Abstract
Although abnormal expression of the long non-coding RNA (lncRNA) MEG3 has been reported in multiple cancer types, the role of MEG3 in the pathobiology of hepatocellular carcinoma (HCC) remains unknown. This study evaluated the expression of lncRNA MEG3 and a microRNA (miRNA-10a-5p) implicated in HCC metastasis in cancer and carcinoma adjacent tissues of HCC samples (n=30 each) via in situ hybridization and quantitative RT-PCR. The effects of overexpressing either MEG3 alone, or MEG3 in combination with miRNA-10a-5p, on the proliferation, apoptosis, cell cycle progression, migration, and invasion of HepG2 cells were evaluated using functional assays. Dual luciferase reporter assays and western blotting were employed to delineate the mechanisms of MEG3 and miRNA-10a-5p regulation of key oncogenes and tumor suppressors in HCC cells. Compared to carcinoma-adjacent regions, MEG3 expression was downregulated in cancer regions of HCC samples; by contrast, miRNA-10a-5p was overexpressed in cancer regions compared to tumor-adjacent areas. Furthermore, overexpression of MEG3 (a) decreased proliferation, migration, and invasion of HepG2 cells; (b) enhanced apoptosis and the proportion of HepG2 cells in G1 of the cell cycle; (c) increased the expression of phosphatase and tensin homolog (PTEN), Bcl2-associated X (Bax), and p53 proteins; and (d) decreased the expression of miRNA-10a-5p, AKT, p-AKT, Bcl-2, and the matrix metalloproteinases (MMPs)-2 and -9. Furthermore, miRNA-10a-5p bound the 3-untranslated region of PTEN mRNA and downregulated PTEN protein expression. Taken together, these data suggest that MEG3 regulates the PTEN/AKT/MMP-2/MMP-9 signaling axis and contributes to HCC development by targeting miRNA-10a-5p.
Keywords: lncRNA MEG3, miRNA-10a-5p, HepG2, PTEN, AKT, MMP-2, MMP-9, Bcl-2, Bax, P-53
Introduction
Cancer is the second largest cause of human mortality, with only cardiovascular disease claiming more lives. Globally, liver cancer is one of the five most common malignancies, affecting about 900,000 people. Hepatocellular carcinoma (HCC), which leads to the death of 700,000 people. is the third leading cause of cancer-related deaths worldwide [1]. HCC is difficult to diagnose at an early stage, and this challenge is the main cause of the high mortality from this disease. Long non-coding RNAs (lncRNAs) are a class of transcribed RNAs that are longer than 200 nucleotides, lack a clear open reading frame, do not encode a protein, and were therefore initially considered to be transcriptional “noise” of the genome [2]. Recent studies have indicated that even though lncRNAs do not encode proteins, these molecules are involved in many intracellular signaling processes, including chromatin modification and transcriptional activation and interference, and may also be involved in the development of a variety of diseases, including cancer [4-12]. Thus, lncRNAs have become a hot topic in cancer research. MicroRNAs (miRNAs), on the other hand, are non-coding RNAs that are generally 20-24 nucleotides long and regulate gene expression by binding to the 3’-untranslated regions of specific mRNA targets to either inhibit the translation of the mRNAs or promote their degradation [13]. Studies have shown that miRNA mutations or ectopic expression are associated with a variety of human cancers. Furthermore, miRNAs can function as tumor suppressors or oncogenes and thus play an important role in the development of cancer [14].
In this study, we first evaluated the expression of lncRNA MEG3 and miRNA-10a-5p in clinical samples from HCC patients and examined the relationship between lncRNA MEG3 and miRNA-10a-5p expression. Next, we explored the molecular mechanism by which lncRNA MEG3 expression affects proliferation, migration, and invasion of the HCC-derived cell line HepG2.
Materials and methods
Materials
The HepG2 HCC cell line was purchased from American Type Culture Collection (ATCC). High-glucose Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), TRIzol, and Lipofectamine TM 2000 transfection reagent were purchased from GIBCO Antibiotic-Antimycotic (100×) solution was purchased from Invitrogen, and the qRT-PCR kit was purchased from Takara. Thiazolyl blue (MTT), dimethyl sulfoxide (DMSO), and crystal violet were purchased from Sigma.
Clinical samples
Clinical samples used in this study were derived from 30 HCC patients who were treated in The Third Affiliated Hospital of Sun Yat-sen University Hospital between November 2015 and December 2016. Formalin-fixed paraffin-embedded samples were used for evaluating lncRNA MEG3 and miRNA-10a-5p expression in cancer and carcinoma-adjacent areas via in situ hybridization (ISH) assay. Cancer and carcinoma-adjacent tissue samples were separated prior to freezing for reverse transcriptase (RT)-PCR analysis of lncRNA MEG3 and miRNA-10a-5p expression.
ISH assay
Tissue sections were dewaxed, permeabilized with pepsin, and rinsed in a 37°C water bath for 30 min. The samples were then dehydrated in an alcohol gradient. After pre-hybridization at 37°C for 2 h, the digoxigenin (DIG)-labeled lncRNA MEG3 and miRNA-10a-5p probes were hybridized overnight in a constant-temperature water bath at 42°C. Tissue sections were then incubated with biotinylated mouse anti-DIG antibody in a water bath at 37°C for 1 h, followed by washing with 0.5 moll hosphate-buffered saline (PBS). Chromogenic visualization of probe-target hybrids was performed following the addition of diaminobenzidine (DAB) color solution via observation of the resulting color under the microscope. Finally, sections were counter-stained using hematoxylin, dehydrated, mounted, and imaged using a microscope.
Cell culture
HepG2 cells were cultured in high-glucose DMEM (containing 10% FBS, 100 U/mL penicillin, and 100 mg/L streptomycin) in a humidified incubator at 37°C and 5% CO2. Culture medium was replaced with fresh medium every 1-2 d, when the cell cultures became 80-90% confluent.
Cell treatments and groups
The lncRNA MEG3 sequence was synthesized (add supplier) and cloned into the pcDNA3.1 vector. The empty pcDNA3.1 vector was utilized as the control. The HepG2 cells were divided into four groups: NC group (untransfected cells); BL group (transfected with empty pcDNA3.1 vector DNA); lncRNA group (transfected with plasmid for lncRNA MEG3 overexpression); and lncRNA+miRNA group (transfected with plasmids for ectopic co-expression of lncRNA MEG3 and miRNA-10a-5p). Transfection was performed using the Lipofectamine TM 2000 transfection reagent according to the manufacturer’s instructions. Cells were transfected 24-48 h prior to being harvested for use in subsequent analysis. RNA was extracted from cells for evaluation of the expression levels of lncRNA MEG3 and miRNA-10a-5p using quantitative RT-PCR.
MTT assay
Cells from the NC, BL, lncRNA, and lncRNA+miRNA groups were inoculated into 96-well plates and cultured overnight at 37°C with 5% CO2 and saturated humidity. Thereafter, 20 μL of 5 mg/mL MTT were added to each well, and cells were incubated at 37°C for an additional 4 h. The culture medium in these wells was then discarded, and 150 μL DMSO was added to each well. The plates were placed on a sample oscillator for 10 min, and absorbance was measured at 490 nm.
Transwell invasion assay
Logarithmic growth phase cells from the NC, BL, lncRNA, and lncRNA+miRNA groups were trypisinized and resuspended in serum-free medium at the same cell density. For each experimental group, 100 μL of cell suspension was seeded into the top compartment of the Transwell chamber, and 600 μL of DMEM containing 10% FBS was added into the bottom compartment. Cells were incubated at 37°C for 24 h in an atmosphere containing 5% CO2. The Transwell insert was then removed, and the side of the insert facing the upper compartment was carefully cleansed with a cotton swab to remove culture medium and cells that had not migrated through the insert. Cells that had migrated through the filter pores to the underside of the insert were fixed for 20 min in 4% poly methanol, stained with Giemsa, and then photographed for quantitation of the number of cells at high magnification.
Wound healing assay
Logarithmic growth phase cells from the NC, BL, lncRNA, and lncRNA+miRNA groups were trypsinized, pelleted by centrifugation, resuspended, and seeded at the same cell density into the wells of 6-well plates. The plates were incubated overnight at 37°C in an atmosphere containing 5% CO2. Once the cells reached 80-90% confluency, a sterile tip of a 10 μL pipette was used to gently scratch the cell monolayer to create a “wound”. Care was taken to ensure that the width of the scratch/wound was nearly identical for all four experimental groups. Cells were washed twice with PBS, and the width of the wound was measured at 0 h at low magnification in three different regions of the wound. Wound widths were also measured after culturing the cells for 24 h at 37°C, and wound closure was quantified.
RT-PCR
To determine the expression levels of lncRNA MEG3 and miRNA-10a-5p in cancer and carcinoma-adjacent tissues from HCC patients, total RNA was extracted using the TRIzol reagent. Real-time PCR reactions were carried out in accordance with instructions provided by the manufacturer of the fluorescence-based qRT-PCR kit. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as the endogenous reference gene for the RT-PCR. The sequences of the primers used for RT-PCR were as follows:
MEG3 F: 5’-CTGCCCATCTACACCTCACG-3’ and R: 5’-CTCTCCGCCGTCTGCGCTAGGGGCT-3’; miRNA-10a-5p F: 5’-CGCTACCCTGTAGATCCGAA-3’ and R: 5’-GTGCAGGGTCCGAGGT-3’; and GAPDH F: 5’-AGCCACATCGCTCAGACAC-3’ and R: 5’-GCCCAATACGACCAAATCC-3’.
Western blot assay
Cells from the NC, BL, lncRNA, and lncRNA+miRNA groups were lysed for 30 min on ice using the RIPA lysis buffer (1 mL RIPA+10 μL phenylmethylsulfonyl fluoride [PMSF]+10 μL aprotinin). Total protein concentration in the cell lysates was determined using the Bicinchoninic Acid (BCA) assay, and 10 μg of total protein was loaded into each lane. Proteins were separated via 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) before being transferred onto a polyvinylidene fluoride (PVDF) membrane. Blocking was performed for 1 h using 5% PBS-Tween containing bovine serum albumin (BSA). The membrane was then incubated with the primary antibody at 4°C overnight, followed by 3 washes using Tris-buffered saline (TBS)-Tween (TBST). The membrane was then incubated with the secondary antibody at room temperature on a shaker overnight. The next day, the membrane was washed three times with TBST. Proteins of interest were then detected using the enhanced chemiluminescence (ECL) kit, followed by autoradiography.
Dual luciferase reporter assay
In the dual luciferase reporter assay, the phosphatase and tensin homolog (PTEN)-luc plasmid contained the 3’-untranslated region of the PTEN gene cloned downstream of the luciferase open reading frame. The plasmid for miRNA-10-5p overexpression was co-transfected with PTEN-luc into HepG2 cells using the Lipofectamine TM 2000 transfection reagent and controlled by Lipofectamine 2000 was transfected into miRNA-10a-5p over expression and control of HepG2 cells and PTEN silencing and control HepG2 cells, After 24 h, cells were lysed, and the dual luciferase reporter assay system was used to detect luciferase activity.
Statistical analyses
Data are displayed as means ± standard deviation (SD). All statistical analyses were performed using SPSS 17.0 statistical software (SPSS, Chicago, IL, USA). The Student’s t-test was used to evaluate the difference between groups. A value of P<0.05 was considered statistically significant.
Results
Expression of lncRNA MEG3 is downregulated in HCC
To gain insight into the potential role of lncRNA MEG3 in the pathobiology of HCC, we evaluated the expression of lncRNA MEG3 in cancer and carcinoma-adjacent tissues of 30 HCC patients via ISH. We also performed ISH for miRNA-10a-5p in the same samples and examined the relationship between the expression levels of lncRNA MEG3 and miRNA-10a-5p. The expression of lncRNA MEG3 was significantly downregulated in cancer regions of HCC compared to carcinoma-adjacent regions (Figure 1A). By contrast, the expression of miRNA-10a-5p was significantly higher in cancer tissues compared to that in carcinoma-adjacent tissues (Figure 1B). We obtained concordant results for both lncRNA MEG3 (Figure 1C) and miRNA-10a-5p (Figure 1D) via RT-PCR. Furthermore, the expression of lncRNA MEG3 was negatively correlated with the expression of miRNA-10a-5p in cancer regions of the HCC samples (Figure 1E).
Figure 1.
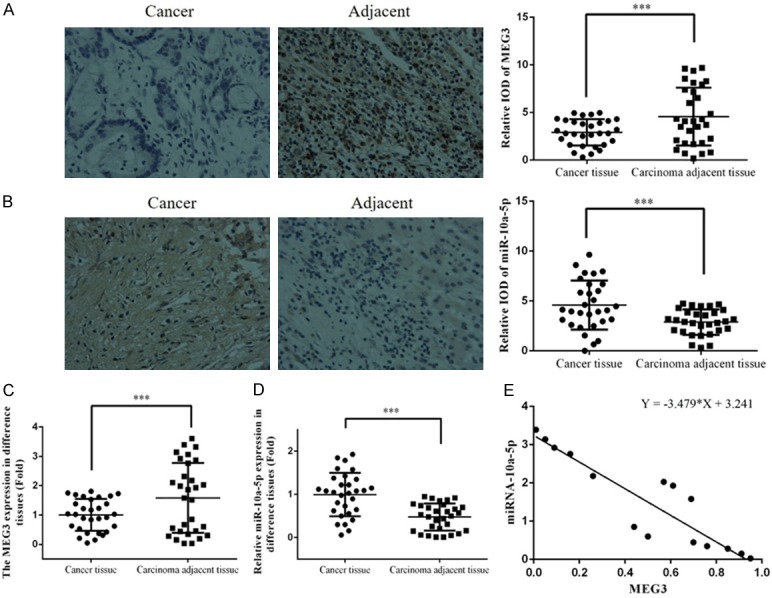
miR-10a-5p and MEG3 expression in clinical tissues. Photomicrographs showing the expression of (A) lncRNA MEG3 and (B) miRNA-10a-5p in cancer and adjacent tissues by ISH. Quantitation of (C) lncRNA MEG3 and (D) miRNA-10a-5p expression in cancer and adjacent tissues by RT-PCR. (E) The correlation between miRNA-10a-5p and MEG3 in cancer tissues. ***: P<0.05. IOD: integrated optical density.
Overexpression of lncRNA MEG3 decreases proliferation of HepG2 cells
To better understand the effects of lncRNA MEG3 expression on the proliferation of HCC cells, we transfected HepG2 cells either with an empty vector (BL group) or with a lncRNA MEG3 overexpression construct (lncRNA group). We also co-transfected HepG2 cells with constructs for lncRNA MEG3 and miRNA-10a-5p overexpression (lncRNA+miRNA group), while a fourth group of untransfected HepG2 cells served as a control (NC group). Cell proliferation in the four groups was evaluated using the MTT assay. The proliferation of the lncRNA group was significantly reduced compared to that of the NC group (P<0.05); however, there were no significant differences in proliferation among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 2).
Figure 2.
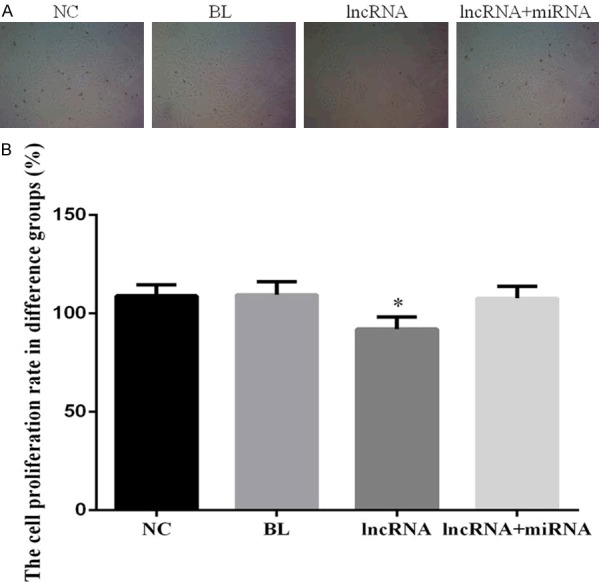
Overexpression of lncRNA MEG3 decreases proliferation in HepG2 cells. A. Photomicrographs showing proliferation in HepG2 experimental groups. B. Bar graphs depicting cell proliferation rate of the HepG2 experimental groups. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
Overexpression of lncRNA MEG3 enhances apoptosis in HepG2 cells
To determine whether expression of lncRNA MEG3 affects apoptosis of HCC cells, we employed flow cytometry to quantify the proportion of cells undergoing apoptosis. Indeed, lncRNA MEG3 overexpression significantly increased apoptosis in HepG2 cells compared to the proportion of apoptotic cells in the NC group (P<0.05; Figure 3). Notably, there were no significant differences in proportion of apoptotic cells among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 3).
Figure 3.
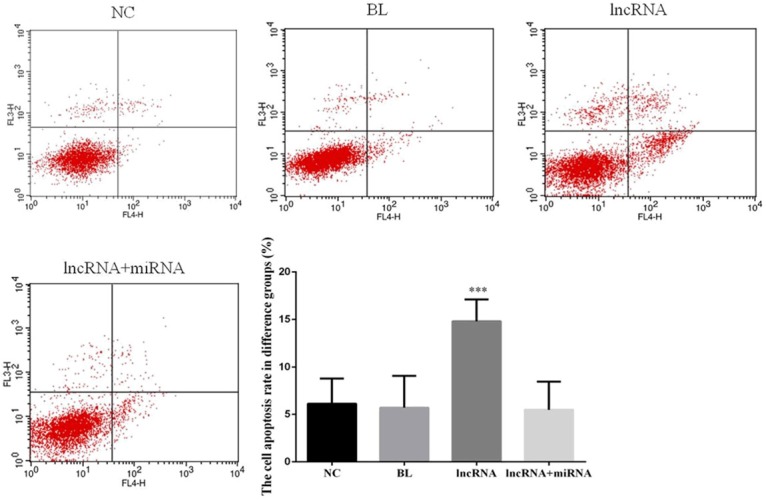
Overexpression of lncRNA MEG3 increases apoptosis in HepG2 cells. The two-parameter (dual color fluorescence) dot plots were obtained via flow cytometric analysis of cells from the indicated experimental groups. Bar graphs depict mean ± SD of 3 times replicates. ***: P<0.05.
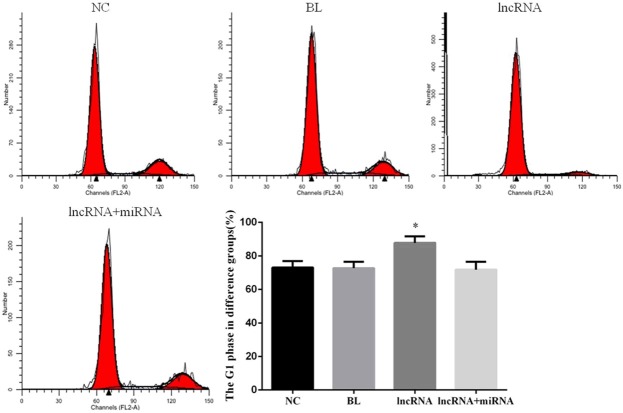
Overexpression of lncRNA MEG3 increases the proportion of cells in G1 phase of the cell cycle
Our data showing that overexpression of lncRNA MEG3 reduces proliferation of HepG2 cells provided impetus for us to evaluate the cell cycle profiles in the four experimental groups via two-color flow cytometry. We observed that the proportion of G1-phase cells in the lncRNA group was significantly higher than that in the NC group (P<0.05; Figure 4). We observed no significant differences in the proportion of G1-phase cells among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 4).
Figure 4.
Overexpression of lncRNA MEG3 increases the proportion of HepG2 cells in G1 phase of the cell cycle. Single parameter histograms depict the cell cycle profiles of cells from the indicated experimental groups. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
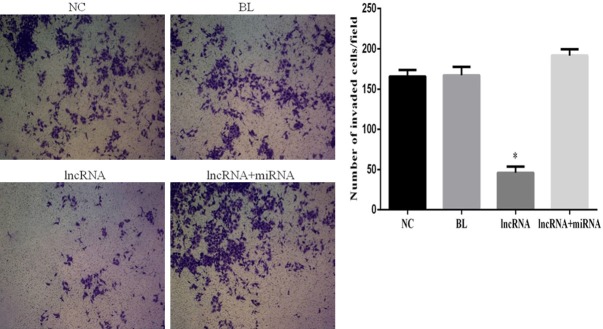
HepG2 cells overexpressing lncRNA MEG3 exhibit diminished cell migration and invasion
To gain a better understanding of the effects of lncRNA expression on invasion and migration of HEPG2 cells, we performed Transwell invasion and wound healing assays, respectively. The lncRNA MEG3-overexpressing cells had a significantly diminished invasion capability compared to the cells in the NC group (P<0.05; Figure 5). Importantly, no significant differences in invasion were noted among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 5). Interestingly, the rate of wound closure in lncRNA MEG3-overexpressing cells was significantly lower than that in untransfected cells in the NC group (P<0.05), while no significant differences in the wound healing rates (and thus, the migration capacity) of cells in the NC, BL and lncRNA+miRNA groups were detected (Figure 6).
Figure 5.
Overexpression of lncRNA MEG3 inhibits invasion of HepG2 cells. Photomicrographs showing results of Transwell cell invasion assay for the indicated experimental groups. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
Figure 6.
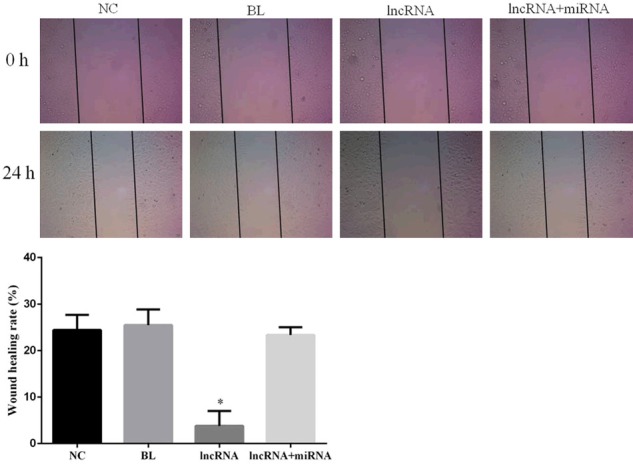
Overexpression of lncRNA MEG3 decreases migration of HepG2 cells. Photomicrographs showing results of wound healing assay. Pictures were taken at 0 and 24 h after cell wounding. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
Overexpression of lncRNA MEG3 results in decreased expression of miRNA-10a-5p
To determine whether lncRNA MEG3 overexpression has any impact on the expression of miRNA-10a-5p and vice versa, we employed quantitative RT-PCR to measure the expression levels of these RNA molecules in our experimental groups. Interestingly, overexpression of lncRNA MEG3 resulted in a striking decrease in the expression of miRNA-10a-5p (Figure 7). Furthermore, the expression levels of lncRNA MEG3 and miRNA-10a-5p in the lncRNA group were significantly different than levels in the NC group (P<0.05). In contrast, no significant differences in the expression levels of lncRNA MEG3 and miRNA-10a-5p were found among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 7).
Figure 7.
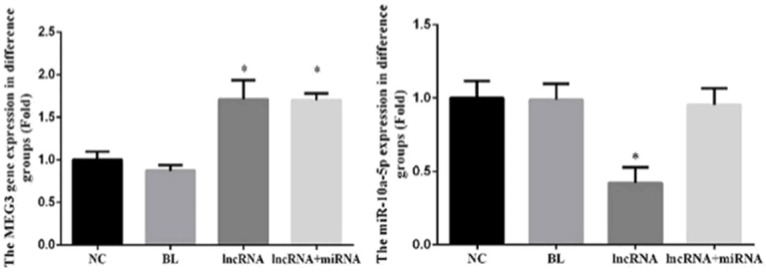
Overexpression of lncRNA MEG3 decreases the expression of miRNA-10a-5p. The expression of MEG3 and miRNA-10a-5p in the indicated experimental groups were assayed by RT-PCR. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
Overexpression of lncRNA MEG3 upregulates expression of PTEN, Bcl-2-associated X (Bax), and p53 proteins in HepG2 cells
Western blotting revealed that the expression of PTEN, Bax, and p53 proteins in the lncRNA group was significantly higher than that in the NC group (P<0.05); however, expression levels of AKT, p-AKT, Bcl-2, matrix metalloproteinase (MMP)-2, and MMP-9 proteins were significantly decreased in the lncRNA group compared to the NC group (P<0.05; Figure 8). The relative expression of these proteins were not significantly different among the NC, BL, and lncRNA+miRNA groups (P>0.05; Figure 8).
Figure 8.
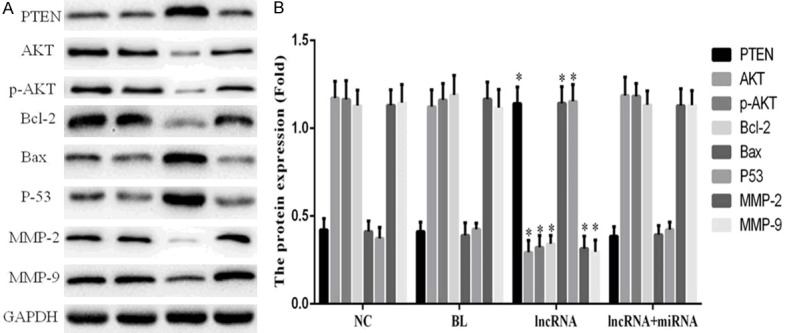
Overexpression of lncRNA MEG3 leads to an upregulation of PTEN, Bax, and p53 proteins and a downregulation of AKT, p-AKT, Bcl-2, MMP-2, and MMP-9 proteins. A. Immunoblots show expression levels of the indicated proteins. B. Bar graphs depict mean ± SD of 3 times replicates. *: P<0.05.
miRNA-10a-5p downregulates expression of PTEN
The 3’-untranslated region of PTEN was cloned into the luciferase reporter vector downstream of the luciferase gene. HepG2 cells transfected with this reporter construct were co-transfected with miRNA-10a-5p mimics. The dual luciferase assay showed that miRNA-10a-5p expression significantly inhibited the activity of the luciferase reporter construct, and this reduction in luciferase expression was abolished when the reporter construct was co-transfected with a construct containing a mutated version of the 3’-untranslated region of PTEN cloned downstream of the luciferase open reading frame (Figure 9). These results suggest that miRNA-10a-5p directly binds to the 3’-untranslated region of PTEN and downregulates PTEN protein expression.
Figure 9.
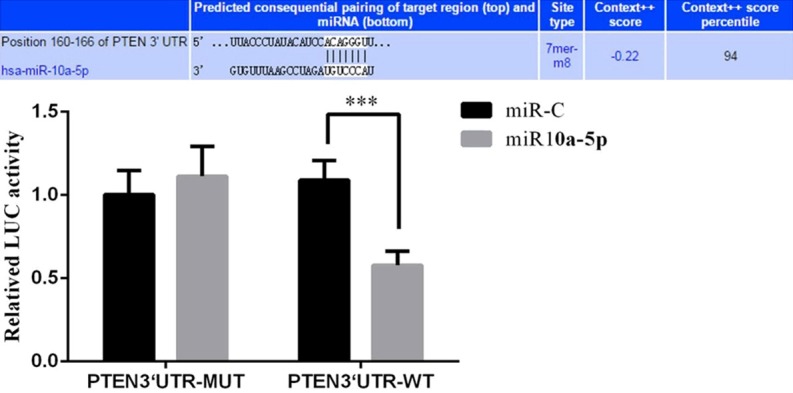
miRNA-10a-5p binds to the 3’-untranslated region of the PTEN mRNA and decreases PTEN protein expression. Bar graphs show the results of the dual-luciferase reporter assay and depict mean ± SD of 3 times replicates. ***: P<0.05.
Discussion
With increasing information about lncRNAs, more studies have indicated that tumor formation is dependent not only on the protein-coding genes but also to many lncRNA genes [15]. Although some lncRNAs have been found to play a key role in the formation and development of tumors, only a small proportion of lncRNAs in our transcriptome have been studied at all, leaving many important issues to be resolved. Here, we have examined the role of lncRNA MEG3 in the development of HCC.
In the present study, the expression of lncRNA MEG3 in HCC tissues was significantly lower than that in normal carcinoma-adjacent tissues. In order to gain a better understanding of how reduced expression of lncRNA MEG3 affects HCC carcinogenesis, we overexpressed lncRNA MEG3 followed by in vitro functional analysis. Our results demonstrate that overexpression of lncRNA MEG3 inhibits the proliferation, migration, and invasion of HCC cells. Together, these findings suggest that lncRNA MEG3 acts as a tumor suppressor gene in HCC and that its absence or decreased expression may drive the development of HCC. Therefore, the lncRNA MEG3 plays a key role in the development of liver cancer and may serve as a potential therapeutic target for the treatment of liver cancer, following further studies.
Over the past few years, many lncRNAs have been discovered, and studies have indicated that lncRNAs regulate gene expression and many other biological processes in a variety of ways [16-19]. Moreover, the expression of these lncRNAs may provide proliferative advantages to cells, leading to uncontrolled cell division [20]. Therefore, lncRNAs were proposed to help fill the gaps previously identified in tumor oncogene and tumor suppressor gene networks. This study revealed that the lncRNA MEG3 regulates the expression of miRNA-10a-5p and plays an effective role in inhibiting the proliferation, migration, and invasion of HepG2 cells.
Recent studies reported that PTEN inhibits the proliferation, migration and invasion of tumor cells [21,22]. PTEN, which is located in the human chromosome 10q23 region, is considered a tumor suppressor gene, and numerous studies have confirmed that PTEN controls tumor cell growth and inhibits tumor invasion and migration by interfering with multiple signaling pathways [23-25]. Downregulation of PTEN expression also results in the upregulation of AKT pathway signaling [26,27], which normally inhibits the proliferation and migration of tumor cells [28-30]. Our study found that lncRNA MEG3 effectively promotes the expression of PTEN and inhibits the activity of AKT, providing insight into a molecular mechanism that may potentially underlie the inhibition of proliferation, migration, and invasion of the HepG2 liver cancer cell line. Bcl-2, Bax, and p53 are downstream genes in the AKT signaling pathway, and their expression directly governs the proliferation and apoptosis of tumor cells. The p53 protein plays vital roles in regulating cell division and apoptosis [31-33]. In this study, we found that the overexpression of the lncRNA MEG3 promotes the expression of Bax and p53 proteins and inhibits the expression of Bcl-2. This result supports the notion that lncRNA MEG3 inhibits HepG2 cell proliferation via the PTEN/AKT/Bcl-2/Bax/p53 signaling pathway, leading to an accumulation of cells in the G1 phase and eventually their death. At the same time, we found that MEG3 inhibits expression of MMP-2 and -9. Studies have confirmed that MMP-2 and -9 are directly involved in the degradation of basement membrane, facilitating tumor metastasis and invasion [34-37]. Therefore, we inferred that lncRNA MEG3 overexpression inhibits HepG2 cell migration and invasion via regulation of PTEN/AKT/MMP-2/9 signaling. Furthermore, the results of our dual luciferase assay confirmed that the 3’-untranslated region of PTEN is the target site of miRNA-10a-5p. We conclude that MEG3 lncRNA plays a role in the PTEN/AKT/MMP-2/9 signaling pathway by targeting miRNA-10a-5p.
The results of this study confirmed that overexpression of lncRNA MEG3 leads to the inhibition of miRNA-10a-5p expression and the PTEN/AKT/Bcl-2/Bax/p53 and PTEN/AKT/MMP-2/9 signaling pathways. Thus, the biology of lncRNA MEG3 in HCC merits further study to evaluate its potential to serve as a therapeutic target for HCC.
Disclosure of conflict of interest
None.
References
- 1.Cristina B, Federica T, Carlo LV. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilusz JE, Sunwoo H, Spector DL. Long nocoding RNAs: functional surprise from the RNA world. Gnes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM. A nocoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HO-TAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, Zhang L, Wang F, Sun SH, Zhou WP. Long nocoding RNA associated with micorvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 10.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long nocording RNA SPRY4-ITI modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 11.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 12.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou Z, Lin S, Dai C, Lu Y, Tian T, Wang M, Liu X, Zheng Y, Xu P, Li S, Sheng Q, Deng Y, Dai Z. Pooling-analysis for diagnostic and prognostic value of MiRNA-100 in various cancers. Chin J Exp Surg. 2015;32:1227–1229. doi: 10.18632/oncotarget.18697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;130:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Hou L, Huang W, Gao Y, Lv X, Tang J. The mechanism of long non-coding RNA MEG3 for neurons apoptosis casused by hypoxia: mediated by miR-181b-12/16-LOX signaling pathway. Front Cell Neurosci. 2016;10:201. doi: 10.3389/fncel.2016.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non-coding RNAs are competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285–2295. [PubMed] [Google Scholar]
- 19.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long nocoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Thrends Cell Biol. 2011;21:351–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increase the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int J Oncol. 2016;10:3751. doi: 10.3892/ijo.2016.3751. [DOI] [PubMed] [Google Scholar]
- 22.Cheng ZY, Guo XL, Yang XY, Niu ZY, Li SH, Wang SY, Chen H, Pan L. PTEN and repamycin inhibiting the growth of K562 cells through regulating mTOR signaling pathway. J Exp Clin Cancer Res. 2008;27:87. doi: 10.1186/1756-9966-27-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Chen Y, Zhao P, Zang L, Zhang Z, Wang X. MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leuk Lymphoma. 2016;10:1–9. doi: 10.1080/10428194.2016.1213827. [DOI] [PubMed] [Google Scholar]
- 24.Lei X, Xu JF, Chang RM, Fang F, Zuo CH, Yang LY. JARID2 promotes invasion and metastasis of hepatocellular carcinoma by facilitating epithelial-mesenchymal transition through PTEN/AKT signaling. Oncotarget. 2016;7:40266–40284. doi: 10.18632/oncotarget.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1647. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 26.Bonnet M, Loosveld M, Montpellier B, Navarro JM, Quilichini B, Picard C, Di Cristofaro J, Bagnis C, Fossat C, Hernandez L, Mamessier E, Roulland S, Morgado E, Formisano-Tréziny C, Dik WA, Langerak AW, Prebet T, Vey N, Michel G, Gabert J, Soulier J, Macintyre EA, Asnafi V, Payet-Bornet D, Nadel B. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood. 2011;117:6650–6659. doi: 10.1182/blood-2011-02-336842. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, Dahlberg S, Neuberg D, Moreau LA, Winter SS, Larson R, Zhang J, Protopopov A, Chin L, Pandolfi PP, Silverman LB, Hunger SP, Sallan SE, Look AT. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Testa JR, Bellacosa A. AKT plays a central role in tumorigenesis. Proc Natl Acad Sci U S A. 2001;98:10983–10985. doi: 10.1073/pnas.211430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura N, Yoshida H, Kitagishi Y, Murakami M, Nishimura Y, Matsuda S. PI3K/AKT/PTEN Signaling as a Molecular Target in Leukemia Angiogenesis. Adv Hematol. 2012;2012:843085. doi: 10.1155/2012/843085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam D, Lee J, Lee SG, Yang WM, Um JY, Sethi G, Ahn KS. Ginkgolic acid inhibits invasion and migration and TGF-β-Induced EMT of lung cancer cells through PI3K/Akt/mTOR inactivation. J Cell Physiol. 2017;232:246–354. doi: 10.1002/jcp.25426. [DOI] [PubMed] [Google Scholar]
- 31.Okumura N, Yoshida H, Kitagishi Y, Murakami M, Nishimura Y, Matsuda S. Berberine protects against 6-OHDA-induced neurotoxicity in PC12 cells and zebrafish through hormetic mechanisms involving PI3K/AKT/Bcl-2 and Nrf2/HO-1 pathways. Redox Biol. 2016;11:1–11. doi: 10.1016/j.redox.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sui Y, Zheng X, Zhao D. Rab31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cell apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumor Biol. 2015;36:8661–8670. doi: 10.1007/s13277-015-3626-5. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Chen C, Hu X, Cai X, Guan Y, Hu H, Wang Q, Chen X, Cai B, Jing X. Suppressing cyclooxygenase-2 prevents nonalcoholic and inhibits apoptosis of hepatocytes that are involved in the Akt/p53 signal pathway. Biochem Biophys Res Commun. 2016;469:1034–1040. doi: 10.1016/j.bbrc.2015.12.096. [DOI] [PubMed] [Google Scholar]
- 34.Chang HR, Chen PN, Yang SF, Sun YS, Wu SW, Hung TW, Lian JD, Chu SC, Hsieh YS. Silibinin inhibits the invasion and migration of renal carcinoma 786-O cells in vitro, inhibits the growth of xenografts in vivo and enhances chemosensitivity to 5-fluorouracil and paclitaxel. Mol Carcinog. 2011;50:811–823. doi: 10.1002/mc.20756. [DOI] [PubMed] [Google Scholar]
- 35.Curran S, Murray GI. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur J Cancer. 2000;36:1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 36.Dong Z, Bonfil RD, Chinni S, Deng X, Trindade Filho JC, Bernardo M, Vaishampayan U, Che M, Sloane BF, Sheng S, Fridman R, Cher ML. Matrix metalloproteinase activity and osteoclasts in experimental prostate cancer bone metastasis tissue. Am J Pathol. 2005;166:1173–1186. doi: 10.1016/S0002-9440(10)62337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rink M, Chun FK, Robinson B, Sun M, Karakiewicz PI, Bensalah K, Fisch M, Scherr DS, Lee RK, Margulis V, Shariat SF. Tissue-based molecular markers for renal cell carcinoma. Minerva Urol Nefrol. 2011;63:293–308. [PubMed] [Google Scholar]