Abstract
Organismal responses to light:dark cycles can result from two general processes: (i) direct response to light or (ii) a free-running rhythm (i.e., a circadian clock). Previous research in cnidarians has shown that candidate circadian clock genes have rhythmic expression in the presence of diel lighting, but these oscillations appear to be lost quickly after removal of the light cue. Here, we measure whole-organism gene expression changes in 136 transcriptomes of the sea anemone Nematostella vectensis, entrained to a light:dark environment and immediately following light cue removal to distinguish two broadly defined responses in cnidarians: light entrainment and circadian regulation. Direct light exposure resulted in significant differences in expression for hundreds of genes, including more than 200 genes with rhythmic, 24-hour periodicity. Removal of the lighting cue resulted in the loss of significant expression for 80% of these genes after one day, including most of the hypothesized cnidarian circadian genes. Further, 70% of these candidate genes were phase shifted. Most surprisingly, thousands of genes, some of which are involved in oxidative stress, DNA damage response, and chromatin modification, had significant differences in expression in the 24 hours following light removal, suggesting that loss of the entraining cue may induce a cellular stress response. Together, our findings suggest that a majority of genes with significant differences in expression for anemones cultured under diel lighting are largely driven by the primary photoresponse rather than a circadian clock when measured at the whole animal level. These results provide context for the evolution of cnidarian circadian biology and help to disassociate two commonly confounded factors driving oscillating phenotypes.
Keywords: Nematostella, circadian, gene expression, photoperiod, transcriptomics
INTRODUCTION
Light is a principal environmental cue that shapes biological communities by influencing the behavior, physiology, and gene expression of individual organisms. For organisms living in photic environments, the presence, duration, and intensity of light is the most predictable cue for shaping time-dependent responses, whether they be at periods of hours, days, or seasons (Bradshaw & Holzapfel, 2007; Dunlap, Loros, & DeCoursey, 2004; Edmunds, 1988). Over evolutionary time, these responses to light have resulted in convergent behavioral and physiological phenotypes including diurnal and nocturnal activity patterns, reproductive windows, and migration patterns, to name a few (A. Brady, Hilton, & Vize, 2009; Gwinner, 1996; Mercier & Hamel, 2010; Tosches, Bucher, Vopalensky, & Arendt, 2014). These light-dependent phenotypes measured at the organismal level are the product of the differential expression of molecular pathways. How light exposure is translated by organisms into these diverse phenotypes has been a central focus for understanding how shifts in the environment can result in different phenotypic outputs through particular changes in gene expression (Cheng, Tsunenari, & Yau, 2009; Fernandes, Fero, Driever, & Burgess, 2013; Roenneberg & Foster, 1997).
Organismal responses to a diel, or daily, light cue can result from two general processes: direct response to the exogenous light or a free-running rhythmic response due to an endogenous time keeper (circadian clock; Berson, Dunn, & Takao, 2002; Miyamoto & Sancar, 1998; D. L. Williams, 2016). Direct responses occur only when an organism responds post-illumination where the light impacts particular cells directly and the response does not continue in the absence of repeated light exposures. Direct light responses occur through ocular (Freedman et al., 1999; Lamb, Collin, & Pugh, 2007) or extra-ocular (Edwards et al., 2008; Porter, 2016) photosensors that may then transmit the light responses to other cells or tissues typically through neural cells. These responses may also occur in any cell exposed to light, which would be particularly common in translucent organisms. On the other hand, circadian clocks generate free-running rhythms as a result of molecular networks that maintain oscillations in phenotype after the entraining cue is removed (Dunlap, 1999; Hardin, 2006). In various animal species, circadian clocks are generally known to be transcription-translation feedback loops that are centrally located in neural cells in the brain or anterior structures (e.g., antennae in some insects) and maintain a free-running period of approximately 24 hours (Dunlap, 1999; Shearman et al., 2000). The central circadian clock can regulate the periodicity of additional tissues through hormonal or other endocrine signaling mechanisms (Gamble, Berry, Frank, & Young, 2014; J. Williams & Sehgal, 2001). Categorization between direct light responses and the endogenous circadian clock can be challenging because these two responses can be causally connected. For example, changes in the timing or duration of the daily light cue can influence the timing of the circadian clock and result in resetting (jet lag; Davis & Mirick, 2006; Mendlewicz, 2009).
Species from early diverging animal phyla have been studied to characterize the mechanisms of photoreception and signal transduction as well as the potential for a circadian clock (Plachetzki, Fong, & Oakley, 2010). Research with sponges has shown that various species have light-dependent behavior (Leys, Cronin, Degnan, & Marshall, 2002) as well as the molecular components for photoreception (cryptochromes; Rivera et al., 2012), components of the classic bilaterian circadian clock (Jindrich et al., 2017; Simionato et al., 2007), and cyclical oscillations in gene expression of these clock genes under diel lighting conditions (Jindrich et al., 2017). Similarly, light has a significant impact on the behavior of ctenophores and previous studies have shown they have various light sensing proteins encoded in their genome (Schnitzler et al., 2012), but the potential for a circadian clock has not yet been studied (Reitzel et al., 2014). Cnidarians have emerged as an informative group of animals to study the evolution of the circadian clock and the role of daily light exposure on behavior, physiology, and gene expression (Hoadley, Vize, & Pyott, 2016; Reitzel, Tarrant, & Levy, 2013). It has been known for decades that light:dark cycles impact the reproduction, movement, and physiology of various cnidarian species (Chalker, Barnes, Dunlap, & Jokiel, 1988). More recently, phylogenomic studies have shown cnidarians have many of the genes that compose the core bilaterian clockwork (Levy et al., 2007; Reitzel, Behrendt, & Tarrant, 2010; Vize, 2009), most of which are expressed in an oscillating pattern under diel lighting conditions (A. K. Brady, Snyder, & Vize, 2011; Oren et al., 2015; Reitzel et al., 2010). Additional studies, primarily with corals, have also shown that hundreds of genes are differentially expressed under light:dark (A. K. Brady et al., 2011; Ruiz-Jones & Palumbi, 2015), and lunar (Oldach, Workentine, Matz, Fan, & Vize, 2017) conditions, many of which appear to dissipate once the entraining cue is removed (A. K. Brady et al., 2011; Peres et al., 2014). It remains unclear how much of the differential gene expression is a product of a direct light response or from an endogenous oscillator (Oldach et al., 2017).
In this study, we utilized comparative transcriptomics to investigate the role of direct light exposure and endogenous circadian oscillations on the gene expression of the starlet sea anemone, Nematostella vectensis (hereafter referred to as just Nematostella). Nematostella has developed into a focal species to determine the potential mechanisms for responses to diel lighting and the circadian clock in cnidarians (Hendricks, Byrum, & Meyer-Bernstein, 2012; Maas, Jones, Reitzel, & Tarrant, 2016). This nocturnal species has clear circadian behavior and physiology with differential activity in diel lighting that is maintained upon removal of the entraining cue. Nematostella has orthologs (bHLH-PAS members Clock and Cycle/Bmal) or homologs (cryptochromes, PAR-bZIP) to genes centrally involved in the bilaterian circadian clock, many of which have oscillating expression under light:dark conditions (Reitzel et al., 2010; Reitzel et al., 2013). In addition, hundreds of genes show differential expression under diel lighting (Leach, Macrander, Peres, & Reitzel, 2018; Oren et al., 2015), but many of these have no evidence for differential expression when animals are cultured under extended periods of darkness (Leach et al., 2018; Peres et al., 2014). Together, these previous studies have shown this species has a diverse transcriptional response to light but the maintenance of these oscillations in gene expression are largely unknown, except after long periods of time (>20 days; Leach et al., 2018; Peres et al., 2014; Reitzel et al., 2010; Reitzel et al., 2013). Understanding how time-dependency of loss of rhythmic expression in genes with removal of the entraining light cue is thus important to discern between direct light effects and those resulting from a free-running circadian clock.
Here, we measure whole organism gene expression changes in Nematostella entrained to a light:dark environment and immediately following light cue removal to distinguish two broadly defined responses in cnidarians: (i): light entrainment and (ii) circadian regulation. By comparing transcriptional patterns before and after exogenous light removal, we report hundreds of cycling light responsive genes including those predicted to be involved in a core clock mechanism, followed by a stress response in constant conditions. Finally, we compared co-expressed genes over time and in each light regime to reveal that light condition, rather than time-of-day, most significantly influences gene expression.
MATERIALS AND METHODS
Culturing and entrainment of Nematostella vectensis
Adult sea anemones from an outbred population of the genome strain (Maryland, USA) were cultured in glass dishes containing 15 parts per thousand (ppt) artificial seawater (ASW). These animals were fed haphazardly three times per week with freshly hatched Artemia nauplii and the water was changed weekly. Animals were maintained at these conditions for ≥1 month in an incubator at 25°C in one of two treatment groups: either a diel light cycle using full spectrum lights (MINGER) or in constant long-term darkness (DD; Fig. S1). Diel conditions were defined as cycles of 12-hour light: 12-hour dark. Zeitgeber time, or ZT = 0/ “lights on” was at 7:00 AM and ZT = 12/ “lights off” was at 7:00 PM.
Light-removal experiment
Individual sea anemones (2–3 cm in length) were sampled from both treatment groups (diel and long-term darkness; Fig. S1) in parallel every 4 hours over a 3-day period, then immediately preserved in RNAlater (Ambion). Four biological replicates were sampled at each time point from both treatment groups for a total of 136 individual samples (n = 68 per treatment group). To measure the time-dependent effects of light removal on gene expression in Nematostella, the light cue was removed from the diel light cycle group after the first 24 hours (at ZT = 0 on the second day) and sampling continued for 44 additional hours. This sampling regime effectively created three treatment subgroups from diel entrained anemones: day 1 of the experiment or ‘light:dark’ (LD), day 2 of the experiment or ‘light removal day 1’ (LR1; i.e., the first day post-light removal), and day 3 of the experiment or ‘light removal day 2’ (LR2; i.e., the second day post-light removal). Thus, samples from the first 24 hours of the experiment will subsequently be referred to as light treatment ‘LD’, samples from the following 24 hours of the experiment will be referred to as LR1, and samples from the last 20 hours of the experiment will be referred to as LR2. Conditions for animals in the long-term darkness (DD) treatment group remained constant during each sampling day (DD1, DD2, and DD3; Fig. S1, Supporting information). We were unable to sample a sixth time at the end of sampling day 3 because of insufficient animals in some treatments, hence why there are only 17 time points over 68 hours rather than a full 72-hour time course (Fig. S1).
Tag-based RNA library preparation, sequencing, and processing
Total RNA was isolated from 136 samples (4 biological replicates * 17 time points * 2 treatment groups) using the RNAqueous kit (Ambion) according to the manufacturer’s protocol. Briefly, after pipetting off and discarding RNAlater from each sample, whole animals were lysed by pipetting in lysis buffer for <2 minutes, washed 2–3 times, and eluted on a column. Genomic DNA was removed using DNA-free kit (Invitrogen), and RNA was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). RNA was shipped for tag-based library preparation at the University of Texas at Austin’s Genomic Sequencing and Analysis Facility (GSAF) as in Meyer et al. (2011) and adapted for Illumina HiSeq 2500. Briefly, total RNA was heat-fragmented and then reverse transcribed into first-strand cDNA. The cDNA was purified using AMPure beads, and PCR-amplified for 18 cycles. Unique Illumina barcodes were added in an additional PCR step for indexing of each sample. After an additional purification step, libraries were pooled, spot checked for quality on a Bioanalyzer (Agilent and Pico), and size-selected using BluePippin (350–550bp fragments). A full version of the library preparation protocol can be found at https://github.com/z0on/tag-based_RNAseq.
Data processing pipeline
Raw sequence data (100 bp, single-end) were delivered from the UT Austin GSAF. Raw reads were trimmed and quality-filtered using the FastX-toolkit (Pearson, Wood, Zhang, & Miller, 1997). Trimmed reads were mapped against the Nematostella Vienna transcriptome (see ‘Data Accessibility’ for link to gene models) using the Bowtie2 aligner (Langmead & Salzberg, 2012) and a read-counts-per-gene file was generated retaining only reads mapping to a single gene (Table S1, Supporting information). Lastly, counts were imported into the R environment for all downstream statistical analysis (R3.5.0, R Core Team 2015).
Identification of cycling genes
Oscillating transcripts were identified with JTK_CYCLEv3.1 in R (Hughes, Hogenesch, & Kornacker, 2010), which determines p-values based on Kendall’s rank correlation coefficient and effectively distinguishes rhythmic and non-rhythmic patterns. JTK_Cycle p-values were Bonferroni-adjusted for multiple testing. Raw counts from 4-hour sampling intervals across all treatment subgroups were used as input data in the in JTK_Cycle script, and the ‘period’ parameter was set to ‘5:7’ to identify genes cycling every 20–28 hours. We compared peak expression times for genes with significantly oscillating expression over multiple days to identify potential shifts in peak transcription with light removal.
Identification of differentially expressed genes (DEGs)
Normalization and differential expression analysis of read counts was performed using a negative binomial generalized linear model in the R package DESeq2 (Fig. S2A–C, Supporting information; Love, Huber, & Anders, 2014). Transcripts with low abundances (mean count <3) were filtered to improve the rate of differential gene discovery as implemented in the DESeq2 pipeline. The arrayQualityMetrics package (Kauffmann, Gentleman, & Huber, 2009) was used to detect outlier transcripts with counts significantly higher than the rest of the total counts, and were discarded from subsequent analysis (Fig. S3A, Supporting information). DESeq2 normalized count data were regularized log transformed using the rlog function. Normalized and rlog transformed counts were used for principal coordinate analysis based on Manhattan distances and significance was evaluated using the vegan package in R (Fig. S2, Supporting information; Dixon, 2003). Gene expression heatmaps were created using the pheatmap package in R with hierarchical clustering of expression patterns (Kolde, 2018). Gene expression graphs were generated with the ggplot2 function in R (Wickham, 2009).
We performed pairwise contrasts between each treatment subgroup, and between each timepoint using Wald tests in DESeq2. P-values were calculated using the Benjamini-Hochberg procedure and false discovery rate (FDR) adjusted for multiple testing. The differentially expressed gene (DEG) lists from each contrast, including adjusted and unadjusted p-values and log2 fold changes, were used for downstream analyses.
Gene ontology (GO) enrichment analysis
Functional summaries of DEGs from each contrast were determined by rank based Gene Ontology (GO) enrichment analysis, using signed, unadjusted log-transformed p-values (positive if up-regulated, negative if down-regulated) with the GO_MWU (https://github.com/z0on/GO_MWU) package. This method utilizes the Mann-Whitney U (MWU) test and measures whether each GO term is significantly enriched in up- or down-regulated genes based on their delta rank (quantitative shift in rank) rather than looking for GO terms among “significant” genes only.
Gene co-expression
We next identified groups (“modules”) of highly correlated genes from each contrast in an unbiased way using weighted gene correlation network analysis (WGCNA; Langfelder & Horvath, 2008). We used genes with an unadjusted p-value <0.1 (5,678 genes) determined by the generalized linear model in DESeq2. The resulting modules were then related to external traits (i.e., sampling day/treatment subgroup, individual, and time) using the eigengene network methodology (Langfelder & Horvath, 2008). This method does not use information regarding how the samples were distributed within experimental conditions to ensure that the module eigengenes correlate with gene expression patterns that reflect biological processes. A sample network identified outlying samples (n = 5, Fig. S3A, Supporting information) with a connectivity score less than −2.5 and were removed from the analysis. A signed co-expression network was built using a soft threshold power of 6 (Fig. S3B, Supporting information) and modules were merged if their eigengene expression correlated with the Pearson correlation coefficient greater than 0.42 (Fig. S3C, Supporting information). Each module’s eigengene expression (the first principal component of all of the genes within that module) was correlated to the day sampling occurred (i.e., treatment subgroup), biological replicate, and time point (Fig. S4, Supporting information). Significant correlations between the membership of the genes in each module and their significance indicates a strong association of the module with a trait (i.e. genes in a particular module are positively or negatively associated with day of sampling, individual, or time; Fig. S5, Supporting information). Module eigengenes were functionally characterized with the GO_MWU package using a transcriptome-wide Fishers exact test for the genes in each module (https://github.com/z0on/GO_MWU).
RESULTS
Transcriptome sequencing produced >396 million raw reads, with an average of 2.9 million reads per sample (each time point had 4 biological replicates, barcoded and sequenced individually). After quality filtering and removal of PCR duplicates, an average of 889,112 reads per sample remained (Table S2, Supporting information). After trimming and deduplication of transcripts, the reads were mapped to the Vienna Nematostella transcriptome (see ‘Data Accessibility’ section for link) with an average mapping efficiency of 74.5%. The mapped reads were converted into a counts per sample table, representing a total of 24,392 genes (Table S1, Supporting information). A generalized linear model in DESeq2 identified unique DEGs normalized with sampling day/treatment subgroup as the covariate. In order to explore gene expression patterns specific to each treatment subgroup, pairwise comparisons between each treatment subgroup were run using Wald statistics to contrast each subgroup to the other two (LD, LR1, LR2). A principal coordinate analysis of the entire rlog transformed dataset separated samples by subgroup and showed distinct clustering of LD, LR1, and LR2 (Fig. 1A). Further, consecutive time points during day and night sampling events tended to cluster within subgroups.
Fig. 1.
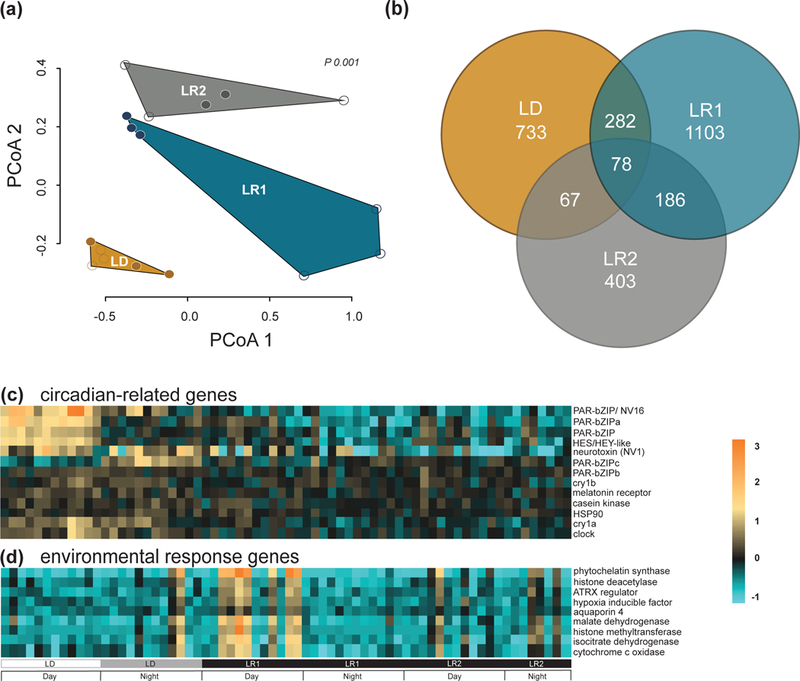
Gene expression of Nematostella vectensis in diel and constant conditions. (A) Principal coordinate analysis based on Manhattan distances. Clusters are grouped by diel treatment subgroups (light:dark or LD, light removal day 1 or LR1, light removal day 2 or LR2) and by time of day (open circles – ‘day’, closed circles – “night”; pPERMANOVA = 0.001). (B) Venn diagram of the total number of differentially expressed genes resulting from each pairwise comparison between the diel treatment sugbroups (LD, LR1, LR2) and control animals kept in long-term constant darkness determined by DESeq2 (Benjamini-Hochberg FDR <0.01). (C) Heatmap of circadian related genes and (D) environmental response genes differentially expressed in light:dark conditions (LD), and after one or two days of light removal (LR1, LR2) determined by DESeq2 (Benjamini-Hochberg FDR <0.01). The experiment key at the bottom identifies the subgroups of the diel light treatment and the ‘day’ and ‘night’ periods of each 24-hour cycle for both C and D. Each row of the heatmaps represent a single annotated gene, and each column represents a single individual in each time point (n = 4 per time point). The color scale is log2 fold change.
Cycling gene expression
To quantify how many genes were cycling on each sampling day, we analyzed the normalized counts by subgroup with non-parametric JTK_Cycle (Table S3, Supporting information). In total, we identified 1,073 cycling genes over the 3-day time course. The number of genes with signatures of cycling differed between subgroups: 228 genes were identified to be significantly cycling (period = 24; p-value <0.01) in LD, 865 genes in LR1, and only 40 genes in LR2. Fifty-two genes were shared between LD and LR1, eight genes were shared between LR1 and LR2, and no genes were shared between LD and LR2 (Fig. S6, Supporting information).
Cycling genes in LD
To explore the genes JTK_Cycle identified as cycling on the first day of the experiment prior to light removal, we isolated genes from the LD subgroup with a significant period of 24 hours (228 genes; p-value <0.01). There were 487 cycling genes at p <0.05, and 35 cycling genes at p <0.001. Only a subset of the 24-hour cycling genes from LD continued to oscillate on the second day and third days, after the light cue was removed (54 genes from subgroups LR1 and LR2). The remaining 174 genes (p-value <0.01) uniquely cycling in LD included light responsive genes (e.g., cryptochromes and rhodopsin) and signal transduction genes (e.g., protein kinase C, and G-protein couple receptor). Several genes previously reported to exhibit rhythmic expression over a diel light cycle in cnidarians (A. K. Brady et al., 2011; Hemond & Vollmer, 2015; Hoadley, Szmant, & Pyott, 2011; Oren et al., 2015) showed expression patterns consistent with a 24-hour rhythm in LD (Fig. 2; Table 1); Clock, Cry1a, Cry1b, PAR-bZIPa, PAR-bZIPc, a Hes/Hey-like transcription factor helt, and a putative clock-interacting circadian pacemaker homolog (CiPC) were expressed with a significant circadian period of 24 hours and all had daytime peaks in expression except PAR-bZIPc (ZT = 18) and CiPC (ZT = 24; Table 1). PAR-bZIPb had a significant period of 28 hours with peak expression in mid-afternoon (Table 1, Fig. 2). The transcription factor helt and the CiPC homolog each had a significant period of 24 hours and were previously identified to have diel expression under LD conditions in cnidarians (Oren et al., 2015; Shoguchi, Tanaka, Shinzato, Kawashima, & Satoh, 2013). Consistent with those earlier studies, helt and CiPC expression peaked mid-morning (ZT = 6) and during subjective night (ZT = 24), respectively (p-value <0.01; Table 1). The subset of significantly cycling genes identified in both the LD and LR1 subgroups included environmental response genes (e.g. peroxiredoxin 5, thioredoxin), genes involved in metabolic processes (e.g., malate dehydrogenase, adenylate cyclase, aspartate aminotransferase) and transcription (e.g., six homeobox). Upon light removal, all but one gene (NVE15806; unidentified protein) phase shifted peak expression. Most of these genes (69%) peaked later in the day, but the remaining genes peaked earlier (28%).
Fig. 2.
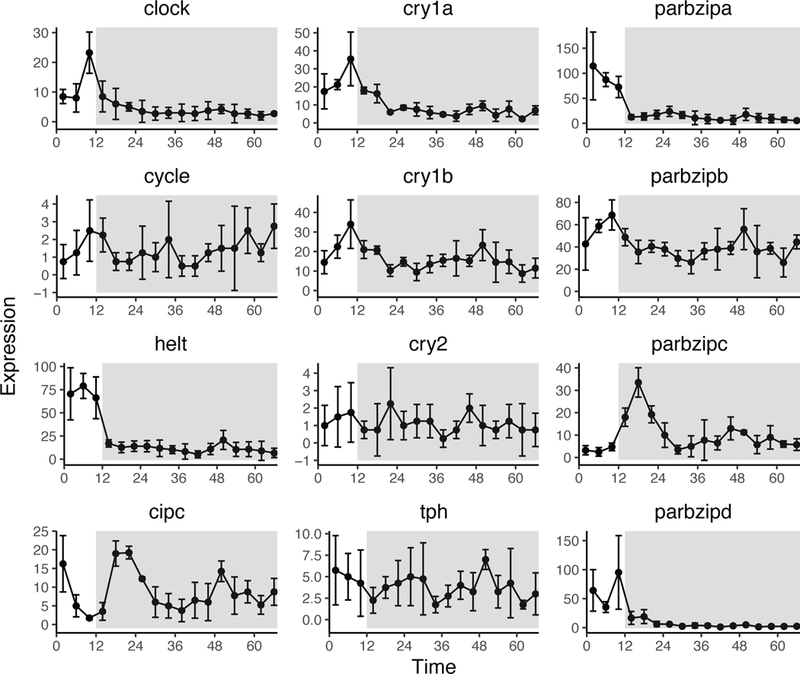
Candidate circadian gene expression profiles over time. Each graph plots a single gene’s expression over the three-day sampling time course for light:dark entrained anemones. Each data point represents the mean of four individually sequenced replicates. Error bars are calculated from the standard deviation for each data point (n=4). The relative expression (y-axis) is shown for each gene over the sampling period (x-axis). The grey shading in each plot indicates light removal.
Table 1:
Cycling and differential expression of candidate circadian clock genes resulting from DESeq2 and JTK_Cycle analysis of subgroups.
| ID | Annotation | DESeq2 p-value* | JTK_Cycle periodicity┼ | Peak Expression⍑ |
||
|---|---|---|---|---|---|---|
| LD | LR1 | LR2 | ||||
| NVE2080 | Clock | 1.20459E-08 | 24 (0.07) | ZT10 | - | - |
| NVE1138 | Cry1a | 1.66397E-18 | 24* | ZT10 | ZT4 | ZT4 |
| NVE24214 | Cry1b | 0.03511226 | 24* | ZT12 | ZT20 | ZT4 |
| NVE14677 | PAR-bZIPa | 1.27133E-12 | 24* | ZT6 | ZT2 | ZT6 |
| NVE20636 | PAR-bZIPb | 0.052867218 | 28* | ZT6 | ZT20 | ZT4 |
| NVE8107 | PAR-bZIPc | 0.007379136 | 24* | ZT18 | ZT22 | ZT6 |
| NVE8679 | helt | 1.40066E-11 | 24* | ZT6 | ZT4 | ZT4 |
| NVE8085 | PAR-bZIPd | 2.2241E-52 | 24* | ZT10 | ZT4 | ZT20 |
| NVE4116 | CiPC | 0.000118936 | 24* | ZT24 | - | - |
Benjamini-Hochberg-adjusted.
Periodicity significance in LD.
Peak expression values in LD correspond to a significant JTK_Cycle period.
Cycling genes in LR1 and LR2
In addition to genes cycling under normal conditions (identified in treatment subgroup LD), we also ran JTK_Cycle on gene counts from the LR1 and LR2 subgroups to reveal expression patterns unique to the first and second days after light removal. During LR1, 1,693 genes were cycling at p <0.05 and 117 genes were cycling at p <0.001. During LR2, 90 genes were cycling at p <0.05 and four genes were cycling at p <0.001. The number of cycling genes at p <0.01 tripled on the first day of light removal compared to LD (Table 1, Fig. S6, Supporting information).
After light cue removal, genes related to signal transduction (e.g., protein phosphatase and protein kinases), metabolism and stress response (e.g., superoxide dismutase, glutathione peroxidase, and HSP70) were uniquely cycling. The bZIP family transcription factors CREB and Maf showed a significant (p <0.01) period of 24 hours during the first day following light cue removal along with DNA regulatory factors (e.g., ARNT and HIF; Table S3, Supporting information). The circadian-associated genes Clock and Cry1a lost signatures of a 24-hour rhythm in the absence of a diel light cue (Clock: period = 0, p-value = 1; Cry1a: period = 2, p-value = 0.33), and the peak expression of Cry1b shifted from ZT = 12 to ZT = 20 in LR1 but was no longer identified by JTK_Cycle to have a significant cycling period (period = 24, p-value = 0.31). PAR-bZIPb maintained a period of 28 hours in the first day following light cue removal (p-value = 0.05), however peak expression shifted from ZT = 6 in LD to ZT = 20 and helt lost evidence of any rhythmicity (period = 0, p-value = 0.52).
Of the previously identified genes hypothesized to be involved in the circadian-clock, only PAR-bZIPb retained a consistent period of 28 hours throughout each treatment subgroup (LD, LR1, LR2; Table 1). However, on the second day after light removal, peak expression of PAR-bZIPb shifted to ZT = 4 (Fig. 2; p-value = 0.01). The remaining 71 uniquely cycling genes during LR2 were sparsely annotated, but included mostly cellular component genes (e.g., ribosomal proteins, solute carrier proteins).
After constant and prolonged exposure to darkness (DD), PAR-bZIPb and PAR-bZIPc were identified by JTK_Cycle to have significant cycling periods of 28 hours (p-value = 0.005 and p-value = 0.02, respectively). The circadian-related tryptophan hydroxylase, or TPH (Peres et al., 2014), did not show evidence of cycling in any diel treatment subgroup (LD, LR1, or LR2) but during DD had a period of 28 hours (p-value = 0.08).
Differential Gene Expression
After comparing genes that show signatures of circadian rhythmicity from each subgroup, we used DESeq2 to analyze differential gene expression of pairwise comparisons between subgroups. These comparisons expose time-dependent transcriptional changes in response to a changing light environment. A total of 2,562 DEGs were differentially expressed over all subgroup comparisons (LD, LR1, LR2; FDR = 0.1, log2 fold-change >1.5). Of these, 350 genes were unique to the contrast of subgroup LD and LR1, 667 genes were unique to the LD v LR2 subgroups, and only 10 genes were shared between the three subgroups. We also compared gene expression between each subgroup and DD. These specific pairwise comparisons establish a baseline for the response of gene expression before and after light cue removal, when compared to a constant condition control (DD). Interestingly, when compared to DD, a nearly 2-fold increase in DEGs was observed in the first 24 hours following light removal (1,649) over the number of DEGs in LD (876). On the second day after light cue removal (LR2) there was a reduction in the number of DEGs (734). This observation is consistent with differential expression between LD and DD (Fig. 1B) and is similar to the pattern of 24-hour cycling genes identified by JTK_Cycle (Fig. S6, Supporting information). Each of these results were in contrast to the patterns of expression for anemones under constant conditions (DD1 v DD2, for example). Pairwise comparisons between each sampling day during DD revealed a total of 66 significant DEGs that were consistently differentially expressed between days (DD1 v DD2, DD1 v DD3, DD2 v DD3; FDR = 0.1, absolute log2 fold-change >1.5).
LD v DD
The contrast of light:dark and long-term darkness allowed us to characterize genes that are differentially expressed during a diel light cycle, before light cue removal. Using a relaxed FDR of 10%, DESeq2 generated 1,160 DEGs (Fig. 1B), predominately comprised of genes up-regulated in LD compared to DD (971 genes). These were genes involved in transcription (e.g., bHLH transcription factors Clock and helt and bZIP transcription factors in the HLF and PAR subfamilies) as well as DNA-photolyase activity (e.g., cryptochromes; Fig. 1C). A GO analysis of genes differentially expressed under diel lighting showed ‘endopeptidase’ and ‘chromatin binding’ as the most enriched terms in up- and down-regulated genes, respectively in the molecular function category (Table 2; Fig. S7, Supporting information). Among biological processes enriched in light:dark conditions, ‘positive regulation of immune system process’ and ‘regulation of immune response’ were upregulated and ‘chromosome organization’ was down-regulated compared to constant conditions (Fig. S7, Supporting information).
Table 2:
Top significantly enriched gene ontology (GO) terms of contrasting treatment subgroups
| Comparison⊥ | Direction | # of DEGs┼ | Top GO Term (# of sig. genes/all genes in category)* |
||
|---|---|---|---|---|---|
| Molecular Function | Biological Process | Cellular Component | |||
| LD v DD | Up | 971 | Endopeptidase (13/150) p < 0.05 | Cellular amide metabolic process (33/367) p < 1e-04 | Cytosolic ribosome (5/79) p < 0.001 |
| Down | 189 | Chromatin binding (19/144) p < 0.05 | Regulation of cell cycle process (51/352) p < 1e-04 | Chromosome (54/389) p < 1e-04 | |
| LR1 v DD | Up | 1391 | ATPase (57/215) p < 1e-04 | Chromosome organization (109/322) p < 1e-09 | Chromosome (126/394) p < 1e-06 |
| Down | 258 | Structural molecule (82/237) p < 1e-04 | Protein localization to ER (20/99) p < 1e-09 | Small ribosomal subunit (30/53) p < 1e-06 | |
| LR2 v DD | Up | 434 | Chromatin binding (135/144) p < 0.05 | Histone modification (121/130) p < 0.001 | Axoneme (53/54) p < 1e-04 |
| Down | 300 | Endopeptidase (134/142) p < 0.05 | NA | Endoplasmic reticulum lumen (57/61) p < 1e-04 | |
| LD v LR1 | Up | 427 | Actin binding (128/135) p < 1e-05 | Actin filament-based process (269/296) p < 1e-05 | Endoplasmic reticulum lumen (60/61) p < 1e-05 |
| Down | 449 | Structural constituent of ribosome (105/111) p < 1e-05 | Translation initiation (111/113) p < 1e-05 | Large ribosomal subunit (61/63) p < 1e-05 | |
| LD v LR2 | Up | 828 | Oxidoreductase (242/266) p < 0.001 | Fatty acid metabolic process (97/102) p < 0.001 | Endoplasmic reticulum lumen (59/61) p < 0.001 |
| Down | 353 | Structural constituent of ribosome (101/111) p < 1e-05 | Spindle elongation (68/69) p < 1e-05 | Cytosolic part (133/139) p < 1e-05 | |
| LR1 v LR2 | Up | 372 | Electron transfer (50/52) p < 0.001 | Mitochondrial ATP synthesis coupled electron transport (32/32) p < 1e-05 | Mitochondrial respiratory chain (33/33) p < 1e-05 |
| Down | 133 | Signaling receptor (136/145) p < 1e-05 | Biological adhesion (212/228) p < 1e-05 | Synapse (242/261) p < 1e-05 | |
Mann-Whitney U test corrected p-value.
Benjamini-Hochberg FDR cutoff of 10%.
Pairwise comparisons using Wald tests in DESeq2.
LD v LR1
The comparison of light:dark conditions and the first day post light cue removal revealed 876 DEGs (449 up-regulated and 427 down-regulated after light removal compared to LD) that exceeded the Benjamini-Hochberg FDR cutoff of 10%. Of these, 125 genes were also cycling (identified by JTK_Cycle, Table S3, Supporting information). A survey of circadian-related genes found Clock, Cry1a, PAR-bZIPa, PAR-bZIPb, and PAR-bZIPc to be down-regulated immediately after light removal compared to diel conditions, along with helt and an additional PAR-bZIPd, previously called NV16 (Fig. 1D; Reinke, Baek, Ashenberg, & Keating, 2013). Genes identified to be up-regulated following light cue removal compared to diel conditions include several environmental response genes, particularly factors involved in the oxidative stress pathway (e.g., hypoxia inducible factor and one cytochrome P450), and heavy metal detoxification (e.g., one phytochelatin synthase; Fig. 1C). Genes involved in metabolic pathways, specifically central enzymes in the citric acid cycle (e.g., malate dehydrogenase and isocitrate dehydrogenase) were also significantly up-regulated after light removal compared to LD. Other essential gene regulatory enzymes, primarily those involved in chromatin organization, were up-regulated in LR1 compared to LD (e.g., histone methyltransferase (HMT), histone deacetylase (HDAC), and transcriptional regulator of ATRX; Fig. 1C, Fig. 3).
Fig. 3.
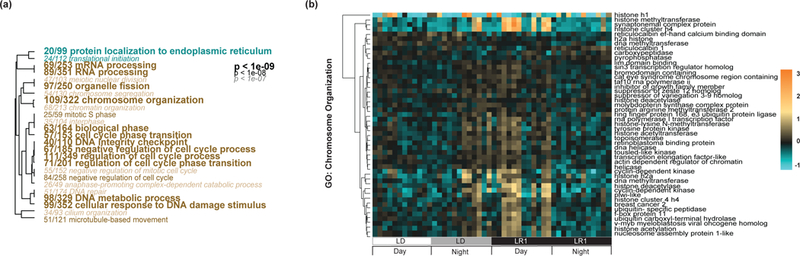
Gene ontology analysis of differently expressed genes after light removal. (A) Significantly up (tan) or down (cyan) -regulated genes related to ‘biological process’ based on a Mann-Whitney U test of the pairwise comparison between the first day following light removal and constant conditions. The font size corresponds to smaller FDR-adjusted p-values, the smallest font is equivalent to a p-value <1e-07, the largest font is equivalent to a p-value <1e-09. Dendrograms represent hierarchical clustering of GO terms based on shared genes in this data set and the ratios in front of each GO term represent the number of genes from that specific GO term in this data set over all genes belonging to that GO term. (B) Clustered heatmap of the top genes (log2 fold change >1.5, DESeq2 p-value <0.001) from the GO term ‘chromosome organization’ (GO:0006325) during light:dark (LD) and one day post light removal (LR1). The color scale is log2 fold change. The experiment key at the bottom identifies the subgroup and the ‘day’ and ‘night’ periods of each 24-hour cycle.
Gene ontology (GO) enrichment analysis of DEGs between LD and LR1 found significantly enriched terms in genes up-regulated after light removal to be ‘structural constituent of ribosome’ in the molecular function category and ‘cellular respiration’ in the biological process category (Table 2). The most significantly enriched terms in genes down-regulated after light removal was ‘actin binding’ of molecular function and ‘lipid metabolic process’ of the biological process category (Table 2). We also compared LR1 to DD, where GO analysis revealed ‘chromosome organization’, ‘cellular response to DNA damage stimulus’, and ‘DNA metabolic process’ as significantly enriched terms from the biological process category for genes up-regulated after light removal compared to long-term darkness (Fig. 3).
LD v LR2
DESeq2 identified 1,181 DEGs passing the Benjamini-Hochberg FDR cutoff of 10% in the contrast between light:dark and the second day after light removal (353 up-regulated and 828 down-regulated after two days of darkness compared to LD). GO enrichment analysis of DEGs between LD and LR2 identified the most significantly enriched GO term in up-regulated genes after two days of light removal as ‘structural constituent of ribosome’ in the molecular function category, and ‘RNA catabolic process’ of biological processes. The most significantly enriched GO term in the molecular function category of down-regulated genes after two days of light removal was ‘oxidoreductase’, and in the biological process category ‘fatty acid metabolism’ was the most enriched. Additionally, comparing DEGs between LR2 and DD, ‘chromatin binding’ and ‘endopeptidase’ were the most enriched GO terms in up- and down-regulated genes, respectively, in the molecular function category (Table 2, Fig. S9, Supporting information).
Weighted gene co-expression network analysis (WGCNA)
After identifying differently expressed gene patterns between subgroups, we performed a weighted gene co-expression network analysis (WGCNA; Langfelder & Horvath, 2008) to isolate groups of genes that show correlated expression across samples without the consideration of experimental conditions. Two modules were significantly and uniquely correlated to light:dark (LD: brown - Pearson’s R2 = 0.35, p-value <4e-05; salmon - Pearson’s R2 = 0.81, p-value <1e-08; Fig 4; Fig. S5, Supporting information). The brown module (1,201 genes) showed GO enrichment for ‘chromatin binding’ in the molecular function category, and ‘chromatin’ in the cellular component category, which were different genes than those with higher expression in the comparison of LR1 and LD. The salmon module (33 genes), also unique to LD, showed significant GO enrichment for ‘transcription factor, RNA polymerase II’ (Fig. 4). One module was significantly correlated to the first day of light removal (LR1: green - Pearson’s R2 = −0.2, p-value <0.02; Fig. S5, Supporting information). GO enrichment analysis of the green module (170 genes) showed enrichment for ‘cytoskeletal protein binding’ and ‘actin binding’ of the molecular function category and ‘cell-to-cell junction’ in the cellular component category. The remaining three modules were significantly and uniquely correlated to the second day of light removal (LR2: turquoise - Pearson’s R2 = 0.22, p-value <0.01; red - Pearson’s R2 = 0.31, p-value <3e-04; purple - Pearson’s R2 = 0.31, p-value <4e-04; Fig. S5, Supporting information). GO analysis of the turquoise module (2,766 genes) showed enrichment for ‘respiratory electron transport chain’ and ‘cellular respiration’ in the biological process category. The red (156 genes) and purple modules (91 genes) were not enriched for any GO terms.
Fig. 4.
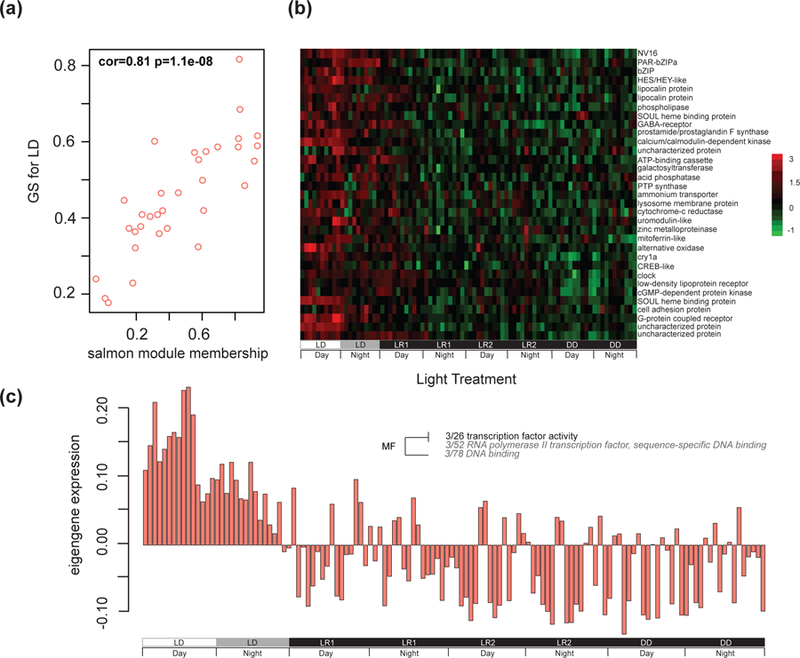
Eigengene expression across all treatment subgroups for light responsive WGCNA salmon module. (A) Scatterplot of the salmon module contains genes co-expressed and positively correlated with light:dark conditions determined by WGCNA analysis and illustrates the genes module membership score along the x-axis and the gene’s significance (GS) for the light treatment trait along the y-axis. A high correlation (cor = 0.81) between these measures indicates a strong association of the module with the trait (i.e. genes in the salmon module are strongly associated with light:dark conditions). (B) Heatmap of genes in the salmon module across each treatment subgroup. The experiment key at the bottom identifies the subgroups and the ‘day’ and ‘night’ periods of each 24-hour cycle. Each row of the heatmap represents a single annotated gene, and each column represents a single individual in each time point (n = 4 per time point). The color scale is log2 fold change. (C) Eigengene expression across all treatment subgroups and long-term darkness with corresponding Gene Ontology. Each bar represents a single individual. The experiment key at the bottom identifies the subgroups and the ‘day’ and ‘night’ periods of each 24-hour cycle. Positive eigengene expression values indicate positive correlation and negative eigengene expression values indicate negative correlation of the module to the light treatment trait. A fisher’s exact test was used to identify significantly enriched GO terms (presence or absence) of the eigengene and categories enriched for molecular function (MF) were assigned. The size and color of the font increases as significance increases, as shown in the inset.
DISCUSSION
Our quantitative analysis of transcriptomes for anemones during consistent light:dark cycles and after removal of the lighting cue revealed unique gene expression profiles over 24-hour periods in the presence of light, after 24 hours of removal, and after longer periods of light removal. Consistent with previous analyses of candidate genes or the whole transcriptome, long-term culturing in all dark conditions resulted in near loss of any differential gene expression over a 24-hour period. The light dependency of the differentially expressed gene sets suggests that many genes under diel lighting are direct response genes and not the product of a circadian timekeeper, at least when measured at the whole individual level. Upon light removal, we measured a large number of genes that were uniquely expressed when the cue was absent for 24 or 44 hours, which appears to be a type of stress response given the types of genes with increased expression.
‘Circadian gene’ expression dependent on light cues
A central finding from our analyses is that the expression of the candidate “circadian clock genes” identified in previous studies is strongly dependent on consistent light:dark cycling, at least when measured in whole animals. Earlier studies by Reitzel et al. (2010), Peres et al. (2014), Oren et al. (2015), and Leach et al. (2018) had shown that Nematostella orthologs to genes central to the circadian clock of bilaterians have oscillating expression in light:dark conditions. Our transcriptome comparisons are consistent with these earlier studies where the bHLH-Pas gene Clock and the bZIP transcription factors in the PAR family had rhythmic expression. Another transcription factor previously identified in corals to be differentially expressed in diel lighting, eyes absent (eya), showed differential expression following light removal (A. K. Brady et al., 2011). Our analyses identified additional PAR-bZIP genes that fit a diel expression pattern, particularly PAR-bZIPd (called NV16 in Reinke et al., 2013) with robust expression in the light period, with peak expression at the beginning of the photoperiod (ZT = 2; Fig. 1C, Fig. 2). This particular PAR-bZIP is a heterodimer partner with other PAR-bZIP proteins from Nematostella previously identified by Reitzel et al. (2013) with different peak expression periods, suggesting the potential for complex gene regulation over a diel period, similar to Drosophila (Cyran et al., 2003). After the removal of the light cue, PAR-bZIPd maintained significant differences throughout light treatments; however, its expression dampened each day following light removal, suggesting light dependency rather than true circadian regulation (Fig. 2). The remaining hypothesized cnidarian circadian genes were not differentially expressed in the absence of a light cue (Fig. 2). Interestingly though, after light cue removal a few of these candidates shifted peak expression time. PAR-bZIPb continued to cycle every 28 hours, but peak expression was phase shifted by 12 hours. Cryptochromes previously identified to have differential expression in response to diel lighting [NvCry1a and NvCry1b in Reitzel et al. (2010)] also experienced peak shifts after light removal. Consistent with previous studies, the hypothesized repressive Type 2 cryptochrome, Nvcry2, showed no response to diel lighting in Nematostella, similar to insect and mammal clocks (Fig. 2; Griffin, Staknis, & Weitz, 1999; Kume et al., 1999; Reitzel et al., 2010; Zhu et al., 2008). Presently, it is unclear what role cryptochromes and PAR-bZIP transcription factors play in the clock of cnidarians; thus, future mechanistic experiments would provide more insight to the potential suppressive role of these proteins and regulatory role of these transcription factors, respectively.
Changes in light condition results in stress and changes to chromatin structure
The large and unique set of differentially expressed genes after one or two days of light removal are broadly consistent with an environmental stress response that involves a number of genes related to cellular stress and chromatin remodeling (Fig. 3, Fig. S8–S9, Supporting information). Removal or time-shifting of entraining cues is broadly known to disrupt physiology and behavior for various animals (Davis & Mirick, 2006; Garaulet & Madrid, 2010; Rhoades, Nayak, Zhang, Sehgal, & Weljie, 2018). Unlike in light:dark conditions, genes involved in cellular and aerobic respiration and cellular response to DNA damage were differentially expressed upon removal of the light cue (Fig. 3). Additionally, Nematostella sampled in constant conditions upregulate hypoxia inducible factor (HIF), cytochrome c oxidase, monoamine oxidase, and aquaporin 4 (Fig. 1D). Genes related to chromatin remodeling that were significantly up regulated after light removal include histone deacetylase and histone methyltransferase (Fig. 4). Broadly, these enzymes regulate gene expression by making modifications to the chromatin structure, ultimately increasing compaction within DNA and reducing transcription factor activity and thus, gene expression.
Photoresponse versus circadian clock
Previous studies in cnidarians have typically relied on a comparison of consistent light:dark cycles and a single day of all darkness immediately after in which to determine if genes are likely “circadian”. The period of free-running behavior or physiology varies between organisms with well-described circadian oscillators but typically last for days or weeks with removal of the entraining cue. Our gene expression results with Nematostella that showed large shifts in the transcriptional profile with removal of light differ from the consistency of a free-run period previously reported for locomotion (Hendricks et al., 2012; Oren et al., 2015) and physiology (Maas et al., 2016). At present, we hypothesize the cause for this discrepancy is the use of whole animals for our sample material when the mechanisms driving cyclic phenotypes are conceivably restricted to a subset of cells, likely neurons. Combining gene expression information from multiple tissues in one sample has the potential to diminish oscillating gene expression signals if present in a small number of cells or if tissues have rhythmic gene expression in different phases, as is known in vertebrates (Albrecht, 2012). Nematostella, like other cnidarians, has a complex but diffuse nervous system without a centralized concentration of neurons (Marlow, Srivastava, Matus, Rokhsar, & Martindale, 2009), which presumably arose in a later common ancestor (Arendt, Tosches, & Marlow, 2015). Recent work (Sebé-Pedrós et al., 2018) has revealed the complex transcriptional differences of the more than eight broad cells types of Nematostella. Moving forward, these cell-type specific analyses of oscillating gene expression will be useful to identify what cells in heterogeneous cell populations may be driving the circadian phenotypes of cnidarians.
Supplementary Material
Supporting Information
Fig. S1: Experimental design of (1) diel and (2) long-term darkness treated Nematostella run in parallel.
Fig. S2: DESeq2 analysis of Nematostella transcriptomes pre- and post-light removal.
Fig. S3: WGCNA analysis of differentially expressed genes across all light treatment subgroups, individuals, and time.
Fig. S4: Module-trait relationship heatmap and significance values of each module eigengene correlated to external traits.
Fig. S5: Gene significance scatter plots for each light treatment subgroup.
Fig. S6: Venn diagram of cycling genes from each light treatment subgroup generated by JTK_Cycle.
Fig. S7: Gene ontology (GO) analysis of differentially expressed genes enriched between light:dark and dark:dark.
Fig. S8: Gene ontology (GO) analysis of differentially expressed genes enriched between light removal day 1 and dark:dark.
Fig. S9: Gene ontology (GO) analysis of differentially expressed genes enriched between light removal day 2 and dark:dark.
Table S1: Raw counts table for the samples reported in this study.
Table S2: Reads per sample for the samples reported in this study.
Table S3: JTK_Cycle analysis of Nematostella transcriptomes pre- and post-light removal.
Acknowledgements
We would like to thank members of the Reitzel and Tarrant (WHOI) laboratories for discussions during the course of this study. We would also like to thank Misha Matz (UT Austin) for assistance with gene expression analyses.
Funding
This research was supported by NIH grant R15GM114740.
Footnotes
Data Accessibility
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Leach et al., 2019) and are accessible through GEO Series accession number GSE132202 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132202). A full detailed protocol of library sequencing preparation and scripts for bioinformatic analysis can be found at https://github.com/z0on/tag-based_RNAseq. The annotation and reference gene model files are available at https://figshare.com/articles/Nematostella_vectensis_transcriptome_and_gene_models_v2_0/807696. Raw data files have been deposited into dryad and are available at: https://datadryad.org/review?doi=doi:10.5061/dryad.pp5qk07.
References
- Albrecht U (2012). Timing to perfection: The biology of central and peripheral circadian clocks. Neuron, 74(2), 246–260. doi: 10.1016/j.neuron.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Arendt D, Tosches MA, & Marlow H (2015). From nerve net to nerve ring, nerve cord and brain — evolution of the nervous system. Nature Reviews Neuroscience, 17, 61. doi: 10.1038/nrn.2015.15 [DOI] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, & Takao M (2002). Phototransduction by retinal ganglion cells that set the circadian clock. Science, 295(5557), 1070–1073. doi: 10.1126/science.1067262 [DOI] [PubMed] [Google Scholar]
- Bradshaw WE, & Holzapfel CM (2007). Evolution of animal photoperiodism. Annual Review of Ecology Evolution and Systematics, 38, 1–25. doi: 10.1146/annurev.ecolsys.37.091305.110115 [DOI] [Google Scholar]
- Brady A, Hilton J, & Vize P (2009). Coral spawn timing is a direct response to solar light cycles and is not an entrained circadian response. Coral Reefs, 28(3), 677–680. [Google Scholar]
- Brady AK, Snyder KA, & Vize PD (2011). Circadian cycles of gene expression in the coral, Acropora millepora. PLoS ONE, 6(9), e25072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker BE, Barnes DJ, Dunlap WC, & Jokiel PL (1988). Light and reef-building corals. Interdisciplinary Science Reviews, 13(3), 222–237. doi: 10.1179/isr.1988.13.3.222 [DOI] [Google Scholar]
- Cheng N, Tsunenari T, & Yau KW (2009). Intrinsic light response of retinal horizontal cells of teleosts. Nature, 460(7257), 899–U139. doi: 10.1038/nature08175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, … Blau J (2003). Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell, 112(3), 329–341. [DOI] [PubMed] [Google Scholar]
- Davis S, & Mirick D (2006). Circadian disruption, shift work and the risk of cancer: A summary of the evidence and studies in Seattle. Cancer Causes & Control, 17(4), 539–545. doi: 10.1007/s10552-005-9010-9 [DOI] [PubMed] [Google Scholar]
- Dixon P (2003). VEGAN, a package of R functions for community ecology. Journal of Vegetation Science, 14(6), 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- Dunlap JC (1999). Molecular bases for circadian clocks. Cell, 96(2), 271–290. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, & DeCoursey PJ (2004). Chronobiology: biological timekeeping: Sinauer Associates. [Google Scholar]
- Edmunds LN Jr. (1988). Cellular and molecular bases of biological clocks. Models and mechanisms for circadian timekeeping Springer. [Google Scholar]
- Edwards SL, Charlie NK, Milfort MC, Brown BS, Gravlin CN, Knecht JE, & Miller KG (2008). A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. Plos Biology, 6(8), 1715–1729. doi: 10.1371/journal.pbio.0060198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AM, Fero K, Driever W, & Burgess HA (2013). Enlightening the brain: Linking deep brain photoreception with behavior and physiology. Bioessays, 35(9), 775–779. doi: 10.1002/bies.201300034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, & Foster R (1999). Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science, 284(5413), 502–504. doi: 10.1126/science.284.5413.502 [DOI] [PubMed] [Google Scholar]
- Gamble KL, Berry R, Frank SJ, & Young ME (2014). Circadian clock control of endocrine factors. Nature Reviews Endocrinology, 10, 466. doi: 10.1038/nrendo.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet M, & Madrid JA (2010). Chronobiological aspects of nutrition, metabolic syndrome and obesity. Advanced Drug Delivery Reviews, 62(9–10), 967–978. doi: 10.1016/j.addr.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Griffin E, Staknis A, & Weitz CJ (1999). Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science, 286, 768–771. [DOI] [PubMed] [Google Scholar]
- Gwinner E (1996). Circadian and circannual programmes in avian migration. Journal of Experimental Biology, 199(1), 39–48. [DOI] [PubMed] [Google Scholar]
- Hardin PE (2006). Essential and expendable features of the circadian timekeeping mechanism. Current Opinion in Neurobiology, 16(6), 686–692. [DOI] [PubMed] [Google Scholar]
- Hemond EM, & Vollmer SV (2015). Diurnal and nocturnal transcriptomic variation in the Caribbean staghorn coral, Acropora cervicornis. Molecular Ecololgy, 24(17), 4460–4473. doi: 10.1111/mec.13320 [DOI] [PubMed] [Google Scholar]
- Hendricks WD, Byrum CA, & Meyer-Bernstein EL (2012). Characterization of circadian behavior in the starlet sea anemone, Nematostella vectensis. PLoS ONE, 7(10), e46843. doi: 10.1371/journal.pone.0046843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KD, Szmant AM, & Pyott SJ (2011). Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE, 6(5), e19755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KD, Vize PD, & Pyott SJ (2016). Current understanding of the circadian clock within Cnidaria. In Goffredo S & Dubinsky Z (Eds.), The Cnidaria, Past, Present and Future: The world of Medusa and her sisters (pp. 511–520). Cham: Springer International Publishing. [Google Scholar]
- Hughes ME, Hogenesch JB, & Kornacker K (2010). JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms, 25(5), 372–380. doi: 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindrich K, Roper KE, Lemon S, Degnan BM, Reitzel AM, & Degnan SM (2017). Origin of the animal circadian clock: diurnal and light-entrained gene expression in the sponge Amphimedon queenslandica. Frontiers in Marine Science, 4(327). doi: 10.3389/fmars.2017.00327 [DOI] [Google Scholar]
- Kauffmann A, Gentleman R, & Huber W (2009). arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics (Oxford, England), 25(3), 415–416. doi: 10.1093/bioinformatics/btn647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R (2018). Pretty Heatmaps Comprehensive R Archive Network. [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, … Reppert SM (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98(2), 193–205. [DOI] [PubMed] [Google Scholar]
- Lamb TD, Collin SP, & Pugh EN Jr. (2007). Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nature reviews. Neuroscience, 8(12), 960–976. doi: 10.1038/nrn2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, & Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 13. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–U354. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach WB, Macrander J, Peres R, & Reitzel AM (2018). Transcriptome-wide analysis of differential gene expression in response to light:dark cycles in a model cnidarian. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 26, 40–49. doi: 10.1016/j.cbd.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, & Hoegh-Guldberg O (2007). Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science, 318(5849), 467–470. doi: 10.1126/science.1145432 [DOI] [PubMed] [Google Scholar]
- Leys SP, Cronin TW, Degnan BM, & Marshall JN (2002). Spectral sensitivity in a sponge larva. Journal of Comparative Physiology A, 188(3), 199–202. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12). doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas AE, Jones IT, Reitzel AM, & Tarrant AM (2016). Daily cycle in oxygen consumption by the sea anemone Nematostella vectensis Stephenson. Biology Open, 5(2), 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, & Martindale MQ (2009). Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Developmental Neurobiology, 69. doi: 10.1002/dneu.20698 [DOI] [PubMed] [Google Scholar]
- Mendlewicz J (2009). Disruption of the circadian timing systems. CNS Drugs, 23(2), 15–26. doi: 10.2165/11318630-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Mercier A, & Hamel JF (2010). Synchronized breeding events in sympatric marine invertebrates: role of behavior and fine temporal windows in maintaining reproductive isolation. Behavioral Ecology and Sociobiology, 64(11), 1749–1765. doi: 10.1007/s00265-010-0987-z [DOI] [Google Scholar]
- Meyer E, Aglyamova GV, & Matz MV (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Molecular Ecology, 20(17), 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, & Sancar A (1998). Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proceedings of the National Academy of Sciences of the United States of America, 95(11), 6097–6102. doi: 10.1073/pnas.95.11.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldach MJ, Workentine M, Matz MV, Fan TY, & Vize PD (2017). Transcriptome dynamics over a lunar month in a broadcast spawning acroporid coral. Molecular Ecology, 26(9), 2514–2526. doi: 10.1111/mec.14043 [DOI] [PubMed] [Google Scholar]
- Oren M, Tarrant AM, Alon S, Simon-Blecher N, Elbaz I, Appelbaum L, & Levy O (2015). Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Scientific Reports, 5, 11418. doi:10.1038/srep1141810.1038/srep11418https://www.nature.com/articles/srep11418#supplementary-informationhttps://www.nature.com/articles/srep11418#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, & Miller W (1997). Comparison of DNA sequences with protein sequences. Genomics, 46(1), 24–36. doi: 10.1006/geno.1997.4995 [DOI] [PubMed] [Google Scholar]
- Peres R, Reitzel AM, Passamaneck Y, Afeche SC, Cipolla-Neto J, Marques AC, & Martindale MQ (2014). Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone Nematostella vectensis. Evodevo, 5, 26. doi: 10.1186/2041-9139-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Fong CR, & Oakley TH (2010). The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proceedings of the Royal Society B-Biological Sciences, 277(1690), 1963–1969. doi: 10.1098/rspb.2009.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ML (2016). Beyond the eye: molecular evolution of extraocular photoreception. Integrative and Comparative Biology, 56(5), 842–852. doi: 10.1093/icb/icw052 [DOI] [PubMed] [Google Scholar]
- Reinke AW, Baek J, Ashenberg O, & Keating AE (2013). Networks of bZIP Protein-Protein Interactions Diversified Over a Billion Years of Evolution. Science, 340(6133), 730. doi: 10.1126/science.1233465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Behrendt L, & Tarrant AM (2010). Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS ONE, 5(9), e12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Passamaneck YJ, Karchner SI, Franks DG, Martindale MQ, Tarrant AM, & Hahn ME (2014). Aryl hydrocarbon receptor (AHR) in the cnidarian Nematostella vectensis: comparative expression, protein interactions, and ligand binding. Development Genes and Evolution, 224(1), 13–24. doi: 10.1007/s00427-013-0458-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM, & Levy O (2013). Circadian clocks in the Cnidaria: Environmental entrainment, molecular regulation, and organismal outputs. Integrative and Comparative Biology, 53(1), 118–130. doi: 10.1093/icb/ict024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades SD, Nayak K, Zhang SL, Sehgal A, & Weljie AM (2018). Circadian- and light-driven metabolic rhythms in Drosophila melanogaster. Journal of Biological Rhythms, 33(2), 126–136. doi: 10.1177/0748730417753003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, & Oakley TH (2012). Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. The Journal of Experimental Biology, 215(Pt 8), 1278–1286. doi: 10.1242/jeb.067140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T, & Foster RG (1997). Twilight times: Light and the circadian system. Photochemistry and Photobiology, 66(5), 549–561. doi: 10.1111/j.1751-1097.1997.tb03188.x [DOI] [PubMed] [Google Scholar]
- Ruiz-Jones LJ, & Palumbi SR (2015). Transcriptome-wide changes in coral gene expression at noon and midnight under field conditions. Biological Bulletin, 228(3), 227–241. doi: 10.1086/BBLv228n3p227 [DOI] [PubMed] [Google Scholar]
- Schnitzler CE, Pang K, Powers ML, Reitzel AM, Ryan JF, Simmons D, … Baxevanis AD (2012). Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biology, 10, 107–107. doi: 10.1186/1741-7007-10-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhé M-P, Renno J, … Marlow H (2018). Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell, 173(6), 1520–1534. 10.1016/j.cell.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng BH, … Reppert SM (2000). Interacting molecular loops in the mammalian circadian clock. Science, 288(5468), 1013–1019. doi: 10.1126/science.288.5468.1013 [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Tanaka M, Shinzato C, Kawashima T, & Satoh N (2013). A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene, 515(2), 426–431. doi: 10.1016/j.gene.2012.12.038 [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, … Vervoort M (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evolutionary Biology, 7(1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosches MA, Bucher D, Vopalensky P, & Arendt D (2014). Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell, 159(1), 46–57. doi: 10.1016/j.cell.2014.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD (2009). Transcriptome analysis of the circadian regulatory network in the coral Acropora millepora. Biological Bulletin, 216(2), 131–137. [DOI] [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2
- Williams DL (2016). Light and the evolution of vision. Eye, 30(2), 173–178. doi: 10.1038/eye.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, & Sehgal A (2001). Molecular components of the circadian system in Drosophila. Annual Review of Physiology, 63(1), 729–755. doi:doi: 10.1146/annurev.physiol.63.1.729 [DOI] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, & Reppert SM (2008). Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biology, 6(1), e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Fig. S1: Experimental design of (1) diel and (2) long-term darkness treated Nematostella run in parallel.
Fig. S2: DESeq2 analysis of Nematostella transcriptomes pre- and post-light removal.
Fig. S3: WGCNA analysis of differentially expressed genes across all light treatment subgroups, individuals, and time.
Fig. S4: Module-trait relationship heatmap and significance values of each module eigengene correlated to external traits.
Fig. S5: Gene significance scatter plots for each light treatment subgroup.
Fig. S6: Venn diagram of cycling genes from each light treatment subgroup generated by JTK_Cycle.
Fig. S7: Gene ontology (GO) analysis of differentially expressed genes enriched between light:dark and dark:dark.
Fig. S8: Gene ontology (GO) analysis of differentially expressed genes enriched between light removal day 1 and dark:dark.
Fig. S9: Gene ontology (GO) analysis of differentially expressed genes enriched between light removal day 2 and dark:dark.
Table S1: Raw counts table for the samples reported in this study.
Table S2: Reads per sample for the samples reported in this study.
Table S3: JTK_Cycle analysis of Nematostella transcriptomes pre- and post-light removal.


