschaus EF Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wold MS Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Alabert C et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29, 585–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKearin DM & Spradling AC bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev 4, 2242–51 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Sogo JM, Stahl H, Koller T & Knippers R Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol 189, 189–204 (1986). [DOI] [PubMed] [Google Scholar]
- 44.Leffak IM, Grainger R & Weintraub H Conservative assembly and segregation of nucleosomal histones. Cell 12, 837–45 (1977). [DOI] [PubMed] [Google Scholar]
- 45.Riley D & Weintraub H Conservative segregation of parental histones during replication in the presence of cycloheximide. Proc Natl Acad Sci U S A 76, 328–32 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weintraub H Cooperative alignment of nu bodies during chromosome replication in the presence of cycloheximide. Cell 9, 419–22 (1976). [DOI] [PubMed] [Google Scholar]
- 47.Annunziato AT Assembling chromatin: the long and winding road. Biochim Biophys Acta 1819, 196–210 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Szenker E, Ray-Gallet D & Almouzni G The double face of the histone variant H3.3. Cell Res 21, 421–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henikoff S & Smith MM Histone variants and epigenetics. Cold Spring Harb Perspect Biol 7, a019364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin C & Felsenfeld G Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21, 1519–29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pomerantz RT & O’Donnell M What happens when replication and transcription complexes collide? Cell Cycle 9, 2537–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiengwe C et al. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep 2, 185–97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGlynn P, Savery NJ & Dillingham MS The conflict between DNA replication and transcription. Mol Microbiol 85, 12–20 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Yarosh W & Spradling AC Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev 28, 1840–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krude T, Christov CP, Hyrien O & Marheineke K Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci 122, 2836–45 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Lebofsky R & Bensimon A DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol 25, 6789–97 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanojcic S et al. Single-molecule analysis of DNA replication reveals novel features in the divergent eukaryotes Leishmania and Trypanosoma brucei versus mammalian cells. Sci Rep 6, 23142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin-Parras L, Hernandez P, Martinez-Robles ML & Schvartzman JB Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol 220, 843–53 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Marheineke K, Hyrien O & Krude T Visualization of bidirectional initiation of chromosomal DNA replication in a human cell free system. Nucleic Acids Res 33, 6931–41 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munden A et al. Rif1 inhibits replication fork progression and controls DNA copy number in Drosophila. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalgaard JZ & Klar AJ A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15, 2060–8 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivessa AS, Zhou JQ & Zakian VA The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100, 479–89 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Sasaki T, Sawado T, Yamaguchi M & Shinomiya T Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol Cell Biol 19, 547–55 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buck SW, Sandmeier JJ & Smith JS RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111, 1003–14 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Hand R Regulation of DNA replication on subchromosomal units of mammalian cells. J Cell Biol 64, 89–97 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huberman JA & Tsai A Direction of DNA replication in mammalian cells. J Mol Biol 75, 5–12 (1973). [DOI] [PubMed] [Google Scholar]
- 67.Palmigiano A et al. PREP1 tumor suppressor protects the late-replicating DNA by controlling its replication timing and symmetry. Sci Rep 8, 3198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soumillon M et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3, 2179–90 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Parisi M et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol 5, R40 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references:
- 70.Feng L, Shi Z & Chen X Enhancer of polycomb coordinates multiple signaling pathways to promote both cyst and germline stem cell differentiation in the Drosophila adult testis. PLoS Genet 13, e1006571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hime GR, Brill JA & Fuller MT Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci 109 ( Pt 12), 2779–88 (1996). [DOI] [PubMed] [Google Scholar]
- 72.Kolb HC, Finn MG & Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl 40, 2004–2021 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Moses JE & Moorhouse AD The growing applications of click chemistry. Chem Soc Rev 36, 1249–62 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Koster DA, Crut A, Shuman S, Bjornsti MA & Dekker NH Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell 142, 519–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang JC Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3, 430–40 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Kuzminov A When DNA Topology Turns Deadly - RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super)Twists in the Replication-Transcription Conflict. Trends Genet 34, 111–120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.LaMarr WA, Yu L, Nicolaou KC & Dedon PC Supercoiling affects the accessibility of glutathione to DNA-bound molecules: positive supercoiling inhibits calicheamicin-induced DNA damage. Proc Natl Acad Sci U S A 95, 102–7 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ljungman M & Hanawalt PC Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci U S A 89, 6055–9 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Techer H et al. Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol 425, 4845–55 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Rezaei Poor Kardost R, Billing PA & Voss EW Jr. Generation and characterization of three murine monoclonal nucleotide binding anti-ssDNA autoantibodies. Mol Immunol 19, 963–72 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data behind graphs shown in Fig. 1d, Fig. 2b,d and Suppl. Fig. 1b are available in Suppl.Tables 1 and 2. Other data that support the findings of this study are available from the corresponding author upon request.