Abstract
To better understand the pathomechanism of psoriasis, a comparative proteomic analysis was performed with non-lesional and lesional skin from psoriasis patients and skin from healthy individuals. Strikingly, 79.9% of the proteins that were differentially expressed in lesional and healthy skin exhibited expression levels in non-lesional skin that were within twofold of the levels observed in healthy and lesional skin, suggesting that non-lesional skin represents an intermediate stage. Proteins outside this trend were categorized into three groups: I. proteins in non-lesional skin exhibiting expression similar to lesional skin, which might be predisposing factors (i.e., CSE1L, GART, MYO18A and UGDH); II. proteins that were differentially expressed in non-lesional and lesional skin but not in healthy and lesional skin, which might be non-lesional characteristic alteration (i.e., CHCHD6, CHMP5, FLOT2, ITGA7, LEMD2, NOP56, PLVAP and RRAS); and III. proteins with contrasting differential expression in non-lesional and lesional skin compared to healthy skin, which might contribute to maintaining the non-lesional state (i.e., ITGA7, ITGA8, PLVAP, PSAPL1, SMARCA5 and XP32). Finally, proteins differentially expressed in lesions may indicate increased sensitivity to stimuli, peripheral nervous system alterations, furthermore MYBBP1A and PRKDC were identified as potential regulators of key pathomechanisms, including stress and immune response, proliferation and differentiation.
Subject terms: Protein-protein interaction networks, Psoriasis
Introduction
To date, all therapies available for psoriasis only manage symptoms. Understanding alterations that cause the disease is highly important for developing new therapies to better manage the disease.
Our skin connects, and at the same time separates internal the external environment. It is constantly subjected to many different stimuli that requires proper response, through which the skin can influences the function of other organs, like the brain and the endocrine system in a mutual way1,2. In psoriasis, the macroscopically healthy looking non-lesional skin harbors alterations that might cause symptoms3. One of the most characteristic properties of non-lesional skin is an altered response to mechanical stress or injury4 leading to barrier disruption5, which leads to an elevated innate immune response6,7. Alterations in non-lesional skin are not restricted to keratinocytes. Angiogenesis is also among those mechanisms that is already affected in non-lesional skin, resulting in altered quantity and quality of microvessels8. In addition, it is becoming clear that some adaptive immune responses are also altered9. Abnormalities in the dermal extracellular matrix composition — such as elevated expression of the oncofetal splice variant of fibronectin10, due to altered splicing events11 indicate the involvement of dermal fibroblasts3. Several matrix metalloproteinases (MMPs), such as MMP-9, previously thought to be increased only in lesions, are now known to be elevated in non-lesional skin compared to healthy skin12. There is also evidence for mechanisms in non-lesional skin that contribute to the maintenance of its state. The PRINS long non-coding RNA is induced by stress and nucleic acids, and it is anticipated to have a protective function in psoriasis. PRINS in the non-lesional skin not only decreases inflammatory responses13 by inhibiting IL-6 and CCL-5 mRNA translation, but also influences anti-apoptotic mechanisms14. Elevated expression of the anti-inflammatory regulator caspase recruitment domain family member 18 (CARD18) in non-lesional skin compared to healthy skin was found to aid the inhibition of inflammatory events15. These mechanisms, among many others, highlight the relevance of comparing non-lesional skin to healthy skin.
One of the most effective ways to study different diseases with such a high complexity and to elucidate related mechanisms is to perform a comparative proteomic analysis of protein extracts derived from affected tissues. Previous large-scale treatises including genomic, transcriptomic and proteomic studies have identified psoriasis-related markers playing key roles in the pathomechanism, such as AKR1B1016, CSTA17, FABP518, PI319, SCCA220, STAT116, STAT321, S100A718–20, S100A819,22 and S100A919,20,22,23, among others, thereby contributing greatly to the better understanding of the disease. However, none of the full scale proteomic studies17,18,22,24–27 to the best of our knowledge, compared lesional and non-lesional psoriatic full thickness skin regions, with the inclusion of biopsies from healthy individuals as a reference in the comparison. The inclusion of healthy skin could provide several important additional information. I. Alterations that are similar in non-lesional and lesional skin, but differ from healthy skin, can be detected and used to identify potential novel disease markers or predisposing factors already present in the non-lesional skin. II. The comparison of non-lesional skin to healthy skin might facilitate the identification of inherent characteristics of psoriatic patients that are already present in their healthy-looking skin prior to lesion development. III. Information could be gained about the extent to which the non-lesional skin is affected in respect to lesional alterations. IV. Altered processes in the non-lesional skin that are contrary to the changes of lesional skin could be identified, some of which may contribute to the maintenance of the non-lesional state and serve as novel intervention points for disease management. We aimed to extend previous proteomic studies, in order to get more information regarding the putative alterations mentioned above. Therefore, a complex comparison was performed, where in addition to non-lesional and lesional skin, samples from healthy skin were also included, in a label-free, semi-quantitative proteomic analysis.
Results
Proteomic workflow and information on involved donors
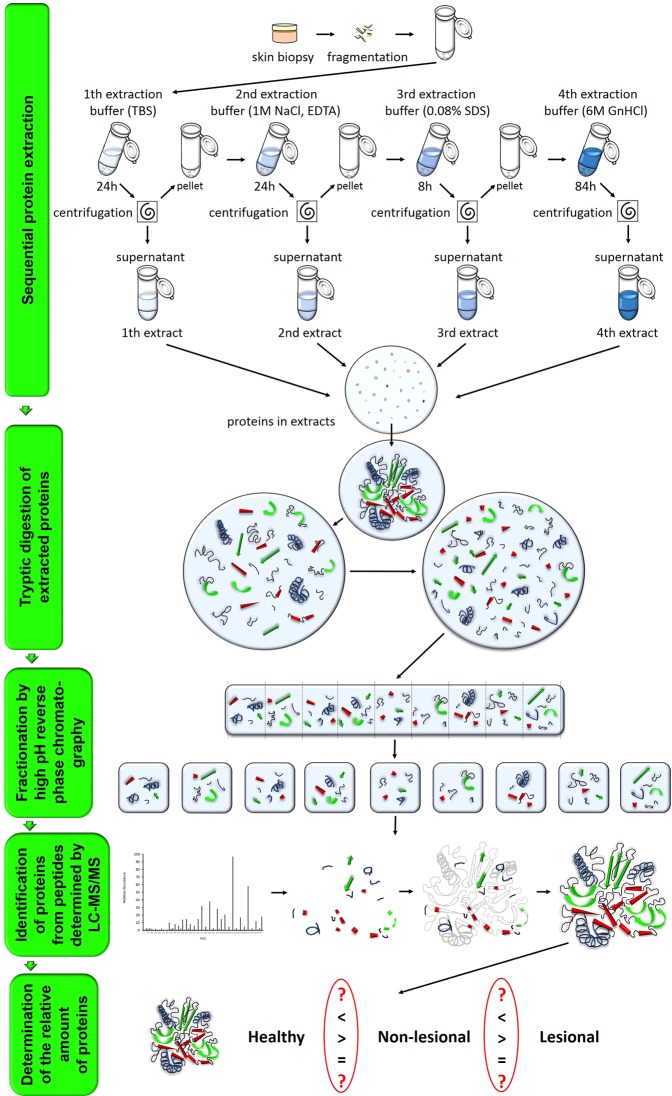
Three biological replicas of our proteomic approach were performed following sequential protein extraction of total skin biopsies. Each proteomic replica contained samples from three healthy donors as well as non-lesional and lesional biopsies from three psoriatic patients. The schematic overview of the applied proteomic strategy is summarized in Fig. 1 (also see Supplementary Information: Materials and Methods), and basic demographic and clinical characteristics of psoriatic patients and healthy donors are listed in Table 1. (Criteria for inclusion of patients in the study and skin sample collection are described at Supplementary Material: Materials and Methods section).
Figure 1.
Schematic illustration of the applied proteomic workflow.
Table 1.
Basic demographic and clinical characteristics of donors involved in the proteomic analysis. (H: healthy donor, P: plaque-type psoriatic patient).
| Proteomic experiment | Group of donors | Donors | Age | Gender | PASI score |
|---|---|---|---|---|---|
| No. 1. | Healthy | H I. | 46 | Male | n/a |
| H II. | 59 | ||||
| H III. | 51 | ||||
| Plaque type psoriasis | P I. | 65 | 17.1 | ||
| P II. | 63 | 9.9 | |||
| P III. | 50 | 5.5 | |||
| No. 2. | Healthy | H IV. | 23 | Female | n/a |
| H V. | 48 | ||||
| H VI. | 51 | ||||
| Plaque type psoriasis | P IV. | 25 | 9.2 | ||
| P V. | 62 | 21.5 | |||
| P VI. | 70 | 17.5 | |||
| No. 3. | Healthy | H VII. | 37 | Male | n/a |
| H VIII. | 39 | ||||
| H IX. | 61 | ||||
| Plaque type psoriasis | P VII. | 49 | 22.4 | ||
| P VIII. | 55 | 12.1 | |||
| P IX. | 61 | 12 |
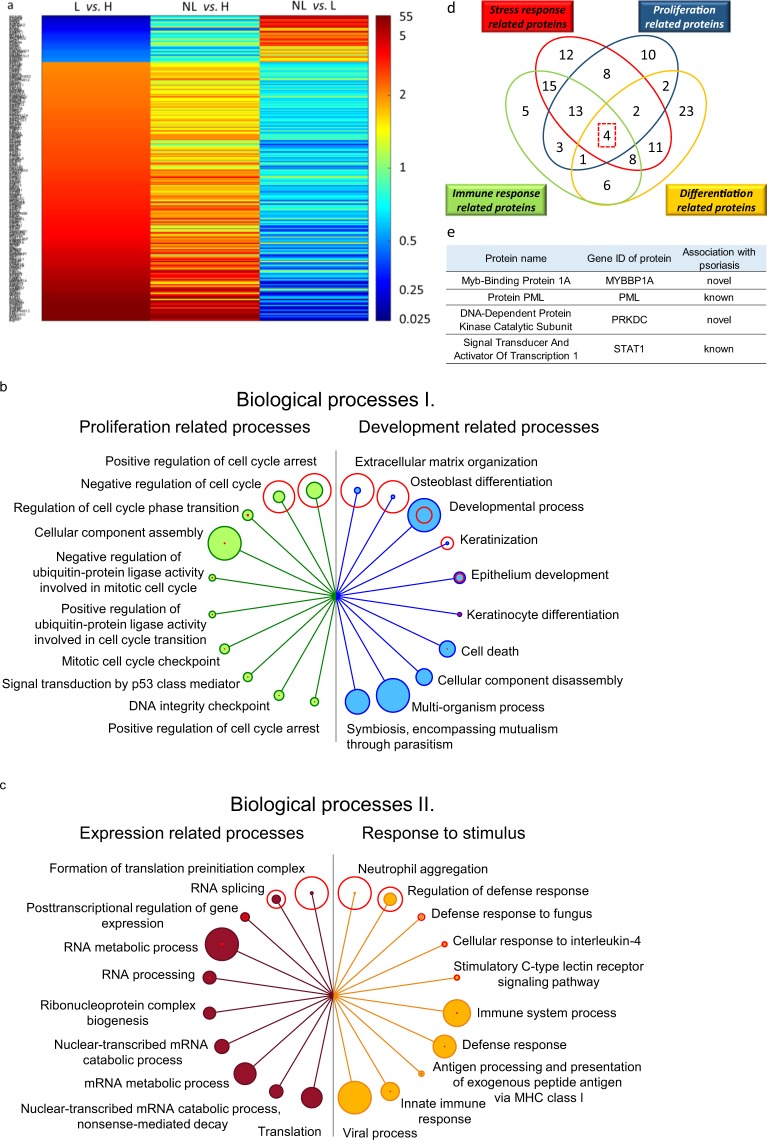
Biological processes associated with differential expression in healthy and lesional skin
As an initial step, proteomic results of lesional and healthy skin samples were compared and the relative abundance of 249 proteins was found to be different (Fig. 2a and Supplementary Table 1). A protein–protein interaction-based enrichment analysis was performed with these proteins. We screened for interaction networks and biological processes related to the observed differences in expression using Gene Ontology (GO) analysis of the STRING database (version 10.5). Based on the GO nomenclature and protein composition, the identified biological processes could be classified into the following categories: development, proliferation, regulation of expression and response to stimulus related processes. The ten most significantly different biological processes of each category are listed in Fig. 2b,c and Supplementary Table 2.
Figure 2.
Characterization of altered protein expression of lesional (L) skin compared to healthy (H) skin. Heatmap of relative expression of proteins differentially expressed in L and H skin (a, left column), and their expression in non-lesional (NL) and L skin (a, middle column) and NL and H skin (a, right column) (a). Biological processes for which proteins were differentially expressed in L and H are listed. The top ten processes are depicted for proliferation (b left, green circles), development (b right, blue circles), expression (c left, filled red circles) and response to stimulus (c right, orange circles). False detection rate (FDR) values are indicated with unfilled red circles around the filled circles for the various biological processes. The size of each circle is proportional to FDR values (unfilled circles) or to the number of proteins (filled circles). Four proteins differentially expressed in H and L skin are believed to participate in all four mechanisms of stress, immune response, proliferation and differentiation (d) and are listed in (e). (*Significant difference in relative protein expression at least by two-fold in L and H comparison).
Since the major characteristics of psoriatic alterations include altered stress and immune responses as well as dysregulation of proliferation and differentiation, we screened among proteins expressed differentially in lesion compared to healthy skin for central regulators participating in all four of these mechanisms (Fig. 2d). As a result, four central proteins — MYBBP1A, PML, PRKDC and STAT1 — were identified (Fig. 2e).
Differential protein expression in non-lesional and lesional skin and the biological processes associated with these proteins
Comparison of non-lesional and lesional skin proteomes led to the identification of 56 proteins exhibiting at least 2-fold differences in relative abundances. Of these proteins, 32 exhibited higher protein abundance in non-lesional skin compared to lesions, whereas 24 exhibited lower abundance (Fig. 3a and Supplementary Table 3). Functional enrichment analysis of these 56 proteins revealed several biological processes identified in psoriasis pathomechanism, including development, and response to stimulus (Fig. 3b and Supplementary Table 4).
Figure 3.
Differential protein expression in lesional (L) and non-lesional (NL) skin and affected biological processes. Heatmap of relative expression for proteins differentially expressed in L and NL skin (a, left column) and the relative expression of these proteins NL and L skin (middle column) and L and healthy (H) skin (right column) (a). Biological processes for which proteins were differentially expressed in L and NL are listed. The top ten processes depicted to be affected in response to stimulus (b left, filled orange circles) and development (b right, filled blue circles). False detection rate (FDR) values are indicated with unfilled red circles around the filled circles for the various biological processes. The size of each circle is proportional to FDR values (unfilled red circles) or to the number of proteins (filled circles) (b). Proteins differentially expressed in L and NL but not in H and L are listed (c). Proteins for which the changes in NL and L compared to H are in different directions (increased vs. decreased and vice versa) are listed (d). Proteins that exhibited altered expression only in lesions (potentially trigger proteins) with known (e) and novel (f) association with psoriasis are listed. (*Significant difference in relative protein expression at least by two-fold in L and NL comparison).
We also found a subset of proteins to be differentially expressed in non-lesional and lesional skin that were not differentially expressed in healthy skin and lesions (Fig. 3c).
The levels of eight proteins were greater in non-lesional skin and lower in lesional skin compared to the levels in healthy skin (non-lesional < healthy < lesional), and one protein exhibited the opposite trend (non-lesional > healthy > lesional). Although the non-lesional and lesional differences in the abundance of these proteins were not statistically significant when compared to healthy skin, the difference in abundance between non-lesional and lesional samples differed significantly by more than two-fold (Fig. 3d).
We also identified 44 proteins that had altered expression only in the comparison of lesional skin to either non-lesional or healthy skin; it is anticipated that these proteins play a role in manifestation and/or maintenance of lesions. The results of a computer-aided, keyword-based literature search suggests that, of these 44 proteins, 23 are already associated with the disease (Fig. 3e), whereas 21 have not yet been associated with psoriasis pathogenesis (Fig. 3f).
Proteins without previous association to psoriasis that exhibited decreased expression in lesions compared to expression in to both non-lesional and healthy skin include modulators of apoptosis, signaling, endothelial cell proliferation, neurite outgrowth, migration, resistance to mechanical stress, cell–cell and extracellular matrix interactions, myelination of peripheral nerves, osmotic and membrane-potential regulation (Supplementary Table 5). In contrast, proteins with increased expression in lesions compared to both non-lesional and healthy skin are involved in cell death, cell proliferation, transcription and translation, calcium sensing (neuronal) and processing of class I MHC peptides (Supplementary Table 5). To further elucidate the significance in psoriasis of the differential expression of these latter three groups of proteins (Fig. 3c,d,f), a detailed automated literature analysis was conducted for associated known functions.
Comparison of protein expression in non-lesional skin compared to healthy skin
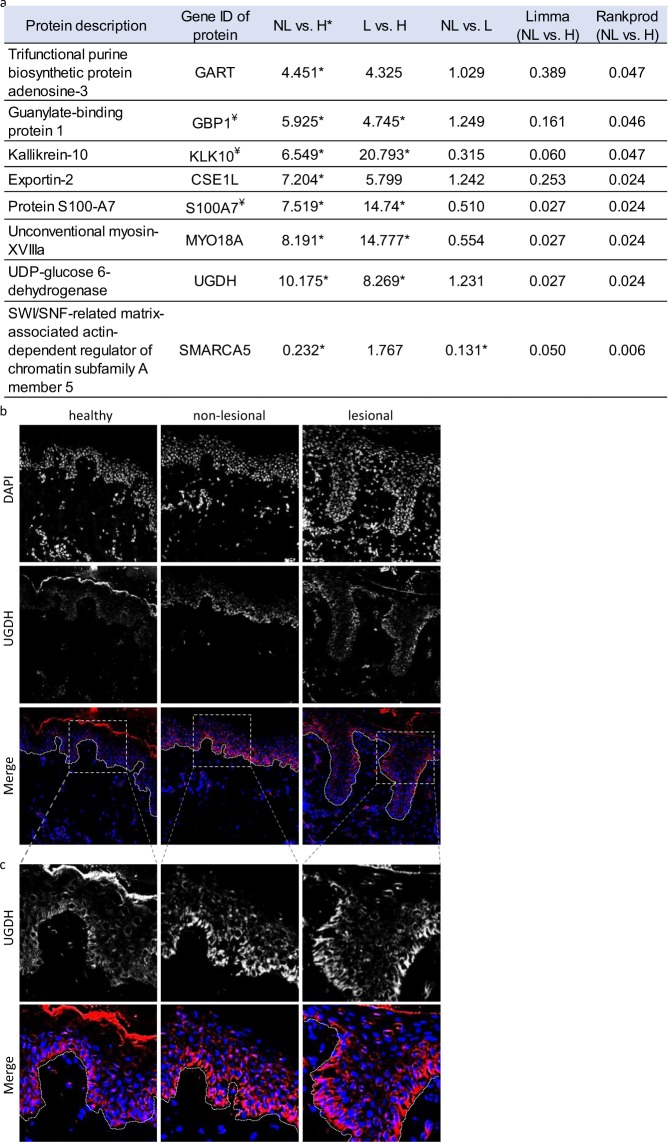
Proteins that were differentially expressed in non-lesional skin compared to healthy were also identified. Seven proteins exhibited higher expression levels in non-lesional skin compared to healthy skin and one with lower expression (Fig. 4a). Among these, the relative amount of four proteins (GART, CSE1L, GBP1 and UGDH) was similar in the non-lesional and lesional skin samples. Out of the eight proteins that are differentially expressed in non-lesional skin compared to healthy GBP1, KLK10 and S100A7 have already been associated with psoriasis pathogenesis; the other five are potential novel, early markers of the disease.
Figure 4.
Differentially expressed proteins in non-lesional (NL) and healthy (H) skin. Proteins with expression that differs by at least 2-fold in non-lesional skin and healthy skin are listed (a). UGDH protein expression is similarly increased in NL and lesional psoriatic skin, compared to H controls. The highest difference in expression for NL and H was seen with immunohistochemical characterization of UGDH (n = 10), which indicated similar patterns of distribution in the three sample types. The strongest staining was observed in basal keratinocytes, and weaker staining was observed in the upper parts of the epidermis. Higher intensity staining UGDH was observed in non-lesional and lesional skin compared to healthy skin (b). A higher magnification of the epidermis is provided (c). (In merged figures, DAPI nuclear staining and UGDH are shown in blue and red, respectively; *: indicates statistical significance, ¥: indicates proteins with known association with psoriasis).
To verify our proteomic results, immunofluorescent staining was performed to gain additional information regarding protein localization, deposition and distribution. UGDH had the largest expression differences in non-lesional and healthy skin. As UGDH has not been linked to psoriasis previously, this protein was chosen for further analysis. UGDH staining showed similar epidermal distribution in all three sample types, with the highest protein levels detected in basal keratinocytes (n = 10 different individuals in each group, listed in Supplementary Table 6). Despite the similarities in the UGDH localization pattern, clear differences in staining intensities were observed. The non-lesional and lesional psoriatic samples displayed more robust intensities compared to that of healthy samples, confirming our proteomic results (Fig. 4b,c).
To determine which lesional alterations and to what extent are manifest in non-lesional skin, we selected the 249 proteins that exhibited differential expression in healthy and lesional skin and their expression levels was compared to those in non-lesional skin. In non-lesional skin, the expression of 199 (79.9%) of the proteins differed from the expression in healthy and lesional skin by less than two-fold. Therefore, this category was termed as intermediate, as they may represent a discrete step in the healthy-to-lesional transition (Supplementary Table 7).
Psoriatic biomarkers, biological functions, canonical pathways and annotation of diseases associated with the detected alterations in protein amounts
To examine the validity of our experimental approach, we further screened our proteomic dataset (lesional vs. healthy) for known, major biomarkers characteristic for psoriasis. These were identified previously in by large scale genomic, transcriptomic and/or proteomic studies. Out of these biomarkers AKR1B10, CSTA, FABP5, PI3, SCCA2, STAT1, STAT3 and members of the S100 family, including S100A7, S100A8, S100A9 were also found in our study. These molecules exhibited elevated expression levels in psoriatic lesions, compared to healthy control skin (Table 2).
Table 2.
Detected expressional differences of classic protein biomarkers for psoriasis.
| Gene ID of protein | L vs. H | NL vs. H | NL vs. L |
|---|---|---|---|
| AKR1B10 | 32.769* | 25.318 | 0.773 |
| CSTA | 2.335* | 1.752 | 0.75 |
| FABP5 | 15.076* | 4.678 | 0.31* |
| PI3 | 52.616* | 4.105 | 0.078 |
| S100A2 | 5.878* | 2.527 | 0.43 |
| S100A7 | 14.74* | 7.519 | 0.51 |
| S100A8 | 20.639* | 5.234 | 0.254* |
| S100A9 | 19.679* | 3.306 | 0.168* |
| SCCA2 | 35.468* | 9.221 | 0.26* |
| STAT1 | 20.504* | 14.478 | 0.706 |
| STAT3 | 3.766* | 2.309 | 0.613 |
Further analysis was performed to identify the cellular mechanisms that may be associated with the proteins that were detected in altered amounts in a proteomic approach, using the Ingenuity Pathway Analysis software (IPA). Diseases annotation revealed ‘psoriasis’ as the first hit when lesional and healthy (Table 3), or lesional and non-lesional differences (Table 3) were compared.
Table 3.
Disease and biological function annotation of differentially expressed proteins.
| Categories (L vs. H) | Diseases or disease related processes | p-value | Predicted Activation State | Activation z-score | Number of Proteins |
|---|---|---|---|---|---|
| Disease annotation of protein expressional differences between lesional (L) and healthy (H) skin | |||||
| Dermatological Diseases and Conditions, Organismal Injury and Abnormalities | Psoriasis | 6.89E-32 | — | — | 61 |
| Chronic psoriasis | 2.15E-23 | — | — | 27 | |
| Cancer, Cell Death and Survival, Organismal Injury and Abnormalities, Tumor Morphology | Cell death of osteosarcoma cells | 4.04E-22 | Decreased | −4.899 | 24 |
| Dermatological Diseases and Conditions, Organismal Injury and Abnormalities | Chronic skin disorder | 7.47E-22 | — | — | 28 |
| Cancer, Cell Death and Survival, Organismal Injury and Abnormalities, Tumor Morphology | Cell death of cancer cells | 4.24E-15 | Decreased | −4.64 | 33 |
| Infectious Diseases | Viral Infection | 1.89E-12 | Increased | 3.883 | 69 |
| Replication of virus | 3.58E-11 | — | 1.819 | 34 | |
| Replication of RNA virus | 1.93E-10 | — | 1.799 | 31 | |
| Dermatological Diseases and Conditions, Organismal Injury and Abnormalities | Plaque psoriasis | 4.05E-10 | — | — | 15 |
| Dermatological Diseases and Conditions, Immunological Disease, Inflammatory Disease, Organismal Injury and Abnormalities | Lichen planus | 4.86E-10 | — | — | 13 |
| Categories (NL vs. L) | Diseases or disease related processes | p-value | Predicted Activation State | Activation z-score | Number of Proteins |
| Disease annotation of protein expressional differences between non-lesional (NL) and lesional (L) skin | |||||
| Dermatological Diseases and Conditions, Organismal Injury and Abnormalities | Psoriasis | 3.72E-15 | — | — | 21 |
| Plaque psoriasis | 1.78E-11 | — | — | 10 | |
| Chronic skin disorder | 3.06E-10 | — | — | 10 | |
| Chronic psoriasis | 8.58E-10 | — | — | 9 | |
| Immunological Disease | Allergy | 8.57E-08 | — | — | 12 |
| Immunological Disease | Hypersensitive reaction | 1.30E-07 | — | — | 12 |
| Immunological Disease | Immediate hypersensitivity | 4.94E-07 | — | — | 10 |
| Dermatological Diseases and Conditions, Inflammatory Disease, Inflammatory Response, Organismal Injury and Abnormalities | Dermatitis | 9.38E-07 | — | −1.067 | 11 |
| Cardiovascular Disease, Organismal Injury and Abnormalities, Renal and Urological Disease | Ischemic acute renal failure | 2.99E-06 | — | — | 3 |
| Organismal Injury and Abnormalities, Reproductive System Disease | Endometriosis | 3.13E-06 | — | — | 10 |
| Categories (L vs.H) | Biological Function | p-value | Predicted Activation State | Activation z-score | Number of Proteins |
| Biological function annotation of protein expressional differences between lesional (L) and healthy (H) skin | |||||
| Protein Synthesis | Initiation of translation of protein | 3.11E-46 | — | — | 42 |
| Translation | 3.28E-40 | — | 0.737 | 57 | |
| Translation of protein | 1.09E-38 | — | 0.555 | 55 | |
| Synthesis of protein | 3.18E-36 | Increased | 2.691 | 64 | |
| Expression of protein | 6.76E-36 | — | 0.527 | 57 | |
| RNA Damage and Repair | Nonsense-mediated mRNA decay | 3.06E-35 | — | — | 32 |
| Protein Synthesis | Metabolism of protein | 6.39E-31 | Increased | 2.92 | 85 |
| Cell Death and Survival | Necrosis | 6.99E-18 | Decreased | −2.168 | 109 |
| RNA Post-Transcriptional Modification | Processing of RNA | 4.39E-14 | — | −0.577 | 32 |
| Cellular Movement | Migration of cells | 1.1E-12 | Increased | 2.067 | 83 |
| Categories (NL vs. L) | Biological Function | p-value | Predicted Activation State | Activation z-score | Number of Proteins |
| Biological function annotation of protein expressional differences between non-lesional (NL) and lesional (L) skin | |||||
| Cell Death and Survival | Killing of Staphylococcus aureus | 8.53E-10 | — | −0.655 | 5 |
| Cellular Movement,Immune Cell Trafficking | Leukocyte migration | 7.62E-08 | — | −0.509 | 18 |
| Cell Death and Survival | Killing of bacteria | 1.10E-07 | — | −1.608 | 6 |
| Cell-To-Cell Signaling and Interaction, Reproductive System Development and Function | Binding of gonadal cell lines | 1.19E-07 | — | 1.964 | 6 |
| Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking | Cell movement of leukocytes | 1.55E-07 | — | −0.429 | 16 |
| Cell Death and Survival | Necrosis | 2.20E-07 | — | −1.927 | 30 |
| Antimicrobial Response, Inflammatory Response | Antimicrobial response | 2.42E-07 | Decreased | −2 | 10 |
| Cell Death and Survival | Killing of Staphylococcus aureus subsp. aureus | 5.03E-07 | — | — | 3 |
| Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking, Inflammatory Response | Cell movement of phagocytes | 5.87E-07 | — | −0.902 | 13 |
| Cellular Compromise, Inflammatory Response | Degranulation of cells | 6.21E-07 | — | −0.87 | 13 |
Annotation of biological functions by IPA highlighted ‘initiation of protein translation’ (Table 3) and ‘killing of Staphylococcus aureus’ as the main functions likely to be affected, respectively. Ingenuity canonical pathway screening identified the ‘role of IL-17A in psoriasis’ among the top ten most significant canonical pathways, when either lesional, or non-lesional protein expression was compared to healthy samples (Table 4). In addition, several cancer, neurological and neuromuscular canonical pathways were also highlighted.
Table 4.
Canonical pathways predicted to be affected in psoriasis based on detected expressional differences of proteins.
| Ingenuity Canonical Pathways (L vs. H) | −log (p-value) |
|---|---|
| EIF2 Signaling | 3.69E + 01 |
| Regulation of eIF4 and p70S6K Signaling | 1.95E + 01 |
| mTOR Signaling | 1.16E + 01 |
| FAT10 Signaling Pathway | 4.22E + 00 |
| tRNA Charging | 4.01E + 00 |
| Role of IL-17A in Psoriasis | 3.32E + 00 |
| RAN Signaling | 2.96E + 00 |
| Intrinsic Prothrombin Activation Pathway | 2.80E + 00 |
| Polyamine Regulation in Colon Cancer | 2.62E + 00 |
| Neuroprotective Role of THOP1 in Alzheimer’s Disease | 2.56E + 00 |
| Ingenuity Canonical Pathways (NL vs. L) | −log(p-value) |
| Caveolar-mediated Endocytosis Signaling | 8.77E + 00 |
| Paxillin Signaling | 5.79E + 00 |
| EIF2 Signaling | 5.35E + 00 |
| Virus Entry via Endocytic Pathways | 4.51E + 00 |
| IL-8 Signaling | 4.41E + 00 |
| Integrin Signaling | 4.31E + 00 |
| Agrin Interactions at Neuromuscular Junction | 4.11E + 00 |
| Regulation of eIF4 and p70S6K Signaling | 3.88E + 00 |
| NF-κB Activation by Viruses | 3.68E + 00 |
| mTOR Signaling | 3.35E + 00 |
| Ingenuity Canonical Pathways (NL vs. H) | −log(p-value) |
| UDP-D-xylose and UDP-D-glucuronate Biosynthesis | 3.10E + 00 |
| 5-aminoimidazole Ribonucleotide Biosynthesis I | 2.92E + 00 |
| Tetrahydrofolate Salvage from 5,10-methenyltetrahydrofolate | 2.70E + 00 |
| Purine Nucleotides De Novo Biosynthesis II | 2.36E + 00 |
| Role of IL-17A in Psoriasis | 2.29E + 00 |
| Colanic Acid Building Blocks Biosynthesis | 2.25E + 00 |
| RAN Signaling | 2.17E + 00 |
| Intrinsic Prothrombin Activation Pathway | 1.79E + 00 |
| SPINK1 Pancreatic Cancer Pathway | 1.66E + 00 |
| MSP-RON Signaling Pathway | 1.54E + 00 |
Discussion
To expand knowledge about the pathomechanism of psoriasis, many extensive, large-scale comparative proteomic approaches have been performed17,24,26. However, the comparison of healthy, non-lesional and lesional skin at the proteomic level has been missing from these studies. To fill this gap, our comparative proteomic analysis included healthy skin as well as non-lesional and lesional psoriatic samples.
In order to check the validity of our proteomic approach we compared major known psoriatic biomarkers published in previous genetic (genome-wide association studies)21, transcriptomic19,21,23 and proteomic studies17,18,20,22 with our proteomic dataset. Known psoriatic lesional biomarkers also found in our study includes AKR1B1016, CSTA17, FABP518, PI319, SCCA220, STAT116, STAT321, S100A718–20, S100A819,22 and S100A919,20,22,23. Moreover, annotation of diseases resulted in the identification of psoriasis with the strongest correlation based on differentially expressed proteins in either lesional vs. healthy or in lesional vs. non-lesional comparison. Canonical pathway analysis of non-lesional differences compared to healthy skin resulted in the identification of ‘Role of IL-17A in Psoriasis’. However, these annotations also highlighted cancer, neurological, neuromuscular or muscular disease-related mechanism, suggesting their potential involvement in disease pathomechanism, or some similarities between these diseases.
Since our proteomic and in silico analysis cannot distinguish between cell-types, and provide information whether mechanistically linked alterations are taking place within the same, or different cell types, further experiments are required in this direction to clarify the exact relevance of these predicted connections to psoriasis pathomechanism.
We performed literature a search for known functions of proteins found to be altered in amounts in our study to suggest mechanism through which they may potentially participate in the pathomechanism of the disease. The detected differences in the expression of proteins in healthy and lesional skin highlighted involvement in psoriasis of cell proliferation28, development29, response to stimulus30, expression31 related processes. In the comparison of non-lesional and lesional skin, we identified 56 proteins with differential expression, which represents only 22.5% of the number of proteins which showed altered expression in the comparison of healthy and lesional skin (56 vs. 249). This highlights the importance of studying healthy skin in comparisons using patient samples for pinpointing disease-associated alterations. Qualitative literature-based analysis of these 56 proteins led to the identification of several mechanism for which association with psoriasis has already been described, including processes related to development29, response to stimulus26 and expression31.
Further analysis focused on gaining insight about the extent to which alterations are manifest in lesions and in non-lesional skin. Strikingly, nearly 80% of the 249 proteins exhibiting differential expression in lesional and healthy skin exhibited an intermediate expression level in the non-lesional skin, suggesting the possible presence of early, lesional-like alterations in non-lesional skin. Divergence from this trend was only observed for two small protein groups. Ten proteins — CHCHD6, CHMP5, COLEC12, FLOT2, ITGA7, LEMD2, NOP56, PLVAP, RRAS and SMARCA5 — differed in relative protein amounts in non-lesional and lesional skin, but the amounts of these proteins were similar in healthy and lesional samples. These ten proteins are likely to represent a group of non-lesional characteristic alteration. For nine proteins — CD207, COLEC12, CTSV, ITGA7, ITGA8, PLVAP, PSAPL1, SMARCA5 and XP32 — the direction of the expressional changes was different in non-lesional and lesional samples compared to healthy skin and might represent proteins that contribute to maintaining the non-lesional state.
Next, with the proteins in these two groups, we performed an extensive literature search to suggest potential mechanisms by which they may influence disease pathogenesis. Interestingly, all the identified proteins may play a role in signaling at different levels starting from the cell surface all away to the nucleus or mitochondria. The identified cell surface receptors include two integrins (ITGA7 and ITGA8) that are important in external signal recognition. Decreased ITGA7 levels — as observed in lesional vs. non-lesional skin — could be associated with delayed autophagy32, differentiation33 and increased migration34, all known to be affected in psoriatic lesions. In contrast, elevation of ITGA7 may induce growth suppression35. However, ITGA7 is characteristically expressed mainly by smooth muscle cells36 in the skin, suggesting that its involvement in keratinocyte related events are at least limited, or none. Instead, may suggest alterations in (vascular) smooth muscle cell adhesion-related processes37. Alternatively, ITGA7 may influence neurite outgrowth38. Therefore, further studies are required to confirm the observed TGA7 expression alteration and to identify the cell-types of source. Another identified cell-surface molecule, MYO18A, through recognizing microorganism lipopolysaccharides, may increase innate immune responses39 promoting cytokine production towards Th1 direction40; known to be important in psoriasis41.
FLOT242, CHMP543 and COLEC1244 participate in endocytic pathways, regulating the levels of cell-surface receptors and, thereby, signaling. FLOT2 is a known component of the raft microdomain complex that represents the major unit regulating STAT signaling pathways according to the raft-STAT signaling hypothesis45. The alteration of FLOT2 expression could suggest a high relevance since STAT3 is a key regulator in psoriasis46. Reduction of the scavenger receptor COLEC12 could trigger psoriasis by trastuzumab treatment47. Moreover, COLEC12 may influence the mitochondrial respiratory chain48, and this property is in agreement with the decreased level of mitochondrial MICOS complex subunit CHCHD649 observed in lesions compared to non-lesional skin. CHCHD6 regulates oxygen consumption and thereby may influence cell growth50. Reduced levels of CHCHD6 were shown previously to lead to a shift from oxidative metabolism to glycolytic metabolism51 that negatively influences keratinocyte differentiation52, and both types of mechanisms are known to be affected in psoriasis53,54.
The altered expression of LEMD2 may suggest that signal transduction is also altered at the level of the nucleus. LEMD2, located in the inner nuclear membrane, regulates nuclear import/export processes55 and thereby intranuclear signaling50. During this regulation, LEMD2 is associated with the same complex as CHMP5, which was also identified in our studies. In the nucleus, the STAT-regulated protein NOP5656, a core protein of the box C/D small nucleolar ribonucleoprotein (snoRNP) complex, participates in the biogenesis of rRNAs57. Increased rRNA biogenesis is suggested to be necessary for high proliferation rate58, a process that is crucial for the development of psoriatic lesions.
Abnormal proliferation28, differentiation52 and, thereby, skin barrier function are key processes during psoriatic plaque formation. SMARCA5 is a component of the nucleosome remodeling factor complex59. Decreasing SMARCA5 levels are required for basal keratinocytes to shift form proliferation toward differentiation60. XP32 is also a component of the epidermal differentiation complex61 and associated with skin barrier function62. The observed contrasting expressional differences of these two proteins in the non-lesional and lesional skin may contribute to our understanding of lesion formation and how non-lesional skin maintains its state.
Overall, our results indicate that dysregulation of cellular signaling — from signal detection, through endocytosis of receptors and transduction of signal from the cell surface to the nucleus — may be affected during the disease. The alteration of these systems is likely to lead to increased reaction to external signals that could contribute to the maintenance of psoriatic plaques.
By comparing non-lesional and healthy skin, differential expression was observed for eight proteins (CSE1L, GART, GBP1, KLK10, MYO18A, S1007A, SMARCA5 and UGDH). Four of these proteins (CSE1L, GART, GBP1, UGDH) might be predisposing factors, as their expression was similar in non-lesional and lesional skin, and their significance would have been missed in comparisons for which healthy samples were not included. Of these, UGDH was detected with the highest relative difference. UGDH has not been highlighted previously in association with psoriasis. We therefore decided to analyze it further. Immunohistochemical analysis confirmed our proteomic results: higher UGDH levels were found in non-lesional and lesional skin compared to healthy skin, that was mainly associated with keratinocytes. Elevated UGDH levels may increase chondrocytes proliferation indirectly, likely through increased hyaluronan production that binds different cytokines63. However, in vitro downregulation of UGDH and consequently decreased hyaluronan amounts did not influence keratinocyte proliferation64. These results are in line with our observation, suggesting that elevated UGDH levels observed in non-lesional keratinocytes are not sufficient to modify their proliferation.
Proteins for which expression was affected only in lesions are often considered “trigger” proteins, as changes in the expression of these proteins are linked to the shift of the disease state. The proteins that have not previously been associated with psoriasis were categorized into two groups. The first group of proteins might contribute to the mechanosensitivity of the tissue (SGCD65, SYNM66, MYH1167, ATP1B168). The second group functions within the nervous system (MPZ, PRX69, CSPG470, CNTN1 and ITGA871, ATP1B172), could suggest the involvement of the peripheral nerve system in psoriasis73.
Finally, we searched for potentially central proteins in disease pathogenesis participating in key mechanisms of psoriasis including regulation of stress and immune response, proliferation and differentiation. Some of the identified proteins, such as PML74 and STAT175, have already been linked to psoriasis. We also identified two proteins, PRKDC and MYBBP1A, which have not previously been highlighted in context with the disease. The PRKDC may plays a role in the detection and repair of breaks in double-stranded DNA76 and mediates the phosphorylation of c-MYC77 and p5378 suggesting a potentially important role in psoriasis. The suggested altered expression by our results of the transcription factor MYBBP1A may also be among the potentially important proteins in psoriasis implicated in the pathogenesis since it functions as a co-repressor of NF-κB that may regulate responses to stress and cytokines79.
Taken together, our comparative proteomic approach of healthy, non-lesional and lesional skin led to the identification of various proteins which may function in psoriasis pathogenesis, providing a strong base for future studies. Proteins exhibiting opposite expression changes in lesional and non-lesional samples compared to healthy skin may function in the maintenance of the non-lesional stage and may represent future targets for therapeutic purposes.
Materials and Methods
Ethics
Skin biopsy collection from donors, the procedure of collection and all experimental protocols were approved by the Regional and Institutional Research Ethics Committee and by the Human Investigation Review Board of the University of Szeged (SOEDAFN-002, IF-562-5/2016 and IF-15056/2015; 157/2015; 3638 and 2799, 3517), strictly following the guidelines and regulations of the Declaration of Helsinki. Prior to surgical intervention and following a detailed description of the skin biopsy donation procedure, participants provided written informed consent. No donor under the age of 18 was included in our study.
Criteria for inclusion of patients in the study and skin sample collection
To identify alterations that are general in chronic plaque psoriasis and keep the number of volunteers for skin biopsy collection to a minimum, for our proteomic approach, we randomly engaged individuals (I) of different age to minimize possible age-related differences; (II) with various Psoriasis Area Severity Index (PASI) scores between 5 and 25, since the score for an individual patient varies over time and with relapse; (III) of both genders to avoid possible gender-associated differences; and (IV) with both early and late onset. A total of 9 (3 × 3) patients suffering from chronic plaque psoriasis and the same number of healthy donors were involved in our study. The data of individuals involved in the study are summarized in Table 1. All psoriatic patients had not received any kind of treatment for the condition for at least 6 months. The 6 mm skin punch biopsies containing the epidermis and the dermis were collected from an area of the upper-middle gluteal region that is not exposed to sunlight. Both lesional and non-lesional samples were collected from patients. Non-lesional samples were taken at least 7 cm from the edge of the lesion subjected for biopsy. The presence of psoriasis was clinically verified for all patients, and clinical as well as demographic data of the donors are presented in Table 1.
Comprehensive and comparative proteomics of healthy, non-lesional and lesional skin
Sample preparation from skin biopsy and sequential protein extraction
Samples were cut with a razor blade. Skin proteins were extracted sequentially in four consecutive solubility-based extraction steps. Extraction buffers were used in increasing order of their solubilizing properties for a better separation of proteins. Samples were initially incubated in extraction buffer I. (0.15 M NaCl, 50 mM Tris-HCl, pH 7.4) for 24 h at 4 °C in the presence of protease inhibitors. Protein extracts were then clarified by centrifugation and separated from the pellet. This step was repeated by resuspending the pellet in extraction buffer II, which contained 1 M NaCl, 25 mM EDTA, 50 mM Tris-HCl, pH 7.4. Following extraction with 250 mM SDS-containing extraction buffer III (8 h at room temperature), guanidine hydrochloride containing extraction buffer IV (4 M GuHCl, 10 mM EDTA, 50 mM Tris-HCl, pH 7.4) was applied for 48 h at 4 °C. The same protein extracts of the three donors were pooled in each investigated group (healthy, non-lesional, lesional). Extraction procedure was carried out three times and each contained extracted proteins of three donors following the pooling of the samples which were than subjected for downstream proteomic analysis.
Protein identification by 2D LC-MSMS
A total 35 µg protein from each sample was applied for mass spectrometry analysis. A modified filter-aided sample preparation method was used for tryptic digestion of the protein extracts80. High-pH reversed-phase chromatography was performed on a C18 column (Phenomenex, Kinetex 5 µ EVO C18 100 A, 2.1 × 100 mm; cat. no. 00D-4622-AN, flow rate: 150 µl/min). Forty-eight fractions were collected from 1 to 25 minutes (half minute/fraction) and 4-4 fractions were combined (1,13,24,37; 2,14,25,38 and so on) to get 12 final fractions. Each fraction was subjected to nano LC-MSMS analysis on an Orbitrap Elite hybrid mass spectrometer (Thermo) coupled with a Waters nanoAcquity UPLC system, using a gradient elution after trapping the samples onto the trap column. Data-dependent analyses were applied; the 20 most intense peaks were selected for ion-trap collision-induced dissociation after each survey scan measured in the Orbitrap. Proteome Discoverer (ver.: 1.3) was used to generate MS/MS peak-list files and our in-cloud ProteinProspector (ver.: 5.16.0) database search engine was used for protein identification against the human sequences from the UniProtKB.2015.12.14.random.concat (149781/55820795 entries searched) database. Detailed protocols and applied counting for semiquantitative analysis is described as Supplementary Information.
Immunofluorescence staining of skin sections for UGDH
For immunofluorescence analysis, 5 µm sections of frozen embedded skin biopsies from psoriatic patients (non-lesional and lesional skin) and healthy individuals were used. After fixation and permeabilization (Foxp3 staining buffer set, fixation/permeabilization kit, Miltenyi Biotec, used according to the description of the manufacturer), samples were blocked in Tris-buffered saline (TBS) containing 1% bovine serum albumin (BSA) and 1% normal goat serum (NGS) for 1 h at room temperature. Samples were incubated overnight at 4 °C in TBS with 1% NGS and primary antibodies against UGDH (rabbit polyclonal antibody, ab155005, Abcam), diluted to 1:100. Following washing in TBS, AF546 secondary antibodies (Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546, A-11035, Invitrogen), diluted in TBS containing 1% NGS to 1:500, were applied for 1 h at room temperature.
Literature search to identify novel psoriasis-associated proteins
To identify proteins not yet linked with the pathomechanism of psoriasis, literature mining was carried out using protein names or the encoding gene’s HUGO Gene Nomenclature Committee (HGNC) symbol(s), applying the following strategy: each protein or gene name was searched together with “psoriasis” as a keyword using the RISmed R package.
Statistical analysis
To compare protein abundance from healthy, lesional and non-lesional skin extracts, significant differences were determined based on relative peptide ion chromatograms and spectrum counting and evaluated using two different approaches: (1) modified t-test (limma) and (2) rank product test (as described by Schwämmle et al.81) following t-test. We considered a protein amount to be different between two samples if at least one of the three tests were significant (test <0.05) and the absolute fold change was at least two or higher.
Supplementary information
Acknowledgements
The MTA Cloud (https://cloud.mta.hu/) and Peter Baker are acknowledged for the use of ProteinProspector database search, which significantly helped us achieving the results published in this paper. The study was supported by NKFI (formal OTKA) PD116992, K111885 and GINOP-2.2.1-15-2016-00007, GINOP-2.3.2-15-2016-00020 research grants and co-financed by the European Social Fund in the framework of TAMOP-4.2.4.A/2-11-1/2012-0001 “National Excellence Program” A2-SZGYA-FOK-13-0001. K.S. is a recipient of the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, and also supported by the UNKP-18-4 New National Excellence Program of the Ministry of Human Capacities. Publication costs were financed by the ‘University of Szeged Open Access Fund’ (Grant number: 4151).
Author Contributions
G.G. conceived the study; G.G., Z.B.C. and L.K. supervised the project; É.H.-G. and G.G. designed the experiments; E.S., R.B., É.H.-G. and G.G. performed the experiments; statistical analysis was done by M.M.; E.S., R.B., M.M. and G.G. analyzed the proteomic data; experiments were performed in the laboratory of Z.B.C., K.L. and K.S.; E.S., K.S. and G.G. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47774-5.
References
- 1.Slominski AT, Zmijewski MA, Plonka PM, Szaflarski JP, Paus R. How UV Light Touches the Brain and Endocrine System Through Skin, and Why. Endocrinology. 2018;159:1992–2007. doi: 10.1210/en.2017-03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slominski, A. T. et al. Sensing the environment: regulation of local and global homeostasis by the skin’s neuroendocrine system. Adv. Anat. Embryol. Cell Biol. 212, v, vii, 1–115 (2012). [DOI] [PMC free article] [PubMed]
- 3.Gubán B, et al. Abnormal regulation of fibronectin production by fibroblasts in psoriasis. Br. J. Dermatol. 2016;174:533–541. doi: 10.1111/bjd.14219. [DOI] [PubMed] [Google Scholar]
- 4.Eyre RW, Krueger GG. Response to injury of skin involved and uninvolved with psoriasis, and its relation to disease activity: Koebner and ‘reverse’ Koebner reactions. Br. J. Dermatol. 1982;106:153–159. doi: 10.1111/j.1365-2133.1982.tb00924.x. [DOI] [PubMed] [Google Scholar]
- 5.Ye L, et al. Abnormal epidermal barrier recovery in uninvolved skin supports the notion of an epidermal pathogenesis of psoriasis. J. Invest. Dermatol. 2014;134:2843–2846. doi: 10.1038/jid.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudjonsson JE, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J. Invest. Dermatol. 2009;129:2795–2804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabó K, et al. Regulatory networks contributing to psoriasis susceptibility. Acta Derm. Venereol. 2014;94:380–385. doi: 10.2340/00015555-1708. [DOI] [PubMed] [Google Scholar]
- 8.Henno A, et al. Altered expression of angiogenesis and lymphangiogenesis markers in the uninvolved skin of plaque-type psoriasis. Br. J. Dermatol. 2009;160:581–590. doi: 10.1111/j.1365-2133.2008.08889.x. [DOI] [PubMed] [Google Scholar]
- 9.Chiricozzi A, et al. Increased expression of interleukin-17 pathway genes in nonlesional skin of moderate-to-severe psoriasis vulgaris. Br. J. Dermatol. 2016;174:136–145. doi: 10.1111/bjd.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting KM, et al. Overexpression of the oncofetal Fn variant containing the EDA splice-in segment in the dermal-epidermal junction of psoriatic uninvolved skin. J. Invest. Dermatol. 2000;114:706–711. doi: 10.1046/j.1523-1747.2000.00871.x. [DOI] [PubMed] [Google Scholar]
- 11.Szlavicz E, et al. Splicing factors differentially expressed in psoriasis alter mRNA maturation of disease-associated EDA+ fibronectin. Mol. Cell. Biochem. 2017;436:189–199. doi: 10.1007/s11010-017-3090-1. [DOI] [PubMed] [Google Scholar]
- 12.Lee SE, Lew W. The Increased Expression of Matrix Metalloproteinase-9 Messenger RNA in the Non-lesional Skin of Patients with Large Plaque Psoriasis Vulgaris. Ann. Dermatol. 2009;21:27–34. doi: 10.5021/ad.2009.21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danis J, Göblös A, Bata-Csörgő Z, Kemény L, Széll M. PRINS Non-Coding RNA Regulates Nucleic Acid-Induced Innate Immune Responses of Human Keratinocytes. Front. Immunol. 2017;8:1053. doi: 10.3389/fimmu.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szegedi K, et al. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp. Dermatol. 2010;19:269–278. doi: 10.1111/j.1600-0625.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- 15.Göblös A, et al. Keratinocytes express functional CARD18, a negative regulator of inflammasome activation, and its altered expression in psoriasis may contribute to disease pathogenesis. Mol. Immunol. 2016;73:10–18. doi: 10.1016/j.molimm.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Suárez-Fariñas M, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J. Invest. Dermatol. 2012;132:2552–2564. doi: 10.1038/jid.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundberg KC, et al. Proteomics of skin proteins in psoriasis: from discovery and verification in a mouse model to confirmation in humans. Mol. Cell. Proteomics MCP. 2015;14:109–119. doi: 10.1074/mcp.M114.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson JC, et al. A proteomics approach to the identification of biomarkers for psoriasis utilising keratome biopsy. J. Proteomics. 2013;94:176–185. doi: 10.1016/j.jprot.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Gudjonsson JE, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 2010;130:1829–1840. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piruzian E, et al. Integrated network analysis of transcriptomic and proteomic data in psoriasis. BMC Syst. Biol. 2010;4:41. doi: 10.1186/1752-0509-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schonthaler HB, et al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39:1171–1181. doi: 10.1016/j.immuni.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Oestreicher JL, et al. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J. 2001;1:272–287. doi: 10.1038/sj.tpj.6500067. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. Identification of unique proteomic signatures in allergic and non-allergic skin disease. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2017;47:1456–1467. doi: 10.1111/cea.12979. [DOI] [PubMed] [Google Scholar]
- 25.Kang H, et al. Exploration of candidate biomarkers for human psoriasis based on gas chromatography-mass spectrometry serum metabolomics. Br. J. Dermatol. 2017;176:713–722. doi: 10.1111/bjd.15008. [DOI] [PubMed] [Google Scholar]
- 26.Méhul B, et al. Noninvasive proteome analysis of psoriatic stratum corneum reflects pathophysiological pathways and is useful for drug profiling. Br. J. Dermatol. 2017;177:470–488. doi: 10.1111/bjd.15346. [DOI] [PubMed] [Google Scholar]
- 27.Ryu J, et al. Proteomic analysis of psoriatic skin tissue for identification of differentially expressed proteins: up-regulation of GSTP1, SFN and PRDX2 in psoriatic skin. Int. J. Mol. Med. 2011;28:785–792. doi: 10.3892/ijmm.2011.757. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M, et al. TGFβ/SMAD/microRNA-486-3p Signaling Axis Mediates Keratin 17 Expression and Keratinocyte Hyperproliferation in Psoriasis. J. Invest. Dermatol. 2017;137:2177–2186. doi: 10.1016/j.jid.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Mali JW. Psoriasis: a dynamic disease. Br. J. Dermatol. 1979;101:725–730. doi: 10.1111/j.1365-2133.1979.tb05655.x. [DOI] [PubMed] [Google Scholar]
- 30.Schopf RE, Straussfeld E. Stimulus-dependent increased generation of oxygen intermediates in monocytes and polymorphonuclear leukocytes in psoriasis. J. Invest. Dermatol. 1985;84:73–76. doi: 10.1111/1523-1747.ep12274844. [DOI] [PubMed] [Google Scholar]
- 31.Dou J, et al. Integrative analyses reveal biological pathways and key genes in psoriasis. Br. J. Dermatol. 2017;177:1349–1357. doi: 10.1111/bjd.15682. [DOI] [PubMed] [Google Scholar]
- 32.Tang Y, et al. ART1 promotes starvation-induced autophagy: a possible protective role in the development of colon carcinoma. Am. J. Cancer Res. 2015;5:498–513. [PMC free article] [PubMed] [Google Scholar]
- 33.Ozeki N, et al. Bone morphogenetic protein-induced cell differentiation involves Atg7 and Wnt16 sequentially in human stem cell-derived osteoblastic cells. Exp. Cell Res. 2016;347:24–41. doi: 10.1016/j.yexcr.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Laszlo V, et al. Epigenetic down-regulation of integrin α7 increases migratory potential and confers poor prognosis in malignant pleural mesothelioma. J. Pathol. 2015;237:203–214. doi: 10.1002/path.4567. [DOI] [PubMed] [Google Scholar]
- 35.Tan L-Z, Song Y, Nelson J, Yu YP, Luo J-H. Integrin α7 binds tissue inhibitor of metalloproteinase 3 to suppress growth of prostate cancer cells. Am. J. Pathol. 2013;183:831–840. doi: 10.1016/j.ajpath.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson E. Alpha 7 beta 1 integrin: putting the brakes on smooth muscle cell proliferation. Circ. Res. 2007;101:651–653. doi: 10.1161/CIRCRESAHA.107.161877. [DOI] [PubMed] [Google Scholar]
- 37.de Rezende FF, et al. Integrin α7β1 is a redox-regulated target of hydrogen peroxide in vascular smooth muscle cell adhesion. Free Radic. Biol. Med. 2012;53:521–531. doi: 10.1016/j.freeradbiomed.2012.05.032. [DOI] [PubMed] [Google Scholar]
- 38.Plantman S, et al. Integrin-laminin interactions controlling neurite outgrowth from adult DRG neurons in vitro. Mol. Cell. Neurosci. 2008;39:50–62. doi: 10.1016/j.mcn.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, et al. SP-R210 (Myo18A) Isoforms as Intrinsic Modulators of Macrophage Priming and Activation. PloS One. 2015;10:e0126576. doi: 10.1371/journal.pone.0126576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hohlfeld JM, Erpenbeck VJ, Krug N. Surfactant proteins SP-A and SP-D as modulators of the allergic inflammation in asthma. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2002;70:287–292. doi: 10.1159/000070744. [DOI] [PubMed] [Google Scholar]
- 41.Jiang W, et al. A Toll-like receptor 7, 8, and 9 antagonist inhibits Th1 and Th17 responses and inflammasome activation in a model of IL-23-induced psoriasis. J. Invest. Dermatol. 2013;133:1777–1784. doi: 10.1038/jid.2013.57. [DOI] [PubMed] [Google Scholar]
- 42.Solis GP, et al. Reggies/flotillins regulate E-cadherin–mediated cell contact formation by affecting EGFR trafficking. Mol. Biol. Cell. 2012;23:1812–1825. doi: 10.1091/mbc.e11-12-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim J-H, et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham SA, et al. Identification of neutrophil granule glycoproteins as Lewis(x)-containing ligands cleared by the scavenger receptor C-type lectin. J. Biol. Chem. 2011;286:24336–24349. doi: 10.1074/jbc.M111.244772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalazar G, et al. Beta-glycoglycosphingolipid-induced alterations of the STAT signaling pathways are dependent on CD1d and the lipid raft protein flotillin-2. Am. J. Pathol. 2009;174:1390–1399. doi: 10.2353/ajpath.2009.080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calautti, E., Avalle, L. & Poli, V. Psoriasis: A STAT3-Centric View. Int. J. Mol. Sci. 19 (2018). [DOI] [PMC free article] [PubMed]
- 47.Kim DH, et al. Psoriasis induced by trastuzumab (herceptin®) Ann. Dermatol. 2013;25:229–231. doi: 10.5021/ad.2013.25.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degenhardt F, et al. Genome-wide association study of serum coenzyme Q10 levels identifies susceptibility loci linked to neuronal diseases. Hum. Mol. Genet. 2016;25:2881–2891. doi: 10.1093/hmg/ddw134. [DOI] [PubMed] [Google Scholar]
- 49.Ott C, Dorsch E, Fraunholz M, Straub S, Kozjak-Pavlovic V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PloS One. 2015;10:e0120213. doi: 10.1371/journal.pone.0120213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.An J, et al. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J. Biol. Chem. 2012;287:7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chella Krishnan K, et al. Integration of Multi-omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-alcoholic Fatty Liver Disease. Cell Syst. 2018;6:103–115.e7. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monteleon CL, et al. Lysosomes Support the Degradation, Signaling, and Mitochondrial Metabolism Necessary for Human Epidermal Differentiation. J. Invest. Dermatol. 2018;138:1945–1954. doi: 10.1016/j.jid.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambert S, Swindell WR, Tsoi LC, Stoll SW, Elder JT. Dual Role of Act1 in Keratinocyte Differentiation and Host Defense: TRAF3IP2 Silencing Alters Keratinocyte Differentiation and Inhibits IL-17 Responses. J. Invest. Dermatol. 2017;137:1501–1511. doi: 10.1016/j.jid.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, et al. Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat. Med. 2018;24:617–627. doi: 10.1038/s41591-018-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu M, et al. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA. 2017;114:E2166–E2175. doi: 10.1073/pnas.1613916114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowling VH, Turner SA, Cole MD. Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene. 2014;33:3519–3527. doi: 10.1038/onc.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lykke-Andersen S, Ardal BK, Hollensen AK, Damgaard CK, Jensen TH. Box C/D snoRNP Autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA. Mol. Cell. 2018;72:99–111.e5. doi: 10.1016/j.molcel.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 58.Marcel V, et al. Expression Profiling of Ribosome Biogenesis Factors Reveals Nucleolin as a Novel Potential Marker to Predict Outcome in AML Patients. PloS One. 2017;12:e0170160. doi: 10.1371/journal.pone.0170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 60.Mulder KW, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat. Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- 61.Zhao XP, Elder JT. Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics. 1997;45:250–258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- 62.Toulza E, et al. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007;8:R107. doi: 10.1186/gb-2007-8-6-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarkin CE, et al. Regulation of UDP-glucose dehydrogenase is sufficient to modulate hyaluronan production and release, control sulfated GAG synthesis, and promote chondrogenesis. J. Cell. Physiol. 2011;226:749–761. doi: 10.1002/jcp.22393. [DOI] [PubMed] [Google Scholar]
- 64.Malaisse J, et al. Hyaluronan Does Not Regulate Human Epidermal Keratinocyte Proliferation and Differentiation. J. Biol. Chem. 2016;291:6347–6358. doi: 10.1074/jbc.M115.661348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goehringer C, et al. Prevention of cardiomyopathy in delta-sarcoglycan knockout mice after systemic transfer of targeted adeno-associated viral vectors. Cardiovasc. Res. 2009;82:404–410. doi: 10.1093/cvr/cvp061. [DOI] [PubMed] [Google Scholar]
- 66.Izmiryan A, et al. Different expression of synemin isoforms in glia and neurons during nervous system development. Glia. 2006;54:204–213. doi: 10.1002/glia.20378. [DOI] [PubMed] [Google Scholar]
- 67.Smith AS, et al. Myosin IIA interacts with the spectrin-actin membrane skeleton to control red blood cell membrane curvature and deformability. Proc. Natl. Acad. Sci. USA. 2018;115:E4377–E4385. doi: 10.1073/pnas.1718285115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mladinov D, Liu Y, Mattson DL, Liang M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase β1. Nucleic Acids Res. 2013;41:1273–1283. doi: 10.1093/nar/gks1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S, et al. Schwann Cell O-GlcNAc Glycosylation Is Required for Myelin Maintenance and Axon Integrity. J. Neurosci. Off. J. Soc. Neurosci. 2016;36:9633–9646. doi: 10.1523/JNEUROSCI.1235-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schäfer Michael K.E., Tegeder Irmgard. NG2/CSPG4 and progranulin in the posttraumatic glial scar. Matrix Biology. 2018;68-69:571–588. doi: 10.1016/j.matbio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 71.Müller U, Bossy B, Venstrom K, Reichardt LF. Integrin alpha 8 beta 1 promotes attachment, cell spreading, and neurite outgrowth on fibronectin. Mol. Biol. Cell. 1995;6:433–448. doi: 10.1091/mbc.6.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johar K, Priya A, Wong-Riley MTT. Regulation of Na(+)/K(+)-ATPase by neuron-specific transcription factor Sp4: implication in the tight coupling of energy production, neuronal activity and energy consumption in neurons. Eur. J. Neurosci. 2014;39:566–578. doi: 10.1111/ejn.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu TH, et al. The Role of the Nervous System in the Pathophysiology of Psoriasis: A Review of Cases of Psoriasis Remission or Improvement Following Denervation Injury. Am. J. Clin. Dermatol. 2016;17:257–263. doi: 10.1007/s40257-016-0183-7. [DOI] [PubMed] [Google Scholar]
- 74.Stoppe M, et al. Cerebellar manifestation of PML under fumarate and after efalizumab treatment of psoriasis. J. Neurol. 2014;261:1021–1024. doi: 10.1007/s00415-014-7311-1. [DOI] [PubMed] [Google Scholar]
- 75.Shi X, et al. IL-17A upregulates keratin 17 expression in keratinocytes through STAT1- and STAT3-dependent mechanisms. J. Invest. Dermatol. 2011;131:2401–2408. doi: 10.1038/jid.2011.222. [DOI] [PubMed] [Google Scholar]
- 76.Lee S-H, Kim C-H. DNA-dependent protein kinase complex: a multifunctional protein in DNA repair and damage checkpoint. Mol. Cells. 2002;13:159–166. [PubMed] [Google Scholar]
- 77.An J, et al. DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Mol. Cancer. 2008;7:32. doi: 10.1186/1476-4598-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gurley KE, Ashley AK, Moser RD, Kemp CJ. Synergy between Prkdc and Trp53 regulates stem cell proliferation and GI-ARS after irradiation. Cell Death Differ. 2017;24:1853–1860. doi: 10.1038/cdd.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owen HR, et al. MYBBP1a is a novel repressor of NF-kappaB. J. Mol. Biol. 2007;366:725–736. doi: 10.1016/j.jmb.2006.11.099. [DOI] [PubMed] [Google Scholar]
- 80.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 81.Schwämmle V, León IR, Jensen ON. Assessment and improvement of statistical tools for comparative proteomics analysis of sparse data sets with few experimental replicates. J. Proteome Res. 2013;12:3874–3883. doi: 10.1021/pr400045u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.