Abstract
In this study, we investigated whether radiomic features of CT image data can accurately predict HMGA2 and C-MYC gene expression status and identify the patient survival time using a machine learning approach in pancreatic ductal adenocarcinoma (PDAC). A cohort of 111 patients with PDAC was enrolled in our study. Radiomic features were extracted using conventional (shape and texture analysis) and deep learning approaches following to segmentation of preoperative CT data. To predict patient survival time, significant radiomic features were identified using a log-rank test. After surgical resection, level of HMGA2 and C-MYC gene expressions of PDAC tumor regions were classified using a support vector machines method. The model was evaluated in terms of accuracy, sensitivity, specificity, and area under the curve (AUC). Besides, inter-reader reliability analysis was used to demonstrate the robustness of the proposed features. The identified features consistently achieved good performance in survival prediction and classification of gene expression status, on images segmented by different radiologists. Using CT data from 111 patients, six features in the segmented region of images were highly correlated with survival time. Using extracted deep features of excised lesions from 47 patients, we observed an average AUC score of 0.90 with an accuracy of 95% in C-MYC prediction (sensitivity: 92% and specificity: 98%). In HGMA2 group, using shape features, the average AUC score was measured as 0.91 with an accuracy of 88% (sensitivity: 89% and specificity: 88%). In conclusion, the radiomic features of CT image can accurately predict the expression status of HMGA2 and C-MYC genes and identify the survival time of PDAC patients.
Keywords: Genomics, machine learning, pancreatic ductal adenocarcinoma, patient survival prediction, texture analysis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive malignant tumor with a poor prognosis [1] and expected to be rated as 3rd for the cause of cancer-related death by 2030 [2]. Even for patients suitable for surgical resection, the average survival time is no more than 18 months [3,4]. Therefore, developing reliable prognostic biomarkers is urgently needed. Previous studies have provided clinical and experimental evidence supporting the oncogenic role of HMGA2 and C-MYC genes in PDAC [5-7]. The high-mobility group AT-hook2 (HMGA2) protein is encoded by the HMGA2 gene on 12q14.3 which is expressed at high levels during the embryonic stage. HMGA2 contains an AT-hook which binds to AT-rich DNA regions and regulates replication, transcription, and repair [8]. Several studies demonstrated the reflection of metastasis in renal carcinoma, esophageal carcinoma, colorectal cancer and PDAC with over-expression of HMGA2 [1,2,5,6]. On the other hand, the oncogene C-MYC is located on chromosome 8q24 and C-MYC protein is a transcription factor that plays a crucial role in cell proliferation, apoptosis, differentiation and metabolism [7]. The studies showed that dysregulated C-MYC is involved in the development and progression of PDAC disease and may contribute to the dedifferentiation of tumor cells, tumor development, and histological tumor grade [9-11]. Therefore, identification of HMGA2 and C-MYC gene expression status requires serious attention for PDAC prognosis.
Tissue biopsy is the crucial step for obtaining the gene expression for diseases, but it is highly invasive and prone to sampling bias due to tissue heterogeneity. Besides, several studies pointed out potential impurity after contamination with healthy tissue in the tumor microenvironment after genomics analysis [12]. Therefore, a non-invasive prognosis approach that reflects the gene expression is severely required.
Computed tomography (CT) is routinely used as a noninvasive imaging approach for diagnosis and staging of PDAC in clinical practice [3,4]. However, information obtained through a routine CT image analysis may include only fundamental properties of tumors e.g. gross area, volume, and simple shape descriptors. On the other hand, texture analysis can provide more complex characteristics of the tissues not observed by human eye [13]. Therefore, the popularity of radiomics analysis which describes the tissue characteristics with high dimensional features has improved during recent years [14,15]. Earlier studies demonstrated the potential of radiomic analysis for prediction of tumor genotypes, grade, and prognosis of the tumor [14,15]. On the other hand, recent studies have investigated the CT image texture extracting image characteristics using deep learning approach for diagnosis purpose [16,17]. Although machine learning has been utilized to classify cancer according to the gene expression patterns, the association between radiomic and genetic features to predict the overall survival of the patients is still understudied [18].
In this study, we focused to evaluate the strengths of radiomic analysis extracting conventional and deep learning features of CT images for prediction of HMGA2 and C-MYC genes expression status for diagnosis of PDAC and overall survival time of the PDAC patients.
Materials and methods
Patients
This retrospective study was approved by the Ethics Committee of Southwest Hospital (Chongqing, China) (No. KY201802). The study cohort consisted of 61 male and 50 female patients with histologically proven PDAC, who underwent surgical resection from 2008 to 2011 at our institution. The average age for the patients was 57 years with 10 months of overall survival time.
Tissue samples and immunohistochemistry
The cross sections of the excised lesions for 47 patients among the study cohort were available for HMGA2 and C-MYC expression analysis. The tissue samples were embedded in paraffin and stained for immunohistochemistry (IHC) studies for C-MYC and HMGA2 genes. After IHC procedure, tissue was quantified using a composite score obtained by multiplying the staining intensity level (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) (Supplementary Figure 1) with the percentage of positive cells (0, 0%; 1, 1-10%; 2, 10-50%; 3, > 50%). The score for each specimen was between 0 and 9, corresponding to the level of protein expression.
CT image acquisition and segmentation
All 111 patients received a preoperative pancreas protocol CT scan with following acquisition parameters: 0.6 mm section width and 2 mm reconstruction interval, 120 kVp and 210-300 mAs. All images were reconstructed with a high-kernel (b30) and a high-resolution matrix (512 × 512). Non-enhanced images from the upper aspect of the liver to the iliac crest were obtained. Omnipaque-350 contrast was then injected intravenously at a rate of 3.5 ml/sec based on total volume weight. Abdominal aortic tracking was used, and the threshold was set to 180 Hu. The arterial phase image was obtained 3 seconds after the trigger, and the venous phase images were obtained 25 seconds after the trigger.
The pancreatic lesion was segmented from the venous phase images, where they were generally more conspicuous. A slice with maximal in-plane tumor diameter was manually chosen as a representative section of the tumor lesion. The region of interest (ROI) of the lesion was defined manually by two radiologists using an in-house developed computer-aided segmentation tool for imaging analysis [19] and inter-reader reliability of segmentation was evaluated in term of intersection-over-union (IoU) measure, denoted as (Equation 1):
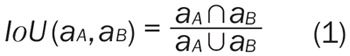 |
Where aA and aB represent the segmented areas of the same patient’s CT data by clinician A and B, respectively.
Radiomic features
All radiomic features (conventional and deep features) were extracted from segmented ROIs on CT data using Matlab® (version 9.1.0, MathWorks, Natick, Massachusetts) and MatConvNet toolbox [20].
Conventional features
The conventional features combines 27 shape (eccentricity, extent, perimeter, orientation, centroid, major axis length, area, solidity, extrema, equivalent-diameter and minor axis length) and 43 texture features (mean gray-level intensity and uniformity) extracted with multi-level of LoG band-pass filter with the parameter σ ranged between 0.5 and 2.5 [21,22].
Deep convolutional features
In recent studies, implementation of convolutional neural networks (CNN) in medical image processing resulted in superior performance [23]. Due to the requirement of large data set to train a new CNN model, the transfer learning approach is commonly preferred while working with relatively small datasets [24]. The recent studies have shown that shallow layers in CNN contain more general information and can be used as feature extractors to obtain common medical image features [23,25,26]. Therefore, we utilized a pre-trained CNN model to extract image characteristics from CT images for diagnosis and prognosis of PDAC [27]. Prior to feature extraction, a sigmoid function normalization was performed to enhance details in the ROIs. Then, masked images were fed into the first three convolutional layers in the CNN model and 256 features were extracted.
Classification
The patients with histology images were labeled according to the level of gene existence as low (staining score < 6) and high expression (staining score ≥ 6) based on staining score. The labels were utilized while generating a classification model using a support vector machine approach with k-fold and leave-one-out cross-validation methods while using all the radiomic features. The performance of the classifier was evaluated in term of accuracy, sensitivity, specificity, and AUC based on the results of cross-validation. The optimal cut-off threshold values were determined at the point on the ROC curve where the positive likelihood ratio (true positive fraction/false positive fraction) was maximal. After that, we calculated the corresponding sensitivity and specificity. The selected ROC cut-offs of the radiomic signature in the training dataset were then applied to the validation dataset to calculate the sensitivity and specificity.
To improve the classifier, we employed an exhaustive feature selection approach to determine the significant features [28]. The number of features is increased in each iteration as long as the performance of the classifier is improved. The pipeline of the method is shown in Figure 1.
Figure 1.
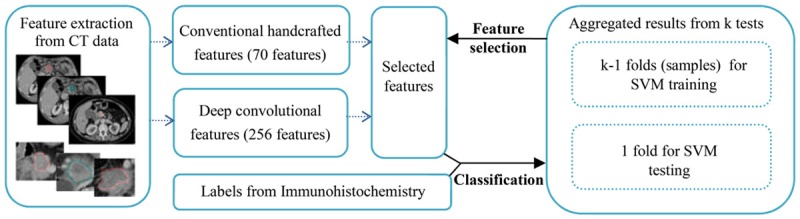
The pipeline of the algorithm used in gene classification. For each preoperative CT image, 326 radiomic features were extracted after manually segmenting the CT images. Following surgical resection pancreatic tumor, HMGA2 and C-MYC expressions of the pancreatic tumor were classified using support vector machine with the selected radiomic features. K-fold cross-validation is used to evaluate the classification model.
Performance
We divided the test into three groups which utilize the conventional features (with shape and texture features), deep features, and all the features. Then, we performed feature selection method and reported the performance with the AUC, accuracy specificity and, sensitivity scores. To obtain more robust results, we use 10-fold, 20-fold, 30-fold, 40-fold, and leave-one-out cross-validation to assess the classification efficacy, which means the scores are averaged over 147 tests.
Statistics
All 111 patients were included in the survival prediction study. To control the confounding effects from age or gender, the patients were grouped into two groups by age or gender, where we divided them by either above or below the median age of 59 [29]. 326 conventional and deep features were extracted from each patient’s CT data. For each feature, we divided the patients into two groups by the median value of the feature.
To examine the effects of gender, age and radiomic features on survival, we utilized log-rank tests to do univariate survival analysis. Besides, we used Kaplan-Meier estimate analysis to assess survival. In the log-rank test, the Chi-squared test was used to compare distributions of categorical variables. The features that showed discrimination with respect to the survival time were shown as the Supplementary Figure 2. P value larger than 0.05 was considered statistically significant.
Results
Segmentation result
Two experienced radiologists blindly segmented CT slices with maximal in-plane tumor diameter (Figure 2) and inter-reader reliability was performed for evaluation of segmented regions by the radiologists. An average IoU score of 0.70 was obtained for 111 patients (Supplementary Figure 3), which was higher than the commonly used threshold of 0.5 to judge inter-reader reliability [30].
Figure 2.
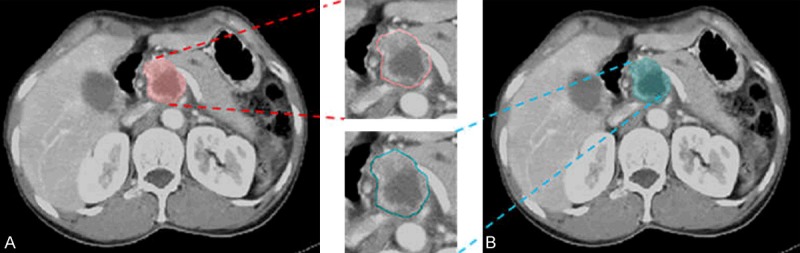
Segmented tumor images from the same patient’s CT data. Different colors indicate the segmentation from the different radiologists. (A) The ROI segmented by Dr. A; (B) The ROI segmented by Dr. B.
Correlation of survival prediction and radiomic features
The characteristics of patients were presented in Supplementary Table 1 with multivariate Cox regression analysis. The results showed that survival time was independent of age and gender. The comparison of the log-rank test was shown in Supplementary Figure 2. In two doctors’ log-rank test, we obtained 39 and 37 significant features for patient survival time, respectively. However, due to variability between different doctors, not all the selected features performed consistently well in different doctor segmented images. Thus, there were only six common features with chi-square values over 3.8 as significant for survival time (Supplementary Figure 4). The feature IDs were 2 (shape feature: extent), 75 (deep convolutional feature), 112 (deep convolutional feature), 156 (deep convolutional feature), 164 (deep convolutional feature), and 284 (deep convolutional feature). The values of those features were presented in Table 1 while the survival curves were shown as Supplementary Figure 2, which indicated that the models built with these 6 features could predict the survival time of patients well.
Table 1.
The values of the selected features
| Feature ID | 2 | 75 | 112 | 156 | 164 | 284 |
|---|---|---|---|---|---|---|
| Dr. A | 0.70 ± 0.08 | 35.58 ± 18.11 | 2.75 ± 6.56 | 1.16 ± 3.32 | 0.75 ± 3.35 | 1.16 ± 3.32 |
| Dr. B | 0.67 ± 0.09 | 36.76 ± 16.55 | 3.93 ± 7.44 | 1.17 ± 3.77 | 0.63 ± 2.57 | 1.17 ± 3.77 |
Note: Six common features with chi-square values over 3.8 are significant for survival time. The feature IDs are 2 (shape feature: extent), 75 (deep convolutional feature), 112 (deep convolutional feature), 156 (deep convolutional feature), 164 (deep convolutional feature), 284 (deep convolutional feature).
The correlation between radiomic features and immunohistochemistry
The classification performance with the best-selected features for C-MYC was presented in Supplementary Figure 5. The selected deep features had much stronger classification power and the performance increased with the size of training data while the test with a bigger k (folds) had better results. Therefore, the best performance was obtained with a leave-one-out test, where AUC, accuracy, specificity, and sensitivity were 0.96, 98%, 100%, and 96%, respectively. Note that, combining deep features with conventional features did not improve the performance, observed from the average values shown in Table 2. This shows that using selected features from deep features will be enough for classification. The classification performance with the best-selected features for HMGA2 was shown in Supplementary Figure 6. The average values of testing results with k-fold cross-validation were shown in Table 2. In terms of AUC, the selected features perform generally well. However, the performance was very sensitive to the size of the training data. This can be observed that AUC scores in 10-fold were much lower than other folds. Note that even the AUCs scores were acceptable in 10-fold, but sensitivity was extremely low due to imbalanced data (much more positive data than negative data). The same holds true for C-MYC, the better performance was obtained by leave-one-out, where the best AUCs was 0.99 with an accuracy of 98%, the specificity of 100%, and sensitivity of 93%.
Table 2.
Values aggregated from all (folds) experiments with respect to AUC, Accuracy, Specificity, Sensitivity scores in C-MYC and HMGA2
| C-MYC | HMGA2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| AUC | Accuracy | Specificity | Sensitivity | AUC | Accuracy | Specificity | Sensitivity | |||||||||
|
|
||||||||||||||||
| Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | Dr. A | Dr. B | |
| Conventional features | 0.72 | 0.74 | 73% | 72% | 86% | 79% | 60% | 66% | 0.81 | 0.81 | 81% | 81% | 85% | 92% | 71% | 57% |
| Deep features | 0.90 | 0.87 | 89% | 86% | 89% | 86% | 88% | 85% | 0.84 | 0.86 | 82% | 85% | 95% | 90% | 53% | 71% |
| All features | 0.90 | 0.87 | 89% | 87% | 89% | 89% | 89% | 85% | 0.84 | 0.86 | 82% | 85% | 95% | 92% | 53% | 69% |
C-MYC, C-MYC protein; HMGA2, high-mobility group AT-hook2.
Finally, we used previously selected features to classify C-MYC and HGMA2 with different doctor segmented images, i.e. fifteen groups (features) from C-MYC and HMGA2 classification test segmented by Dr. A, fifteen groups (features) from C-MYC and HMGA2 classification test segmented by Dr. B. Another 240 additional experiments in total were conducted to assess our selected radiomic features (Figure 3).
Figure 3.
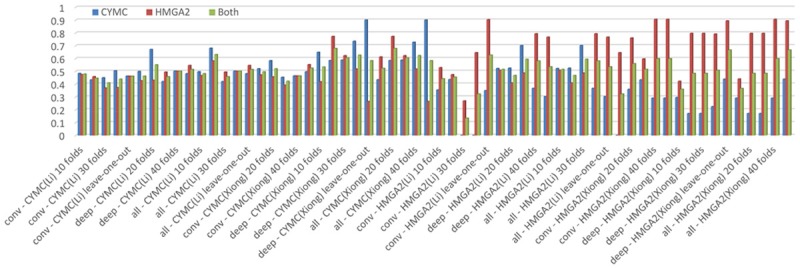
The AUC scores in C-MYC (blue bars) and HGMA2 (red bars) classification using selected features with different doctor segmented images in different K-folds validations. Green bars indicate the mean AUC scores in both C-MYC and HGMA2 classification. In x-axis label, “conv” means using selected conventional features, “deep” means using selected deep convolutional features, while “all” means using both selected conventional features and deep convolutional features. “A” indicates that the ROIs are segmented by Dr. A, and “B” indicates that the ROIs are segmented by Dr. B.
In C-MYC test, we could find the features selected from Dr. B could obtain consistently good performance with segmented images, where average AUC was about 0.90 with an accuracy of 95%, a sensitivity of 92%, and a specificity of 98% (feature IDs = 147 and 148, deep features). Note pure deep features contributed to the good performance. On the other hand, we identified features selected from Dr. B again provided good performance with segmented images for HMGA2 test while obtaining an AUC of 0.91 and an accuracy of 88%. Besides, the features demonstrated a sensitivity of 89% and a specificity of 88% for shape feature (extrema points in the region). We presented the ROC figures using the best-performed features selected in the experiment for C-MYC and HMGA2 classification in Figure 4.
Figure 4.
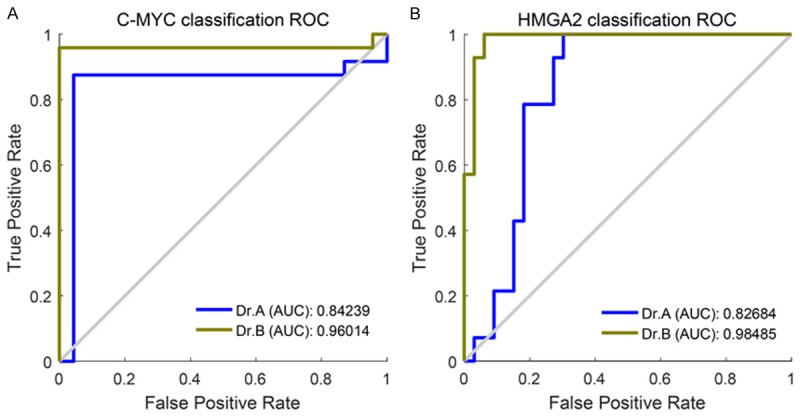
The ROCs with AUC scores in C-MYC and HMGA2 classification: (A) ROCs in C-MYC classification with best-performed features, namely features 147 and 148 (deep features); (B) ROCs in HMGA2 classification with best-performed feature 14 (shape feature: extrema points in the region).
Discussion
In our study, we showed a significant relationship between textural features and expression of C-MYC and HMGA2 proteins. C-MYC was correlated with two deep convolutional features (No. 147 and 148) while HMGA2 was directly expressed with single shape feature (extrema points in the region). Our results also indicate that a total of 6 texture features (conventional and deep convolutional) were significantly associated with survival of PDAC patients. The radiomic features of CT image were well correlated with HMGA2 and C-MYC gene expression status and also showed high accuracy in predicting survival time of the patients.
In oncology, radiomic data is in a mineable form to build a quantitative characteristic model relating image features to phenotypes or gene-protein signatures [31]. The core hypothesis of radiomics is that texture may provide valuable diagnostic, prognostic or predictive information as revealing complex patterns [32]. Hanania et al utilized radiomic features to evaluate the malignant potential of intraductal papillary mucinous neoplasm (IPMNs) [33]. 14 features were selected from 360 features of 53 patients. The most predictive marker differentiated low-grade and high-grade lesions with an AUC of 82% with a sensitivity of 85% and specificity of 68%. Hou et al reported that radiomic analysis in contrast-enhanced CT could well differentiate responders from nonresponses to chemoradiotherapy in esophageal carcinoma [34]. Five features were used to construct classifier models using ANN and SVM methods which resulted in accuracies of 97% and 89% for ANN and SVM, respectively. Yang et al conducted a study to investigate the correlation between radiomic features and KRAS/NRAS/BRAF mutations in colorectal cancer, extracting radiomic features from CT images of 117 patients with 10-fold cross-validation [35]. The AUC, sensitivity, and specificity for predicting KRAS/NRAS/BRAF mutations were measured as 0.83, 69%, and 86% in the validation cohort, respectively. There was a thorough intra-/inter-reader agreement evaluation. Besides, 10-fold cross-validation was repeated for 1000 times to prevent overfitting in training and to select the model with the best performance. Huang et al reported that radiomics nomogram for preoperative prediction of lymph node (LN) metastasis in patients with colorectal cancer and validation of the radiomics nomogram was performed [36]. Eilaghi et al reported that CT texture features were promising prognostic imaging biomarkers of overall survival in PDAC for 30 patients included in the study [37]. CT imaging is preferably used for initial exams of pancreas despite the ability to obtain more detailed information with other imaging modalities e.g. US and MRI [38]. In this study, we investigated the CT image texture of 111 patients with PDAC disease to identify quantitative imaging biomarkers which are significantly correlated with overall survival. We performed segmentation by two experienced radiologists and evaluated the segmentation results (average IoU score of 0.70 (Supplementary Table 2) to improve the robustness of the study and prevent potential bias caused by segmentation. The segmentation error of our in-house software was within an acceptable range, which provides a prerequisite for future multi-center research. We also demonstrated that deep convolutional neural networks (CNNs) could automatically learn well-performed features and achieve superior performance in medical image processing. The radiomic features provided high accuracy in predicting survival time of the patients. To the best of our knowledge, we are the first one to systematically evaluate the proposed prognostic factors, i.e., conventional features and deep features, in PDAC under different cross-validation criteria with inter-reader reliability analysis.
Our study had several limitations. First, it was lack of a patient cohort for external validation where verification of the proposed prediction models requires large cohorts of patients. We performed a retrospective single-institution study and the study has all limitations inherent to retrospective studies and accompanying chances of bias. Additionally, we only used the regions with the maximum tumor area, which cannot reflect the overall characteristics and heterogeneity of the tumor. In the next step, we will adopt a 3D segmentation method. Finally, we only cross-checked the included samples. More results need to be verified in subsequent randomized controlled trials.
In conclusion, our study showed that CT radiomic features of PDAC can accurately predict HMGA2 and C-MYC gene expression levels and identify the overall survival time of the PDAC patients. Our findings may provide potential clinical value for evaluation of PDAC disease with a noninvasive approach.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics for hispanics/latinos, 2012. CA Cancer J Clin. 2012;62:283–298. doi: 10.3322/caac.21153. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Mayo SC, Gilson MM, Herman JM, Cameron JL, Nathan H, Edil BH, Choti MA, Schulick RD, Wolfgang CL, Pawlik TM. Management of patients with pancreatic adenocarcinoma: national trends in patient selection, operative management, and use of adjuvant therapy. J Am Coll Surg. 2012;214:33–45. doi: 10.1016/j.jamcollsurg.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, Herman JM, Cameron JL, Yeo CJ, Halushka MK, Eshleman JR, Raben M, Klein AP, Hruban RH, Hidalgo M, Laheru D. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mito JK, Agoston AT, Dal Cin P, Srivastava A. Prevalence and significance of HMGA2 expression in oesophageal adenocarcinoma. Histopathology. 2017;71:909–917. doi: 10.1111/his.13310. [DOI] [PubMed] [Google Scholar]
- 6.Na N, Si T, Huang Z, Miao B, Hong L, Li H, Qiu J, Qiu J. High expression of HMGA2 predicts poor survival in patients with clear cell renal cell carcinoma. Onco Targets Ther. 2016;9:7199–7205. doi: 10.2147/OTT.S116953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mai S, Mushinski JF. c-Myc-induced genomic instability. J Environ Pathol Toxicol Oncol. 2003;22:179–199. doi: 10.1615/jenvpathtoxoncol.v22.i3.30. [DOI] [PubMed] [Google Scholar]
- 8.Strell C, Norberg KJ, Mezheyeuski A, Schnittert J, Kuninty PR, Moro CF, Paulsson J, Schultz NA, Calatayud D, Lohr JM, Frings O, Verbeke CS, Heuchel RL, Prakash J, Johansen JS, Ostman A. Stroma-regulated HMGA2 is an independent prognostic marker in PDAC and AAC. Br J Cancer. 2017;117:65–77. doi: 10.1038/bjc.2017.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hessmann E, Schneider G, Ellenrieder V, Siveke JT. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35:1609–1618. doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- 10.Schleger C, Verbeke C, Hildenbrand R, Zentgraf H, Bleyl U. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol. 2002;15:462–469. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- 11.Zhang M, Fan HY, Li SC. Inhibition of c-Myc by 10058-F4 induces growth arrest and chemosensitivity in pancreatic ductal adenocarcinoma. Biomed Pharmacother. 2015;73:123–128. doi: 10.1016/j.biopha.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, Rubin DL, Napel S, Plevritis SK. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol. 2008;15:1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Zhang Z, Chen YC, Zhao ZY, Yin XD, Jiang HB. A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Transl Oncol. 2019;12:292–300. doi: 10.1016/j.tranon.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts HJWL. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nian Y, Li M, Cui H, Hu X, Xie B, Li K, Xiong X, Xiao J, Chen W. Graph-based unsupervised segmentation for lung tumor CT images; The 3rd IEEE international Conference on Computer and Communications (ICCC); 2017. [Google Scholar]
- 20.Vedaldi A, Lenc K. MatConvNet: convolutional neural networks for MATLAB. 2014:689–692. [Google Scholar]
- 21.Miles KA, Ganeshan B, Griffiths MR, Young RC, Chatwin CR. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology. 2009;250:444–452. doi: 10.1148/radiol.2502071879. [DOI] [PubMed] [Google Scholar]
- 22.Mookiah MRK, Rohrmeier A, Dieckmeyer M, Mei K, Kopp FK, Noel PB, Kirschke JS, Baum T, Subburaj K. Feasibility of opportunistic osteoporosis screening in routine contrast-enhanced multi detector computed tomography (MDCT) using texture analysis. Osteoporos Int. 2018;29:825–835. doi: 10.1007/s00198-017-4342-3. [DOI] [PubMed] [Google Scholar]
- 23.Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 24.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–1359. [Google Scholar]
- 25.Antropova N, Huynh B, Giger M. SU-D-207B-06: predicting breast cancer malignancy on DCE-MRI data using pre-trained convolutional neural networks. Medical Physics. 2016;43:3349. [Google Scholar]
- 26.Oktay O, Bai W, Guerrero R, Kamnitsas K, Caballoro J, de Marvao A, Cook S, O’Regan D, Rueckert D. Multi-input cardiac image super-resolution using convolutional neural networks; International Conference on Medical Image Computing and Computer-Assisted Intervention; 2016. [Google Scholar]
- 27.Chatfield K, Simonyan K, Vedaldi A, Zisserman A. Return of the devil in the details: delving deep into convolutional nets. Computer Science. 2014 [Google Scholar]
- 28.James F. The elements of statistical learning: data mining, inference and prediction. Neural Netw. 2009;22:958–69. [Google Scholar]
- 29.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Critical Care. 2004;8:389. doi: 10.1186/cc2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everingham M, Gool LV, Williams CKI, Winn J, Zisserman A. The pascal visual object classes (VOC) challenge. International Journal of Computer Vision. 2010;88:303–338. [Google Scholar]
- 31.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanania AN, Bantis LE, Feng Z, Wang H, Tamm EP, Katz MH, Maitra A, Koay EJ. Quantitative imaging to evaluate malignant potential of IPMNs. Oncotarget. 2016;7:85776–85784. doi: 10.18632/oncotarget.11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou Z, Ren W, Li S, Liu J, Sun Y, Yan J, Wan S. Radiomic analysis in contrast-enhanced CT: predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget. 2017;8:104444–104454. doi: 10.18632/oncotarget.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, Zhang H, Ying J, Zhao X, Tian J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28:2058–2067. doi: 10.1007/s00330-017-5146-8. [DOI] [PubMed] [Google Scholar]
- 36.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 37.Eilaghi A, Baig S, Zhang Y, Zhang J, Karanicolas P, Gallinger S, Khalvati F, Haider MA. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma - a quantitative analysis. BMC Med Imaging. 2017;17:38. doi: 10.1186/s12880-017-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee ES, Lee JM. Imaging diagnosis of pancreatic cancer: a state-of-the-art review. World J Gastroenterol. 2014;20:7864. doi: 10.3748/wjg.v20.i24.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


