Abstract
Biochemical mechanisms emerged and were integrated into the metabolic plan of cellular life long before molecular oxygen accumulated in the biosphere. When oxygen levels finally rose, they threatened specific types of enzymes: those that use organic radicals as catalysts, and those that depend upon iron centers. Nature has found ways to ensure that such enzymes are still used by contemporary organisms. In some cases they are restricted to microbes that reside in anoxic habitats, but in others they manage to function inside oxygen-filled cells. In the latter case, it is frequently true that the ancestral enzyme has been modified to fend off poisoning. In this review we survey a range of protein adaptations that permit radical-based and low-potential iron chemistry to succeed in oxic environments. In many cases, accessory domains shield the vulnerable radical or metal center from oxygen. In others, the structures of iron cofactors evolved to less oxidizable forms, or alternative metals replaced iron altogether. The overarching view is that some classes of biochemical mechanism are intrinsically incompatible with the presence of oxygen. The structural modification of target enzymes is an under-recognized response to this problem.
Keywords: hydrogen peroxide, superoxide, obligate anaerobiosis, iron-sulfur clusters, radical enzymes
Background.
Life evolved in a world devoid of molecular oxygen. The first organisms that arose were probably methanogens and acetogens, which generated energy by transferring electrons from hydrogen and sulfide to carbon dioxide [1]. Subsequently bacteria developed the capacity for cyclic photosynthesis, which enables growth even in the absence of favorable redox substrates. Fermentative microbes then evolved to degrade dead autotrophs. This cohort of microbes thrived, refined their enzymology, and grew to dominate the planet.
A billion years later a second photosynthetic apparatus appeared: photosystem II, which harvests biosynthetic electrons from water. It was a boon for life. However, a problem was in the offing: the by-product of water oxidation is molecular oxygen. In early stages it diffused out of cells and was quickly consumed through its reduction by compounds in the environment. This period is documented in the geological record as the conversion of soluble ferrous iron to ferric precipitate and the solubilization of sulfide salts as sulfate. And once such reduced species had been titrated from the environment, about 2.5 Gyr ago, molecular oxygen began to accumulate in the atmosphere [2]. Levels held steady at about 1% of contemporary concentrations for the next billion years; then, with the full-scale photosynthetic action of plant life, they rose to the 0.2 atm, where they remain today.
This history is important if one wants to understand how the biota reacted to this threat. Molecular oxygen appeared very late in the history of metabolic evolution. It attacked only a few particular biochemical mechanisms—albeit ones that were fundamental to the lifestyles of the existing microbes. The metabolic schemes that these mechanisms underlay could not easily be walked back and replaced. And so extant life forms, committed to types of biochemistry that had made good sense in an anoxic world, struggled to cope with the toxic effects of oxygen. Because oxygen accumulated gradually, adaptations emerged in incremental layers. Most famously, enzymes appeared that could scavenge the most reactive forms of reduced oxygen. The focus of this review, however, is upon the tweaks and adjustments that have accrued in the classes of enzymes that oxidants directly attack. These modifications say interesting things about how oxygen attacks them and about the facility with which evolution can address such problems.
The problem with molecular oxygen.
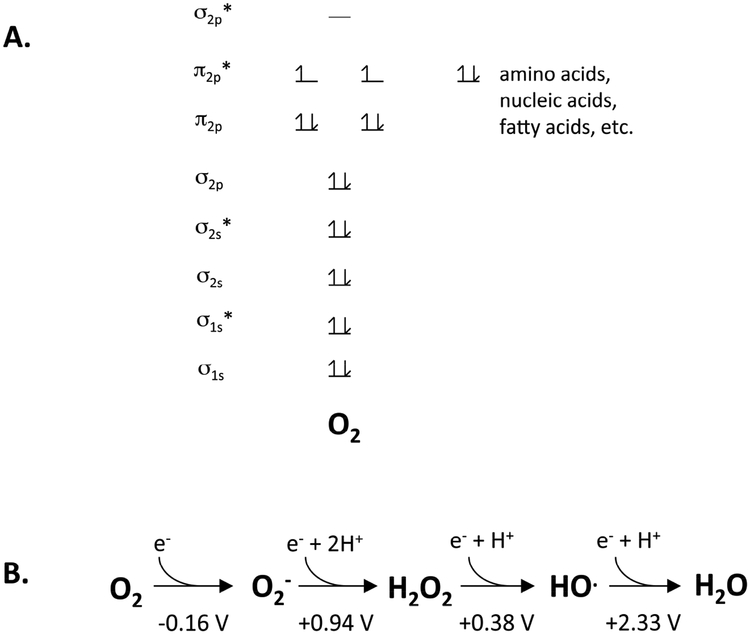
Two distinctive features guide the chemical behavior of molecular oxygen. First, despite having an even number of electrons, it is a diradical, with its two outermost electrons in spin-paired orbitals (Fig. 1A) [3]. That arrangement means that it rapidly forms adducts with other radical species—a serious problem for the radical-based enzymes that had arisen in the anoxic world.
Figure 1. The chemical nature of molecular oxygen.
(A) Molecular oxygen is a di-radical. This arrangement favors rapid reaction with other radicals but permits only single-electron transfers from spin-paired biomolecules. (B) The reduction potential of oxygen indicates that it is a mediocre univalent oxidant, in contrast to superoxide, hydrogen peroxide, and the hydroxyl radical.
The fact that these electrons are unpaired also means that molecular oxygen can abstract only one electron at a time from the familiar spin-paired biomolecules that comprise the structural moieties of living cells: amino acids, nucleic acids, lipids, and carbohydrates. This constraint has an enormous impact: molecular oxygen has a much more negative univalent reduction potential (Eo’ = −0.16 V; Fig. 1B) than those molecules (e.g., Eo’ of deoxyguanosine radical cation = +1.29 V), and so it cannot directly oxidize them at meaningful rates. Biologists will recognize that these molecules are stable in aerobic buffers. However, oxygen can oxidize the metal centers, flavins, and quinones that catalyze univalent electron transfers in low-potential redox processes. When oxygen does so inside cells, it is converted to reduced species (ROS) like superoxide and hydrogen peroxide. These ROS have much more potent oxidizing potentials (Eo’ = + 0.94 and +0.38 V, respectively) than does oxygen, and we will see that they quickly oxidize metalloenzymes with which oxygen itself reacts sluggishly.
Finally, the orbital structure of oxygen provides one oxidizing advantage that superoxide and hydrogen peroxide lack: it can oxidize strong reductant partners from a distance. The underlying reason is that outer-sphere electron transfers to oxygen do not require bond-breaking, as in the case of hydrogen peroxide, and electron acceptance by oxygen is not impaired by anionic status, as for superoxide. Low-potential metal centers near enzyme surfaces can be oxidized by molecular oxygen even which it cannot contact them directly.
Therefore, the accumulation of oxygen threatened both the radical and iron-based metalloenzymes that underpinned extant biochemistry. It is widely recognized that the biota responded by evolving scavenging enzymes that suppress the toxic actions of superoxide and hydrogen peroxide. The purpose of this review is to consider another form of evolutionary adaptation: the modifications of erstwhile oxidative target enzymes so they remain functional upon occasional or even constant oxygen exposure. The discussion is not a comprehensive survey; instead, it aims to convey key ideas through illustrative examples.
Enzymes that perform radical-based chemistry.
Glycyl-radical enzymes (I): pyruvate:formate lyase.
Two core enzymes of the pre-oxic world were pyruvate:formate lyase (PFL) and ribonucleotide reductase (RNR). Both catalyze reactions that cannot easily be achieved by standard acid-base mechanisms; they solve this problem by resorting to radical-based chemistry. In their resting states, these enzymes maintain a glycyl radical on a peptide strand near the enzyme surface; when substrate binds, an electron moves to the glycyl radical from an active-site cysteine [4](Fig. 2A). The resultant thiyl radical initiates the chemistry by abstracting a hydrogen atom from the substrate, and upon completion, the radical returns to the glycyl residue. The mechanisms and the energetics of these reactions have been reviewed [4]. This discussion focuses upon the problem arises when cells containing such enzymes are exposed to dissolved oxygen. Even in committed anaerobes such an event is likely commonplace: without exception they sport full sets of antioxidant enzymes, including scavengers of superoxide and hydrogen peroxide, in testimony to the fact that no environment or lifestyle is reliably oxygen-free. Glycyl-radical enzymes are extremely sensitive to molecular oxygen in vitro: even at O °C, aeration inactivates PFL within 10 sec [5]. Oxygen adds directly to the radical, producing a glycyl-peroxyl radical species that ultimately cleaves the polypeptide immediately adjacent to the glycine residue [6]. Activity is lost. The question is: Have organisms that use such enzymes, and that might encounter oxygen, made any tactical adjustments in order to minimize this problem?
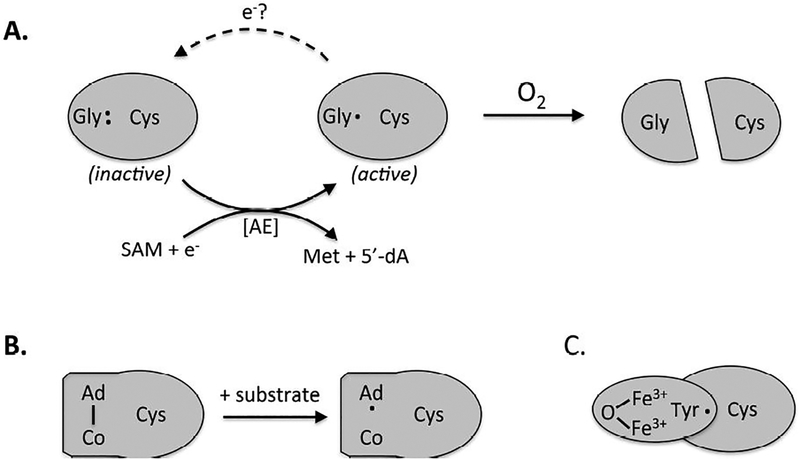
Figure 2. The evolution of oxygen-tolerant radical enzymes.
All ribonucleotide reductases maintain similar active sites, but they employ distinct mechanisms to generate the catalytic cysteinyl radical. The ancestral enzyme (A) features a resting-state glycyl radical near the protein surface, which is itself generated by a radical SAM activating enzyme (AE). Glycyl radical enzymes are rapidly inactivated by oxygen and are found only in contemporary anaerobic microbes. (B) The binuclear isozymes maintain a buried tyrosyl radical in an accessory subunit; the radical is physically insulated from the approach of oxygen. (C) In B12-dependent isozymes, an initiating adenosyl radical is generated by homolytic cleavage of the cofactor only when substrate binds, thus minimizing opportunity for reaction with O2.
A whole series of such adjustments have been revealed in the model bacterium E. coli. This bacterium dwells in sometimes-oxic, sometimes-anoxic boundary [7] near the epithelial wall of the mammalian intestine. The role of PFL is to cleave pyruvate without producing NADH when the microbe grows in anoxic microhabitats; this strategy is advantageous because the use of acetyl-CoA to reoxidize NADH diminishes the economy of anaerobic fermentations. When E. coli enters oxic environments, however, PFL is ably replaced by NADH-forming pyruvate dehydrogenase (PDH). This shift makes sense not only because PFL is incapable of oxic activity but because the oxidation of NADH by the aerobic respiratory chain makes a virtue of NADH formation. Transcription of pfl is reduced by an order of magnitude [8]. In extension of the same logic, bacteria that live in reliably oxic environments have completely dispensed with PFL.
It might seem odd that some residual PFL synthesis still occurs when E. coli is in the presence of oxygen—but, interestingly, under this condition the enzyme is not fully matured to the radical form that oxygen would instantly cleave [9]. Formation of the PFL glycyl radical requires an S-adenosylmethionine-dependent activating system (see below)—and this accessory enzyme creates the PFL glycyl radical only when oxygen levels are very low. The mechanism of this control is not clear. The residual synthesis of unactivated PFL becomes immediately useful when oxygen levels drop, since rapid PFL activation restores activity without any delay for protein synthesis.
The preceding describes a PFL control tactic that evolved to optimize its performance when cells transit from oxic to anoxic environments. What about the opposite scenario? Consider erstwhile anoxic cells that are suddenly confronted with an influx of oxygenated waters: the expectation would be that oxygen would immediately destroy the extant glycyl-radical enzymes, making them unavailable when anoxia is restored. However, workers reported that upon cellular aeration the PFL glycyl radical is quickly reduced to its original glycine form—thereby avoiding oxidative cleavage. The mechanism is mysterious. Two groups reported that such deactivation could be catalyzed in vitro by alcohol dehydrogenase [10–12]; another group was unable to reproduce the phenomenon, despite significant effort [13]. This issue awaits resolution.
Finally, investigators have discovered that even if PFL is cleaved, all is not lost. The small protein GrcA can adhere to oxygen-cleaved glycyl-radical enzymes, including PFL; GrcA itself is then activated to a glycyl-radical form that replaces the lost section of the native enzyme [14]. Authors have speculated that this ad hoc solution may enable the functions of glycyl-radical enzymes to be quickly recovered in post-aeration situations, pending the de novo synthesis of new PFL. The upshot is that in this post-anoxic world, a host of adaptations have arisen to minimize and tolerate the quenching of PFL by oxygen. Sitting at the center of metabolism, PFL is an abundant and indispensable enzyme, and so these adjustments play a key role in bacteria that live at oxic/anoxic interfaces or that transit between such habitats.
Glycyl-radical enzymes (II): ribonucleotide reductase.
The adaptation story is completely different for ribonucleotide reductase (RNR), another enzyme that employs radical chemistry to catalyze a challenging reaction. Contemporary anaerobes employ a glycyl-radical RNR [15]. Like PFL, which it resembles [16], it employs a glycyl radical to initiate a cysteinyl-radical attack on its substrate. Just as for PFL, such an enzyme cannot work in an oxic environment—but the difference is that radical-based ribonucleotide reduction must somehow continue.
Accordingly, most committed aerobes have replaced the glycyl mechanism with a second subunit in which iron (or manganese) and oxygen collaborate to generate a resting tyrosyl radical [4, 17](Fig. 2B). Unlike the glycyl radical of the anaerobic enzyme, the tyrosyl radical is buried away from the protein surface and thereby is shielded from inappropriate reactions with oxygen. When substrate binds, the tyrosyl radical abstracts the electron from the active-site cysteine, and the reaction commences. This element of the mechanism resembles that of the glycyl-radical RNR, which is ancestral to it. The evolution of oxygen-tolerant RNRs constitutes one of the clearest examples of enzymic adaptation to oxygen; indeed, the presence or absence of the glycyl-radical- and tyrosyl-radical-based enzymes constitutes definitive evidence of whether a microbe replicates in the presence or absence of oxygen.
B12-dependent enzymes.
A third family of ribonucleotide reductase generates its catalytic radical using a B12-dependent mechanism [4, 17](Fig. 2C). Cobalamin is perhaps the most complex cofactor in nature, and it is therefore assumed to be a late addition. In B12 enzymes radical formation begins with the binding of substrate, which conformationally strains the cofactor and prompts homolytic scission of its adenosyl-Co(III) covalent bond [18]. The resultant adenosyl radical pivots towards the catalytic cysteine residue, enabling electron abstraction and subsequent cysteinyl radical assault upon the substrate. After completion of the catalytic cycle, the adenosyl radical reappears and then condenses with the cobalt ion to regenerate the non-radical resting state. Thus the radical species is transient, a feature that minimizes its availability for reaction with oxygen. Accordingly, the B12-dependent ribonucleotide reductase can operate in both oxic and anoxic environments.
Why doesn’t oxygen poison B12 enzymes during the course of the reaction? Such enzymes owe some of their aerobic stability to the shielding of the adenosyl radical by the bound substrate. This inference is drawn from studies of glycerol and diol dehydratases. These enzymes dehydrate substrates that are not amenable to standard dehydration mechanisms, as they lack carbonyl groups to serve as electron sinks; therefore, radical mechanisms are used. As with RNRs, some diol dehydratases are glycyl-radical enzymes and work only in anoxic cells, while others use B12 and can operate when oxygen is present [19].
Yet in the test tube B12-based diol dehydratases are inactivated by oxygen with half-times as short as 3 minutes [20], which is at odds with their ability to function in oxic cells. Study revealed that the rate of inactivation of the substrate-free enzyme correlates with the facility of spontaneous cleavage of the adenosyl-Co bond [20]. The first well-defined reaction product is Co(III) plus 5’-peroxyadenosine, indicating that, in the absence of substrate, molecular oxygen adds directly to the adenosyl radical. The resultant peroxyl radical apparently oxidizes the adjacent Co(II), forming a non-radical dead-end product. Importantly, the inactivation rate is far lower in the presence of substrate, suggesting that substrate either physically shields the catalytic radical or outcompetes oxygen for it. Presumably the presence of substrate inside cells is a key factor that lessens the rate of this inactivating process.
The importance of cofactor shielding was reinforced by examination of methylmalonyl-CoA mutase, which has a half-life in oxic buffers of 3 hours [21]. This value drops to 3 minutes, however, in a mutant where the close packing of residues near the B12 has been disrupted, presumably allowing access of molecular oxygen.
Even if the B12 is oxidized, there may be a final safety net: the operons that encode the B12-dependent dehydratases also encode “reactivating enzymes.” Upon the occasion of B12 oxidation, the reactivating enzymes extract the damaged cofactor from the active site of the enzyme, and they enable binding of a new, undamaged B12 molecule so that full activity is restored [22]. Thus it appears that the evolution of B12 mechanisms has substantially suppressed, if not fully extinguished, the problems that oxygen poses for radical-based biochemistry.
Non-redox enzymes with exposed iron cofactors.
Mononuclear iron enzymes.
Some bacterial enzymes use a single Fe(II) cofactor to bind substrates and activate them for non-redox reactions. Examples include ribulose-5-phosphate 3-epimerase (RPE, in the pentose phosphate pathway) [23] and peptide deformylase (PDF, which deformylates the N-terminal methionine residue of nascent proteins) [24, 25]. The role of the Fe(II) is to bind the substrate and subsequently to stabilize an anionic reaction intermediate (Fig. 3A). In vitro these enzymes gradually lose their activities in oxic buffers, because molecular oxygen oxidizes the iron to Fe(III), which dissociates. How do these enzymes fare in aerated cells? In general they do well: these enzymes continue to function when cells grow in air-saturated media. Substrate protection may slow their rate of oxidation, and the straightforward remetallation of apoprotein helps to counteract whatever oxidation does occur.
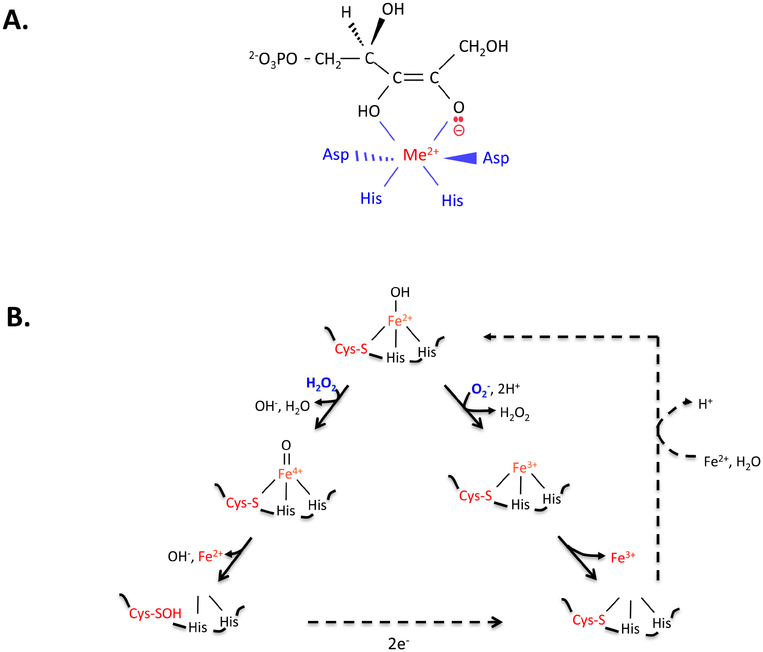
Figure 3. Non-redox iron-dependent enzymes are inactivated by hydrogen peroxide and superoxide.
(A) Mononuclear enzymes (here, ribose-5-phosphate 3-epimerase, RPE) employ an active-site divalent metal (Me2+) to stabilize anionic reaction intermediates. (B) In the substrate-free enzyme (here, peptide deformylase, PDF), the solvent exposure of the catalytic iron atom leaves it vulnerable to oxidation by H2O2 (left branch) and O2- (right). Dissociation of ferric iron inactivates the enzyme. If cysteine coordinates the metal, the ferryl radical generated by H2O2 is quickly quenched, and activity can be restored by reduction of the cysteine sulfenate residue and remetallation. Thioredoxins are likely mediators of the reduction. Some organisms employ manganese or zinc in place of iron, conferring full resistance to ROS.
However, the inactivation of these enzymes becomes a major issue whenever cells experience high levels of intracellular superoxide or hydrogen peroxide [23, 26, 27]. Both of these species are more-potent univalent oxidants than is molecular oxygen itself (Fig. 1B). Unlike oxygen, they must directly bind iron atoms to oxidize them—but the special vulnerability of these iron enzymes is that their iron atom must be solvent-exposed so that it can directly bind substrate. Therefore, the metal is unavoidably accessible to these oxidants. They bind the iron and oxidize it, leading to metal dissociation (Fig. 3B). Strikingly, studies with mutants of E. coli have shown that the cell processes that are most sensitive to H2O2 are all driven by mononuclear Fe(II) enzymes. For example, as little as 0.5 micromolar intracellular H2O2 is enough to block aromatic biosynthesis through its inactivation of DAHP synthase [28].
The iron oxidation by H2O2 also creates a second problem: the Fenton reaction generates a hydroxyl-like ferryl radical, which is a powerful oxidant that can covalently damage the active-site polypeptide. Consequently, some mononuclear enzymes—including Rpe—are irreversibly inactivated upon H2O2 exposure [23]. Others, however, feature a metal-binding arrangement that quenches the radical. PDF is an example [26]. One of its ligands to iron is a cysteine residue, which is not the most avid binder of iron but is an efficient reductant of strong oxidants. When H2O2 reacts with the bound iron atom, cysteine immediately transfers an electron to the nascent ferryl radical, preempting the release of a hydroxyl radical and thereby avoiding the oxidation of other residues (Fig. 3B). Cysteine sulfenate is formed, which is then restored to the thiol form by cellular reductive processes. Remetallation restores activity. From this standpoint, it is plausible that cysteine coordination arose in these enzymes as a defensive tactic to avoid the irreversible oxidation of other protein residues.
In oxic environments, cells continuously generate ROS through adventitious electron transfers from redox enzymes to oxygen [29]. The combination of repair processes and scavenging enzymes ensure that mononuclear enzymes maintain adequate steady-state activities under routine conditions.
However, problems can still arise when forcing circumstances cause ROS levels to rise. For example, mononuclear enzymes fail when the obligate anaerobe Bacteroides thetaiotaomicron encounters significant oxygen, because its rate of intracellular superoxide formation is extremely high [30]. The enzymes can also lose activity even in air-tolerant bacteria, if the bacteria enter environments that contain micromolar H2O2 [23, 26]. Hydrogen peroxide accumulates in some habitats through its direct excretion by lactic acid bacteria, the chemical oxidation of reduced metals and sulfur species at oxic/anoxic interfaces, the action of phagosomal NADPH oxidase, and incidental photochemistry.
For years workers have known that oxidative stress can be relieved if manganese is included in the medium. Studies with E. coli revealed how this treatment protects mononuclear enzymes [26, 28]. When H2O2 enters E. coli, it activates the transcription factor OxyR; OxyR then stimulates the transcription of a variety of genes, including ones that encode Dps, a mini-ferritin that sequesters loose iron [31–33], and MntH, an importer that delivers high levels of manganese into the cell [34, 35]. Their combined efforts enable the iron cofactor of non-redox mononuclear enzymes to be replaced by manganese. Manganese is not quite as good as iron at activating these enzymes—but it has the virtue of not being oxidized by H2O2. By switching metal cofactors E. coli restores the function of these enzymes during periods of H2O2 exposure.
This metal-replacement strategy extends far beyond this bacterium. E. coli is a facultative organism that spends most of its life in the hypoxic intestine, and its exposure to oxidants must only be occasional. In contrast, some obligate aerobes might be expected to contend with oxidants almost constantly. Lactic acid bacteria, for example, generate H2O2 as a stoichiometric by-product of their main fermentative strategy. They are famous for their ability to continue growing even in laboratory cultures that have accumulated millimolar H2O2—a level 10,000 times what would poison E. coli [36]. The difference is that lactic acid bacteria maintain very high intracellular pools of manganese [37], and they apparently employ it as their routine cofactor for enzymes like Rpe and Pdf. In effect, they constitutively engage the H2O2 defense that E. coli employs only under duress.
Bacterial pathogens have evolved to withstand the oxidative burst of neutrophils and macrophages—and a big part of the picture might be the alternative metallation strategies that they use for their mononuclear enzymes. The PDF of Borrelia burgdorferi has somehow evolved to work well with zinc [38]. Interestingly, its active-site structure is not obviously different from that of the E. coli enzyme, which utterly fails with zinc. The guess is that some second-sphere tinkering has tweaked the active-site geometries and polarities in subtle ways. The PDF of Staphylococcus aureus is completely resistant to oxidants [39]. The crystal structure revealed unprecedented iron ligation by cysteine sulfinic acid rather than cysteine, leading the investigators to surmise that the sulfinicferric complex is catalytically active [40]. Mycobacterium tuberculosis also employs an oxidant-resistant PDF, though in this case it appears that a closed-off active site effectively excludes oxidants from the ferrous cofactor [41]. Presumably a gating mechanism must operate to allow substrate entry. In sum, the fact that mononuclear enzymes are the weak links during oxidative stress has imposed successful selection for modification. It is striking that evolution repeatedly succeeded by retrofitting the extant metalloenzymes, rather than by replacing them wholesale with novel proteins that use a different catalytic strategy.
Iron-sulfur dehydratases.
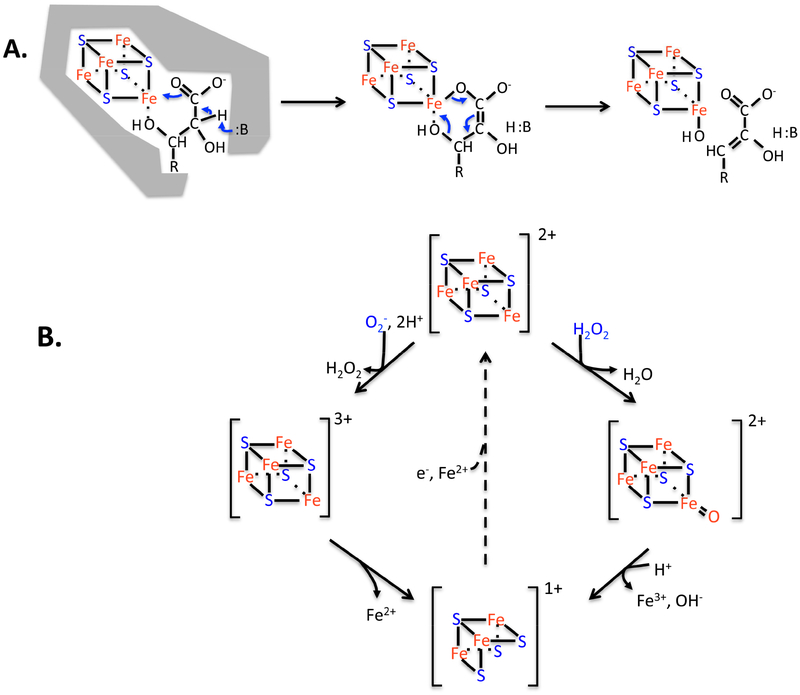
Superoxide and hydrogen peroxide attack one other class of enzymes: dehydratases that use active-site [4Fe-4S] clusters to bind their substrates. Aconitase is the founding member of this family [42]. These enzymes use a catalytic strategy analogous to that of mononuclear Fe(II) enzymes. One iron atom of the cluster projects into the active site and is under-coordinated so that it can form bidentate bonds to substrate (Fig. 4A). The binding of the iron activates the substrate for deprotonation because it electrostatically stabilizes the resultant anion. The same iron atom then acts as a Lewis acid to withdraw the hydroxide leaving group, thereby completing the dehydration reaction. Such non-redox surface chemistry is facile, and turnover numbers are high. The ROS problem for such enzymes resembles that of mononuclear enzymes: O2- and H2O2 enter the active site, directly bind the exposed iron atom of the cluster, and univalently oxidize it to create an unstable [4Fe-4S]3+ valence. The key iron atom dissociates, leaving behind a catalytically inactive [3Fe-4S]+ cluster (Fig. 4B). During periods of oxidative stress, the inactivation of these enzymes is responsible for failures in TCA cycling and amino acid biosynthesis [43–46]. If ROS stress is mild and oxidation of these enzymes is slow, the damage can be substantially countervailed by cluster repair that restores activity [47]. However, when oxidant levels are especially high, this dynamic equilibrium shifts towards the inactive form, metabolism fails, and growth ceases. These phenotypes occur in eukarya as well [48].
Figure 4. Dehydration by [4Fe-4S] clusters is elegant, but the enzymes are vulnerable to ROS.
(A) The unique iron atom of the solvent-exposed cluster provides an electrostatic partner that activates substrate for deprotonation, and then it serves as a Lewis acid for dehydroxylation. (B) Hydrogen peroxide and superoxide (not shown) can bind and oxidize the cluster, destabilizing it. The steady-state cluster status in vivo represents the dynamic balance between oxidation and repair. The repair process (dotted line) has been observed but has not been genetically defined. The evolution of [2Fe-2S] and cluster-free dehydratases enables some enzymes to completely resist oxidation.
Once again, nature has evolved modified forms of these enzymes that can withstand this stress. When E. coli encounters natural redox-cycling antibiotics, which fill its cytoplasm with superoxide [49], it replaces its [4Fe-4S] fumarase with an analogue that employs charged amino acid residues rather than a cluster to position and activate the substrate [50]. Thus oxidants have no impact upon its activity. Strikingly, higher organisms evolved to use the same cluster-free isozyme as their housekeeping enzyme; humans lack any cluster-containing fumarase. One interpretation is that higher eukaryotes have fully converted to the oxidant-resistant isozyme because our mitochondria are more frequently stressed than are facultative bacteria. Conversely, one might wonder why E. coli does not make the same choice: Why does it express the oxidant-resistant enzyme only when stress is detected? The obvious hypothesis is that the cluster-containing enzyme is simply a more efficient catalyst, and some kinetic analyses support this idea [51, 52].
A different story has emerged for dihydroxyacid dehydratase (DHAD), an iron-sulfur dehydratase in the pathway of branched-chain biosynthesis in microbes and plants. Oxidants rapidly poison the [4Fe-4S]2+ enzyme of E. coli, but they do not impair the homolog found in spinach chloroplasts [43, 53]. The key is that the latter enzyme uses a [2Fe-2S]2+ cluster. Whereas the iron atoms of [4Fe-4S]2+ dehydratases are formally in mixed Fe(III) and Fe(II) valences, those of the [2Fe-2S] clusters are already both in the ferric form, which likely makes the cluster refractory to further oxidation. An additional feature is that the substrate-free [2Fe-2S] cluster may be loosely coordinated by an accessory amino acid residue, as resonance Raman spectroscopy indicated ligation by an oxygen atom but EPR analysis apparently excluded water and hydroxide as the ligand [54]. The [2Fe-2S] DHAD is less active than the [4Fe-4S] enzyme, indicating that catalytic efficiency has been compromised for the sake of stability [55]. This is a recurring pattern among oxidant-resistant enzymes.
The DHAD enzymes of most obligate aerobes, including eukarya, are also predicted to employ [2Fe-2S] clusters [56]. It is striking that [2Fe-2S] isozymes have not been found for other [4Fe-4S] dehydratases, leaving some organisms with a counterintuitive a mixture of oxidant-sensitive and –resistant enzymes. For example, although yeasts possess a cluster-free fumarase and a [2Fe-2S] DHAD, those two enzymes lie in pathways that retain oxidant-sensitive [4Fe-4S] aconitase and isopropylmalate isomerase. In stressful habitats, oxidation of the latter enzymes creates crippling metabolic bottlenecks [48]. The logic of this arrangement awaits explication.
Redox enzymes with low-potential metal centers.
Pyruvate:ferredoxin oxidoreductase (PFOR).
The non-redox iron-sulfur clusters discussed above are vulnerable to superoxide and H2O2, which must directly bind iron to oxidize it. In contrast, these ROS cannot damage clusters that are buried in polypeptide—which is true of most clusters that are used in electron-transfer reactions. For example, the cluster-rich respiratory enzymes remain fully active in ROS-stricken cells [46].
However, molecular oxygen can oxidize clusters without binding them directly. While oxygen is not as thermodynamically potent as other ROS, it carries the distinction of being able to accept electrons by outer-sphere transfer—if the reduction potential of the cluster is low enough. One apparent example is pyruvate:ferredoxin oxidoreductase (PFOR), which terminates glycolysis in many anaerobic bacteria. This enzyme is an analogue of pyruvate dehydrogenase—it cleaves pyruvate into acetyl-CoA plus CO2. The key distinction is that rather than transfer an electron pair to NAD+, it transfers the pair through a series of iron-sulfur clusters to the enzyme surface, where the electrons hop to ferredoxin (Fig. 5A). The reduced ferredoxin delivers the electrons to hydrogenase, and the redox balance is achieved by hydrogen evolution. Thus PFOR, like PFL, plays a central role in the economy of many anaerobic fermentations.
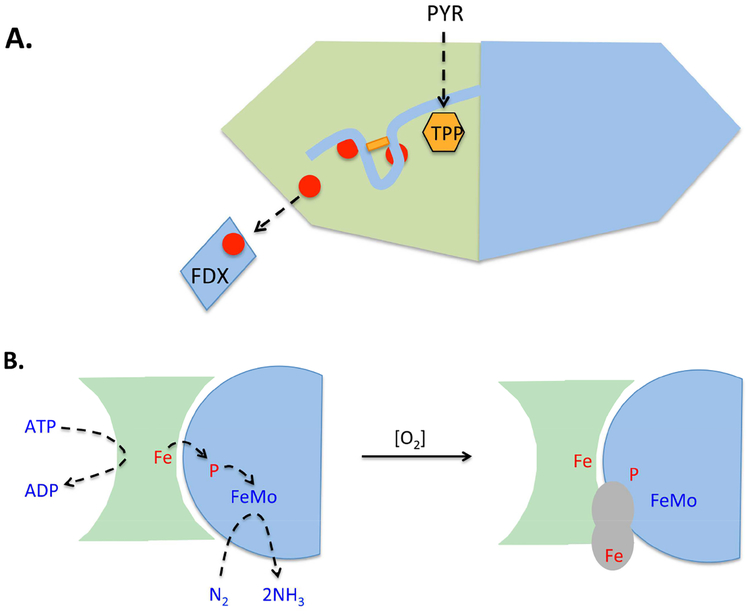
Figure 5. Protection of low-potential electron-transfer chains by accessory protein domains.
(A) The pyruvate:formate lyase of Desulfovibrio africanus features an unusual C-terminal extension [61]. Electron flow is from the thiamine pyrophosphate cofactor (TPP) to ferredoxin. Upon exposure to oxygen, a newly formed disulfide bond (rectangle) locks the extension over the iron-sulfur clusters. (B) Protection of nitrogenase from molecular oxygen. (Left) ATP-driven electrons move from the [4Fe-4S] cluster (Fe) of the FeSII protein through the P cluster to the molybdenum cofactor of the FeMo protein, where N2 is reduced. Full reduction requires multiple cycles of FeSII-FeMo association, reduction, and dissociation. (Right) When oxygen is present, Shethna protein (gray) forms a complex with the FeSII and FeMo proteins, interrupting activity but stabilizing their cofactors. The binding activity of the Shethna protein is likely triggered by oxidation of its [2Fe-2S] cluster (Fe) [66].
But if anaerobes are aerated, PFOR activity disappears over the next 20 minutes or so [57]. The deactivation is suspected of involving the over-oxidation and consequent destruction of one or more of the clusters, since cluster bleaching occurs concomitant with the loss of activity. The +2/+1 operating potentials of these clusters must be low enough (−540 to −390 mV [58] in Desulfovibrio africanus) to favor electron flow to resident ferredoxins (−410 to −385 mV [59, 60]) and, ultimately, to enable the downstream reduction of protons to molecular hydrogen (Eo’ = −414 mV). It seems plausible that the +3/+2 potential is relatively low—and therefore within the reach of oxidation by molecular oxygen.
PFOR inactivation appears to be a key reason that metabolism ceases when many anaerobes are exposed to oxygen [57]. Strikingly, several bacteria maintain relatively stable forms of PFOR. The best-studied of these belongs to Desulfovibrio species. This enzyme possesses an extra C-terminal extension containing two cysteine residues, which are quickly oxidized during oxygen exposure. Since oxygen is not a good direct oxidant of cysteine residues, it seems likely that their oxidation is mediated by the metal centers. The resultant disulfide bond closes the domain over the thiamine-proximal cluster of the electron wire [61](Fig. 5A). This shielding action is expected to suppress electron hopping to molecular oxygen in the bulk solution, since electron-transfer rates vary inversely with the log of distance [62]. Indeed, deletion of the terminal domain leaves a protein that shows anoxic activity but is fully vulnerable to oxygen [63]. In vitro, full activity is revived when the disulfide bond is reduced by thiol agents. Evidence indicates that a thioredoxin reactivates the oxidized enzyme when anoxia is restored in vivo [64].
PFOR apparently manifests a common pattern: that nature has learned to stabilize low-potential enzymes by occluding their metal centers. The following section extends this theme to the case of nitrogenase.
Nitrogenase.
The reduction of the triple bond of dinitrogen is thermodynamically challenging, and so electrons are conveyed to it at low potential. As was manifested by PFOR, oxygen and low-potential redox centers are a bad mix. Indeed, when nitrogenase is exposed to oxygen in vitro, both components of this bipartite enzyme—the Fe and FeMo proteins—are quickly and irrevocably damaged.
Yet some nitrogen-fixing bacteria are obligate aerobes, which seems like an impossible situation. In fact, these bacteria use a variety of tactics to keep their internal oxygen levels low: the excretion of high-viscosity biofilms, the formation of specialized cells with thick walls, rapid uncoupled internal respiration, the synthesis of oxygen-binding globins, and so on [65]. These strategies are expensive, and resort to them reflects the truth that life has failed to evolve emphatically oxygen-resistant forms of the enzyme.
However, the bacterium Azotobacter vinelandii does have a closely studied mechanism that transiently stabilizes the nitrogenase complex for the duration of oxygen stress, albeit in a non-active form [66]. If oxygen levels rise, the 13 kDa Shethna protein binds to the nitrogenase, forming a three-member Shethna/Fe protein/MoFe protein complex (Fig. 5B). When hypoxia is restored, Shethna protein dissociates, and nitrogenase turnover resumes. The apparent effect of Shethna is to physically shield the redox centers of the other two proteins so that oxygen cannot approach near enough to oxidize them. Shethna contains a [2Fe-2S] ferredoxin cluster, poised at −340 mV, whose role is presumably to sense a threatening rise in oxygen [67]. The obvious model is that oxidation of the cluster triggers a domain movement that enables it to bind nitrogenase. The same movement uncovers the Shethna cluster, so that when oxygen levels fall, it is accessible for quickly re-reduction by cellular reductants. Reduction would trigger back-movement of the domain and dissociation from nitrogenase. The overarching principle—that a protein senses oxygen and then occludes a redox center to prevent its oxidation—is the same as that which protects D. africanus PFOR. The difference is that in its shielded form PFOR retains some activity, whereas nitrogenase is stable but inactive.
Hydrogenases.
As mentioned earlier, many carbohydrate- and amino-acid-fermenting bacteria employ hydrogenase reactions to consume electrons and thereby sustain the redox balance of their fermentation schemes.
| (1) |
Reduced ferredoxins transfer electrons to a multi-cluster electron wire that terminates at a binuclear iron site. Protons are reduced at the distal iron atom. When oxygen is introduced, turnover ceases. Analyses indicate that oxygen binds to the distal iron atom, and continued delivery of electrons reduce it to iron-coordinated superoxide, peroxyl, and ferryl (hydroxyl) species [68]. If electron delivery is fast enough, full reduction can be achieved, water is released, and the enzyme remains competent to form hydrogen. However, if electron in-flow is slow, oxygen binding causes irreversible damage through a mechanism that is incompletely understood. One proposal is that intermediate ROS species, such as superoxide or H2O2, are long-lived enough that they dissociate from the iron and diffuse to the adjacent [4Fe-4S] sub-cluster, which they then oxidize and fatally destabilize. An alternative is that the ferryl—or hydroxyl—intermediate oxidizes an adjacent cysteine residue to a sulfenic acid, ending its function in proton conduction. An empty bi-nuclear site, degraded sub-cluster, or oxidized cysteine have all been observed in oxygen-inactivated hydrogenases [68–70]; the uncertainty lies in which injury occurs first, and under what conditions. No di-iron hydrogenase is known that can withstand oxygen.
The reverse reaction—electron transfer from molecular hydrogen to cellular electron carriers—is typically catalyzed by an evolutionarily distinct hydrogenase with a nickel-iron binuclear center (Fig. 6). Nickel is the site of hydrogen binding; its avidity for hydrogen minimizes the chance that oxygen will bind in its place. Still, if oxygen levels rise high enough, as in air-saturated cells, most of these enzymes are poisoned [71]. As with the di-iron hydrogenases, if reverse electron flow to the active site is rapid, it fully reduces oxygen to a hydroxide form, and after its dissociation further electron input through the wire can restore full activity within seconds. However, if electron inflow is slower, the catalytic center is trapped in a complex with an oxygen intermediate that is tentatively proposed to be a peroxide species. Resolution of this species is extremely slow and does not occur under physiological conditions. Thus the standard Ni-Fe hydrogenases, like Fe-Fe hydrogenases, cannot function in oxic environments.
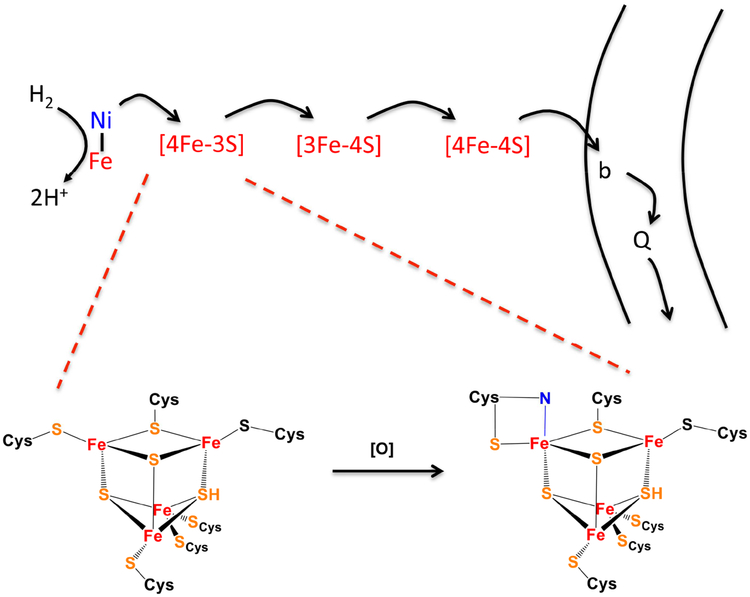
Figure 6. An unusual [4Fe-3S] cluster stabilizes membrane-bound NiFe hydrogenases against oxidation.
In soluble NiFe hydrogenases, oxygen may bind the reduced enzyme and pull electrons toward it, forming a dead-end peroxide complex that cannot be reactivated. In the membrane-bound enzymes, physiological electron flow is from hydrogen through the binuclear center and the iron-sulfur clusters to the membrane-bound b-type cytochrome and quinone pool. If oxygen binds the reduced enzyme, the back-flow of electrons to oxygen is amplified by oxidation of the proximal cluster, enabling complete oxygen reduction to water. The unusual structure of the cluster (bottom) [72] prevents disintegration of its super-oxidized form, and subsequent enzyme reduction restores full activity. In the oxidized cluster one iron atom receives bidentate coordination from the thiolate and amide nitrogen of a single cysteine residue.
Yet thermodynamic circumstance has prompted nature to evolve oxygen-resistant forms of hydrogen-oxidizing hydrogenase. Oxic/anoxic interfaces exist in sediments and at the periphery of the intestinal lumen, and at those interfaces hydrogen- and oxygen-containing fluids mix. This situation provides a wonderful opportunity for enterprising bacteria to exploit the strongly exergonic oxidation of hydrogen. Indeed, a variety of bacteria, including E. coli, carry Ni-Fe hydrogenases that adhere to the outer face of the cytoplasmic membrane; these enzymes transfer electrons from periplasmic hydrogen to their respiratory quinones, which ultimately deliver the electrons to a cytochrome oxidase, thereby capturing the redox energy as a membrane potential. What trick enables these hydrogenases to withstand the oxygen?
The core iron-nickel site of these hydrogenases is conventional, but the structure of the iron-sulfur cluster proximal to it is unprecedented [72] (Fig. 6). One of the usual inorganic sulfur atoms is replaced by a bridging cysteine thiolate, and an adjacent iron atom is coordinated by two rather than one cysteine residue. The key is that if oxygen binds to the reduced nickel site, the oxidation event also includes the abstraction of two electrons from this cluster. Cluster oxidation is permissible because the superoxidized cluster is stabilized through additional coordination by a deprotonated backbone amide nitrogen atom. The benefit is that the extra electrons provided by cluster ensure the full reduction of nickel-bound oxygen to water, thereby avoiding the putative inhibitory peroxyl state. Turnover is unabated. The protein modifications that solved the oxygen problem in this way are strikingly modest: the polypeptide thread is essentially conserved, but it has gained the two key cysteine residues that allow the unusual cluster structure [73]. This evolutionary parsimony resembles the generation of oxidant-resistant DHAD clusters. Another parallel is notable: the oxygen-resistant Ni-Fe hydrogenases appear to be less efficient than the oxygen-sensitive homologs.
Why are radical SAM enzymes stable in air-saturated cells?
The most common radical-based mechanisms in nature start with the splitting of S-adenosylmethionine (SAM) to produce an adenosyl-radical species [4]. Theses radicals initiate a wide range of challenging chemistry. Radical SAM enzymes are notoriously difficult to work with in vitro, requiring full anoxia. But this observation is strange, in that the same enzymes are routinely used by organisms that grow in oxygenated environments. E. coli, for example, relies upon such enzymes to synthesize lipoate, thiamine, and biotin—all of which must happen in oxic as well as anoxic habitats. How do the enzymes escape the damage that occurs in vitro?
Radical SAM enzymes employ [4Fe-4S] clusters to transfer single electrons from flavodoxins to SAM, triggering its cleavage to generate the catalytic radical (Fig. 7). This process is driven by substrate binding, so the resultant radical has a momentary lifetime in a shielded environment, much like B12-driven radicals. Indeed, it turns out to be the cluster, rather than the transitory catalytic radical species, that is at risk in oxic buffers. Because the cluster must directly bind incoming SAM, in the resting enzyme it sits in a solvent-exposed orientation—and presumably oxygen can easily approach it. When it does so, oxygen over-oxidizes the cluster to an unstable +3 state, which then degrades [74]. Both [3Fe-4S] and [2Fe-2S] products are observed; these forms are catalytically incompetent. These events parallel those which degrade the clusters of dehydratases. However, the reaction rate of radical SAM enzymes with oxygen far exceeds that of dehydratase clusters: in oxic buffers radical SAM enzymes lose activity within minutes. It seems plausible that, given the extremely low potential of the catalytic +1/+2 cycling (as low at −0.55 V [75]), the cluster +2/+3 potential is also relatively low. This might enable facile oxidation even though in most contexts molecular oxygen (−0.16 V) is not a compelling oxidant of [4Fe-4S] clusters. This idea is recalls the oxygen sensitivities of the low-potential metal cofactors of nitrogenase and PFOR.
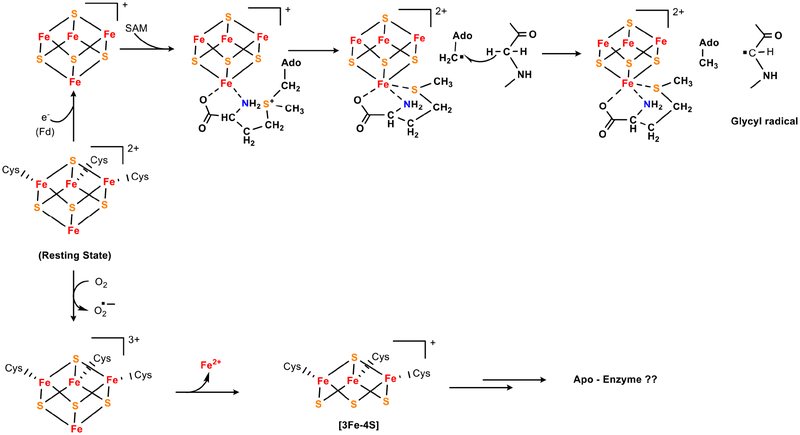
Figure 7. Radical SAM enzymes remain functional in oxic cells despite featuring a low-potential cluster.
(Top) The many members of this enzyme class cleave S-adenosylmethionine by electron transfer from the coordinating cluster to SAM. The resultant adenosyl radical then operates upon substrate (in this example, activating a glycyl-radical enzyme). (Bottom) The cluster is rapidly degraded when the enzyme is exposed to oxygen in vitro, but such enzymes remain functional inside air-saturated cells. One possibility is that the cluster is shielded in vivo by SAM or other metabolites.
Such instability would not be tolerable in vivo—and the mechanism by which these enzymes are protected is an important unresolved question. One possibility is that the constant delivery of electrons keeps the steady-state cluster in a +1 valence, whose oxidation can be tolerated, rather than allowing it to linger in the vulnerable +2 valence. But whole-cell EPR data do not support this idea: overproduced radical SAM enzyme tends to sit in the +2 valence even in oxic media [74], without rapid decomposition. Therefore it seems more likely that substate physically shields the cluster from oxygen. Three cysteine residues ligand the cluster, leaving a unique iron atom under-coordinated in the SAM-free enzyme. In the reaction cycle this iron atom binds nitrogen and carboxylate moieties of SAM in bidentate fashion, filling the iron ligand sphere and blocking direct ligation by oxygen. By keeping oxygen at a distance, this feature seems likely to help stabilize the cluster against oxidation. Indeed, the analogous [4Fe-4S] clusters in dehydratase-class enzymes are strongly stabilized when substrate binds.
This model of protection requires that SAM occupy the resting enzymes in vivo. Direct evidence is lacking, but some data support this idea. In E. coli the steady-state SAM level might be inferred from the affinities for SAM of the proteins that control its synthesis. The transcription factor MetJ drives transcription of MetK, the methionine adenosyltransferase that generates SAM. MetJ is inhibited by SAM, with a dissociation constant of 180 micromolar [76]; this observation suggests that E. coli maintains a level of SAM far higher than the 10 micromolar Kd, for example, of the SAM radical enzyme that activates RNR [15]. On the activity level, turnover of MetK itself is controlled by competition between SAM and ATP for its substrate binding site; the respective Kd values are 10 micromolar and 100 micromolar [77]. Because the concentration of enzyme-accessible ATP is ~ 3 mM in the cell [78], the upshot is that half-inhibition of MetK cannot happen until SAM levels approach 300 micromolar. Thus both the transcriptional and activity controls upon SAM synthesis are calibrated to maintain pools that far exceed the binding constant of the SAM enzyme, which would seem to ensure that its cluster will be fully coordinated—and minimally exposed to oxygen—in vivo.
Interestingly, whole-cell EPR analysis of one SAM radical enzyme suggested that other metabolites, possibly including AMP, predominantly bound the cluster of the overproduced enzyme [74]. It seems unworkable for the enzyme to be occupied by AMP in vivo, and one wonders whether SAM levels were deficient under the non-growing conditions that were used in that experiment.
Summary points.
In general, aerobes did not jettison the oxygen-sensitive pathways that they inherited from their anaerobic ancestors; instead, they modified the vulnerable enzymes so they could withstand the stress.
Radical-based mechanisms enable chemistry that acid-base catalysis cannot replicate—but they can be rapidly poisoned by oxygen through a radical-radical addition reactions. As organisms moved to oxic environments, some enzymes with exposed glycyl radicals were replaced by B12-dependent isozymes, in which the radical species are transitory and shielded.
The [4Fe-4S] clusters of dehydratases are oxidatively degraded by superoxide and hydrogen peroxide. Some of these enzymes have now given way to oxidant-resistant [2Fe-2S] paralogs or cluster-free analogs. The latter enzymes are less catalytically efficient, which is a common theme of oxidant-resistant isozymes.
Non-redox mononuclear enzymes appear to work best with iron, perhaps due to its superiority at ligand-exchange behavior. However, ROS inactivate these enzymes. Alternative metallation with manganese, zinc, or novel iron complexes allows activity in microbes that are chronically exposed to oxidants.
Oxygen can oxidize and destabilize the low-potential electron wires of nitrogenase and pyruvate:ferredoxin oxidoreductase. In some species, metal-based sensors trigger the movement of polypeptide over the vulnerable wire, blocking any further approach of oxygen—albeit with diminution or temporary inhibition of enzyme function.
Radical SAM enzymes present a puzzle that is not yet solved: They are very oxygen-sensitive in vitro but not in vivo. One possibility is that metabolic controls ensure the enzymes are saturated with SAM, which would occlude and stabilize their clusters.
Acknowledgment.
This work was supported by grants GM49640 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- [1].Schonheit P; Buckel W; Martin WF On the origin of heterotrophy. Trends Microbiol 24:12–25; 2016. [DOI] [PubMed] [Google Scholar]
- [2].Anbar AD Elements and evolution. Science 322:1481–1483; 2008. [DOI] [PubMed] [Google Scholar]
- [3].Naqui A; Chance B Reactive oxygen intermediates in biochemistry. Ann Rev Biochem 55:137–166; 1986. [DOI] [PubMed] [Google Scholar]
- [4].Shibata N; Toraya T Molecular architectures and functions of radical enzymes and their (re)activating proteins. J. Biochem 158:271–292; 2015. [DOI] [PubMed] [Google Scholar]
- [5].Knappe J; Neugebauer FA; Blaschkowski HP; Ganzler M Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc. Natl. Acad. Sci. USA 81:1332–1335; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang W; Wong KK; Magliozzo RS; Kozarich JW Inactivation of pyruvate formate-lyase by dioxygen: defining the mechanistic interplay of glycine 734 and cysteine 419 by rapid freeze-quench EPR. Biochemistry 40:4123–4130; 2001. [DOI] [PubMed] [Google Scholar]
- [7].Espey MG Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Rad. Biol. Med 55:130–140; 2013. [DOI] [PubMed] [Google Scholar]
- [8].Sawers G Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol 10:737–747; 1993. [DOI] [PubMed] [Google Scholar]
- [9].Reddy SG; Wong KK; Parast CV; Peisach J; Magliozzo RS; Kozarich JW Dioxygen inactivation of pyruvate formate-lyase: EPR evidence fo the formation of protein-based sulfinyl and peroxyl radicals. Biochemistry 37:558–563; 1998. [DOI] [PubMed] [Google Scholar]
- [10].Kessler D; Leibrecht I; Knappe J Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Letts 281:59–63; 1991. [DOI] [PubMed] [Google Scholar]
- [11].Kessler D; Herth W; Knappe J Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzymic AdhE protein of Escherichia coli. J. Biol. Chem 267:18073–18079; 1992. [PubMed] [Google Scholar]
- [12].Asanuma N; Yoshii T; Hino T Molecular characteristics and transcription of the gene encoding a multifunctional alcohol dehydrogenase in relation to the deactivation of pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Arch. Microbiol 181:122–128; 2004. [DOI] [PubMed] [Google Scholar]
- [13].Nnyepi MR; Peng Y; Broderick JB Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. Arch. Biochem. Biophys 459:1–9; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wagner AF; Schultz S; Bomke J; Pils T; Lehmann WD; Knappe J YfiD of Escherichia coli and Y061 of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvatet formate-lyase. Biochem. Biophys. Res. Commun 285:456–462; 2001. [DOI] [PubMed] [Google Scholar]
- [15].Fontecave M; Mulliez E; Logan DT Deoxyribonucleotide synthesis in anaerobic microorganisms: the class III ribonucleotide reductase. Prog. Nucl. Acid Res 72:95–127; 2002. [DOI] [PubMed] [Google Scholar]
- [16].Stubbe J Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol 10:731–736; 2000. [DOI] [PubMed] [Google Scholar]
- [17].Nordlund P; Reichard P Ribonucleotide reductases. Annu. Rev. Biochem 75:681–706; 2006. [DOI] [PubMed] [Google Scholar]
- [18].Dowling DP; Croft AK; Drennan CL Radical use of Rossmann and TIM barrel architectures for controlling coenzyme B12 chemistry. Annu. Rev. Biophys 41:403–427; 2012. [DOI] [PubMed] [Google Scholar]
- [19].Hao J; Lin R; Zheng Z; Liu H; Liu D Isolation and characterization of microorganisms able to produce 1,3-propanediol under aerobic conditions. World J. Microbiol. Biotechnol 24:1731–1740; 2008. [Google Scholar]
- [20].Schwartz PA; Frey PA Dioldehydrase: an essential role for potassium ion in the homolytic cleavage of the cobalt-carbon bond in adenosylcobalamin. Biochemistry 46:7293–7301; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Thoma NH; Evans PR; Leadlay PF Protection of radical intermediates at the active site of adenosylcobalamin-dependent methylmalonyl-CoA mutase. Biochemistry 39:9213–9221; 2000. [DOI] [PubMed] [Google Scholar]
- [22].Toraya T; Tanokuchi A; Yamasaki A; Nakamura T; Ogura K; Tobimatsu T Diol dehydratase-reactivase is essential for recycling of coenzyme B12 in diol dehydratase. Biochemistry 55:69–78; 2015. [DOI] [PubMed] [Google Scholar]
- [23].Sobota JM; Imlay JA Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. USA 108:5402–5407; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rajagopalan PTR; Pei D Oxygen-mediated inactivation of peptide deformylase. J. Biol. Chem 273:22305–22310; 1998. [DOI] [PubMed] [Google Scholar]
- [25].Becker A; Schlichting I; Kabsch W; Groche D; Schultz S; Wagner AF Iron center, substrate recognition and mechanism of peptide deformylase. Nat Struct Biol 5:1053–1058; 1998. [DOI] [PubMed] [Google Scholar]
- [26].Anjem A; Imlay JA Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J. Biol. Chem 287:15544–15556; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gu M; Imlay JA Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol. Microbiol 89:123–134; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sobota JM; Gu M; Imlay JA Intracellular hydrogen peroxide and superoxide poison 3-deoxy-D-arabinoheptulosonate 7-phosphate synthase, the first committed enzyme in the aromatic biosynthetic pathway of Escherichia coli. J. Bacteriol 196:1980–1991; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Imlay JA The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol 11:443–454; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lu Z; Sethu R; Imlay JA Endogenous superoxide is a key effector of the oxygen sensitivity of a model obligate anaerobe. Proc. Natl. Acad. Sci. USA 115:e3266–e3275; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Altuvia S; Almiron M; Huisman G; Kolter R; Storz G The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol 13:265–272; 1994. [DOI] [PubMed] [Google Scholar]
- [32].Ilari A; Ceci P; Ferrari D; Rossi G; Chiancone E Iron incorporation into E. coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem 277:37619–37623; 2002. [DOI] [PubMed] [Google Scholar]
- [33].Park S; You X; Imlay JA Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA 102:9317–9322; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kehres DG; Janakiraman A; Slauch JM; Maguire ME Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184:3151–3158; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Anjem A; Varghese S; Imlay JA Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72:844–858; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pericone CD; Park S; Imlay JA; Weiser JN Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185:6815–6825; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Archibald FS; Fridovich I Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol 145:442–451; 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nguyen KT; Wu J-C; Boylan JA; Gherardini FC; Pei D Zinc is the metal cofactor of Borrelia burgdorferi peptide deformylase. Arch. Biochem. Biophys 468:217–225; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baldwin ET; Harris MS; Yem AW; Wolfe CL; Vosters AF; Curry KA; Murray RW; Bock JH; Marshall VP; Cialdella JI; Merchant MH; Choi G; M R Deibel J Crystal structure of type II peptide deformylase from Staphylococcus aureus. J. Biol. Chem 277:31163–31171; 2002. [DOI] [PubMed] [Google Scholar]
- [40].Kreusch A; Spraggon G; Lee CC; Klock H; McMullan D; Ng K; Shin T; Vincent J; Warner I; Ericson C; Lesley SA Structure analysis of peptide deformylases from Streptococcus pneumoniae, Staphylococcus aureus, Thermotoga maritima and Pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase. J. Mol. Biol 330:309–321; 2003. [DOI] [PubMed] [Google Scholar]
- [41].Kumar S; Kanudia P; Karthikeyan S; Chakraborti PK Identification of crucial amino acids of bacterial peptide deformylase affecting enzymatic activity in response to oxidative stress. J. Bacteriol 196:90–99; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lauble H; Kennedy MC; Beinert H; Stout CD Crystal structures of aconitase with isocitrate and nitroisocitrate bound. Biochemistry 31:2735–2748; 1992. [DOI] [PubMed] [Google Scholar]
- [43].Kuo CF; Mashino T; Fridovich I α,β-dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem 262:4724–4727; 1987. [PubMed] [Google Scholar]
- [44].Gardner PR; Fridovich I Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem 266:19328–19333; 1991. [PubMed] [Google Scholar]
- [45].Flint DH; Tuminello JF; Emptage MH The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem 268:22369–22376; 1993. [PubMed] [Google Scholar]
- [46].Jang S; Imlay JA Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem 282:929–937; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gardner PR; Fridovich I Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J. Biol. Chem 267:8757–8763; 1992. [PubMed] [Google Scholar]
- [48].Wallace MA; Liou L-L; Martins J; Clement MHS; Bailey S; Longo VD; Valentine JS; Gralla EB Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis: Cross-compartment protection by CuZnSOD. J. Biol. Chem 279:32055–32062; 2004. [DOI] [PubMed] [Google Scholar]
- [49].Babu BN; Brown OR Quantitative effects of redox-cycling chemicals on the oxidant-sensitive enzyme dihydroxy-acid dehydratase. Microbios 82:157–170; 1995. [PubMed] [Google Scholar]
- [50].Liochev SI; Fridovich I Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA 89:5892–5896; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Woods SA, Schwartzbach SD, and Guest JR Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 954:14–26; 1988. [DOI] [PubMed] [Google Scholar]
- [52].Flint DH Initial kinetic and mechanistic characterization of Escherichia coli fumarase A. Arch. Biochem. Biophys 311:509–516; 1994. [DOI] [PubMed] [Google Scholar]
- [53].Flint DH; Emptage MH Dihydroxyacid dehydratase: isolation, characterization as Fe-S proteins, and sensitivity to inactivation by oxygen radicals N. Y.: VCH Publishers; 1990. [Google Scholar]
- [54].GAo H; Azam T; Randeniya S; Couturier J; Rouhier N; Johnson MK Function and maturation of the Fe-S center in dihydroxyacid dehydratase from Arabidopsis. J. Biol. Chem 293:4422–4433; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Flint DH; Allen RM Iron-sulfur proteins with nonredox functions. Chem. Rev 96:2315–2334; 1996. [DOI] [PubMed] [Google Scholar]
- [56].Muhlenhoff U; Richter N; Pines O; Pierik AJ; Lill R Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem 286:41205–41216; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pan N; Imlay JA How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron? Mol. Microbiol 39:1562–1571; 2001. [DOI] [PubMed] [Google Scholar]
- [58].Pieulle L; Guigliarelli B; Asso M; Dole F; Bernadac A; Hatchikian EC Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta 1250:49–59; 1995. [DOI] [PubMed] [Google Scholar]
- [59].Hagtchikian EC; Cammack R; Patil DS; Robinson AE; Richards AJM; George S; Thomson AJ Spectroscopic characterization of ferredoxins I and II from Desulfovibrio africanus. Biochim. Biophys. Acta 784:40–47; 1984. [Google Scholar]
- [60].Armstrong FA; Cammack R; George S; Hatchikian EC; Thomson AJ Electrochemical and spectroscopic characterization of the 7Fe form of ferredoxin III from Desulfovibrio africanus. Biochem. J 264:265–273; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chabriere E; Charon MH; Volbeda A; Pieulle L; Hatchikian EC; Fontecilla-Camps JC Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol 6:182–190; 1999. [DOI] [PubMed] [Google Scholar]
- [62].Moser CC; Keske JM; Warncke K; Farid RS; Dutton PL Nature of biological electron transfer. Nature 355:796–802; 1992. [DOI] [PubMed] [Google Scholar]
- [63].Pieulle L; Magro V; Hatchikian EC Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J Bacteriol 179:5684–5692; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pieulle L; Stocker P; Vinay M; Nouailler M; Vita N; Brasseur G; Garcin E; Sebban-Kreuzer C; Dolla A Study of the thiol/disulfide redox systems of the anaerobe Desulfovibrio vulgaris points out pyruvate:ferredoxin oxidoreductase as a new target for thioredoxin 1. J. Biol. Chem 286:7812–7821; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fay P Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev 56:340–373; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schlesier J; Rohde M; Gerhardt S; Einsle O A conformational switch triggers nitrogenase protection from oxygen damage by Shethna protein II (FeSII). J. Am. Chem. Soc 138:239–247; 2015. [DOI] [PubMed] [Google Scholar]
- [67].Milton RD; Cai R; Sahin S; Abdellaoui S; Alkotaini B; Leech D; Minteer SD The in vivo potential-regulated protective protein of nitrogenase in Azotobacter vinelandii supports aerobic bioelectrochemical dinitrogen reduction in vitro. J. Am. Chem. Soc 139:9044–9052; 2017. [DOI] [PubMed] [Google Scholar]
- [68].Kubas A; Orain C; Sancho DD; Sensi LS; Gauquelin C; Meynial-Salles I; Soucaille P; Bottin H; Baffert C; Fourmond V; Best RB; Blumberger J; Leger C Mechanism of O2 diffusion and reduction in FeFe hydrogenases. Nat. Chem 9:88–95; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Swanson KD; Ratzloff MW; Mulder DW; Artz JH; Ghose S; Hoffman A; White S; Zadvornyy OA; Broderick JB; Bothner B; King PW; Peters JW [FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster. J. Am. Chem. Soc 137; 2015. [DOI] [PubMed] [Google Scholar]
- [70].Stripp ST; Goldet G; Brandmayr C; Sanganas O; Vincent KA; Haumann M; Armstrong FA; Happe T How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. USA 106:17331–17336; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Stiebritz MT; Reiher M Hydrogenases and oxygen. Chem. Sci 3:1739–1751; 2012. [Google Scholar]
- [72].Goris T; Wait AF; Saggu M; Fritsch J; Heidary N; Stein M; Zebger I; Lendzian F; Armstrong FA; Friedrich B; Lenz O A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase. Nat. Chem. Biol 7:310–318; 2011. [DOI] [PubMed] [Google Scholar]
- [73].Shomura Y; Yoon K-S; Nishihara H; Higuchi Y Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479:253–257; 2011. [DOI] [PubMed] [Google Scholar]
- [74].Yang J; Naik SG; Ortillo DO; Garcia-Serres R; Li M; Broderick WE; Huynh BH; Broderick JB The iron-sulfur cluster of pyruvate formate-lyases activating enzyme in whole cells: cluster interconversion and a valence-localized [4Fe-4S]2+ state. Biochemistry 48:9234–9241; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mulliez E; Padovani D; Atta M; Alcouffe C; Fontecave M Activation of class III ribonucleotide reductase by flavodoxin: a protein radical-driven electron transfer to the iron-sulfur center. Biochemistry 40:3730–3736; 2001. [DOI] [PubMed] [Google Scholar]
- [76].Marti-Arbona R; Teshima M; Anderson PS; Nowak-Lovato KL; Hong-Geller E; Unkefer CJ; Unkefer PJ Identification of new ligands for the methionine biosynthesis transcriptional regulator (MetJ) by FAC-MS. J. Mol. Microbiol. Biotech 22:205–214; 2012. [DOI] [PubMed] [Google Scholar]
- [77].Markham GD; Hafner EW; Tabor CW; Tabor H S-adenosylmethionine synthetase from Escherichia coli. J. Biol. Chem 255:9082–9092; 1980. [PubMed] [Google Scholar]
- [78].Schneider DA; Gourse RL Relationship between growth rate and ATP concentration in Escherichia coli. A bioassay for available cellular ATP. J. Biol. Chem 279:8262–8268; 2004. [DOI] [PubMed] [Google Scholar]