Abstract
To detect the expression pattern of microRNA-765 in breast cancer (BCa) and its regulatory effect on the disease progression. Expression level of microRNA-765 in 66 paired BCa tissues and matched normal tissues was detected by qRT-PCR. The relationship between microRNA-765 level and clinical data of BCa patients was analyzed. Subsequently, microRNA-765 level in BCa cell lines was examined as well. Changes in proliferative, migratory and invasive abilities in MCF-7 and SKBR3 cells either overexpressing microRNA-765 or not were evaluated. Furthermore, expression level of EZH1 in BCa tissues and cell lines was determined. The regulatory interaction between microRNA-765 and EZH1 was identified. Finally, the role of microRNA-765/EZH1 axis in the progression of BCa was assessed. MicroRNA-765 was downregulated in BCa tissues relative to matched normal ones. BCa patients expressing low expression of microRNA-765 presented higher tumor stage, higher metastatic rate and worse overall survival. Overexpression of microRNA-765 attenuated proliferative, migratory and invasive abilities in MCF-7 and SKBR3 cells. In addition, EZH1 was upregulated in BCa tissues and cell lines. EZH1 level was negatively regulated by microRNA-765 in BCa. Overexpression of EZH1 reversed the inhibitory effects of microRNA-765 on malignant progression of BCa. MicroRNA-765 is downregulated in BCa and closely correlated to tumor stage, lymphatic metastasis, distant metastasis and poor prognosis of BCa patients. Overexpression of microRNA-765 attenuates the malignant progression of BCa through negatively regulating EZH1.
Keywords: MicroRNA-765, EZH1, breast cancer, proliferation, metastasis
Introduction
Breast cancer (BC) is the most prevalent malignancy in women, and occasionally occurs in men [1,2]. The incidence of BCa ranks first among female malignant tumors in the developed countries, such as Europe and the United States. Its incidence also ranks first in Chinese female population, and exerts an increasing trend year by year [3,4]. BCa has become the number one killer threatening female physical and mental health [5]. With the improvement of early-stage diagnosis and health screening of BCa, individualized treatment is preferred and improves the survival of BCa patients [6,7]. Tumorigenesis is a complex process involving multiple genes, signaling pathways and biological processes [8]. A great number of factors are related to the tumorigenesis of BCa, including age at menarche, age of menopause, fertility, breastfeeding, genetics, lifestyle, endocrine status, etc. [9,10]. Currently, explorations on the occurrence and development of tumors at molecular and genetic levels have been well concerned [11].
MiRNAs are a class of endogenous, short, non-coding RNAs that regulate gene expressions by complementary pairing with the 3’UTR of the target mRNA [12,13]. In addition, miRNAs are also able to inhibit translation of the target mRNAs in an incomplete pairing way [14]. MiRNAs have tissue-specific characteristics, which participate in cellular behaviors and tumor progression [15]. Abnormally expressed miRNAs influence the downstream genes and further alter the phenotypes of tumor cells [16]. So far, targeted therapy and individualized treatment of tumors have already achieved great strides in tumor treatment. It is of significance to develop novel hallmark targeting BCa that contributes to improve the clinical outcome [17,18]. It is reported that microRNA-765 is downregulated in many types of tumors, serving as a tumor suppressor in the pathological progression [19,20].
Identification the role of miRNAs in regulating the downstream targets helps to determine its biological function in cellular behavior mediation [14-16]. It is necessary to discover functional signaling pathways that a single miRNA is involved in [16]. This study explored the regulatory effects of microRNA-765 and its downstream gene EZH1 on the progression of BCa. Our conclusion may provide new ideas for clinical treatment of BCa.
Methods
Patients and BC samples
Paired BCa tissues and matched adjacent tissues were surgically resected from 66 BCa patients. None of them received preoperative anti-tumor therapies. Clinical indexes and follow-up data of them were collected for further analyses. Patients and their families in this study have been fully informed. This study was approved by Ethics Committee of The Second Affiliated Hospital of Nanchang University.
Cell lines and reagents
Mammary epithelial cell line (MCF-10A) and BCa cell lines (MDA-MB-231, MCF-7 and SKBR3) were provided by ATCC, USA. Cells were cultured in Dulbecco’s modified eagle medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS) (Life Technologies, USA) and maintained in a 37°C, 5% CO2 incubator. Medium was replaced every 2-3 days. Cell passage was conducted at 90% of confluence.
Transfection
Transfection plasmids were provided by GenePharma, Shanghai. Cells were pre-seeded in the 6-well plates and transfected using Lipofactamine 2000 at 70% of confluence. Transfected cells at 48 h were harvested for subsequent experiments.
Cell proliferation assay
Cells were seeded in the 96-well plate with 2 × 103 cells per well. At the appointed time points, absorbance (A) at 450 nm of each sample was recorded using the CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) for depicting the viability curve.
Colony formation assay
Cells were seeded in the 6-well plate with 2.5 × 103 cells per well and cultured for 2 weeks. Subsequently, cells were subjected to 15-min fixation in 4% paraformaldehyde and 10-min staining in Giemsa solution. After removing the staining solution, colonies were washed, air dried and observed under a microscope.
Transwell migration and invasion assay
Transfected cells for 48 h were adjusted to the dose of 2.0 × 105/mL. 200 μL/well suspension was applied in the upper side of Transwell chamber (Millipore, MA, USA) pre-coated with Matrigel. In the bottom side, 700 μL of medium containing 10% FBS was applied. After 48 h of incubation, cells invaded to the bottom side were subjected to fixation in methanol for 15 min, crystal violet staining for 20 min and cell counting using a microscope. Penetrating cells were counted in 5 randomly selected fields per sample. Migration assay was performed in the same way except for Matrigel pre-coating.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), purified by DNase I treatment, and reversely transcribed into cDNA using Primescript RT Reagent (Takara, Otsu, Japan). The obtained cDNA was subjected to qRT-PCR using SYBR®Premix Ex Taq™ (Takara, Japan). Glyceraldheyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal references. Each sample was performed in triplicate, and relative level calculated by 2-ΔΔCt was analyzed by iQ5 2.0.
Western blot
Total protein was extracted from cells or tissues using radioimmunoprecipitation assay (RIPA) (Beyotime, Shanghai, China) and loaded for electrophoresis. After transferring on a polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) membrane, it was blocked in 5% skim milk for 2 hours, and subjected to incubation with primary antibodies at 4°C overnight and secondary antibodies for 2 hours. Bands were exposed by electrochemiluminescence (ECL) (Pierce, Rockford, IL, USA) and analyzed by Image Software.
Statistical analysis
SPSS 22.0 (SPSS IBM, Armonk, NY USA) was used for data analyses. Data were expressed as mean ± standard deviation. Intergroup differences were analyzed by the t-test. Kaplan-Meier curves were introduced for assessing the prognostic value of microRNA-765 on BCa patients. Spearman regression test was performed to evaluate the relationship between two genes. P < 0.05 was considered as statistically significant.
Results
MicroRNA-765 was lowly expressed in BCa
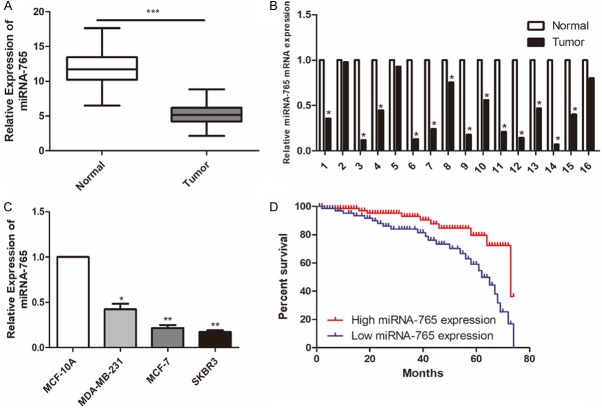
Compared with matched normal tissues, microRNA-765 was downregulated in BCa tissues (Figure 1A, 1B). Identically, microRNA-765 was lowly expressed in BCa cell lines relative to mammary epithelial cell line (Figure 1C). Kaplan-Meier curves revealed worse survival in BCa patients presenting low expression of microRNA-765 (Figure 1D).
Figure 1.
MiRNA-765 was lowly expressed in BCa. A. Relative level of miRNA-765 in BCa tissues and matched normal tissues (n = 66). B. Relative level of miRNA-765 in 16 paired BCa tissues and matched normal tissues. C. Relative level of miRNA-765 in mammary epithelial cell line (MCF-10A) and BCa cell lines (MDA-MB-231, MCF-7 and SKBR3). D. Kaplan-Meier curves revealed the overall survival in BCa patients with high and low expression of miRNA-765.
MicroRNA-765 expression was correlated with tumor stage and metastasis in BC patients
Enrolled BCa patients were divided into high expression and low expression groups based on their microRNA-765 levels. The data showed that microRNA-765 level was negatively correlated to tumor stage, lymphatic metastasis and distant metastasis, rather than age and gender of BCa patients (Table 1). It is suggested that microRNA-765 may serve as a prognostic marker of BCa.
Table 1.
Association of miRNA-765 expression with clinicopathologic characteristics of breast cancer
| Parameters | Number of cases | miRNA-765 expression | P-value | |
|---|---|---|---|---|
|
| ||||
| High (%) | Low (%) | |||
| Age (years) | 0.399 | |||
| < 60 | 35 | 19 | 16 | |
| ≥ 60 | 31 | 20 | 11 | |
| Gender | 0.122 | |||
| Male | 32 | 22 | 10 | |
| Female | 34 | 17 | 17 | |
| T stage | 0.021 | |||
| T1-T2 | 38 | 27 | 11 | |
| T3-T4 | 28 | 12 | 16 | |
| Lymph node metastasis | 0.006 | |||
| No | 40 | 29 | 11 | |
| Yes | 26 | 10 | 16 | |
| Distance metastasis | 0.019 | |||
| No | 55 | 36 | 19 | |
| Yes | 11 | 3 | 8 | |
Knockdown of microRNA-765 attenuated proliferative, migratory and invasive abilities of BCa cells
To explore the biological function of microRNA-765 in BCa progression, microRNA-765 mimics was first constructed. Transfection of microRNA-765 mimics markedly upregulated microRNA-765 level in MCF-7 and SKBR3 cells (Figure 2A). CCK-8 assay showed that transfection of microRNA-765 mimics attenuated the viability in BCa cells (Figure 2B). Colony formation assay revealed the decreased number of colonies in BCa cells overexpressing microRNA-765, suggesting the inhibited proliferative ability (Figure 2C). Moreover, numbers of migratory and invasive cells were reduced after transfection of microRNA-765 in MCF-7 and SKBR3 cells, indicating the attenuated migratory and invasive abilities (Figure 2D).
Figure 2.
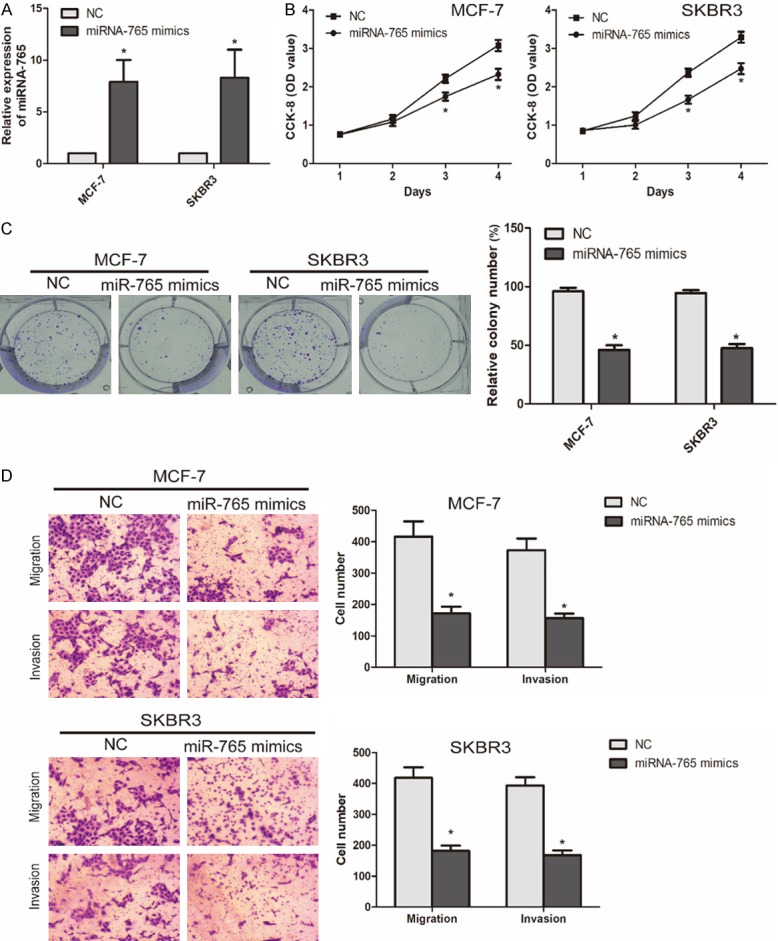
Knockdown of miRNA-765 attenuated proliferative, migratory and invasive abilities of BCa cells. A. Transfection efficacy of miRNA-765 mimics in MCF-7 and SKBR3 cells. B. CCK-8 assay showed viability in MCF-7 and SKBR3 cells transfected with NC or miRNA-765 mimics. C. Colony formation assay showed colony number in MCF-7 and SKBR3 cells transfected with NC or miRNA-765 mimics (*P < 0.05, compared with NC group). D. Transwell assay showed migratory and invasive cells in MCF-7 and SKBR3 cells transfected with NC or miRNA-765 mimics (magnification 20 ×) (*P < 0.05, compared with NC group).
EZH1 was highly expressed in BCa
EZH1 was screened out to be the downstream gene of microRNA-765 (data not shown). We thereafter examined EZH1 level in BCa. As qRT-PCR data showed, EZH1 was upregulated in BCa tissues and cell lines (Figure 3A, 3B). Furthermore, a negative correlation was observed between expression levels of EZH1 and microRNA-765 in BCa tissues (Figure 3C). Transfection of microRNA-765 mimics remarkably downregulated EZH1 level in MCF-7 and SKBR3 cells (Figure 3D).
Figure 3.
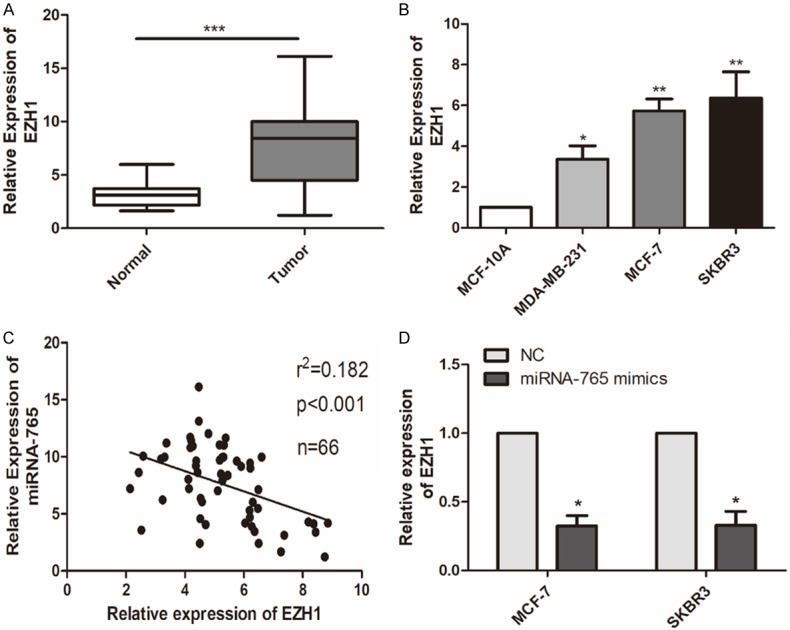
EZH1 was highly expressed in BCa. A. Relative level of EZH1 in BCa tissues and matched normal tissues (n = 66) (***P < 0.001, compared with normal group). B. Relative level of EZH1 in mammary epithelial cell line (MCF-10A) and BCa cell lines (MDA-MB-231, MCF-7 and SKBR3) (*P < 0.05, compared with MCF-10A group **P < 0.01, compared with MCF-10A group; ***P < 0.001, compared with MCF-10A group). C. A negative correlation between expression levels of miRNA-765 and EZH1 in 66 cases of LCa tissues. D. Relative level of EZH1 in MCF-7 and SKBR3 cells transfected with NC or miRNA-765 mimics (*P < 0.05, compared with NC group).
EZH1 modulated microRNA-765 expression in BCa cells
It is speculated that EZH1 may be involved in microRNA-765-mediated BCa progression. We established pcDNA-EZH1 and tested its transfection efficacy in BCa cells. Transfection of pcDNA-EZH1 greatly upregulated mRNA and protein levels of EZH1 in BCa cells. Interestingly, the downregulated EZH1 in BCa cells overexpressing microRNA-765 was reversed by co-transfection of pcDNA-EZH1 (Figure 4A, 4B). Overexpression of EZH1 in MCF-7 cells enhanced the numbers of colonies, migratory cells and invasive cells. It is noteworthy that the decreased numbers of colonies, migratory cells and invasive cells were partially reversed by EZH1 overexpression (Figure 4C, 4D). Therefore, it is concluded that overexpression of microRNA-765 alleviated the malignant progression of BCa through negatively regulating EZH1.
Figure 4.
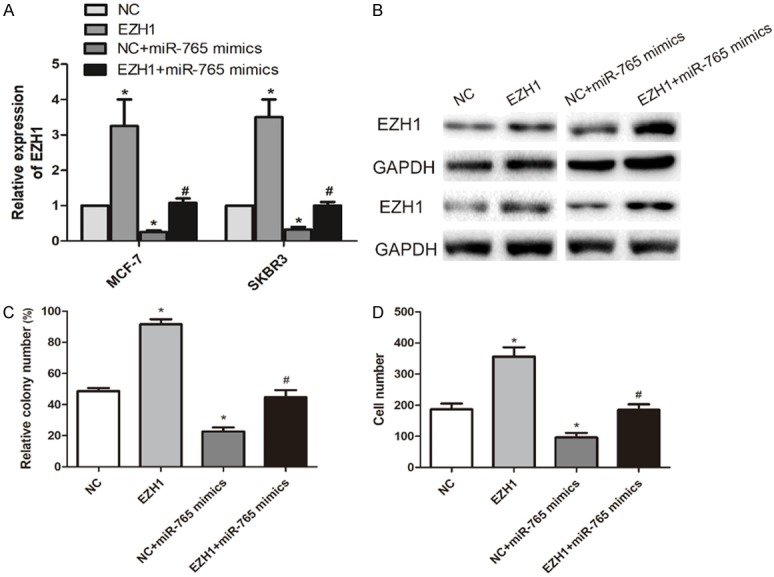
EZH1 modulated miRNA-765 expression in BCa cells. A. Relative level of EZH1 in MCF-7 and SKBR3 cells transfected with NC, pcDNA-EZH1, NC + miR-765 mimics or pcDNA-EZH1 + miR-765 mimics (*P < 0.05, compared with NC group; #P < 0.05, compared with pcDNA-EZH1 group). B. Protein level of EZH1 in MCF-7 and SKBR3 cells transfected with NC, pcDNA-EZH1, NC + miR-765 mimics or pcDNA-EZH1 + miR-765 mimics. C. Relative colony number in MCF-7 cells transfected with NC, pcDNA-EZH1, NC + miR-765 mimics or pcDNA-EZH1 + miR-765 mimics (*P < 0.05, compared with NC group; #P < 0.05, compared with pcDNA-EZH1 group). D. Relative migratory cell number in MCF-7 cells transfected with NC, pcDNA-EZH1, NC + miR-765 mimics or pcDNA-EZH1 + miR-765 mimics (*P < 0.05, compared with NC group; #P < 0.05, compared with pcDNA-EZH1 group).
Discussion
With the progress of the Human Genome Project, researches on molecular level have been extensively conducted [12-15]. Since the discovery of miRNAs, they have been recognized to widely participate in regulating cellular behaviors [12,13]. Furthermore, abnormally expressed miRNAs in tumors are well concerned [14-17]. Functionally, miRNAs target on the corresponding mRNAs, and thus degrade them or inhibit their translation [12,13]. It is estimated that over 30% of human genes and cellular processes are regulated or controlled by miRNAs. MiRNAs exert important effects on tumor growth, invasion and apoptosis [13].
Current researches on BCa-related miRNAs mainly focus on three aspects: miRNA profiling of BCa tissues; function of BCa-related miRNAs; and diagnostic, therapeutic and prognostic applications of miRNAs in BCa [17,18]. A miRNA is differentially expressed in normal tissues and tumor tissues with different degrees of differentiation [19]. A great number of studies have identified the carcinogenic or tumor-inhibiting role of miRNAs in tumor diseases [15]. The completely contrary effect of miRNAs in tumors depends on the downstream genes they regulate [14-17]. Moreover, miRNA level is considered to be related to tumor metastasis [13,20]. Previous studies have shown the downregulated microRNA-765 in tumors [19,20].
Consistently, our results also identified that microRNA-765 was downregulated in BCa tissues and cell lines. Its level was negatively correlated to tumor stage and metastatic rates of BCa, indicating its tumor-suppressor effect. In vitro experiments suggested that overexpression of microRNA-765 attenuated proliferative, migratory and invasive abilities of BCa cells. To further illustrate the molecular mechanism of microRNA-765 in regulating BCa progression, we screened out EZH1 as the downstream gene of microRNA-765. The data showed that EZH1 was upregulated in BCa tissues and cell lines. Besides, EZH1 level was negatively correlated to microRNA-765 level in BCa. Interestingly, EZH1 overexpression reversed the inhibited proliferative and metastatic abilities in BCa cells overexpressing microRNA-765. To sum up, microRNA-765 overexpression alleviated the malignant phenotypes of BCa cells via negatively regulating EZH1.
Conclusions
MicroRNA-765 is downregulated in BCa and closely correlated to tumor stage, lymphatic metastasis, distant metastasis and poor prognosis of BCa patients. Overexpression of microRNA-765 attenuates the malignant progression of BCa through negatively regulating EZH1.
Disclosure of conflict of interest
None.
References
- 1.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganz PA, Goodwin PJ. Breast cancer survivorship: where are we today? Adv Exp Med Biol. 2015;862:1–8. doi: 10.1007/978-3-319-16366-6_1. [DOI] [PubMed] [Google Scholar]
- 3.Yuan X, Liu J, Ye X. Effect of miR-200c on the proliferation, migration and invasion of breast cancer cells and relevant mechanisms. J Buon. 2019;24:61–67. [PubMed] [Google Scholar]
- 4.Li T, Mello-Thoms C, Brennan PC. Descriptive epidemiology of breast cancer in China: incidence, mortality, survival and prevalence. Breast Cancer Res Treat. 2016;159:395–406. doi: 10.1007/s10549-016-3947-0. [DOI] [PubMed] [Google Scholar]
- 5.Bromley HL, Petrie D, Mann GB, Nickson C, Rea D, Roberts TE. Valuing the health states associated with breast cancer screening programmes: a systematic review of economic measures. Soc Sci Med. 2019;228:142–154. doi: 10.1016/j.socscimed.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Zhuang X, Wang J. Correlations of MRP1 gene with serum TGF-beta1 and IL-8 in breast cancer patients during chemotherapy. J Buon. 2018;23:1302–1308. [PubMed] [Google Scholar]
- 7.McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical diagnosis and management of breast cancer. J Nucl Med. 2016;57(Suppl 1):9S–16S. doi: 10.2967/jnumed.115.157834. [DOI] [PubMed] [Google Scholar]
- 8.Chow KH, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwan ML, Kroenke CH, Sweeney C, Bernard PS, Weltzien EK, Castillo A, Factor RE, Maxfield KS, Stijleman IJ, Kushi LH, Quesenberry CJ, Habel LA, Caan BJ. Association of high obesity with PAM50 breast cancer intrinsic subtypes and gene expression. BMC Cancer. 2015;15:278. doi: 10.1186/s12885-015-1263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji ZC, Han SH, Xing YF. Overexpression of miR-3196 suppresses cell proliferation and induces cell apoptosis through targeting ERBB3 in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:8383–8390. doi: 10.26355/eurrev_201812_16536. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz B, Wienert J, Bethge M. Development and implementation of work-related medical rehabilitation in cancer patients using organizational ethnography and action research methodology. Int J Occup Med Environ Health. 2019;32:217–228. doi: 10.13075/ijomeh.1896.01250. [DOI] [PubMed] [Google Scholar]
- 12.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 13.Tutar L, Ozgur A, Tutar Y. Involvement of miRNAs and pseudogenes in cancer. Methods Mol Biol. 2018;1699:45–66. doi: 10.1007/978-1-4939-7435-1_3. [DOI] [PubMed] [Google Scholar]
- 14.Andres-Leon E, Cases I, Alonso S, Rojas AM. Novel miRNA-mRNA interactions conserved in essential cancer pathways. Sci Rep. 2017;7:46101. doi: 10.1038/srep46101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu N, Yong S, Kim HK, Choi YL, Jung Y, Kim D, Seo J, Lee YE, Baek D, Lee J, Lee S, Lee JE, Kim J, Kim J, Lee S. Identification of tumor suppressor microRNAs by integrative microRNA and mRNA sequencing of matched tumor-normal pairs in lung adenocarcinoma. Mol Oncol. 2019 doi: 10.1002/1878-0261.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham VV, Zhang J, Liu L, Truong B, Xu T, Nguyen TT, Li J, Le TD. Identifying miRNA-mRNA regulatory relationships in breast cancer with invariant causal prediction. BMC Bioinformatics. 2019;20:143. doi: 10.1186/s12859-019-2668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filella X, Foj L. miRNAs as novel biomarkers in the management of prostate cancer. Clin Chem Lab Med. 2017;55:715–736. doi: 10.1515/cclm-2015-1073. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W, Chen Y, Fan Y, Qi L, Yin Y, Zhu W, Shu Y, Liu P. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188–196. doi: 10.1158/1055-9965.EPI-16-0607. [DOI] [PubMed] [Google Scholar]
- 19.Leung YK, Chan QK, Ng CF, Ma FM, Tse HM, To KF, Maranchie J, Ho SM, Lau KM. Correction: Hsa-miRNA-765 as a key mediator for inhibiting growth, migration and invasion in fulvestrant-treated prostate cancer. PLoS One. 2019;14:e214184. doi: 10.1371/journal.pone.0214184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung YK, Chan QK, Ng CF, Ma FM, Tse HM, To KF, Maranchie J, Ho SM, Lau KM. Hsa-miRNA-765 as a key mediator for inhibiting growth, migration and invasion in fulvestrant-treated prostate cancer. PLoS One. 2014;9:e98037. doi: 10.1371/journal.pone.0098037. [DOI] [PMC free article] [PubMed] [Google Scholar]