Abstract
Metabolomics is an emerging science that can inform pathogenic mechanisms behind clinical phenotypes in COPD. We aimed to understand disturbances in the serum metabolome associated with respiratory outcomes in ever-smokers from the SPIROMICS cohort. We measured 27 serum metabolites, mostly amino acids, by 1H-nuclear magnetic resonance spectroscopy in 157 white ever-smokers with and without COPD. We tested the association between log-transformed metabolite concentrations and one-year incidence of respiratory exacerbations after adjusting for age, sex, current smoking, body mass index, diabetes, inhaled or oral corticosteroid use, study site and clinical predictors of exacerbations, including FEV1% predicted and history of exacerbations. The mean age of participants was 53.7 years and 58% had COPD. Lower concentrations of serum amino acids were independently associated with 1-year incidence of respiratory exacerbations, including tryptophan (β = −4.1, 95% CI [−7.0; −1.1], p = 0.007) and the branched-chain amino acids (leucine: β = −6.0, 95% CI [−9.5; −2.4], p = 0.001; isoleucine: β = −5.2, 95% CI [−8.6; −1.8], p = 0.003; valine: β = −4.1, 95% CI [−6.9; −1.4], p = 0.003). Tryptophan concentration was inversely associated with the blood neutrophil-to-lymphocyte ratio (p = 0.03) and the BODE index (p = 0.03). Reduced serum amino acid concentrations in ever-smokers with and without COPD are associated with an increased incidence of respiratory exacerbations.
Subject terms: Chronic obstructive pulmonary disease, Predictive markers, Translational research
Introduction
Chronic obstructive pulmonary disease (COPD) is a prevalent condition typically caused by exposure to smoking or other noxious particles and characterized by chronic respiratory symptoms and irreversible airflow obstruction1. COPD is a highly heterogeneous disorder; some patients experience frequent respiratory exacerbations while other do not2,3, not all patients progress towards severe emphysema regardless of the severity of airflow obstruction, and women tend to have more rapid disease progression than men4. This complex heterogeneity cannot be fully captured by lung function measurement alone or by any single biomarker.
Metabolomics is an emerging science that involves the simultaneous measurement of endogenous low molecular weight compounds (≤1,500 Daltons) in a biological specimen5,6. Not only does the metabolome interact with and reflect the activity of the genome, epigenome and proteome, but it is also influenced by one’s diet, lifestyle and medications5. Therefore, given its capture of gene function, enzyme activity and physiologic changes, the study of metabolomics has the potential to inform the clinical heterogeneity of COPD phenotypes. While human studies conducted to date have showed that metabolomics analysis of the serum, plasma, urine and exhaled breath condensate can discriminate between never smokers, smokers with COPD and smokers without COPD7–11, the relationship between metabolic profiles and the clinical heterogeneity of COPD has not yet been fully characterized.
A key outcome in the natural history of COPD is respiratory exacerbations which are associated with accelerated lung function decline12 and increased mortality13. It has been established that plasma metabolomics become significantly disturbed during acute exacerbation events14. We hypothesized that an altered serum metabolome among ever-smokers in the stable state would be associated with an increased risk of future respiratory exacerbations. To test this hypothesis, we conducted a prospective analysis of the relationship between the baseline serum metabolome measured by 1H-nuclear magnetic resonance (NMR) spectroscopy and the incidence of exacerbations over one year of follow-up in participants of the Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS) cohort15. We also explored cross-sectional associations between serum metabolomics and other pulmonary parameters such as lung function, extent of emphysema on chest computed tomography (CT) and exertional capacity.
Results
Description of participants and metabolites
The 157 participants included in this analysis had a mean age of 53.7 years, with 56.8% being current smokers and 58.0% having COPD confirmed on spirometry (Table 1). The prevalence of diabetes mellitus in our cohort was 7.1%, and 30.5% of participants were on inhaled corticosteroids. Among the 138 participants for whom information on prospective respiratory exacerbations was available, 109 reported no exacerbations, while 29 experienced at least one exacerbation over one year of follow-up. Participants in the exacerbation group were more likely to have COPD (especially with spirometry grade Global Initiative for Chronic Obstructive Lung Disease (GOLD) 3–4 severity), more emphysema on chest CT and diabetes mellitus, and were more likely to be using an inhaled corticosteroid (Table 1). NMR spectroscopy reliably identified and quantified 27 serum metabolites in all participants. Measured metabolites were mostly amino acids or amino acid derivatives, but also included glucose, choline and three organic acids (2-hydroxybutyrate, citrate and lactate).
Table 1.
Baseline characteristics of participants.
| All participants (n = 157)* | No exacerbation (n = 109) | ≥1 exacerbation (n = 29) | p-value** | |
|---|---|---|---|---|
| Demographics | ||||
| Age | 53.7 (4.8) | 53.7 (4.8) | 54.4 (4.5) | 0.50 |
| Female | 79 (50.3%) | 52 (47.7%) | 18 (62.1%) | 0.17 |
| Smoking history | ||||
| Smoking pack-years | 45.8 (35.1) | 45.2 (39.5) | 50.3 (25.6) | 0.40 |
| Current smoking | 88 (56.8%) | 59 (55.1%) | 17 (58.6%) | 0.74 |
| Pulmonary parameters | ||||
| FEV1/FVC < 0.7 | 91 (58.0%) | 58 (53.2%) | 26 (89.7%) | 0.0004 |
| GOLD spirometry grade 1–2 | 62 (39.5%) | 45 (41.3%) | 14 (48.3%) | 0.50 |
| GOLD spirometry grade 3–4 | 29 (18.5%) | 13 (11.9%) | 12 (41.4%) | 0.0003 |
| 6MWD (m) | 445.0 (138.5) | 460.6 (136.1) | 403.1 (153.1) | 0.051 |
| % emphysema on CT | 5.2 (9.0) | 3.8 (6.7) | 9.2 (12.7) | 0.04 |
| Exacerbation predictors | ||||
| FEV1% predicted | 78.7 (27.4) | 83.1 (24.9) | 57.8 (23.2) | <0.0001 |
| ≥1 exacerbation during previous year | 30 (19.1%) | 16 (14.7%) | 14 (48.3%) | 0.0001 |
| SGRQ score | 35.0 (22.2) | 30.0 (20.4) | 57.3 (13.1) | <0.0001 |
| WBC count (thousands) | 7.7 (2.5) | 7.6 (2.4) | 8.6 (3.0) | 0.07 |
| GERD | 55 (35.0%) | 39 (35.8%) | 16 (55.2%) | 0.07 |
| Medication use | ||||
| Inhaled corticosteroids | 47 (30.5%) | 25 (23.4%) | 18 (62.1%) | <0.0001 |
| Oral corticosteroids | 2 (1.3%) | 1 (0.9%) | 1 (3.4%) | 0.32 |
| Metabolic parameters | ||||
| BMI (kg/m2) | 27.7 (4.9) | 27.9 (5.0) | 28.1 (4.6) | 0.79 |
| Diabetes mellitus | 11 (7.1%) | 5 (4.7%) | 5 (17.2%) | 0.02 |
Data are expressed as mean (SD) for continuous variables and count (percentage) for categorical ones. FEV1, forced expiratory volume in the first second; FVC, forced vital capacity; GOLD, Global Initiative for Chronic Obstructive Lung Disease; 6MWD, 6-minute walking distance; CT, computed tomography; SGRQ, St. George’s Respiratory Questionnaire; WBC, white blood cell; GERD, gastroesophageal reflux disease; BMI, body mass index.
*Exacerbation data missing in 19 participants.
**p-value comparing characteristics between group with no exacerbation and group with ≥1 exacerbation.
Association of serum metabolites with incident respiratory exacerbations
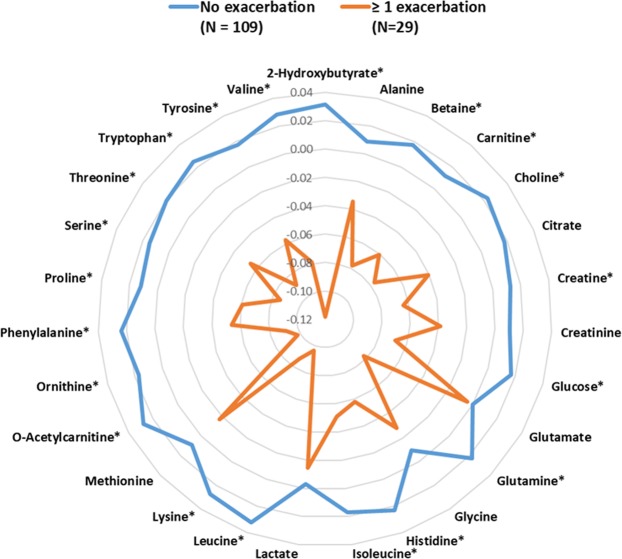
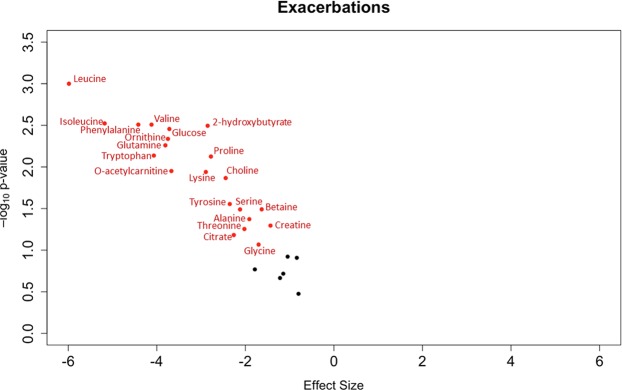
Compared to participants without exacerbations, those with at least one exacerbation had significantly lower serum concentrations for 20 of the 27 identified metabolites (Fig. 1 and Supplementary Table S1). This negative association between baseline metabolite concentrations and incident respiratory exacerbations persisted in adjusted analyses (Fig. 2) where a total of 21 metabolites were statistically significant after correction for multiple comparisons, including tryptophan (β = −4.1, 95% CI [−7.0; −1.1], p = 0.007) and the branched-chain amino acids (leucine: β = −6.0, 95% CI [−9.5; −2.4], p = 0.001; isoleucine: β = −5.2, 95% CI [−8.6; −1.8], p = 0.003; and valine: β = −4.1, 95% CI [−6.9; −1.4], p = 0.003). Multiple imputation results for missing concentrations of metabolites yielded similar results except for histidine which reached statistical significance in this sensitivity analysis (Supplementary Fig. S2).
Figure 1.
Radar plot showing mean-centered and range-scaled log-transformed metabolite concentrations by exacerbation group. Asterisks (*) denote statistical significance at the 10% false discovery rate.
Figure 2.
Volcano plot showing the adjusted association between log-transformed metabolite concentrations and incident respiratory exacerbations. Red dots represent metabolites that are statistically significant at the 10% false discovery rate. Black dots represent metabolites that are not statistically significant.
Association of serum metabolites with pulmonary parameters
In multivariable models, none of the metabolites were significantly associated with FEV1% predicted, % emphysema on chest CT or 6-minute walking distance (6MWD) after correction for multiple comparisons. Most serum metabolites were positively associated with the forced expiratory volume during the first second (FEV1) % predicted (Fig. 3A) and negatively associated with % emphysema (Fig. 3B). No trend was found when 6MWD was the outcome of interest (Fig. 3C). Notably, tryptophan had considerable associations with all pulmonary parameters: FEV1% predicted (β = 11.87; 95% CI [1.38, 22.36]; p = 0.03), % emphysema (β = −3.77; 95% CI [−7.17, −0.36]; p = 0.03) and 6MWD (β = 66.02; 95% CI [6.59, 125.45]; p = 0.03).
Figure 3.
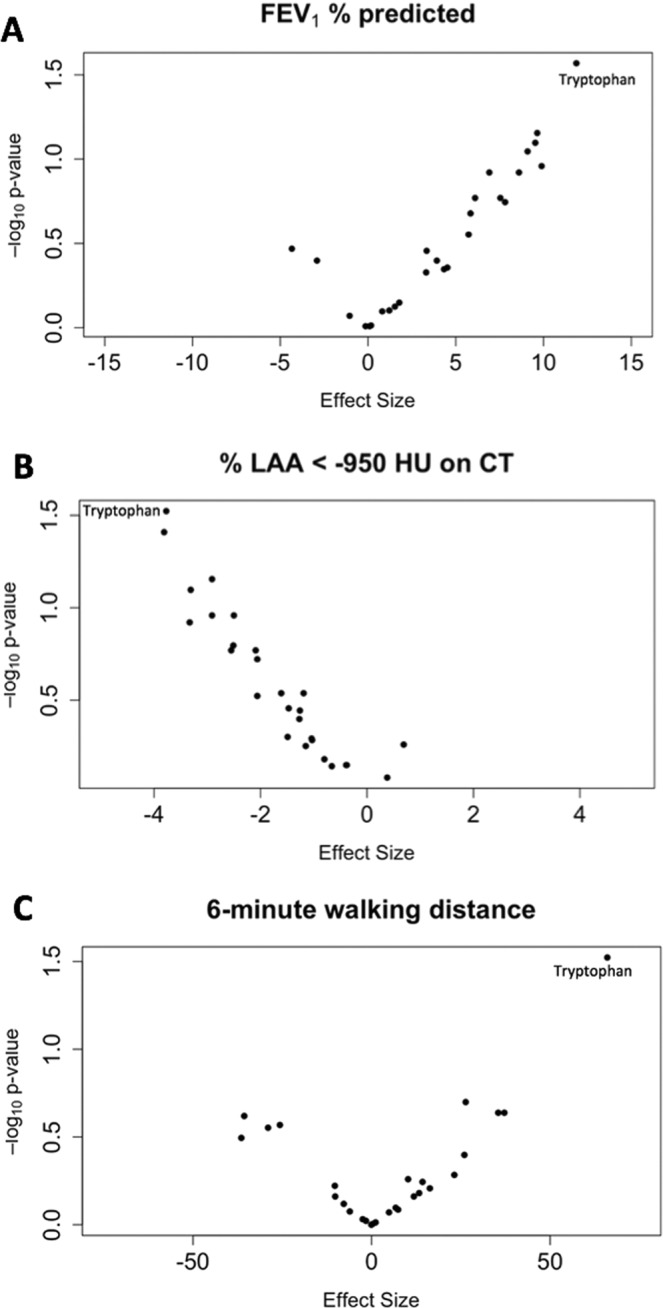
Volcano plots showing the association between log-transformed metabolite concentrations and FEV1% predicted (A), % low attenuation area (LAA) <−950 Hounsfield Units (HU) on chest computed tomography (CT) (B), and 6-minute walking distance (C). Black dots represent individual metabolites.
Association of tryptophan with inflammatory and prognostic indices
Because tryptophan was consistently related to all outcomes, we tested its association with both blood neutrophil-to-lymphocyte ratio (NLR) and the Body mass index, airflow Obstruction, Dyspnea, Exercise capacity (BODE) index. After adjusting for relevant clinical covariates, tryptophan was inversely associated with NLR (β = −0.02; 95% CI [−0.03, −0.001]; p = 0.03) in all participants and with BODE (β = −0.04; 95% CI [−0.08, −0.003]; p = 0.03) in those with COPD.
Discussion
In this NMR metabolomics analysis of current and former smokers in the SPIROMICS cohort, we found that reduced serum concentrations of amino acids are associated with a higher incidence of respiratory exacerbations. In addition, we show consistent and substantial associations between decreased tryptophan concentration and worse lung function, % emphysema on chest CT and exertional capacity. These preliminary findings support the concept of COPD as a systemic disorder and offer new insights into potential pathogenic pathways.
Circulating amino acids are involved in multiple physiologic functions, including protein synthesis, gluconeogenesis, immunity, and cell signaling16. Prior studies have described decreased serum concentrations of many amino acids in individuals with COPD compared to healthy subjects17,18. Our study further extends this knowledge by showing that lower amino acid concentrations are independently associated with worse clinical outcomes in ever-smokers. An untargeted liquid chromatography mass spectrometry (LC-MS)-based plasma metabolomics study of smokers from the COPDGene cohort also showed an inverse association between conccentrations of some amino acids (including tryptophan, tyrosine and glutamine) and exacerbation frequency19. This reduction in amino acids could be due to a consumptive process at the level of either or both the immune and musculoskeletal systems that rely on amino acids for their function, as described below. We cannot exclude a contribution from altered uptake of dietary sources as gut-resident bacteria can limit availability of amino acids, especially tryptophan20. However, given the tight regulation of levels of most serum amino acids in health, we consider this alternative explanation less likely, though meriting investigation.
The essential amino acid tryptophan regulates physiologic processes within multiple organs and systems. The amount of tryptophan that contributes to protein synthesis or to serotonin production is minimal as more than 95% is converted into kynurenine and related molecules, initially by indoleamine 2,3-dioxygenase (IDO), the rate-limiting extra-hepatic enzyme in the kynurenine pathway20. Kynurenine can reduce the activity of natural killer cells, antigen-presenting cells, and T cells. Conversely, immune cells themselves produce enzymes regulating the kynurenine pathway in response to pro-inflammatory cytokines and pathogens20,21. Gulcev and colleagues found decreased plasma tryptophan levels and increased IDO activity in COPD patients experiencing an acute exacerbation compared to COPD controls in the stable state14. One possible explanation of our results is that increased serum kynurenine dampens immune defenses to respiratory pathogens, predisposing to subsequent exacerbation events. Furthermore, tryptophan metabolites are major endogenous ligands of the aryl hydrocarbon receptor (AhR), a transcription factor with important immunomodulatory effects in the respiratory tract that is expressed in lung epithelial cells and a variety of immune cells22. Support for a generalized inflammatory milieu contributing to higher exacerbation risk in susceptible hosts comes from our finding of an inverse association between tryptophan and blood NLR, a well-studied predictor of COPD progression and outcomes23. Because all amino acids, not only tryptophan, are necessary for successful immune responses24, it is noteworthy that we found significant associations between concentrations of many of them and incident respiratory exacerbations, even after adjusting for relevant clinical covariates, including prior exacerbation history which is one of the strongest predictors of future exacerbations25.
Beyond their role in the immune system, amino acids are also major determinants of muscle mass, implying that body composition might mediate the association between decreased amino acid concentrations and poor pulmonary outcomes. Muscle dysfunction is prevalent in COPD and lower muscle mass is correlated with increased exacerbation risk as well as worse lung function, respiratory symptoms, exertional capacity and emphysema26. The branched-chain amino acids (leucine, isoleucine, valine) are particularly important contributors to muscle health, as they modulate oxidative use of glucose by muscles27. Serum levels of branched-chain amino acids have consistently been found to be lower in individuals with COPD17,28,29. Notably, one study showed increased quadriceps-to-plasma leucine content in COPD subjects compared to controls, suggesting increased muscle uptake as part of a consumptive process30. None of the metabolites identified in our study had a significant association with 6MWD; it is unclear whether the high baseline exertional capacity (mean 6MWD 445 meters) of our participants attenuated a potential association. In a small randomized placebo-controlled trial of COPD patients on long-term oxygen therapy, dietary supplementation with creatine and CoQ10 resulted in decreased dyspnea and improved functional performance11. Although amino acid supplementation holds promise, it still needs to be validated on a broad scale.
We found an inverse relationship between concentrations of most metabolites and severity of emphysema, although statistical significance was not met. Our results complement a report showing decreased tryptophan levels in COPD patients with predominantly emphysema versus those with predominantly airways disease17. Importantly, whether tryptophan or other amino acids mechanistically contribute to emphysema progression or lung function decline, remains unknown. Nonetheless, we showed a significant association between tryptophan concentrations and the BODE index, a validated and widely used predictor of mortality in individuals with COPD31.
Beyond identification of the serum amino acid metabolome as a biomarker of worse clinical parameters and outcomes in COPD, this metabolomics analysis provides important preliminary data to study some of the mechanisms involved in COPD pathogenesis. A molecular understanding of the disturbances affecting the metabolome may lead to a better understanding of COPD as a systemic condition. This is because of prevalent altered metabolic and body composition parameters in COPD, including cachexia, sarcopenia and osteoporosis in patients with emphysema, and overweight/obesity, diabetes mellitus and the metabolic syndrome in patients with chronic bronchitis. Given the complex heterogeneity of COPD, it is possible that different metabolic pathways are associated with specific clinical phenotypes19. Therefore, a comprehensive approach involving mechanistic studies, pathway analyses and integration of metabolomics data with data from other -omics sciences has the potential to identify specific pathogenic metabolic pathways. Once such pathways are identified, their individual components (e.g. gene expression, protein/enzyme activity, metabolite concentration) can be the subject of further studies to determine the etiology behind the observed alterations. This knowledge may lead to new therapeutic targets which are currently sorely needed in COPD.
Our study has a number of strengths, including its well-characterized and relatively large number of participants as well as the prospective collection of exacerbation events. We also acknowledge several limitations. Only white participants 40–60 years of age were included and less than 20% had a GOLD spirometry grade of ≥3. Therefore, our results are not generalizable to all smokers, including those with severe emphysema who may be cachexic and have substantially altered metabolomes. We could not assess the effects of diet and body composition beyond the body mass index (BMI) on the concentrations of serum metabolites, as data from food questionnaires and measurements of muscle mass and fat-free body mass were not available for our participants. This is particularly important because muscle mass could be a confounder underlying our findings, i.e. low muscle mass may lead to lower metabolite concentrations and also increase the risk of exacerbations. Furthermore, while we adjusted our models for the presence of diabetes mellitus, we did not have data available on the severity of diabetes and the use of insulin or oral hypoglycemic agents, which are factors that can affect systemic metabolism and therefore influence the association between metabolomics and exacerbation incidence. The use of NMR mostly detected amino acids and their derivatives; other classes of metabolites, especially lipids, could not be evaluated. In addition, 3-hydroxybutyrate and acetate were not included in this analysis due to aberrantly high concentrations in a subset of participants. Finally, we measured metabolites at a single point in time, precluding assessment of the association between temporal changes in the metabolome and longitudinal outcomes.
In summary, we showed that a reduction in serum amino acid concentrations is independently associated with an increased risk of respiratory exacerbations in ever-smokers with and without COPD. This association may be related to underlying systemic inflammation, immune dysregulation and/or skeletal muscle dysfunction. Further studies are needed to confirm our preliminary findings and tie them to results from other –omics sciences to better understand the pathogenic pathways involved.
Methods
Study participants and phenotypic measurements
The design of SPIROMICS, a multicenter longitudinal study funded by the National Health Lung and Blood Institute (ClinicalTrials.gov identifier: NCT01969344) has been previously reported15. In this metabolomics sub-study, participants were selected from 12 different sites to approximately reflect the entire cohort in terms of distribution of sex (equal numbers of males and females) and spirometry severity. To limit the effect of demographic heterogeneity on metabolomics results32,33, only white, ever-smoker (≥20 pack-years) participants, 40–60 years of age, were selected for this pilot analysis. Airflow obstruction was defined as a post-bronchodilator ratio of the forced expiratory volume in the first second to the forced vital capacity (FEV1/FVC) < 0.7. Spirometry tracings were reviewed to confirm adherence to American Thoracic Society and European Respiratory Society standards34. The severity grade of airflow limitation was determined in participants with COPD based on GOLD guidelines1. History of respiratory exacerbations in the year before study enrollment, smoking history, self-reported comorbidities, and medication use were all obtained from participants at the baseline visit. In addition, a 6-minute walk test and a high-resolution CT of the chest were acquired during that visit. Chest CTs were performed and analyzed according to the SPIROMICS quantitative CT lung assessment system protocol35. Extent of emphysema on CT was measured as the percent of lung volume with attenuation <−950 Hounsfield Units (HU) at full inspiration36. Prospective data on exacerbations was captured every three months through a structured telephone questionnaire. Exacerbations were defined as symptomatic respiratory events that necessitated health care utilization (clinic visit, Emergency Department visit or hospitalization) and treatment with antibiotics, systemic corticosteroids, or both. This definition was applied to both smokers with and without COPD as the latter group can experience significant morbidity from exacerbation-like events37,38.
Serum sample processing and quantitative 1H-NMR metabolomics
Blood was collected from fasting participants by direct venipuncture, into no-additive red top tubes (BD Vacutainer®, Franklin Lakes, NJ, USA) in accordance with the SPIROMICS Manual of Operations. Blood was allowed to coagulate at room temperature for 30 minutes, after which it was centrifuged (3000 × g for 15 minutes). The resulting serum was aliquoted and stored at −80 °C. At the time of assay, samples were randomly assigned to a total of 14 batches and were thawed in an ice water bath. Pre-chilled methanol was added to each sample in a 2:1 ratio to remove proteins and macromolecules by precipitation. The aqueous fraction was lyophilized, suspended in deuterated water (D2O) and filtered (3KDa Pall Nanosep omega filters Sigma-Aldrich, St. Louis, MO, USA) to remove residual macromolecules, after which a known amount of formate was added as an internal standard as previously described39,40. On the day of NMR acquisitions, the pH was adjusted to a 6.5–7.5 range, and samples were transferred to NMR tubes. A one-dimensional 1H-NMR spectrum was generated for each sample using a 500 MHz spectrometer (Agilent Inc., Santa Clara, CA, USA) equipped with a 5-mm Agilent “One-probe” at the University of Michigan NMR Biochemical Core Laboratory. Spectra were acquired by 32 scans of the first increment of a 1H, 1H-NOESY pulse sequence41 and were analyzed using Chenomx NMR Suite 8.3 (Chenomx Inc., Edmonton, Alberta, Canada). We identified metabolites using the compound library of the Chenomx software and determined their concentrations relative to the internal standard. A member of the NMR Laboratory cross-checked the quality of each NMR spectrum, and 157 subjects with interpretable NMR spectra were included in this analysis (Supplementary Fig. S3).
Statistical analysis
We reported baseline characteristics of participants as mean and standard deviation for continuous variables and as count and percentage for categorical ones. We assessed differences in characteristics between participants who had at least one respiratory exacerbation and those who did not experience any exacerbations over one year of follow-up using the two-sample t-test for continuous variables and the Chi-square test for categorical ones. We used a two-sample t-test to assess differences in metabolite concentrations between the two exacerbation groups. Results of this analysis were displayed on a radar plot by exacerbation group after the log-transformed concentrations of all metabolites were mean-centered and range-scaled (plot generated in Microsoft Excel 2016). To test the association between log-transformed metabolite concentrations and incidence of respiratory exacerbations, we built a multivariable logistic regression model for each metabolite adjusting for demographics (age and sex), study site, clinical factors that can affect metabolism (smoking status, BMI, diabetes mellitus, and current use of inhaled or oral corticosteroids)42, and clinical predictors of exacerbations (FEV1% predicted, history of respiratory exacerbations during the preceding year, white blood cell count, St. George’s Respiratory Questionnaire score and presence of gastroesophageal reflux disease)25,38. In addition to the multivariable logistic regression models above that dropped individuals with missing metabolite concentrations, we performed a multiple imputation sensitivity analysis that filled in missing metabolite concentration values (which comprised only 0.6% of all expected values) using the PROC MI function in SAS (Cary, NC, USA). The resulting 100 logistic regression outputs were combined using PROC MI ANALYZE which provided multiple imputation parameter estimates, confidence intervals and p-values. To test the association between log-transformed metabolite concentrations and each of FEV1% predicted, % emphysema on chest CT and 6MWD, we built multivariable linear regression models using the same covariates as above except for clinical predictors of exacerbations. Given that associations were tested for multiple metabolites, we corrected p-values for a false discovery rate (FDR) of 10% using the Benjamini-Hochberg method43. We used volcano plots to display the associations found in the multivariable models between metabolite concentrations and clinical parameters with effect size (beta coefficient for each metabolite) on the x-axis and −log10 of p-value on the y-axis (plots generated in R v3.4.0). We finally tested the association between a metabolite of interest (tryptophan) and each of the blood NLR, a known inflammatory biomarker in smokers23, and the BODE index, a well-validated prognostic tool in COPD31, after adjusting for age, sex, study site, smoking status, diabetes, inhaled or oral corticosteroid use, history of respiratory exacerbations during the preceding year, FEV1% predicted and BMI (the latter two variables were not included in the model for BODE given their inclusion in the outcome variable). All statistical analyses were performed in SAS v9.4 except when indicated.
Ethical approval and informed consent
All methods were performed in accordance with the relevant guidelines and regulations. All participants gave written informed consent. The SPIROMICS protocol was approved by the institutional review boards of all participating institutions (Columbia University IRB 2, University of Iowa IRB-01, Johns Hopkins IRB-5, University of California Los Angeles Medical IRB 1, University of Michigan IRBMED B1 Board, National Jewish Health IRB, University of California San Francisco IRB Parnassus Panel, Temple University IRB A2, University of Alabama at Birmingham IRB #2, University of Illinois IRB #3, University of Utah IRB Panel Review Board 5, Wake Forest University IRB #5, University of North Carolina at Chapel Hill Non-Biomedical IRB).
Supplementary information
Acknowledgements
The authors thank the SPIROMICS participants and participating physicians, investigators and staff for making this research possible. More information about the study and how to access SPIROMICS data is at www.spiromics.org. We would like to acknowledge the following current and former investigators of the SPIROMICS sites and reading centers: Neil E Alexis, MD; Wayne H Anderson, PhD; Mehrdad Arjomandi, MD; Igor Barjaktarevic, MD, PhD; R Graham Barr, MD, DrPH; Lori A Bateman, MSc; Surya P Bhatt, MD; Eugene R Bleecker, MD; Richard C Boucher, MD; Russell P Bowler, MD, PhD;; Stephanie A Christenson, MD; Alejandro P Comellas, MD; Christopher B Cooper, MD, PhD; David J Couper, PhD; Gerard J Criner, MD; Ronald G Crystal, MD; Jeffrey L Curtis, MD; Claire M Doerschuk, MD; Mark T Dransfield, MD; Brad Drummond, MD; Christine M Freeman, PhD; Craig Galban, PhD; MeiLan K Han, MD, MS; Nadia N Hansel, MD, MPH; Annette T Hastie, PhD; Eric A Hoffman, PhD; Yvonne Huang, MD; Robert J Kaner, MD; Richard E Kanner, MD; Eric C Kleerup, MD; Jerry A Krishnan, MD, PhD; Lisa M LaVange, PhD; Stephen C Lazarus, MD; Fernando J Martinez, MD, MS; Deborah A Meyers, PhD; Wendy C Moore, MD; John D Newell Jr, MD; Robert Paine, III, MD; Laura Paulin, MD, MHS; Stephen P Peters, MD, PhD; Cheryl Pirozzi, MD; Nirupama Putcha, MD, MHS; Elizabeth C Oelsner, MD, MPH; Wanda K O’Neal, PhD; Victor E Ortega, MD, PhD;; Sanjeev Raman, MBBS, MD; Stephen I. Rennard, MD; Donald P Tashkin, MD;; J Michael Wells, MD; Robert A Wise, MD; and Prescott G Woodruff, MD, MPH. The project officers from the Lung Division of the National Heart, Lung, and Blood Institute were Lisa Postow, PhD, and Lisa Viviano, BSN. SPIROMICS was supported by contracts from the NIH/NHLBI (HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C, HHSN268200900019C, HHSN268200900020C), and a grant from the NIH/NHLBI (U01 HL137880), and supplemented by contributions made through the Foundation for the NIH and the COPD Foundation from AstraZeneca/MedImmune; Bayer; Bellerophon Therapeutics; Boehringer-Ingelheim Pharmaceuticals, Inc.; Chiesi Farmaceutici S.p.A.; Forest Research Institute, Inc.; GlaxoSmithKline; Grifols Therapeutics, Inc.; Ikaria, Inc.; Novartis Pharmaceuticals Corporation; Nycomed GmbH; ProterixBio; Regeneron Pharmaceuticals, Inc.; Sanofi; Sunovion; Takeda Pharmaceutical Company; Theravance Biopharma; and Mylan.
This analysis was also supported by National Institutes of Health grants T32HL007749 (WWL), R01GM111400 (KAS), R01HL122438 (MKH), K24HL138188 (MKH); by Merit Review award I01 CX000911 from the Clinical Laboratory Research and Development Service, Department of Veterans Affairs (JLC); and by funding from the University of Michigan Health System-Peking University Health Science Center Joint Institute for Translational and Clinical Research.
Author Contributions
W.W.L., T.J.S., K.A.S. and M.K.H. designed the study. K.A.S. and L.Y. acquired metabolomics data. W.W.L., T.G., S.M., K.A.S. and M.K.H. performed the statistical analysis. W.W.L., S.K.M., J.L.C., T.J.S., K.A.S. and M.K.H. prepared the manuscript. All other authors revised the manuscript and approved the final draft.
Data Availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
WWL, TG, SM, JLC, APC, LY, REK, RP, MLNM, SPP, WKO, WD, BH, TJS and KAS declare no competing interests. RPB serves on the advisory boards of GSK, BI and Mylan, and received research grants from GSK and BI. RGB reports grants from NIH during the conduct of the study; grants from Alpha1 Foundation, Foundation for the NIH, and COPD Foundation and personal fees from UpToDate outside the submitted work. N. N. H. serves on the advisory board of AstraZeneca, GlaxoSmithKline, and Mylan, and has received research grants fromAstraZeneca, Boehringer-Ingelheim Pharmaceuticals, Inc, Forest Research Institute, Inc, and GlaxoSmithKline. CBC reports grants from Equinox Health Clubs, personal fees from Equinox Health Clubs, grants from Amgen, personal fees from PulmonX, other from GlaxoSmithKline and part-time employment in scientific engagement for the GlaxoSmithKline Global Respiratory Franchise. IB reports personal fees from Astra Zeneca, grants from GE Healthcare, personal fees from Grifols, personal fees from CSL Behring, grants from Amgen, grants from PCORI. JAK reported serving on a data monitoring committee for Sanofi and receiving grants from the National Institutes of Health and the Patient-Centered Outcomes Research Institute. PGW reports personal fees from AstraZeneca, Theravance, Regeneron, Sanofi, Genentech, and Novartis. FJM reports personal fees and non-financial support from American College of Chest Physicians, AstraZeneca, Boehringer Ingelheim, Continuing Education, ConCert, Genentech, GlaxoSmithKline, Inova Fairfax Health System, Miller Communications, National Association for Continuing Education, Novartis, Pearl Pharmaceuticals, PeerView Communications, Prime Communications, Puerto Rican Respiratory Society, Chiesi, Roche, Sunovion, and Theravance; non-financial support from ProterrixBio; personal fees from Columbia University, Haymarket Communications, Integritas, Inthought Research, MD Magazine, Methodist Hospital Brooklyn, New York University, Unity, UpToDate, WebMD/MedScape, Western Connecticut Health Network, and American Thoracic Society. MKH reports personal fees from GSK, personal fees from BI, personal fees from AZ, other from Novartis, other from Sunovion.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kathleen A. Stringer and MeiLan K. Han jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47761-w.
References
- 1.Vogelmeier CF, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report. GOLD Executive Summary. Am J Respir Crit Care Med. 2017;195:557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 2.Han MK, et al. Frequency of exacerbations in patients with chronic obstructive pulmonary disease: an analysis of the SPIROMICS cohort. Lancet. Respir Med. 2017;5:619–626. doi: 10.1016/S2213-2600(17)30207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst JR, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363:1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 4.Coxson HO, et al. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1:129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 5.Bowler RP, et al. New Strategies and Challenges in Lung Proteomics and Metabolomics. An Official American Thoracic Society Workshop Report. Ann Am Thorac Soc. 2017;14:1721–1743. doi: 10.1513/AnnalsATS.201710-770WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1:a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, et al. Serum Metabolite Biomarkers Discriminate Healthy Smokers from COPD Smokers. PLoS One. 2015;10:e0143937. doi: 10.1371/journal.pone.0143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Laurentiis G, et al. Separating smoking-related diseases using NMR-based metabolomics of exhaled breath condensate. J Proteome Res. 2013;12:1502–1511. doi: 10.1021/pr301171p. [DOI] [PubMed] [Google Scholar]
- 9.Paige M, et al. Pilot analysis of the plasma metabolite profiles associated with emphysematous Chronic Obstructive Pulmonary Disease phenotype. Biochem Biophys Res Commun. 2011;413:588–593. doi: 10.1016/j.bbrc.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, et al. Metabonomic profiling of serum and urine by (1)H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PLoS One. 2013;8:e65675. doi: 10.1371/journal.pone.0065675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Benedetto F, et al. Supplementation with Qter((R)) and Creatine improves functional performance in COPD patients on long term oxygen therapy. Respir Med. 2018;142:86–93. doi: 10.1016/j.rmed.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Dransfield MT, et al. Acute exacerbations and lung function loss in smokers with and without chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195:324–330. doi: 10.1164/rccm.201605-1014OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soler-Cataluna JJ, et al. Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax. 2005;60:925–931. doi: 10.1136/thx.2005.040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulcev M, et al. Tryptophan catabolism in acute exacerbations of chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2016;11:2435–2446. doi: 10.2147/COPD.S107844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couper D, et al. Design of the Subpopulations and Intermediate Outcomes in COPD Study (SPIROMICS) Thorax. 2014;69:491–494. doi: 10.1136/thoraxjnl-2013-203897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 17.Ubhi BK, et al. Targeted metabolomics identifies perturbations in amino acid metabolism that sub-classify patients with COPD. Mol Biosyst. 2012;8:3125–3133. doi: 10.1039/c2mb25194a. [DOI] [PubMed] [Google Scholar]
- 18.Pouw EM, Schols AM, Deutz NE, Wouters EF. Plasma and muscle amino acid levels in relation to resting energy expenditure and inflammation in stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;158:797–801. doi: 10.1164/ajrccm.158.3.9708097. [DOI] [PubMed] [Google Scholar]
- 19.Cruickshank-Quinn CI, et al. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci Rep. 2018;8:17132. doi: 10.1038/s41598-018-35372-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervenka Igor, Agudelo Leandro Z., Ruas Jorge L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science. 2017;357(6349):eaaf9794. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 21.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 22.Puccetti M, et al. Towards Targeting the Aryl Hydrocarbon Receptor in Cystic Fibrosis. Mediators Inflamm. 2018;2018:1601486. doi: 10.1155/2018/1601486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paliogiannis Panagiotis, Fois Alessandro G., Sotgia Salvatore, Mangoni Arduino A., Zinellu Elisabetta, Pirina Pietro, Negri Silvia, Carru Ciriaco, Zinellu Angelo. Neutrophil to lymphocyte ratio and clinical outcomes in COPD: recent evidence and future perspectives. European Respiratory Review. 2018;27(147):170113. doi: 10.1183/16000617.0113-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, Yin YL, Li D, Kim SW, Wu G. Amino acids and immune function. Br J Nutr. 2007;98:237–252. doi: 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 25.Hurst JR, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363:1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 26.McDonald ML, et al. Quantitative computed tomography measures of pectoralis muscle area and disease severity in chronic obstructive pulmonary disease. A cross-sectional study. Ann Am Thorac Soc. 2014;11:326–334. doi: 10.1513/AnnalsATS.201307-229OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layman DK, Walker DA. Potential importance of leucine in treatment of obesity and the metabolic syndrome. J Nutr. 2006;136:319S–323S. doi: 10.1093/jn/136.1.319S. [DOI] [PubMed] [Google Scholar]
- 28.Ubhi BK, et al. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J. 2012;40:345–355. doi: 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- 29.Hofford JM, et al. The nutritional status in advanced emphysema associated with chronic bronchitis. A study of amino acid and catecholamine levels. Am Rev Respir Dis. 1990;141:902–908. doi: 10.1164/ajrccm/141.4_Pt_1.902. [DOI] [PubMed] [Google Scholar]
- 30.Engelen MP, Wouters EF, Deutz NE, Menheere PP, Schols AM. Factors contributing to alterations in skeletal muscle and plasma amino acid profiles in patients with chronic obstructive pulmonary disease. Am J Clin Nutr. 2000;72:1480–1487. doi: 10.1093/ajcn/72.6.1480. [DOI] [PubMed] [Google Scholar]
- 31.Celli BR, et al. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:1005–1012. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- 32.Patel MJ, et al. Race and sex differences in small-molecule metabolites and metabolic hormones in overweight and obese adults. Omics. 2013;17:627–635. doi: 10.1089/omi.2013.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darst BF, Koscik RL, Hogan KJ, Johnson SC, Engelman CD. Longitudinal plasma metabolomics of aging and sex. Aging (Albany NY) 2019;11:1262–1282. doi: 10.18632/aging.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MR, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 35.Sieren JP, et al. SPIROMICS Protocol for Multicenter Quantitative Computed Tomography to Phenotype the Lungs. Am J Respir Crit Care Med. 2016;194:794–806. doi: 10.1164/rccm.201506-1208PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gevenois PA, de Maertelaer V, De Vuyst P, Zanen J, Yernault JC. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. Am J Respir Crit Care Med. 1995;152:653–657. doi: 10.1164/ajrccm.152.2.7633722. [DOI] [PubMed] [Google Scholar]
- 37.Woodruff PG, et al. Clinical Significance of Symptoms in Smokers with Preserved Pulmonary Function. N Engl J Med. 2016;374:1811–1821. doi: 10.1056/NEJMoa1505971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowler RP, et al. Prediction of acute respiratory disease in current and former smokers with and without COPD. Chest. 2014;146:941–950. doi: 10.1378/chest.13-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagana Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal Chem. 2015;87:706–715. doi: 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McHugh Cora, Flott Thomas, Schooff Casey, Smiley Zyad, Puskarich Michael, Myers Daniel, Younger John, Jones Alan, Stringer Kathleen. Rapid, Reproducible, Quantifiable NMR Metabolomics: Methanol and Methanol: Chloroform Precipitation for Removal of Macromolecules in Serum and Whole Blood. Metabolites. 2018;8(4):93. doi: 10.3390/metabo8040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacy P, et al. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PLoS One. 2014;9:e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinke Stacey N., Gallart-Ayala Héctor, Gómez Cristina, Checa Antonio, Fauland Alexander, Naz Shama, Kamleh Muhammad Anas, Djukanović Ratko, Hinks Timothy S.C., Wheelock Craig E. Metabolomics analysis identifies different metabotypes of asthma severity. European Respiratory Journal. 2017;49(3):1601740. doi: 10.1183/13993003.01740-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.