Abstract
Natural killer (NK) cells play an important role in immune surveillance and protective immunity, mainly through rapid cytokine release and cytolytic activities. But how such responses are negatively regulated remains poorly defined. Here, we demonstrated that the E3 ubiquitin ligase TRIM29 is a crucial regulator of NK cell functions. We found that TRIM29 was not expressed in resting NK cells but was readily up-regulated following activation, especially after interleukin 12 (IL-12) plus interleukin 18 (IL-18) stimulation. The levels of TRIM29 expression were inversely correlated with interferon-γ (IFN-γ) production by NK cells, suggesting that TRIM29 inhibits NK cell functions. Indeed, deficiency of TRIM29 specifically in NK cells resulted in an enhanced IFN-γ production and consequently protected mice from murine cytomegalovirus (MCMV) infection. Mechanistically, we showed that once induced in NK cells, TRIM29 ubiquitinates and degrades the TGF-β activated kinase 1 binding protein 2 (TAB2), a key adaptor protein in IFN-γ production by NK cells. These results identify TRIM29 as a negative regulator of NK cell functions and may have important clinical implications.
Introduction
NK cells are vital to protective innate immunity and immune surveillance; they provide a key defense mechanism against microbial pathogens as well as tumors (1, 2). NK cells also play a regulatory role in adaptive immune responses and are actively involved in autoimmune diseases (3–5). NK cells contribute to host defense primarily through their ability to rapidly secrete cytokines (mainly IFN-γ and TNF-α) and chemokines (e.g., MIP-1α/β), as well as release of cytolytic granules containing granzymes and perforin to kill target cells (6, 7). In particular, production of IFN-γ, a cytokine essential for both innate and adaptive immune responses, is a hallmark of NK cell activation. It is critical for suppressing the proliferation of tumor and virus-infected cells (8, 9). In models of MCMV infections, NK cell production of IFN-γ during early infection is required for viral control (10). IL-12 and IL-18 are powerful inducers of NK cell activation leading to IFN-γ production(4, 11). Upon binding of the IL-12 receptor (IL-12R) consisting of β1 and β2 chains, IL-12 induces phosphorylation and activation of JAK2, TYK2 kinases, and the transcription factor STAT4, which translocates to the nucleus and activates transcription of the Ifng gene (12). IL-18 is structurally related to IL-1β, but by itself, IL-18 is a poor inducer of IFN-γ production by NK cells due to low IL-18R expression on the cell surface (13). However, when combined with IL-12, IL-18 is remarkably synergistic in NK-mediated IFN-γ production, due in part to upregulation of the IL-18R on the cell surface by IL-12 (14, 15). The binding of IL-18 to IL-18Rα and β chains leads to a cascade of signaling events, primarily through MyD88 recruitment of TNF receptor-associated factor 6 (TRAF6), TGF-β activated kinase 1 (TAK1) and their adaptor proteins such as the TGF-β activated kinase 1 binding protein 1 and 2 (TAB1 and TAB2) (16). Furthermore, IL-18 also activates AP-1 via MAP kinases to stabilize IFN-γ mRNA and enhance IFN-γ secretion by NK cells (17). In most cases, an increased level of IFN-γ is protective against acute viral infections, but uncontrolled and excessive production of IFN-γ by NK cells can lead to immune disorders, such as inflammatory bowel disease and atherosclerosis (10, 18, 19). Thus, it is crucial that this IFN-γ production is finely tuned during immune activation where it provides optimal protection against pathogens, while avoiding unwanted inflammation and tissue damage. However, the precise mechanisms that negatively regulate IFN-γ expression after productive NK cell activation have not been carefully explored and are largely unknown.
We previously reported that TRIM29 is a crucial negative regulator of alveolar macrophages and controls macrophage activation in the respiratory tract (20). TRIM29 is an E3 ubiquitin ligase; it employs the B-box domain to catalyze substrate ubiquitination instead of the typical RING domain (21, 22). In general, E3 ligases transfer ubiquitin groups from the E2 to the target proteins. These activities often mediate substrate degradation or other protein activities through either Lys48-linked or Lys63-linked ubiquitination, respectively (23, 24). These post-translational modifications have been involved in diverse signaling events. In this study, we examined the role of TRIM29 in NK cells and demonstrated a novel role for the E3 ligase TRIM29 in regulating NK cell functions. TRIM29 is induced in NK cells by IL-12 and IL-18, binds the TAB2 molecule at the N terminal domain, and promotes proteasome-mediated degradation of TAB2, thus, inhibiting IFN-γ production by activated NK cells. Indeed, deficiency of TRIM29 in NK cells leads to markedly enhanced NK cell functions after IL-12 and IL-18 stimulation. Our data identify TRIM29 as a key checkpoint regulator of IFN-γ production in NK cells.
Materials and Methods
Animals
Trim29f/f mice were designed with loxP sites flanking exon 2 of Trim29. NKp46-iCre mice were provided by Eric Vivier (25), and were bred to Trim29f/f mice to generate Trim29f/fNKp46-iCre mice. All animals were maintained in a specific pathogen free facility at Houston Methodist Research Institute in Houston, Texas. Animal use and care were approved by the Houston Methodist Animal Care Committee in accordance with institutional animal care and use guidelines. Mice at 7–8 wk of age were infected with 5 × 104 PFU (plaque-forming units) of the MCMV Smith strain by intraperitoneal injection (i.p.). In experiments involving IFN-γ neutralization, mice were injected twice i.p. with α-IFN-γ antibody (H22, 200 μg) 1 day and 3 days prior to infections.
Antibodies and reagents
Fluorochrome-conjugated antibodies to mouse CD45.2 (104), CD3ε (145–2C11), CD19 (1D3), NK1.1 (PK136), NKp46 (29A1.4), Ly49H (3D10), Ly49D (4E5), KLRG1 (2F1), CD69 (H1.2F3), CD49a (HMa1), CD49b (DX5), CD11b (M1/70), CD27 (LG.3A10), IFN-γ (XMG1.2), CD107a (1D4B) were purchased from Biolegend. RORγt (B2D) were purchased from eBioscience. Intracellular IFN-γ staining was performed using Intracellular Fixation and Permeabilization Buffer Set (eBioscience). Mouse recombinant mouse IL-15, IL-12, and IL-18 were purchased from Peprotech (Rocky Hill, NJ). The following antibodies were used for immunoblot analysis: anti-TRIM29 (1:1,000) (H-300; Santa Cruz), anti-TAB2 (1:1,000) (C88H10; CST), anti-TRAF6 (1:1,000) (D-10; Santa Cruz), anti-TAK1 (1:1,000) (D94D7; CST), anti-P38 (1:1,000) (D13E1; CST), anti-ERK1/2 (1:1,000) (137F5; CST), anti-JNK (1:1,000) (#9252; CST), anti-STAT4 (1:1,000) (C46B10; CST), anti-ubiquitin (1:1,000) (P4D1; Santa Cruz), K63-specific anti-ubiquitin (1:1,000) (05–1313; Millipore), K48-specific anti-ubiquitin (1:1,000) (05–1307; Millipore), K27-specific anti-ubiquitin (1:1,000) (EPR17034; Abcam), anti-β-actin (1: 10,000) (A2228; Sigma), anti-HA (1: 2,000) (HA-7; Sigma), anti-Myc (1: 2,000) (9E10; Santa Cruz), All HRP-labelled secondary antibodies were from Cell Signaling. Protein A-agarose were from Santa Cruz. Anti-Myc Agarose Affinity Gel antibody produced in rabbit were from Sigma. IP lysis buffer were from Thermofisher. Mouse IFN-γ ELISA MAX Deluxe sets (B168582) were from Biolengend.
Flow cytometric analysis
Single-cell suspensions of bone marrow, blood, spleen, liver and intestine from mice were obtained. Cells were stained with the Zombie Aqua Dye (BioLegend) to exclude dead cells. Cells were then blocked and stained with cell surface markers or mouse IgG1 isotype control in for 30min at 4 °C in the dark. Cells were washed and collected with a FACSCalibur cytometer (BD Immunocytometry Systems, San Jose, CA), and analyzed using FlowJo (Tree Star, Ashland, OR). For apoptosis assay, cells were stained with APC Annexin V apoptosis detection Kit with 7-AAD (BioLegend).
ELISA
Blood was obtained from uninfected control and MCMV-infected mice. Sera were collected and assayed by ELISA for IFN-γ according to the manufacturer’s instructions.
MCMV Quantification by Plaque Assay
Spleen and liver were harvested from day 1.5 post-infection and weighed, homogenized to prepare extracts in 0.5 ml DMEM. Virus titers were quantified as described previously (Lindenberg et al., 2014). Briefly, the serial dilution of homogenates were inoculated on 3T3-NIH cell overlays for 2 hours to allow virus attachment. Cells were covered with pre-warmed 0.75% (w/v) carboxymethylcellulose DMEM medium, cultured for 5 more days, then fixed with formalin and stained with crystal violet.
Cells purification
Mouse Spleens and livers were cut into small pieces and mechanically homogenized in RPMI1640 Glutamax medium (Gibco) supplemented with 10% fetal bovine serum (FBS), 5 mM β-mercaptoethanol (Gibco), and 100 U/mL penicillin/streptomycin (Biochrom). A single-cell suspension was prepared, and red blood cells (RBCs) were lyzed in ACK lysing buffer (ThermoFisher). For NK cell sorting, spleen or liver single-cell suspensions from 3 to 5 mice were pooled and enriched for NK cells using the NK Cell Isolation Kit II (Miltenyi Biotec). Following magnetic separation on an autoMACS Separator (Miltenyi Biotec), spleen NK cells were gated as CD3e–NK1.1+ cells. Liver ILC1 cells were identified and gated as CD45+CD3ε−CD19−TCRβ−NK1.1+CD49b−CD49a+ cells. CD4+ T cells in spleen were sorted from as CD3ε +NK1.1−CD4+ cells. CD8+ T cells in spleen were sorted as CD3ε +NK1.1−CD8+ cells. NKT cells in spleen were sorted as CD3ε + NK1.1+ cells. All cell sorting was performed using the FACSAria II (BD Biosciences).
NK Cell Stimulation in vitro
Sorted splenic NK cells were stimulated in RPMI1640 containing 10% FBS for different time points in the presence of low dose recombinant murine IL-15 (5ng/mL), and stimulated with recombinant mouse IL-12 (2 ng/ml), IL-18 (2 ng/ml). CD107a antibody was added with BD GolgiStop™ (containing Monensin) 4 hours prior to intracellular staining. Cells were first stained for surface markers, followed by fixation and permeabilization with the Fixation and Permeabilization Solution set (eBioscience). Cells were then resuspended in 1× permwash solution and stained with antibodies.
In Vitro Cytotoxicity Assays
The cytolytic activities of NK cells in vitro were determined as following: YAC-1 target cells were labelled with CFSE (2.5 μM, Molecular Probes) and co-cultured in triplicates with magnetically enriched splenic NK cells at different effector to target (E:T) ratios. In some experiments, NK cells were pre-activated with IL-12 (2 ng/ml) and IL-18 (2 ng/ml) for 24 hrs before co-culturing with YAC-1 cells. After 4 hrs co-culture, cells were stained with 7-aminoactinomycin D (7-AAD, eBioscience) and killing of target YAC-1 cells by NK cells was determined by flow cytometry. The cytolytic activities were calculated as % of CFSE+7-AAD+ cells co-cultured with NK cells – % CFSE+7-AAD+ cells in the absence of NK cells.
Co-immunoprecipitations and Western blotting analysis
Forty-eight after transfection, cells were lysed in IP lysis buffer (#87788, Fisher Scientific) with proteases and phosphatase inhibitor cocktail for 30 min. Soluble extracts of cell lysates were collected by centrifugationat 15,000g for 10 min. Pre-cleaning was performed by adding protein A beads into collected lysate and incubated with constant rotation for 1 h at 4°C, followed by another 15,000g centrifugation for 10 min at 4°C. MYC-tagged recombinant proteins in the supernatants were immunoprecipitated by incubation with anti-MYC antibodies with constant rotation overnight at 4°C. After adding the protein A agarose beads suspension, the mixture was further incubated with constant rotation for 4 hrs at 4°C. The precipitates were washed four times by using cold IP lysis buffer. After final washing, the beads were resuspended in sample buffer and boiled for 10 mins. Coimmunoprecipitated samples or the whole cell lysates were analyzed by Western blotting with the antibodies indicated in the figures. The specific bands were visualized using the enhanced chemiluminescence reagents (Thermo Scientific, Waltham, MA). For all immunoblotting experiments, β-actin was used as a loading control.
Real time quantitative RT-PCR analysis
Total RNA was extracted with the RNeasy mini kit (#74106, Qiagen) according to the manufacturer’s introduction. RNA quality was assessed using NanoDrop 2000 and a total of 400ng RNA were used for reversed transcription using High-Capacity cDNA Reverse Transcription Kit (#4375575, ABI) with random primers. Quantitative real-time PCR was performed by BIO-RAD CFX96 real time PCR system with SYBR Green Mix and specific primers (Sigma). Data were normalized to the expression of housekeeping GAPDH gene and the relative abundance of transcripts was calculated by the 2−ΔΔCt method.
Statistical analysis
All graphs were generated with Prism 5 software (GraphPad). A two-tailed Student t test or one-way ANOVA was used to generate p value data, as specified in the figure legends. A p value <0.05 was considered statistically significant.
Results
NK cell production of IFN-γ is dynamic and correlated with TAB2 expression
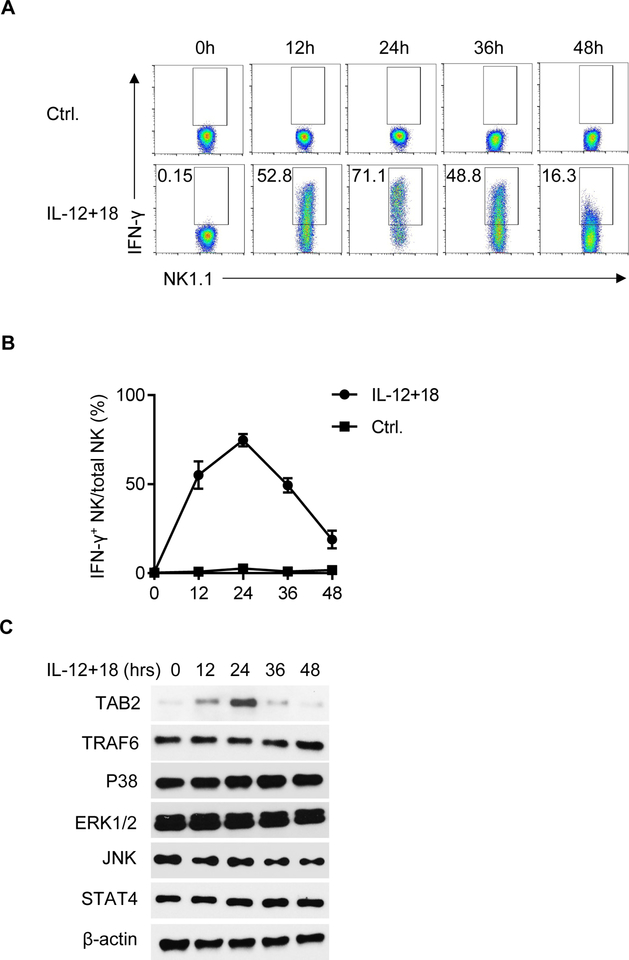
We used IFN-γ as a readout to examine NK cell functions and how the IFN-γ response is regulated. Using intracellular IFN-γ staining and flow cytometry, we measured the IFN-γ expression of resting NK cells, as well as of NK cells stimulated with IL-12 and IL-18 in vitro. Splenic NK cells were purified (Fig. S1) and cultured in the presence of IL-12 and IL-18. We examined the kinetics of IFN-γ expression in NK cells at different time points (0–48 hr) after activation. As compared to unstimulated resting NK cells, IFN-γ was highly induced in NK cells stimulated with IL-12 and IL-18 (Fig. 1A). Kinetic analysis showed that IFN-γ production peaked at 24 hr, and gradually decreased at 48 hr after stimulation under such activation conditions (Fig. 1B). We also evaluated CD107a expression, which is a marker of NK cytotoxic capacity. As shown in Fig. S2A and S2B, we observed comparable levels of CD107a in NK cells with or without IL-12 plus IL-18 (IL-12/18) stimulation, which is consistent with a previous report that IL-12/18 stimulation has a minimal effect on NK cell cytotoxicity (26). The dynamic expression of IFN-γ in NK cells prompted us to determine the feedback regulatory mechanisms in the IL-12/18 induced NK cell activation. We assessed the induction of key molecules downstream of IL-12/18 pathways by immunoblot analysis. We found that TAB2 expression in NK cells showed the most striking correlation to IFN-γ expression in response to IL-12/18 stimulation, which peaked at 24 hr and gradually declined over 48 hr (Fig. 1C). We also examined other signaling molecules besides TAB2 and found that the protein levels of TARF6, P38, ERK1/2, JNK and STAT4 remained constant throughout activation.
Figure 1. The kinetics of IFN-γ expression in NK cells in response to IL-12 and IL-18 stimulation.
(A, B) Splenic NK cells were stimulated with IL-12 (2 ng/ml) and IL-18 (2 ng/ml) in a low concentration of IL-15 (5 ng/ml) for the indicated periods. The expression (A) and the kinetics (B) of IFN-γ were determined by flow cytometry. (C) NK cells were stimulated with IL-12 (2 ng/ml) and IL-18 (2 ng/ml) for 0, 12, 24, 36 or 48 hrs. Cell lysate were prepared for western blot using indicated antibodies. β-actin served as a loading control. Data are representative of 3 independent experiments. Data are mean ± SEM.
TRIM29 is induced in activated NK cells, but not in resting NK cells
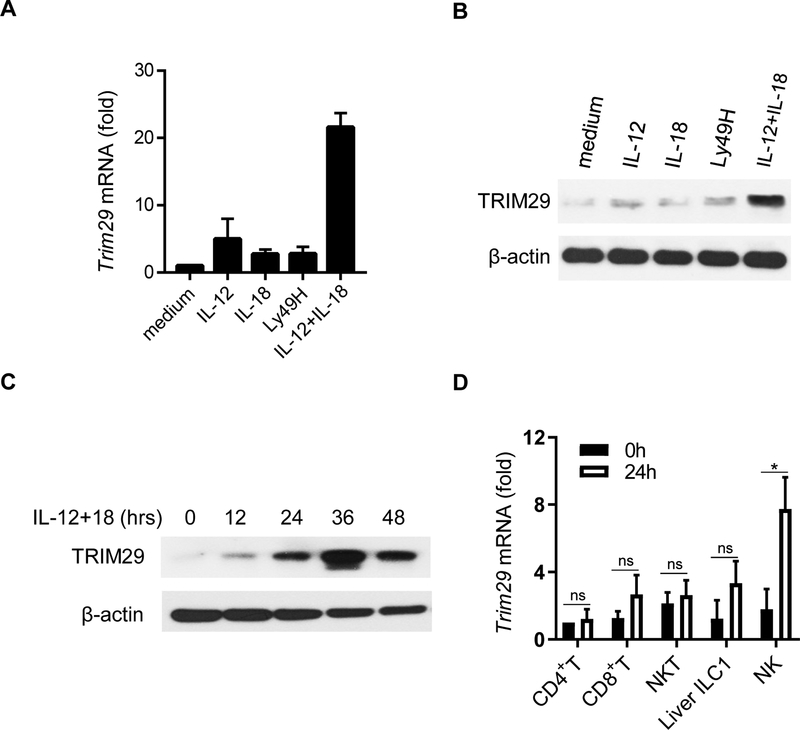
To determine whether TRIM29 plays a role in NK cell activation, we cultured splenic NK cells for 24 hrs under cytokines (IL-12 and IL-18, either alone or combination) or pre-coated antibodies against NK cell receptor Ly49H and measured the induction of TRIM29 by quantitative RT-PCR (qRT-PCR) and immune blot. The expression of TRIM29 was extremely low in resting NK cells, but its expression was markedly induced by IL-12/18 treatment. However, TRIM29 expression was not induced by individual cytokines alone or the ligation of Ly49H (Fig. 2A and 2B). We next examined the kinetics of TRIM29 expression in NK cells in response to IL-12/18. The expression of TRIM29 peaked at 36 hr after stimulation and gradually decreased at 48 hr (Fig. 2C). As controls, we also examined the expression of TRIM29 in liver ILC1 cells (defined as CD45+CD3ε−CD19−TCRβ−NK1.1+CD49b−CD49a+ cells), CD4+ T cells, CD8+ T cells and NKT cells upon IL-12/IL-18 stimulation. As shown in Fig. 2D, TRIM29 expression was primarily induced in NK cells in response to IL-12/18 stimulation, but not in other cell types (Fig. 2D). Taken together, these data suggest that TRIM29 expression in NK cells is an induced event and is likely involved in regulating NK cell activation in response to IL-12/18 stimulation.
Figure 2. The induction of TRIM29 in NK cells in response to IL-12 and IL-18 stimulation.
(A) Real-time PCR quantification of Trim29 mRNA in splenic NK cells at 24 hr in response to various combinations of IL-12, IL-18, and plate-bound anti-Ly49H mAbs, shown as fold expression relative to medium control. (B) TRIM29 protein levels were determined by immunoblot at 24 hr. (C) Splenic NK cells were stimulated with IL-12 and IL-18 for the indicated periods and immunoblotted by TRIM29 antibodies. β-actin was used as a loading control. (D) Real-time PCR quantification of Trim29 mRNA in liver CD45+CD3ε−NK1.1+CD49b−CD49a+ ILC1, splenic CD3ε+CD4+ T cells, CD3ε+CD8+ T cells, CD3ε+NK1.1+ NKT cells and CD3ε−NK1.1+ NK cells in response to IL-12 and IL-18, shown as fold expression relative to naïve CD4+ T cells. Data are representative of 3 independent experiments. Data are mean ± SEM.
**P < 0.01; unpaired Student’s t‐test.
TRIM29 negatively regulates TAB2 and IFN-γ expression in activated NK cells
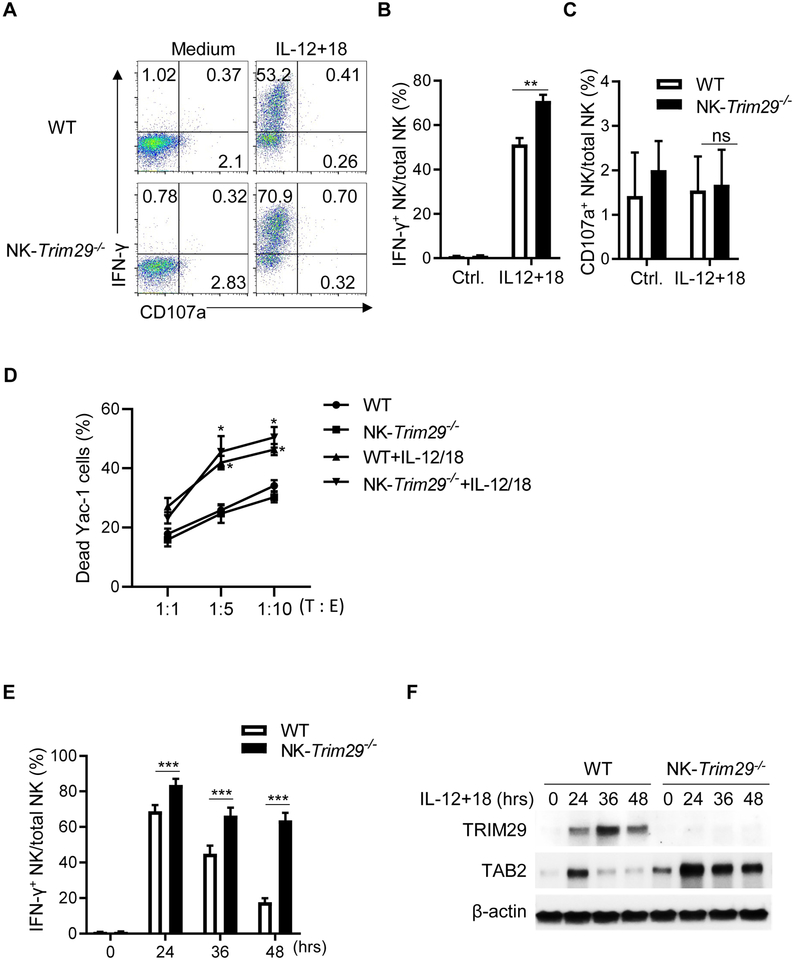
To determine the role of TRIM29 in regulation of NK cells, we deleted TRIM29 specifically in NK cells by crossing TRIM29 floxed mice (Trim29f/f) with NKp46-Cre (Nkp46iCre) mice (25) (Fig. S3A). This cross resulted in Trim29f/fNKp46iCre/+ mice (called ‘NK-Trim29−/− mice hereafter), carrying the conditional deletion of TRIM29 in NKp46+ NK cells (Fig. S3A and S3B). We next examined NK cell numbers, subsets, and phenotypes in various tissues of NK-Trim29−/− and Trim29f/fNKp46+/+ mice (WT hereafter). The frequency of splenic NK cell and NKp46 expression was normal in NK-Trim29−/− mice (Fig. S3C), and absolute number of NK cells from bone marrow, spleen and liver was comparable in NK-Trim29−/− or Trim29f/fNKp46+/+ WT littermates (Fig. 3C). Deletion of TRIM29 has no effect on the expression of maturation markers (i.e., CD27, CD11b, Ly49D, and Ly49H, CD49a, CD49b), activating or inhibitory receptors (i.e., KLRG1, TIGIT, NKG2A/C/E, CD226 and NKG2D) or migration marker CD62L in the spleen (Fig. S3E and S3F). Furthermore, the percentage of NKp46+ liver-resident innate lymphoid cell subset 1 (ILC1) and innate lymphoid cell subset 3 (ILC3) was unaltered in NK-Trim29−/− mice (Fig. S4A and S4B). We further examined whether TRIM29 deficiency affected the production of IFN-γ in NK cells stimulated by IL-12 and IL-18. Compared with WT mice, NK cells from NK-Trim29−/− mice produced higher levels of IFN-γ at 12 hr post IL-12/18 stimulation (Fig. 3A and 3B), while the expression of CD107a was similar in these cells (Fig. 3C). We then determined the cytolytic activities of NK cells against the YAC-1 target cells. As exposure of NK cells to IL-12 and IL-18 is known to boost NK cell cytolytic activities (27), we examined whether TRIM29 would inhibit the effect of IL-12/IL-18 in promoting NK cell cytotoxicity. We treated NK cells from WT and NK-Trim29−/− mice with IL-12 and IL-18 for 24 hrs, followed by incubation with YAC-1 targets. We found that both WT and NK-Trim29−/− NK cells were comparable in killing YAC-1 target cells after IL-12/IL-18 priming in vitro in various effector-to-target (E/T) ratios (Figure 3D). These results suggest that TRIM29 is likely not affecting NK cell cytolytic activities following IL-12/18 activation. IFN-γ production in NK cells peaked at 24 hr upon stimulation then gradually decreased at 36 hr and 48 hr, and knockout of TRIM29 significantly enhanced these IFN-γ production (Fig. 3E). These results suggest that TRIM29 plays a role in suppressing IFN-γ production in NK cells without affecting their cytolytic potential in response to IL-12/18 activation.
Figure 3. Sustained TAB2 expression in TRIM29-deleted NK cells promotes IFN-γ production upon cytokines stimulation in vitro.
(A) Flow cytometry plots of IFN-γ and CD107a expression in WT (Trim29f/fNKp46+/+) and NK-Trim29−/− (Trim29f/fNKp46iCre/+) splenic NK cells after 12 hrs upon IL-12 and IL-18 stimulation in vitro. (B and C) Quantification of percentage of intracellular expression of IFN-γ (B) and CD107a production (C) by splenic NK cells by flow cytometric analysis. (D) In vitro cytotoxicity of freshly isolated NK cells or IL-15 (5ng/ml), IL-12 (2 ng/ml) and IL-18 (2 ng/ml)–activated splenic NK cells from WT (Trim29f/fNKp46+/+) and NK-Trim29−/− (Trim29f/fNKp46iCre/+) mice towards YAC-1 targets at the indicated target–to–effector cell ratios (T : E) as quantified by FACS analysis. (E) Quantification of percentage of intracellular expression of IFN-γ by splenic NK cells at 24, 36 and 48 hr after IL-12 and IL-18 stimulation. (F) Immunoblotting of TRIM29 and TAB2 at various time points after stimulation. Data are representative of 3 independent experiments. Data are mean ± SEM.
*P < 0.05, **P < 0.01, ***P < 0.001; unpaired Student’s t‐test.
TRIM29 binds TAB2 and induces its ubiquination and degradation
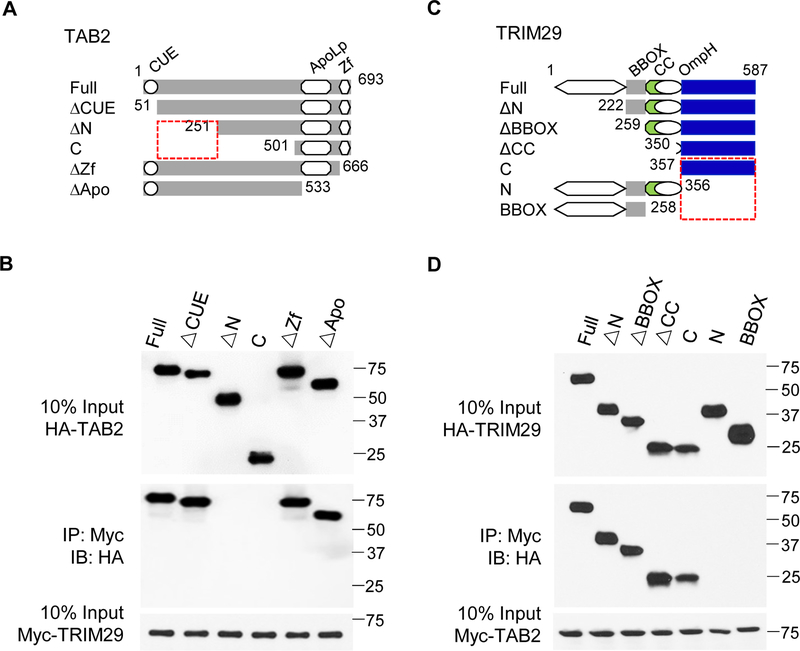
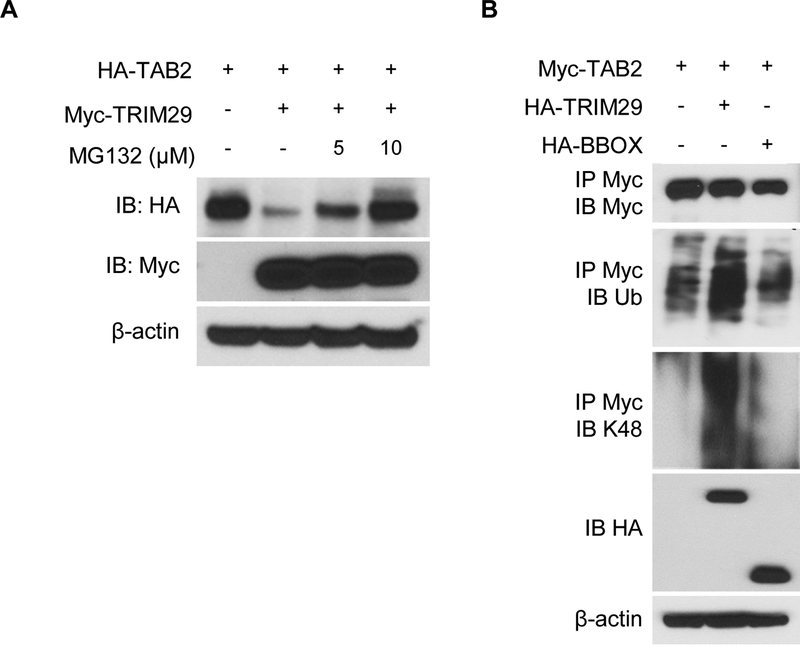
To explore the mechanisms of action of TRIM29, we investigated whether deletion of TRIM29 in NK cells would affect the expression of TAB2. As shown in Fig. 3F, TAB2 was abundantly expressed in both freshly isolated and activated NK cells from TRIM29-deficient mice as compared to that in WT NK cells. Notably, TRIM29-deficient NK cells showed enhanced expression of TAB2 upon activation (24 hr to 48 hr). These data suggest the possibility that TRIM29 may limit the expression of TAB2 in activated NK cells. We next screened potential proteins that could interact with TRIM29 by performing co-immunoprecipitation assays in which the TRIM29 was immunoprecipitated first using a specific TRIM29 mAb, followed by protein sequencing by liquid chromatography–mass spectrometry. We found molecules including TAK1, TAB1, TAB2 and TRAF6 which are the downstream of IL-18 signaling in this TRIM29-binding protein complex (data not shown). To ascertain the physical interactions between TRIM29 and TAB2, we mapped the interaction sites between TRIM29 and TAB2. We transfected HEK 293T cells with plasmids encoding the full-length (FL) or different truncations of TAB2 (ΔCUE, ΔN, C, ΔZf, and ΔApo; Fig. 4A) with a HA tag together with Myc-tagged full-length TRIM29. Myc-tagged TRIM29 was immunoprecipitated and then analyzed by Western blotting with anti-HA antibody to detect binding proteins in the complex. The results showed that both FL TAB2 and the N terminal (ΔCUE, ΔZf, and ΔApo) of TAB2, but not its C terminal ΔN and C domains, could bind TRIM29 (Fig. 4B), suggesting that the N-terminal domain of TAB2 interacts with TRIM29 (Fig. 4B). Next, we analyzed the TRIM29 domains mediating TRIM29–TAB2 interactions. We expressed HA-tagged full-length and truncated TRIM29 (ΔN, ΔBBOX, ΔCC, C, N and BBOX; Fig.4C), co-expressed them with Myc-tagged full-length TAB2 in HEK 293T cells, and examined their binding by Co-IP analysis. As shown in Fig 4D, the full-length TRIM29 and all mutants except its N-terminal domain could interact with TAB2 (Fig. 4D), suggesting that TRIM29 uses its C-terminal domain to bind to TAB2 (Fig. 4D). Because TRIM29 is an E3 ubiquitin ligase, we next determined whether TAB2 is the target of TRIM29 for ubiquitination and degradation. Co-expression of TAB2 with TRIM29 showed that TRIM29 induced degradation of TAB2. Treatment with the proteasome inhibitor MG132 rescued this degradation (Fig. 5A), suggesting that TRIM29 was able to induce proteasome-dependent degradation of TAB2. To determine whether TAB2 undergoes ubiquitination, we co-transfected plasmids expressing Myc-tagged TAB2 with HA-tagged full length TRIM29 or TRIM29-BBOX mutation into HEK293T cells. Immunoblotting analysis demonstrated the full length of TRIM29, but not its mutation, induced the ubiquitination of TAB2 by K48-mediated linkage (Fig. 5B).
Figure 4. Molecular mapping of TRIM29 and TAB2 interactions.
(A) Outline of the constructs of full-length TAB2 and of its truncated mutants: N-terminal coupling of ubiquitin conjugation to ER degradation (CUE), C-terminal zinc finger (ΔZf), and apolipophorin III like (ΔApo). Each construct includes a C-terminal HA tag. (B) Immunoblotting (IB) of purified Myc-tagged TRIM29 with anti-Myc (third blot), purified HA-tagged full-length TAB2 (Full) or its truncations alone (top) or after incubation with Myc-tagged TRIM29 and immunoprecipitation (IP) with anti-Myc (second blot). (C) Outline of the constructs of full-length TRIM29 and of its truncated mutants: the B-box zinc-finger domain (BBOX) and coiled-coil domain (CC). Each construct includes a C-terminal HA tag. (D) Immunoblotting of purified Myc-tagged TAB2 with anti-Myc (third blot), and purified HA-tagged full-length TRIM29 (Full) or its truncations alone (top) or after incubation with Myc-tagged TAB2 and immunoprecipitation with anti-Myc (second blot). Data are representative of 3 independent experiments.
Figure 5.
TRIM29 induces ubiquitination and degradation of TAB2 via K48 linkage. (A) Immunoblotting of HA-tagged TAB2 with anti-HA (top blot) or of Myc-tagged TRIM29 with anti-Myc (second blot) in HEK293T cells transfected with empty vector or expression vector for Myc-tagged TRIM29 and treated for 4 hrs with 5 or 10 μM of MG132. β-actin was used as a loading control (bottom). (B) Immunoblotting with anti-Myc (top blot), anti-total ubiquitination (ub, 2nd blot), anti-K48-linked ubiquitination (K48, 3rd blot) of Myc-tagged TAB2 in HEK293T cells transfected with empty vector or expression vector for HA-tagged TRIM29, TRIM29-BBOX and treated for 4 hrs with 10 μM of MG132, assessed after immunoprecipitation with anti-Myc, and with anti-HA (4th blot) or anti-β-actin (bottom blot) in whole-cell lysates. Data are representative of 3 independent experiments.
Conditional deletion of TRIM29 in NK cells enhances host defense against virus infection in vivo
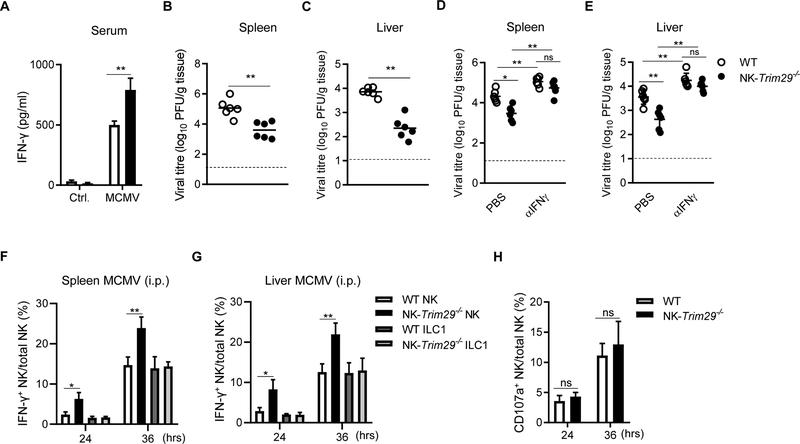
To assess the role of TRIM29 in NK cell functions in vivo, we intraperitoneally infected both NK-Trim29−/− mice and WT mice with MCMV, and monitored both IFN-γ production and viral titers in vivo over time. NK-Trim29−/− mice produced significantly higher IFN-γ production in the serum than did WT mice at 36 hr (Fig. 6A). Viral titer of MCMV in spleen and liver homogenates was also measured by plaque forming assay after infections. We found that NK-Trim29−/− mice had markedly decreased viral titers than those of WT mice at this early time point (Fig. 6B and 6C). To confirm that the control of MCMV in NK-Trim29−/− mice is due to an increase in IFN-γ, we treated WT and NK-Trim29−/− mice with α-IFN-γ prior to MCMV infection. Viral titers were measured in the spleen and liver at 36 hr. (Fig. 6D and E). As expected, NK-Trim29−/− mice showed increased viral titers in the spleen and liver, and both WT and NK-Trim29−/− mice had heightened levels of viral titers after neutralizing IFN-γ as compared to PBS control groups, suggesting that enhanced IFN-γ production due to TRIM29 deficiency is responsible for the control of MCMV replications. During MCMV infection, it is well described that the production of IFN-γ by NK cells in response to pro-inflammatory cytokines stimulation during infection is required in the early defense against MCMV infection. Under certain conditions, ILC1 cells also contribute to early IFN-γ production to aid in MCMV control (28). To clarify whether TRIM29 was also involved in ILC1 cell activation in viral control, we determined the IFN-γ expression in ILC1 cells from infected mice by flow cytometry. We found that despite enhanced number of IFN-γ+ NK-cells in NK-Trim29−/− mice as compared to WT mice at 24hr and 36 hr after infection (Fig. 6F and 6G), the ILC1 cells showed no differences in the spleen or liver in WT and NK-Trim29−/− mice at various time points (Fig. 6F and 6G). These results suggest that the enhanced anti-MCMV response in NK-Trim29−/− mice is through regulation of IFN-γ production by NK cells but not ILC1 cells. Furthermore, as shown in Fig. 6H, NK-Trim29−/− mice had no alteration in the percentage of CD107a+ NK cells. Thus, deletion of TRIM29 exhibited enhanced IFN-γ production in NK cells following acute viral infections and TRIM29 played a key role in modulating the anti-MCMV response. These data suggest that TRIM29 is involved in control of MCMV infection through regulation of NK cells.
Figure 6. TRIM29 deficiency enhances IFN-γ production by NK cells against viral infection.
(A-C) WT (Trim29f/fNKp46+/+) and NK-Trim29−/− (Trim29f/fNKp46iCre/+) mice were infected with MCMV (5 × 104 PFU) intraperitoneally (i.p.). Serum concentration of IFN-γ were measured by ELISA (A), viral titers were measured in spleen (B) and liver (C) at 36 hr post-infection. Dotted line represents the limit of detection for the assay. (D and E) For IFN-γ neutralizing studies, WT and NK-Trim29−/− mice were treated with PBS or α-IFN-γ, followed by infection with MCMV (i.p.). Viral titers were measured in the spleen (D) and liver (E) at 36 hr post-infection. (F and G) Quantification of intracellular of IFN-γ staining of NK cells and ILC1 cells in (F) spleen and (G) liver obtained from WT or NK-Trim29−/− mice following MCMV (i.p.). (H) Quantification of percentage of CD107a in splenic NK cells obtained from WT or NK-Trim29−/− mice following MCMV (i.p.). Data are representative of 3 independent experiments. Data are mean ± SEM.
*P < 0.05, **P < 0.01; unpaired Student’s t‐test.
Discussion
NK cells are a key cell type in innate immunity because of their cytotoxic capacity and their ability to produce high levels of cytokines during viral infections and tumor transformations (29, 30). Resting NK cells are self-tolerant, show limited effector functions, and require prior activation to execute effector activities including IFN-γ production. It has been shown that signals from cytokines, especially IL-12 and IL-18, are synergistic in activating NK cells and in IFN-γ production (31). However, persistent and uncontrolled IFN-γ production can lead to tissue inflammation and tissue damage, resulting in autoimmune disorders. As compared to signals and pathways that activate NK cells, little is known about how NK cells are suppressed after productive activation. In the current study, we studied the regulatory mechanisms that negatively regulate NK activities, using IL-12 and IL-18 as drivers of NK activation and IFN-γ production as a readout. We demonstrated that TRIM29 is readily induced in activated NK cells and acts a feedback regulator of IFN-γ production by NK cells. Mechanistically, we showed that once TRIM29 is induced, it binds to TAB2 and mediates TAB2’s Lys48 linked ubiquitination and proteasome degradation. As TAB2 is a key adaptor protein in IL-12 and IL-18 signaling in NK cells IFN-γ production, degradation of TAB2 abrogates IFN-γ expression in NK cells in response to IL-12 and IL-18. This notion is supported using conditional TRIM29 deficient mice where deletion of TRIM29 specifically in NK cells enhanced IFN-γ production in vitro, and protected mice from MCMV infections in vivo. Thus, our data identify TRIM29 as a key checkpoint regulator of NK activities and may have important clinical implications.
The E3 ubiquitin ligase TRIM29 has recently been shown to be a negative regulator of alveolar macrophages and to play a critical role in suppressing macrophage activation and pro-inflammatory cytokine production (20). The chief function of TRIM29 in alveolar macrophages is suppression of NF-kB activation. The identification of TRIM29 in NK cells is new, and its role in feedback suppression of NK functions is significant. Here, we showed that TRIM29 functions as a negative regulator of IFN-γ production in activated NK cells via the modulation of TAB2 expression. TRIM29 is barely expressed in resting NK cells but highly induced and peaked at 36 hr after IL-12/18 stimulation (Fig. 2C). In addition, the peak of IFN-γ production after IL-12/18 stimulation is at 24 hr (Fig. 1A and 1B) in vitro, thus the induction of TRIM29 in NK cells is a late activation event after early IFN-γ production. As reported previously, NK cells play a key role in the early control of MCMV infection (32). NK cells confer immunity against MCMV through the production of IFN-γ and by directly killing of infected cells. NK cell activation is regulated by various proinflammatory cytokines through crosstalk with DC and macrophages. Among those, IL-12 has been shown to elevate IFN-γ secretion by NK cells. Simultaneously, MCMV-induced IL-18 enhances the secretion of IFN-γ by NK cells while IL-15 has been described to trigger NK cell cytotoxicity, proliferation, and survival (26, 33, 34). In our studies, we found that NK cells from NK-Trim29−/− mice have higher and sustained IFN-γ levels than WT mice upon IL-12/18 stimulation (Fig. 3D) and higher IFN-γ in the serum at day 1.5 post infection (Fig. 6D). On the other hand, we did not find a difference in MCMV-induced NK-cell CD107a production in NK-Trim29−/− mice. Our results suggested that the induction of TRIM29 is specifically regulated by IL-12/18 signaling in NK cells and restricts hyper-activation of NK cells through inhibition of IFN-γ production.
The TRIM family of E3 ligases typically contain the RING domain, and some family members are known to inhibit cytokine production via binding to key signaling components and/or targeting such components for proteasome-mediated degradation through the classic RING domain dependent ubiquitination (35). However, TRIM29 interestingly lacks the RING domain. Instead, TRIM29 has a B-box domain that also has E3 ligase activity. Other studies have demonstrated that TRIM29 is constitutively expressed in macrophages and restricts both the IRF-mediated type I IFN production pathway and the NF-κB-mediated pro-inflammatory signaling pathway (20). Specifically, TRIM29 can interact with the adaptor NEMO in macrophages via the OmpH-OmpH domains, and induce the ubiquitination and degradation of NEMO at Lys183 by K48 linkage (20). Here we found that TRIM29 is induced in NK cells and interacts with TAB2 via C-terminal domain. Thus, TRIM29 uses different domains to regulate different pathways in unique cell types. NK cells, as key effector cells against viral infections and tumors, have established multiple layers of control to ensure effective function while minimize autoimmunity and tissue inflammation. NK cells express MyD88, which is required to transduce DC derived IL-1 family cytokine signals (such as IL-18). As compared with that in WT cells, the production of IFN-γ in MyD88-deficient NK cells was reduced upon MCMV infection. However, how MyD88 signaling is regulated in NK cells has not been clarified. Recent studies report that IL-1R8 on NK cells serves as a checkpoint for NK cell maturation and effector function (36). IL-1R8-deficiency was associated with more efficient control of MCMV infection and lung metastasis. These findings are reminiscent of our own, and the possibility that TRIM29 also regulates the IL-1R8 pathway through TAB2 merits further investigation. Induction of TRIM29 during infection represents an important immune inhibitory pathway, and the role of NK cells in inflammatory and autoimmune conditions associated with TRIM29 deficiency will need to be examined.
In summary, we have identified TRIM29 as a negative regulator of NK cell activities and show how TRIM29 inhibits TAB2 expression and IFN-γ production in NK cells. Our findings demonstrated the importance of TRIM29 in negative regulation of NK cell functions through post-translational modifications of TAB2. These new findings provide mechanistic insights into the molecular mechanisms that control IFN-γ production by NK cells and may have important clinical implications.
Supplementary Material
Key points.
TRIM29 is induced in NK cells and inhibits NK functions.
TRIM29 deletion in NK cells enhances IFN-γ production.
TRIM29 ubiquitinates and degrades TAB2 in activated NK cells.
Acknowledgements
We thank Laurie Minze for excellent operational supports. We thank Preston Arnold for proofread and the flow cytometry core at Houston Methodist Research Institute for excellent services.
This work was supported by grants from the National Institutes of Health (R01AI080779).
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Lanier LL 2005. NK cell recognition. Annu Rev Immunol 23: 225–274. [DOI] [PubMed] [Google Scholar]
- 2.Chiossone L, Dumas PY, Vienne M, and Vivier E. 2018. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol 18: 671–688. [DOI] [PubMed] [Google Scholar]
- 3.Wensveen FM, Jelencic V, Valentic S, Sestan M, Wensveen TT, Theurich S, Glasner A, Mendrila D, Stimac D, Wunderlich FT, Bruning JC, Mandelboim O, and Polic B. 2015. NK cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat Immunol 16: 376–385. [DOI] [PubMed] [Google Scholar]
- 4.Parikh BA, Piersma SJ, Pak-Wittel MA, Yang L, Schreiber RD, and Yokoyama WM. 2015. Dual Requirement of Cytokine and Activation Receptor Triggering for Cytotoxic Control of Murine Cytomegalovirus by NK Cells. PLoS Pathog 11: e1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gur C, Porgador A, Elboim M, Gazit R, Mizrahi S, Stern-Ginossar N, Achdout H, Ghadially H, Dor Y, Nir T, Doviner V, Hershkovitz O, Mendelson M, Naparstek Y, and Mandelboim O. 2010. The activating receptor NKp46 is essential for the development of type 1 diabetes. Nat Immunol 11: 121–128. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL 2008. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 8: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trambas CM, and Griffiths GM. 2003. Delivering the kiss of death. Nat Immunol 4: 399–403. [DOI] [PubMed] [Google Scholar]
- 8.Sun JC, and Lanier LL. 2011. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol 11: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, Sexl V, Jonjic S, Seliger B, Zitvogel L, and Mandelboim O. 2018. NKp46 Receptor-Mediated Interferon-gamma Production by Natural Killer Cells Increases Fibronectin 1 to Alter Tumor Architecture and Control Metastasis. Immunity 48: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberger CM, Clark AE, Treuting PM, Johnson CD, and Aderem A. 2008. ATF3 regulates MCMV infection in mice by modulating IFN-gamma expression in natural killer cells. Proc Natl Acad Sci U S A 105: 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, and Fitzgerald KA. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol 3: 133–146. [DOI] [PubMed] [Google Scholar]
- 13.Akira S 2000. The role of IL-18 in innate immunity. Curr Opin Immunol 12: 59–63. [DOI] [PubMed] [Google Scholar]
- 14.Chaix J, Tessmer MS, Hoebe K, Fuseri N, Ryffel B, Dalod M, Alexopoulou L, Beutler B, Brossay L, Vivier E, and Walzer T. 2008. Cutting edge: Priming of NK cells by IL-18. J Immunol 181: 1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madera S, and Sun JC. 2015. Cutting edge: stage-specific requirement of IL-18 for antiviral NK cell expansion. J Immunol 194: 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah AY, and Cooper MA. 2016. Metabolic Regulation of Natural Killer Cell IFN-gamma Production. Crit Rev Immunol 36: 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mavropoulos A, Sully G, Cope AP, and Clark AR. 2005. Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood 105: 282–288. [DOI] [PubMed] [Google Scholar]
- 18.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, Kita M, Ueda Y, Iwakura Y, Kataoka K, Okanoue T, and Mazda O. 2006. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol 146: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon ML, and Zuckerman SH. 2005. Gamma interferon: a central mediator in atherosclerosis. Inflamm Res 54: 395–411. [DOI] [PubMed] [Google Scholar]
- 20.Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin R, Lu H, Li XC, Liu YJ, and Zhang Z. 2016. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol 17: 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, and Zhang Z. 2017. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun 8: 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X, Du H, and Massiah MA. 2011. Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J Mol Biol 407: 505–520. [DOI] [PubMed] [Google Scholar]
- 23.Xing J, Zhang A, Minze LJ, Li XC, and Zhang Z. 2018. TRIM29 Negatively Regulates the Type I IFN Production in Response to RNA Virus. J Immunol 201: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, Jamieson AM, Langdon WY, Ikeda F, Fededa JP, Cronin SJ, Nitsch R, Schultz-Fademrecht C, Eickhoff J, Menninger S, Unger A, Torka R, Gruber T, Hinterleitner R, Baier G, Wolf D, Ullrich A, Klebl BM, and Penninger JM. 2014. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507: 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, and Vivier E. 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci U S A 108: 18324–18329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puttur F, Francozo M, Solmaz G, Bueno C, Lindenberg M, Gohmert M, Swallow M, Tufa D, Jacobs R, Lienenklaus S, Kuhl AA, Borkner L, Cicin-Sain L, Holzmann B, Wagner H, Berod L, and Sparwasser T. 2016. Conventional Dendritic Cells Confer Protection against Mouse Cytomegalovirus Infection via TLR9 and MyD88 Signaling. Cell Rep 17: 1113–1127. [DOI] [PubMed] [Google Scholar]
- 27.Ni J, Miller M, Stojanovic A, Garbi N, and Cerwenka A. 2012. Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J Exp Med 209: 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, Degli-Esposti MA, Leslie CS, Sun JC, and O’Sullivan TE. 2017. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 171: 795–808 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narni-Mancinelli E, Jaeger BN, Bernat C, Fenis A, Kung S, De Gassart A, Mahmood S, Gut M, Heath SC, Estelle J, Bertosio E, Vely F, Gastinel LN, Beutler B, Malissen B, Malissen M, Gut IG, Vivier E, and Ugolini S. 2012. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science 335: 344–348. [DOI] [PubMed] [Google Scholar]
- 30.Putz EM, Mayfosh AJ, Kos K, Barkauskas DS, Nakamura K, Town L, Goodall KJ, Yee DY, Poon IK, Baschuk N, Souza-Fonseca-Guimaraes F, Hulett MD, and Smyth MJ. 2017. NK cell heparanase controls tumor invasion and immune surveillance. J Clin Invest 127: 2777–2788. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Miyake T, Satoh T, Kato H, Matsushita K, Kumagai Y, Vandenbon A, Tani T, Muta T, Akira S, and Takeuchi O. 2010. IkappaBzeta is essential for natural killer cell activation in response to IL-12 and IL-18. Proc Natl Acad Sci U S A 107: 17680–17685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lisnic B, Lisnic VJ, and Jonjic S. 2015. NK cell interplay with cytomegaloviruses. Curr Opin Virol 15: 9–18. [DOI] [PubMed] [Google Scholar]
- 33.Kroemer A, Xiao X, Degauque N, Edtinger K, Wei H, Demirci G, and Li XC. 2008. The innate NK cells, allograft rejection, and a key role for IL-15. J Immunol 180: 7818–7826. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, and Biron CA. 2002. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol 169: 4279–4287. [DOI] [PubMed] [Google Scholar]
- 35.Chang M, Jin W, and Sun SC. 2009. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol 10: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molgora M, Bonavita E, Ponzetta A, Riva F, Barbagallo M, Jaillon S, Popovic B, Bernardini G, Magrini E, Gianni F, Zelenay S, Jonjic S, Santoni A, Garlanda C, and Mantovani A. 2017. IL-1R8 is a checkpoint in NK cells regulating anti-tumour and anti-viral activity. Nature 551: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.