Abstract
Salivary duct carcinomas (SDC) and Her2/Neu3-overexpressing invasive breast carcinomas (HNPIBC/IBC) are histologically indistinguishable. We investigated whether common histopathologic and immunophenotypic features of SDC and IBC are mirrored by a similar microRNA (miRNA) profile. MiRNA profiling of 5 SDCs, 6 IBCs Her2/Neu3+, and 5 high-grade ductal breast carcinoma in situ (DCIS) was performed by NanoString platform. Selected miRNAs and HOXA1 gene were validated by RT-PCR. We observed similar miRNA expression profiles between IBC and SDC with the exception of 2 miRNAs, miR-10a and miR-142-3p, which were higher in IBC tumors. DCIS tumors displayed increased expression of miR-10a, miR-99a, miR-331-3p and miR-335, and decreased expression of miR-15a, miR-16 and miR-19b compared to SDC. The normal salivary gland and breast tissues also showed similar expression profiles. Interestingly, miR-10a was selectively increased in both IBC and normal breast tissue compared to SDC and normal salivary gland tissue. Moreover, our NanoString and RT-PCR data confirmed that miR-10a was upregulated in IBC and DCIS compared to SDC. Finally, we show downregulation of HOXA1, a miR-10 target, in IBC tumors compared to normal breast tissue. Taken together, our data demonstrates that, based on miRNA profiling, SDC is closely related to HNPIBC. Our results also suggest that miR-10a is differentially expressed in IBC compared to SDC and may have potential utility as a diagnostic biomarker in synchronous or metachronous malignant epithelial malignancies involving both organs. In addition, miR-10a could be playing an important role as a mammary-specific oncogene, involved in breast cancer initiation (DCIS) and progression (IBC), through mechanisms that include modulation of HOXA1 gene expression.
Electronic supplementary material
The online version of this article (10.1007/s12105-018-0971-x) contains supplementary material, which is available to authorized users.
Keywords: Salivary duct carcinoma, Her2/Neu+ breast carcinoma, MicroRNA profiling, MiRNA-10, Oncogene
Introduction
Salivary duct carcinomas (SDCs) are rare, highly aggressive malignancies of salivary gland origin that are histologically similar to invasive breast cancer and account for about 2% of salivary gland malignancies [1]. They occur predominantly in the parotid and less frequently in other salivary glands, mostly in elderly men [2]. SDCs are characterized by frequent early distant metastases and a 10-year mortality rate as high as 65%. A significant percentage of patients die within 5 years of diagnosis of this disease [3]. Phenotypically, SDCs bear a close histopathologic semblance to high-grade Her2/Neu-overexpressing invasive breast carcinomas (HNPIBCs/IBCs), with both tumors frequently expressing androgen receptor (AR) and Her2/Neu by immunohistochemistry [3]. Microscopically, both of these tumors span the same range of histopathologic patterns (cribriform, comedo, apocrine, etc.). Moreover, SDCs exhibit a similar immunohistochemical phenotype to ductal adenocarcinomas of the breast. Indeed, a third of the latter are positive for Her2/Neu, estrogen receptor/progesterone receptor (ER/PR), and AR [4]. HNPIBCs represents approximately 15–20% of breast carcinomas and are characterized by unique expression signatures, poor cellular differentiation, high rates of cellular proliferation, frequent nodal metastases, relative resistance to certain chemotherapy and overall worse prognosis [5]. Indeed, HNPIBC represent an aggressive type of breast carcinoma, which in spite of anti-Her2/Neu agents, still lead to high mortality and toxicity profile [5]. Although histopathologic, immunophenotypical and cytogenetic similarities exist between SDC and IBC [3], comparative analysis of the molecular mechanisms that drive these anatomically distinct tumors are largely unknown.
Molecular profiling has proven to be a powerful tool in the characterization of various tumors. One such method involves the characterization of microRNAs (miRNAs), which are small noncoding RNAs [6] involved in post-transcriptional regulation of gene expression [7]. They have been implicated in numerous biological processes—cell growth and differentiation [8–10]—as well as pathological processes including cancer [11, 12]. Studies on miRNA expression signatures from hematological and solid organ tumors suggest that miRNA profiles are tissue-specific, and they have a remarkable ability to discriminate between different types of cancers with a high degree of accuracy [13, 14]. MiRNA signatures are therefore potentially useful in identifying tumor type, and possible therapeutic options, when histopathologic and immunophenotypic data are unable to provide such information.
MiRNAs have been shown to play a major role in breast cancer development. Indeed, Iorio et al. demonstrated that miR-125b, miR-145, miR-21, and miR-155 were deregulated in breast cancer compared to normal breast tissue [15]. Previous molecular genetic studies of SDC have focused on analysis of a limited gene panel [16, 17]. However, comparative miRNA expression profiles of SDC and HNPIBC have not been previously investigated. In this study, we analyzed the miRNA profiles of 5 cases of SDC and 6 cases of HNPIBC to determine whether the morphological similarities between these two types of cancers are mirrored by similar molecular profiles. In view of the premalignant nature of ductal carcinoma in situ (DCIS), and the potential biologic relatedness between mammary DCIS and IBC [18, 19], we included cases of high grade DCIS (n = 5) in the study to ascertain if there are similar dysregulations of candidate miRNAs between DCIS and IBC. Our results demonstrate that de novo SDCs have a very similar molecular profile to that of HNPIBCs. However, miR-10a is highly expressed in mammary tumors (IBCs and DCIS) but not in SDCs. Thus, we suggest that miR-10a may be a useful biomarker for discriminating HNPIBC from SDC. Furthermore, over-expression of miR-10a leads to downregulation of HOXA1 in breast cancer, which might contribute to the initiation and progression of mammary carcinogenesis.
Materials and Methods
Tumor Characterization and Sampling
SDCs (n = 5), IBCs (n = 6) and DCIS (n = 5) formalin-fixed, paraffin-embedded (FFPE) cases were selected from The Ohio State University Wexner Medical Center archive after approval through the Institutional Review Board of The Ohio State University, Department of Pathology (Institutional Review Board-approved protocol number 2002H0089). All cases were re-reviewed to confirm the diagnosis. In view of the morphologic overlap between the two entities, none of the patients with de novo SDC had concurrent IBC. The diagnosis of SDC was based on established criteria [3]. All IBC cases were Her2/Neu amplified by both immunohistochemistry (3+ positivity) and fluorescence in situ hybridization (two of which were also positive for ER and PR). All DCIS cases were morphologically high-grade and all but 1 case were negative for ER and PR.
The relative stability of miRNAs in FFPE archival tissue has made them an attractive resource for miRNA profiling studies, and indeed such tissue has been shown to be comparable to fresh frozen specimens [20].
Against this background, we had optimized a technique in our recent work using FFPE material in which we used the patient’s non-tumoral tissue as a means to minimize the variability introduced by using normal tissue controls from different patients [21]. We used a tissue microarrayer to core tumor and non-tumoral areas that had been mapped on hematoxylin and eosin-stained (H&E) sections so that the tumor samples were matched with paired controls from the same patient. In this regard, representative H&E-stained slides were identified for each case, and the tumoral and non-tumoral (“normal”) appearing areas were marked and used as a guide for the tissue microarrayer to simultaneously obtain both tumor tissue and normal tissue for each patient as duplicate (paired) samples. Both areas (tumor and non-tumoral) were appropriately marked as a guide for the tissue microarrayer to obtain multiple 1.75 mm cores simultaneously from the corresponding areas on the FFPE blocks.
For HOXA1 validation experiments, a separate cohort of snap-frozen Her2/Neu3+ positive IBC and normal breast specimens (n = 3 per group) were obtained through the Tissue Procurement Service of the Department of Pathology at The Ohio State University Wexner Medical Center.
RNA Extraction
Paraffin-embedded tissue cores from blocks were deparaffinized in xylene before protease digestion. RNAs were extracted by using the Ambion RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Invitrogen, AM1975) according to the manufacturer’s protocol. Samples were eluted in water and cleaned up by glycogen-Na acetate precipitation procedure. Briefly, 1/10 vol of 3 M sodium acetate (pH 5.2), 1 µg glycogen, and 3.5 volumes of ice-cold 95% ethanol was added to each sample’s tube. Samples were incubated on ice for at least 30 min, centrifuged at top speed for 20 min, washed twice with cold 70% ethanol, and finally re-suspended in RNase-free water.
Frozen tissues were extracted by using TRIzol solution (Invitrogen). Tissues were frozen immediately after collection and maintained at − 80 °C until extraction. Each sample was homogenized in 1 ml of TRIzol and extracted following the manufacturer’s protocol.
NanoString nCounter Platform Profiling
The NanoString nCounter Human miRNA Expression Assay Kit (http://www.NanoString.com/) was used to profile human and human-associated viral miRNAs in all samples. 100 ng of total RNA was used as input for nCounter miRNA sample preparation reactions. All sample preparation and hybridization reactions were performed according to manufacturer’s instructions (NanoString Technologies). Data collection was carried out on the nCounter Digital Analyzer (NanoString Technologies) following the manufacturer’s instructions to count individual fluorescent barcodes and quantify target RNA molecules present in each sample. For each assay, a high-density scan (600 fields of view) was performed.
Real Time Polymerase Chain Reaction (RT-PCR)
Reverse transcriptase and real-time PCR (RT-PCR) reactions were performed according to the manufacturer’s protocols. All RT-PCR reactions were run in a GeneAmp PCR 9700 Thermocycler (Applied Biosystems).
Mature miR-10a (Applied Biosystems, assay ID: 000387), miR-21 (Applied Biosystems, assay ID: 000397) and miR-148 (Applied Biosystems, assay ID: 000470) expression were analyzed by Taqman micro-RNA assay and normalized to RNU44 (endogenous control, assay ID: 001094) according to the manufacturer’s protocol. 10 ng of total RNA from all samples was reverse transcribed for each target and endogenous control. Samples were analyzed in triplicate, and the expression of miR-10a, miR-21 and miR-148 was determined using the comparative Ct method (2−ΔCt).
Expression of the target gene HOXA1 was also tested by RT-PCR, using GAPDH and OAZ1 as housekeeping genes for normalization. Targets were assayed using Taqman probes (HOXA1 assay ID: Hs00939046_m1, cat#: 4331182, GAPDH assay ID: Hs02758991_g1, cat# 4351370, OAZ1 assay ID: Hs00427923_m, cat# 4351370) on total cDNA obtained from the same RNA samples previously checked for miRNA expression. Gene expression levels were quantified using the ABI Prism 7900HT Sequence detection system (Applied Biosystems).
Statistical Analysis
Raw NanoString data, which are proportional to copy number, were log-transformed and normalized by the quantile method after application of a manufacturer-supplied correction factor for several miRNAs. Data were filtered to exclude relatively invariant features (IQR 0.5) and features below the detection threshold (defined for each sample by a cutoff corresponding to approximately twice standard deviation of negative control probes plus the mean of them) in at least half of the samples. Using R/Bioconductor and the filtered dataset, linear models for microarray data analysis [22] were employed with a contrast matrix in order to identify deregulated miRNAs. P values were used to rank miRNAs of interest, and correction for multiple comparisons was done by the Benjamini Hocheberg method [23]. Raw data that were above background, as well as the corresponding quantile-normalized data, were also imported into MultiExperiment Viewer [24] to create heat maps. For the RT-PCR data, Student’s t test was employed to determine the statistical significance of values obtained.
Results
SDCs Closely Resemble IBCs Histopathologically
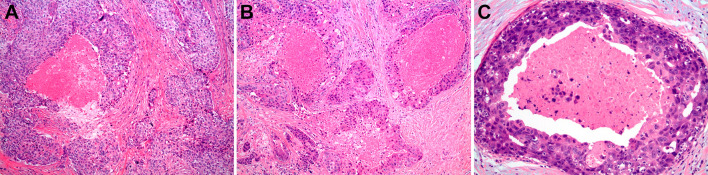
A histopathologic review of five cases of SDC, six cases of IBC and five cases of DCIS showed a striking microscopic similarity (Fig. 1). All three tumor types exhibited ductal proliferation with central necrosis (comedocarcinoma) and high-grade cellular and nuclear pleomorphism. In all three tumors, neoplastic cells were cuboidal to columnar, with nuclei showing prominent nucleoli. SDC and IBC showed such prominent morphologic phenotypic mimicry that they were difficult (if not impossible) to distinguish from each other. Our data highlighted the value of identifying molecular biomarkers to differentiate between these histologically and embryologically similar cancer types.
Fig. 1.
Histopathological photomicrographs of a invasive breast carcinoma (IBC), b salivary duct carcinoma (SDC) and c ductal carcinoma in situ (DCIS) of breast, showing striking histopathologic similarity, rendering IBC and SDC morphologically indistinguishable. The central comedonecrosis and marked cellular and nuclear atypia are prototypic features of all three entities [H&E, × 200]
MiRNA Profiling Shows SDC to be More Closely Related to IBC than DCIS
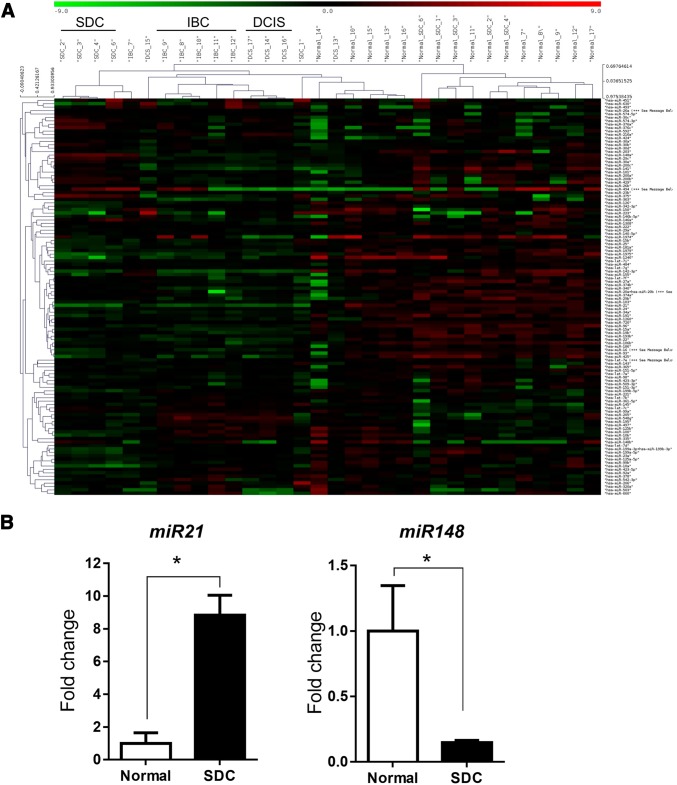
In order to identify distinguishing molecular characteristics of SDS, IBC and DCIS, we performed miRNA profiling of tumors and surrounding normal tissues from the three tumor types (Fig. 2a). We observed an increased expression of miR-21 and miR-155, and a decreased expression of miR-10b and miR-145 in IBCs compared to surrounding normal breast tissue. In DCIS, we observed an increased expression of miR-21 compared to surrounding breast tissue (Table 1). We also found an increased expression of miR-21 and decreased expression of miR-148 in SDC compared to normal salivary tissue (Fig. 2b). Detailed miRNA profiles are provided in Supplementary Tables 1 and 2. Our data suggest that, although there are morphological similarities between these tumors, distinct molecular pathways drive their respective carcinogenic processes.
Fig. 2.
a Global miRNA profiling of normal breast, normal salivary tissues, IBC, DCIS and SDC as determined by NanoString analysis. Heat maps show the distinct microRNA signature of SDC, IBC and DCIS tumors. b RT-PCR validation of miR-21 and miR-148 expression in SDC tumors compared to normal salivary tissue, *p < 0.05
Table 1.
miRNA expression comparisons between normal breast tissue, IBC and DCIS tumors from patients determined by NanoString analysis
| microRNA | Mean | Fold change | p-values | ||||
|---|---|---|---|---|---|---|---|
| Normal | IBC | DCIS | Log twofold change | Linear fold change | Raw p-values | Adj p-values | |
| hsa-miR-21 | 10.87 | 14.84 | 3.97 | 15.70 | 1.49E−07 | 5.09E−06 | |
| hsa-miR-155 | 5.52 | 7.65 | 2.13 | 4.37 | 9.15E−04 | 3.00E−03 | |
| hsa-miR-145 | 11.31 | 9.42 | − 1.90 | 0.27 | 4.59E−04 | 1.64E−03 | |
| hsa-miR-10b | 6.75 | 6.00 | − 0.75 | 0.59 | 1.89E−02 | 3.78E−02 | |
| hsa-miR-21 | 10.87 | 13.75 | 2.88 | 7.39 | 8.12E−05 | 5.99E−04 | |
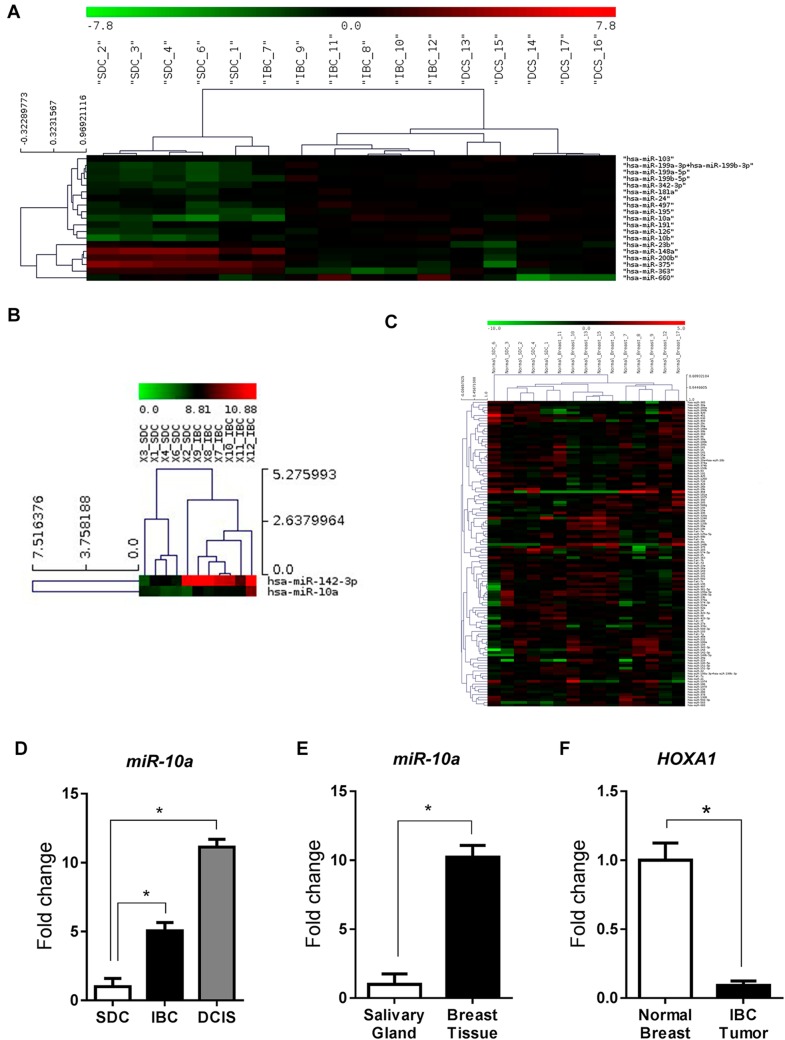
Further comparative analysis of miRNA profiles between SDC, IBC and DCIS identified a distinct miRNA signature for SDC as shown by the hierarchical clustering on the heat map (Fig. 3a). Interestingly, comparisons between SDC and IBC revealed only two miRNAs which were differentially expressed in these two cancers, miR-10a and miR-142-3p, found to be more expressed in IBC than in SDC (Fig. 3b). On the other hand, SDC versus DCIS comparisons showed seven differentially expressed miRNAs: miR-10a, miR-99a, miR-331-3p, miR-335 up-regulated; and miR-15a, miR-16, miR-19b, down-regulated in DCIS compared to SDC. Finally, IBC vs DCIS miRNA profile comparisons show differential miRNA expression in five miRNAs: miR-15a, miR-16, miR-19b, miR-222 and miR-374 which were down-regulated in DCIS compared to IBC. These data are summarized in Table 2. It should be noted that these results were based on analysis of NanoString data alone. Taken together, our data seems to suggest that miRNA expression profiles show SDC to be more closely related to IBC than DCIS.
Fig. 3.
a Heat map showing the NanoString gene expression profiles of SDC, IBC and DCIS. b Heat map comparison between SDC and IBC showing expression of miR-142-3p and miR-10a. c Heat map showing miRNA profiles of normal salivary and normal breast tissues as determined by NanoString analysis. d Gene expression of miR-10a in SDC, IBC and DCIS as determined by RT-PCR. e Gene expression of miR-10a in normal salivary and normal breast tissue as determined by RT-PCR. fHOXA1 gene expression in IBC and normal breast tissue as determined by RT-PCR analysis, *p < 0.05
Table 2.
Selected miRNA expression comparisons between tumors from SDC, IBC and DCIS patients determined by NanoString analysis
| microRNA | Mean | Log fold change | p-values | ||
|---|---|---|---|---|---|
| SDC | IBC | DCIS | |||
| hsa-miR-142-3p | 8.314 | 10.417 | 2.1027 | 0.0375 | |
| hsa-miR-10a | 6.718 | 8.432 | 1.7137 | 0.0016 | |
| hsa-miR-99a | 8.974 | 10.128 | 1.1535 | 0.0167 | |
| hsa-miR-10a | 6.718 | 8.293 | 1.5745 | 0.0099 | |
| hsa-miR-335 | 5.374 | 6.680 | 1.306 | 0.0100 | |
| hsa-miR-331-3p | 5.642 | 6.800 | 1.158 | 0.0002 | |
| hsa-miR-16 | 12.796 | 12.078 | − 0.7185 | 0.0040 | |
| hsa-miR-19b | 9.336 | 7.890 | − 1.446 | 0.0060 | |
| hsa-miR-15a | 10.312 | 9.305 | − 1.007 | 0.0078 | |
| hsa-miR-374a | 7.903 | 6.348 | − 1.5558 | 0.0176 | |
| hsa-miR-19b | 9.288 | 7.890 | − 1.3983 | 0.0038 | |
| hsa-miR-15a | 10.432 | 9.305 | − 1.1267 | 0.0068 | |
| hsa-miR-222 | 8.935 | 7.913 | − 1.0225 | 0.0148 | |
| hsa-miR-16 | 12.968 | 12.078 | − 0.8908 | 0.0025 | |
MiR-10a Is Selectively Expressed in Normal Breast Compared to Salivary Gland Tissue
Real-time PCR validation of SDC, IBC and DCIS tumors showed a significantly increased expression of miR-10a in IBC and DCIS compared to SDC (Fig. 3d). This led us to further investigate whether miR-10a upregulation in IBC and DCIS tumors compared to SDC parallels the same pattern in the surrounding non-tumoral (“normal”) mammary and salivary gland tissue respectively. Our analysis of adjacent non-tumoral tissues from IBC and SDC cases revealed an increased expression of miR-10a in breast tissue compared to salivary tissue (Fig. 3c, e). Our data suggests that, in contrast to SDC, miR-10a could be playing an important role in breast tumors, possibly acting as a mammary specific oncogene, the activation of which might function as an initiator of tumorigenesis.
HOXA1, a Target of miR-10a Is Down-Regulated in IBC
Given the differential expression levels of miR-10a observed between SDC and IBC, we next investigated whether putative targets of miR-10a were differentially regulated in these cancers. One of the many putative miR-10a targets as determined by in silico experiments using Target Scan and PicTar is HOXA1, a gene known to be implicated in a number of cancers [25–27]. HOXA1 regulation by miR-10a has been reported in some studies [28, 29]. We therefore evaluated gene expression levels of HOXA1 in snap-frozen tissues of normal breast and IBC cases from new patients using RT-PCR. Our results showed a significant decrease in HOXA1 expression in IBC compared to normal breast tissue (Fig. 3f). These results correlate with the increased miR-10a expression levels in IBC compared to IDC, supporting the hypothesis that over-expression of miR-10 in IBC leads to downregulation of HOXA1 which might contribute to the initiation and progression of mammary cancers.
Discussion
SDCs are highly lethal salivary gland malignancies characterized by poor prognosis in spite of multimodality treatment, consisting of surgical resection, with or without adjuvant chemoradiation therapy. Despite their frequent expression of Her2/Neu and AR, response to targeted Her2/Neu inhibition and androgen deprivation therapies, respectively, has been disappointingly inconsistent [1, 30, 31]. There is thus an urgent need for a basic understanding of the pathogenesis of these rare tumors, with a view to developing novel therapeutic targets.
Moreover, given the striking histopathologic and immunophenotypical similarities between SDC and HNPIBC, in those exceptionally rare clinical circumstances where synchronous or metachronous tumors occur, distinguishing between the two malignancies may be exceptionally challenging. Because there may be differing implications for staging, prognosis and therapy, the discovery of a specific molecular signature that would discriminate these two tumors could facilitate more accurate diagnostic and prognostic assessments, and provide insights into possible therapeutic strategies.
As regulators of gene expression, miRNAs have increasingly become the focus of extensive research in cancer development as well as in the discrimination of various cancer types. In this study, we used miRNA profiling to determine whether the similarities that exist among SDC, HNPIBC, and high-grade DCIS are reflected in the molecular landscape of these cancers. The goal was to identify specific molecular signatures that discriminate among these histopathologically similar malignancies in order to characterize pathways that potentially could represent new targets for specific therapy. Our results demonstrate that there are striking similarities in the miRNA expression profiles of SDC and IBC. This finding is in accord with previous studies that have used different platforms to compare the two entities [1, 32]. It should be noted that, without RT-PCR validation, miRNA expression as determined by nanostring analysis should be interpreted with caution. Although beyond the scope of our current study, a more extensive validation of the differential miRNA profiles observed between SDC, DCIS and IBC will be required.
Although the role of specific miRNAs in the pathogenesis of SDC is yet to be fully characterized, evidence suggests that they are significant contributors to salivary gland epithelial malignancies. For example, an examination of mucoepidermoid carcinoma (MEC) of the upper aerodigestive tract by Chiosea et al. revealed a significant correlation between the DICER protein, which is involved in miRNA maturation, and advanced stages/high grades of MEC as determined by immunohistochemistry [33]. We found miR-21 to be significantly up-regulated in SDC compared to normal salivary gland and miR-148 to be down-regulated in SDC compared to surrounding normal salivary gland, suggesting that these miRNAs may be functioning as oncomir and tumor suppressor miRNA, respectively, in SDC.
miR-21 is located on chromosome 17q23.2 in humans and has been shown to be the most commonly upregulated miRNA in both solid and hematological malignancies [12, 34]. The mechanism of oncogenesis is varied amongst the different tumor types. For example, in colorectal carcinomas, it has been shown that miR-21 induces “stemness” by down-regulating transforming growth factor receptor beta while it stimulates invasion and metastasis by suppressing PDCD4 [35, 36]. In non-small cell lung cancer, miR-21 enhances oncogenic K-ras activation and modulates tumorigenesis by targeting SPRY2, BTG2, and PDCD4 [37]. MiR-148 is down-regulated in many solid organ malignancies, including breast, colorectal, pancreatic, hepatocellular, endometrial and non-small cell lung cancers [38]. It has been shown that increased expression of miR-148a inhibits both tumor growth and metastasis in a hepatocellular carcinoma tumor stem cell-line by controlling the ACVR1/BMP/Wnt signaling pathway [39]. In the context of our current study, differential expression of miR-21 and miR-148 between normal salivary gland and SDC, suggests these miRNAs may represent potential tumor biomarkers and possible therapeutic targets. Additional studies will be needed to determine the biologic roles of specific miRNAs in SDC pathogenesis.
The differential expression of miR-10a between SDCs and IBCs illustrates the potential utility of miRNAs to distinguish between these histologically similar cancer types. This will be particularly useful in resolving clinical dilemmas, such as one in which an IBC develops synchronously with a secondary nodule in the parotid gland, the latter of which could either represent a metastasis from the breast primary or a de novo SDC. The impact on staging and treatment would be significant, depending on which scenario is correct.
Our miRNA analysis of breast tumors relative to surrounding non-involved normal breast tissue is in agreement with a previous study by Iorio et al. [15], which showed five miRNAs that were dysregulated (miR-10b, miR-125b, and miR-145 down-regulated; and miR-21 and miR-155 up-regulated). In our study, we found four of these miRNAs (miR-10b, miR-145, miR-21 and miR-155) to be dysregulated. Interestingly, miR-10b, which was down-regulated in IBC compared to normal breast tissue (in both Iorio et al. and our study), has been shown to be highly expressed in metastatic breast cancer cells [40]. Indeed, therapeutic targeting of miR-10b inhibits breast cancer metastasis but does not inhibit primary mammary tumor growth in a mouse breast cancer model [41]. This highlights the potential value of miRNA targeting in cancer therapy.
A significant finding in our study was the selectively increased expression of miR-10a, which followed a tissue-specific expression pattern. First, miR-10a displayed increased expression in normal breast tissue compared to the normal salivary gland tissue. Second, miR-10a expression was increased in HNPIBC compared to SDC. These results suggest that miR-10a could be biologically relevant in mammary oncogenesis, at least in a subset of breast carcinomas. This is not surprising, as previous studies have reported an amplification of the miR-10 gene locus in breast cancer patients [42, 43]. Thus, it is plausible that miR-10a could act as mammary tissue oncogene, and its overexpression may be an initiation step in mammary carcinogenesis. Indeed, miR-10a up-regulation might facilitate malignant transformation of normal glandular breast epithelium, and could trigger uncontrolled growth that initially leads to DCIS and then to IBC. Previous research has linked miR-10a to increased chemo-resistance of breast cancer cell-line MCF7 to cisplatin [44]. Hence, miR-10a might not only promote tumorigenesis but may also be involved in chemoresistance, suggesting that this microRNA could be a potential therapeutic target in some breast cancer types. Further studies are warranted to determine the correlation between miR-10a expression and pre-neoplastic ductal breast lesions such as usual ductal hyperplasia (UDH) and atypical ductal hyperplasia (ADH), including columnar cell hyperplasia and flat epithelial atypia. Given these results, it is possible that miR-10a plays a tissue-specific role in breast carcinogenesis compared to the histopathologically similar SDC.
To further define the involvement of miR-10a in breast carcinogenesis, we investigated the expression of the putative miR-10a target HOXA1. HOXA1 is a transcription factor that is part of the homeobox gene cluster A, involved in gene expression, cell differentiation, and temporo-spatial body development [25]. It is believed that the HOX gene family is an important regulator of tumor invasion and metastasis. MiR-10a and miR-10b are both located close to the homeobox genes and in silico studies (Target Scan) show that miR-10a targets HOXA1, which has also been confirmed by luciferase assay experiments [29]. MiR-10a has also been shown to repress transcriptional expression of another member of the HOX gene family, HOXD4, in breast cancer cells [45]. In line with these studies, our analysis of breast and salivary gland tumors showed decreased expression of HOXA1 in IBC compared to SDC, which is consistent with the enhanced miR-10a expression observed in IBC tumors.
MiR-10a expression and concomitant repression of HOXA1 has been shown to be an important regulatory pathway in a number of cancers, including acute myeloid leukemia [46] and pancreatic cancer [28]. In pancreatic cancer, miR-10a over-expression decreases HOXA1 levels, which trigger increased invasiveness of malignant epithelial cells [28]. In another study, retinoic acid receptor antagonists, which inhibit miR-10a expression, prevented the metastatic behavior of pancreatic cancer [47]. Therefore, it is possible that down-regulation of HOXA1 in IBC tumors might prevent the differentiation of ductal epithelial cells and increase their invasiveness, characteristic of high-grade carcinomas. Furthermore, it seems that HOXA1 might be up-regulated in certain types of breast cancers (basal-like, Her2Neu-, ER-) but decreased in others (Her2Neu+, ER+), and its down-regulation might be influenced by miR-10a via 3′UTR direct targeting, or via enhanced HOXA1 promoter methylation. It should be noted that our data did not show a significant difference in the expression of miR-10a between normal breast tissue and IBC. While additional experiments will be required to explain this apparent discrepancy, other studies using in silico and in vitro reporter assays demonstrate that HOXA1 is targeted by other miRNAs including miR-30b, miR-30c, miR-30e, miR-99a miR-100, miR-181c and miR-203 [48–54]. Interestingly, we observed a moderate increase in miR-30e expression in IBCs compared to normal breast tissue (Supplementary Table 2). It is therefore possible that downregulation of HOXA1 is mediated by one or more miRNAs during breast oncogenesis.
In conclusion, our studies reveal similar miRNA expression patterns between SDC and IBC, but with differential expression of miR-10a in IBC compared to SDC. We also suggest that miR-10a could be useful in resolving difficult clinical scenarios of patients with synchronous HNPIBC and SDC, and serve as a potential biomarker in the early detection of a subset of breast cancer. We propose a new tumorigenic mechanism in breast cancers, represented by the over-expression of a breast specific miRNA, miR-10a, that down-regulates HOXA1 and leads to proliferation of less differentiated ductal epithelial cells with increased invasiveness. Conceivably, down-regulation of miR-10a through antisense compounds might prove to be a potentially effective therapy for these types of cancer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge Dr. Carl Allen of the Department of Oral and Maxillofacial Pathology and Radiology, College of Dentistry, The Ohio State University Wexner Medical Center for proof-reading the manuscript and providing editorial assistance.
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
This study was approved by the Institutional Review Board of The Ohio State University, Department of Pathology (Institutional Review Board-approved Protocol No. 2002H0089).
References
- 1.Dalin MG, Desrichard A, Katabi N, Makarov V, Walsh LA, Lee KW, et al. Comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin Cancer Res. 2016;22(18):4623–4633. doi: 10.1158/1078-0432.CCR-16-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grenko RT, Gemryd P, Tytor M, Lundqvist PG, Boeryd B. Salivary duct carcinoma. Histopathology. 1995;26(3):261–266. doi: 10.1111/j.1365-2559.1995.tb01440.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnes L, Eveson JW, Reichart P, Sidransky D. World Health Organization classification of tumours: pathology and genetics of head and neck tumours. Lyon: IARC Press; 2005. [Google Scholar]
- 4.Wick MR, Ockner DM, Mills SE, Ritter JH, Swanson PE. Homologous carcinomas of the breasts, skin, and salivary glands: a histologic and immunohistochemical comparison of ductal mammary carcinoma, ductal sweat gland carcinoma, and salivary duct carcinoma. Am J Clin Pathol. 1998;109(1):75–84. doi: 10.1093/ajcp/109.1.75. [DOI] [PubMed] [Google Scholar]
- 5.Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353(16):1652–1654. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, et al. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18(7):1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 2010;285(13):9383–9389. doi: 10.1074/jbc.M109.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuhara T, Matsuura Y. Role of miR-122 and lipid metabolism in HCV infection. J Gastroenterol. 2013;48(2):169–176. doi: 10.1007/s00535-012-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Hua X, Haubrock M, Wang J, Wingender E. The architecture of the gene regulatory networks of different tissues. Bioinformatics. 2012;28(18):i509–i514. doi: 10.1093/bioinformatics/bts387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Chiosea SI, Williams L, Griffith CC, Thompson LD, Weinreb I, Bauman JE, et al. Molecular characterization of apocrine salivary duct carcinoma. Am J Surg Pathol. 2015;39(6):744–752. doi: 10.1097/PAS.0000000000000410. [DOI] [PubMed] [Google Scholar]
- 17.Ku BM, Jung HA, Sun JM, Ko YH, Jeong HS, Son YI, et al. High-throughput profiling identifies clinically actionable mutations in salivary duct carcinoma. J Transl Med. 2014;12:299. doi: 10.1186/s12967-014-0299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowell CF, Weigelt B, Sakr RA, Ng CK, Hicks J, King TA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol. 2013;7(5):859–869. doi: 10.1016/j.molonc.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To T, Wall C, Baines CJ, Miller AB. Is carcinoma in situ a precursor lesion of invasive breast cancer? Int J Cancer. 2014;135(7):1646–1652. doi: 10.1002/ijc.28803. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 2013;6(286):ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 24.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 25.Chariot A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Commun. 1996;222(2):292–297. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- 26.Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, et al. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007;26(27):3998–4008. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, et al. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J Biol Chem. 2006;281(10):6471–6481. doi: 10.1074/jbc.M512666200. [DOI] [PubMed] [Google Scholar]
- 28.Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19(7):2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 29.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105(10):3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limaye SA, Posner MR, Krane JF, Fonfria M, Lorch JH, Dillon DA, et al. Trastuzumab for the treatment of salivary duct carcinoma. Oncologist. 2013;18(3):294–300. doi: 10.1634/theoncologist.2012-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, Kwon OJ, Park JJ, Seo JH. Salivary duct carcinoma of the parotid gland: is adjuvant HER-2-targeted therapy required? J Oral Maxillofacial Surg. 2014;72(5):1023–1031. doi: 10.1016/j.joms.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Hoang MP, Callender DL, Sola Gallego JJ, Huang Z, Sneige N, Luna MA, et al. Molecular and biomarker analyses of salivary duct carcinomas: comparison with mammary duct carcinoma. Int J Oncol. 2001;19(4):865–871. doi: 10.3892/ijo.19.4.865. [DOI] [PubMed] [Google Scholar]
- 33.Chiosea SI, Barnes EL, Lai SY, Egloff AM, Sargent RL, Hunt JL, et al. Mucoepidermoid carcinoma of upper aerodigestive tract: clinicopathologic study of 78 cases with immunohistochemical analysis of dicer expression. Virchows Arch. 2008;452(6):629–635. doi: 10.1007/s00428-007-0574-5. [DOI] [PubMed] [Google Scholar]
- 34.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y, Nangia-Makker P, Farhana L, Rajendra SG, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Deng X, Zeng X, Peng X. The role of Mir-148a in cancer. J Cancer. 2016;7(10):1233–1241. doi: 10.7150/jca.14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, et al. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61(2):574–584. doi: 10.1002/hep.27543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 41.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lund AH. miR-10 in development and cancer. Cell Death Differ. 2010;17(2):209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127(8):1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 45.Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, et al. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryant A, Palma CA, Jayaswal V, Yang YW, Lutherborrow M, Ma DD. miR-10a is aberrantly overexpressed in Nucleophosmin1 mutated acute myeloid leukaemia and its suppression induces cell death. Mol Cancer. 2012;11:8. doi: 10.1186/1476-4598-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137(6):2136–45, e1. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 48.Ning ZQ, Lu HL, Chen C, Wang L, Cai W, Li Y, et al. MicroRNA-30e reduces cell growth and enhances drug sensitivity to gefitinib in lung carcinoma. Oncotarget. 2017;8(3):4572–4581. doi: 10.18632/oncotarget.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni LY, Zhao JD, Lu YH, Li W, Li BL, Wang XC, et al. MicroRNA-30c suppressed giant-cell tumor of bone cell metastasis and growth via targeting HOXA1. Eur Rev Med Pharmacol Sci. 2017;21(21):4819–4827. [PubMed] [Google Scholar]
- 50.Wang X, Li Y, Qi W, Zhang N, Sun M, Huo Q, et al. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget. 2015;6(32):32737–32747. doi: 10.18632/oncotarget.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma G, Agarwal SM. Identification of critical microRNA gene targets in cervical cancer using network properties. MicroRNA. 2014;3(1):37–44. doi: 10.2174/2211536603666140417214659. [DOI] [PubMed] [Google Scholar]
- 52.Mukherjee A, Shrivastava S, Bhanja Chowdhury J, Ray R, Ray RB. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol. 2014;88(14):7929–7940. doi: 10.1128/JVI.00787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang J, et al. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur J Cancer. 2014;50(8):1541–1554. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, Chen Z, Jin Y, Dragas D, Zhang L, Adjei BS, et al. MicroRNA-99 family members suppress Homeobox A1 expression in epithelial cells. PLoS ONE. 2013;8(12):e80625. doi: 10.1371/journal.pone.0080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.