Abstract
Colon cancer is one of the most common malignancies worldwide, while the molecular mechanism remains largely unknown. miR-223-3p plays an important role in cancer development. Here, we found that miR-223-3p was up-regulated in 30 cases of colon cancer tissues as compared with their adjacent normal tissues. Lentivirus-mediated miR-223-3p over-expression promoted the proliferation, colony formation, migration and invasion of colon cancer cells. Inverse results were observed in miR-223-3p knockdown cells. Epithelial-mesenchymal transition (EMT) was regulated by miR-223-3p. In addition, cell apoptosis was suppressed and enhanced by miR-223-3p over-expression and knockdown, respectively. We further identified PRDM1, a tumor suppressor, was the target of miR-223-3p using microarray and luciferase assay. Our findings suggested that miR-223-3p acts as an oncogenic microRNA in colon cancer through regulating EMT and PRDM1.
Keywords: Colon cancer, miR-223-3p, EMT, PRDM1
Introduction
Colon cancer is the third most frequent malignant tumor and the fourth leading cause of cancer-related death throughout the world [1,2]. The surgery followed by adjuvant therapy is the prior treatment option for colon cancer patients, whereas only a small percentage of the patients benefit [3]. Most of the patients relapse and finally die with a more malignant disease stage. For the past decades, efforts are made to investigate the pathological and molecular process of colon cancer, and some oncogenes and tumor suppressors are found to play important roles in the development of colon cancer [4,5]. However, exploiting these trigger genes as therapeutic targets for colon cancer remains far from satisfactory in clinic. There is still constant need to identify novel molecular contributing to this disease.
MicroRNAs (miRNAs) are a class of endogenous 16-24 nt non-coding RNAs, which negatively regulate the expression of gene at the post-transcriptional level via directly binding with its mRNA [6-8]. Numerous studies have shown that miRNAs modulate cancer development by serving as oncogenes or tumor suppressors [9-11]. The expression and function of miRNAs have also been reported in colon cancer. For example, miRNA-143 suppresses colon cancer cell growth through inhibiting glucose uptake and glucose transporter 1 (GLUT1) expression and glucose uptake [12]. miR-185 is down-regulated in colon cancer tissues and its over-expression inhibits the colon cancer proliferation, growth, invasion and migration [13]. Recently, the role of miR-223-3p in tumor growth has been described. For example, miR-223-3p expression is increased in ovarian cancer specimens. miR-223-3p over-expression promotes the proliferation, migration, and invasion of ovarian cancer cells [14]. Reversely, ectopic expression of miR-223-3p could inhibit the invasion, migration, growth, and proliferation in osteosarcoma cells [15]. Therefore, the involvement of miR-223-3p in other cancers, including colon cancer, should be determined. Here, we aimed to investigate the role of miR-223-3p in colon cancer development. Firstly, miR-223-3p was up-regulated in colon cancer tissues. miR-223-3p positively promoted the proliferation, growth, invasion, migration and epithelial-mesenchymal transition (EMT) of colon cancer cells. Furthermore, PRDM1 served as a direct downstream target of miR-223-3p. Our results suggest the oncogenic role of miR-223-3p in colon cancer.
Material and methods
Clinical samples and TCGA database
The colon cancer and paired adjacent normal tissues were collected from 30 cancer patients, who enrolled in the First Hospital of Shanxi Medical University during May 2016 to July 2017. All the patients were diagnosed by the pathologists. The written informed consent was obtained from each patient. All the procedures were approved by the Ethics Committee of First Hospital of Shanxi Medical University. miR-223-3p expression was analyzed from 30 cases of colon cancer and paired normal tissues, and confirmed by the Cancer Genome Atlas (http://cancergenome.nih.gov) database.
Cell culture
Human normal colon epithelial cells (NCM460) and colon cancer cell lines (RKO, HCT116 and SW620) were obtained from the American Type Culture Collection (ATCC). These cells were cultured in 1640 (Hyclone) or Dulbecco’s Modified Eagle dedium (DMEM, Hyclone), which was supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin solution (Corning). All the cells were maintained at 37°C with 5% CO2.
Total RNA extraction and quantitative PCR
The total RNA was extracted from the cells by Trizol reagent (Invitrogen) or Ultrapure RNA Kit (CWBIO), following the manufacturer’s instructions. 0.5 μg RNA was subjected to reversed-transcriptional reaction using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO). Quantitative PCR reaction was conducted on a Bio-rad IQ5 machine using SYBR qPCR master mixture (Transgen). The primer sequences were as following: PRDM1 forward, 5’-TAAAGCAACCGAGCACTGAGA-3’, PRDM1 reverse, 5’-ACGGTAGAGGTCCTTTCCTTTG-3’; and GAPDH forward, 5’-TGACTTCAACAGCGACACCCA-3’, GAPDH reverse, 5’-CACCCTGTTGCTGTAGCCAAA-3. The relative expression was normalized to GAPDH mRNA expression.
Western blot
The treated cell proteins were extracted using lysis buffer (Beyotime) and measured using BCA protein assay kit (Beyotime). Equal protein was separated on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by transferring to polyvinylidenefluoride membranes (PVDF). The membranes were blocked with 5% skimmed milk at room temperature for 60 minutes and incubated with primary antibodies at 4°C overnight. Antibody against PRDM1 and GAPDH were obtained from the Abcam and Abclone, respectively.
Cell proliferation and growth assay
CCK assay was used to detect the viability of colon cancer cells. Briefly, a total of 3000 miRNA-control and miR-223-3p over-expressed RKO cells, miRNA-control and miR-223-3p silenced HCT 116 cells were seeded in 96-well plates. After 1, 2, 3, 4 and 5 days treatment, the CCK reagent was added into each well and maintained at 37°C for 3 hours. Then, the optical density (OD) value was determined by a micro-plate reader at 450 nm.
For colony formation assay, equal HCT 116 or RKO cells transfected with indicated lentivirus were seeded 6-well plates and maintained at 37°C for 9 days. The colonies were washed and fixed with methanol for 30 minutes. Then, the colonies were stained with crystal violet solution for 20 minutes. The colonies were photographed and the number was calculated.
Cell invasion and migration assays
Transwell assay was used to detect the cell migration. RKO or HCT 116 cells (5 * 104/well) were seeded into the upper chamber of a 24-well insert (8-μm pore size; Corning). The upper compartment contained serum-free 1640 medium and the lower chamber was filled with 1640 medium with 10% FBS. After 48 h of incubation, RKO or HCT 116 cells invaded into the lower surface of the insert were fixed with methanol, stained by crystal violet solution and counted under a light microscope.
Wound-healing assay was used to measure the cell migration. RKO or HCT 116 cells were seeded and cultured in the 6-cm plates. When the cells reached to 90% confluence, the wound at the middle of the plates was produced using the pipette tip. The wound was photographed and measured at 0 h and 24 h.
Microarray
Total RNA from RKO cells was extracted using Trizol reagent (Invitrogen). NanoDrop 2000 and Agilent Bio analyzer 2100 were used to detect the RNA quantity and quality. Affymetrix human GeneChip primeview was used for microarray processing to determine gene expression profile according to the manufacturer’s instructions. Significantly different genes between miRNA-control and miR-223-3p over-expressed RKO cells were identified depending on the following criteria: P < 0.05 and the absolute fold change > 1.5.
Luciferase reporter assay
We constructed plasmid vectors containing the 3’UTR of PRDM1 with either a wild-type (WT) or a mutant (MUT) version of the predicted target site. The regions were amplified and cloned into the psi-Check2 dual-luciferase miRNA vector (Promega). Then, the miR-223-3p mimics and psiCheck2 vector/psiCheck2-PRDM1-WT/MUT were co-transfected into RKO cells. The dual-luciferase reporter assay kit (Promega) was used to detect the luciferase activity at 48 hours after transfection.
Statistical analysis
GraphPad prism software was applied for statistical analysis. Difference between two groups was analyzed by un-paired students’ t tests. One-way ANOVA was used for analyzing the difference when more than two groups. P value of less than 0.05 was considered statistical significance.
Results
miR-223-3p is elevated in colon cancer tissues and cells
We firstly investigated the significance of miR-223-3p in colon cancer. The expression of miR-223-3p was increased in colon cancer tissues as compared with their adjacent tissues (Figure 1A, 1B). Consistently, TCGA database shown that miR-223-3p expression was higher in colon adenocarcinoma tissues than that of in normal tissues (Figure 1C). Furthermore, miR-223-3p was also high expression in colon cancer cell lines (RKO, HCT 116 and SW620), compared with the normal colon epithelial cells (Figure 1D). These results suggested that miR-223-3p was significantly elevated in colon cancer specimens and cell lines.
Figure 1.
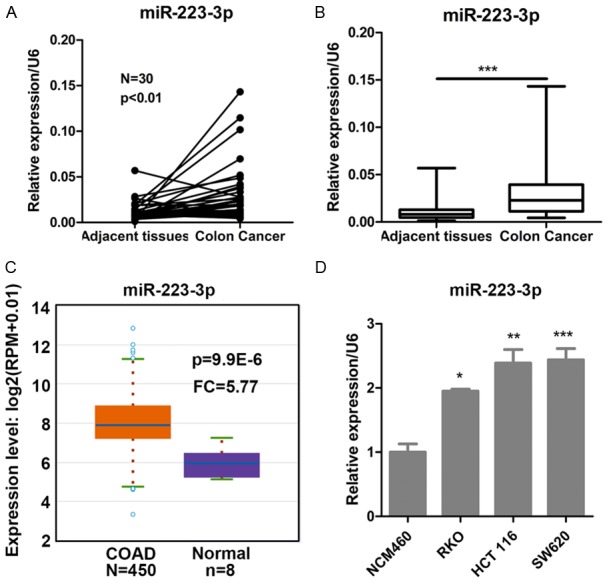
miR-223-3p is elevated in colon cancer tissues and cells. A. Relative expression of miR-223-3p in the cancer and the paired adjacent normal tissues of colon cancer patients. N = 30, P < 0.01 compared with paired adjacent normal tissues. B. Relative expression of miR-223-3p in colon cancer and adjacent normal tissues. ***P < 0.001 compared with adjacent normal tissues. C. TCGA database showed that miR-223-3p was elevated in colon adenocarcinoma (COAD, n = 450) compared with normal tissues (n = 8). P = 9.9E-6 compared with adjacent normal tissues. D. Relative miR-223-3p expression in normal colon epithelial cells (NCM460) and colon cancer cell lines (RKO, HCT116, SW620). *P < 0.05 and **P < 0.01 compared with NCM460 cells.
miR-223-3p enhances the proliferation and reduces the apoptosis in colon cancer cells
To explore the role of miR-223-3p in colon cancer, using lentivirus transfection, we generated the miR-223-3p over-expressed and knocked down in RKO and HCT 116 cells, respectively. qRT-PCR analysis shown that miR-223-3p was efficiently over-expressed in RKO cells (Figure 2A). CCK assay indicated that miR-223-3p increased the proliferation of RKO cells since day 4 (Figure 2B). By contrast, silencing of miR-223-3p reduced the viability of HCT 116 cells (Figure 2C). Then, these cells were subjected to colony formation and apoptosis analysis. We found that miR-223-3p ectopic expression promoted the colony growth of RKO cells, while miR-223-3p silencing suppressed the colony formation of HCT 116 cells (Figure 2D-H). Apoptosis was inhibited by miR-223-3p over-express in RKO cells (Figure 2F). Inversely, miR-223-3p knockdown induced the apoptosis of HCT 116 cells (Figure 2I).
Figure 2.
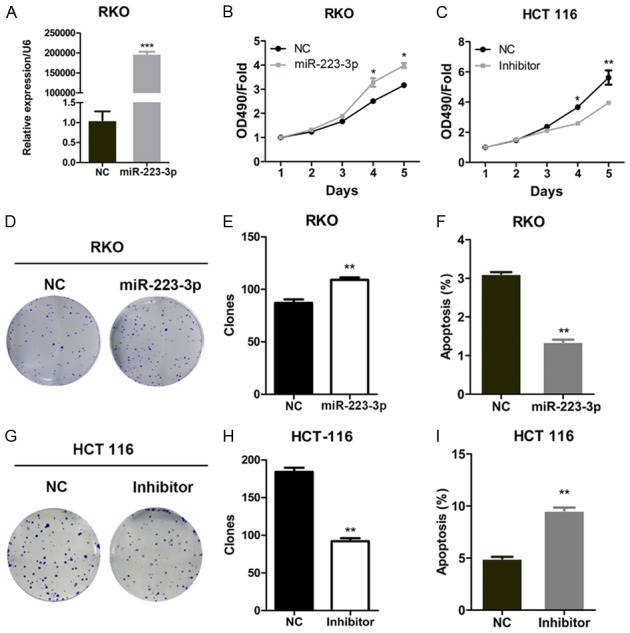
miR-223-3p promotes the proliferation and growth of colon cancer cells. (A and B) Negative control (NC) or miR-223-3p over-expressed RKO cells were subjected to qRT-PCR analysis of miR-223-3p expression (A) and CCK analysis of cell viability (B). *P < 0.05 and ***P < 0.001 compared with NC group. (C) Negative control (NC) or miR-223-3p silenced (Inhibitor) HCT 116 cells were subjected to CCK analysis of cell viability (C). *P < 0.05 and **P < 0.01 compared with NC group. (D and E) Colony formation analysis of NC or miR-223-3p over-expressed RKO cells. Representative images (D) and quantification (E) of colonies. **P < 0.01 compared with NC group. (F) Cells described in (A) were subjected to apoptosis measurement. **P < 0.01 compared with NC group. (G and H) Colony formation analysis of NC or miR-223-3p silenced HCT 116 cells. (G) Representative images (G) and quantification (H) of colonies. **P < 0.01 compared with NC group. (I) Cells described in (C) were subjected to apoptosis measurement. **P < 0.01 compared with NC group.
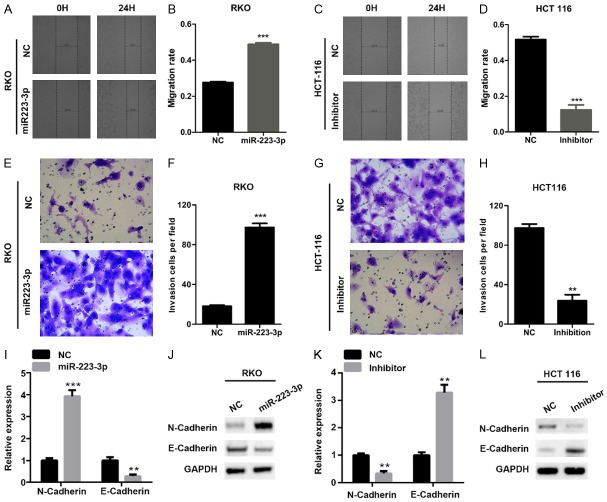
miR-223-3p increases the invasion and migration of colon cancer cells
To investigate the function of miR-223-3p in colon cancer on cell migration and invasion, miR-223-3p over-expressed RKO and silenced HCT 116 cells were subjected to wound-healing and transwell assays. We found that miR-223-3p over-expression promoted the migration of RKO cells, while inverse results were observed in miR-223-3p knockdown HCT 116 cells (Figure 3A-D). Similarly, miR-223-3p ectopic expression increased the invasiveness capability of RKO cells (Figure 3E, 3F). By contrast, miR-223-3p silencing reduced the invasion in HCT 116 cells (Figure 3G, 3H). qRT-PCR and Western blot results shown that miR-223-3p over-expression markedly up-regulated the mesenchymal marker, N-cadherin, and down-regulated the epithelial marker E-cadherin in RKO cells (Figure 3I, 3J), while its knockdown caused an opposite effect in HCT 116 cells (Figure 3K, 3L). Taken together, miR-223-3p promoted the epithelial-mesenchymal transition (EMT), migration and invasion of colon cancer cells.
Figure 3.
miR-223-3p increases the invasion and migration capacity of colon cancer cells. (A and B) The migration of negative control (NC) or miR-223-3p over-expressed RKO cells was determined by wound-healing assay. (A) Representative wound-healing images. (B) Migration rate. ***P < 0.001 compared with NC group. (C and D) The migration of negative control (NC) or miR-223-3p silenced HCT 116 cells was determined by wound-healing assay. (C) Representative wound-healing images. (D) Migration rate. ***P < 0.001 compared with NC group. (E and F) Transwell assay was used to analyze the invasion of RKO cells described in (A). (E) Representative transwell images. (F) Invasion rate. ***P < 0.001 compared with NC group. (G and H) Transwell assay was used to analyze the invasion of HCT 116 cells described in (C). (G) Representative transwell images. (H) Invasion rate. **P < 0.01 compared with NC group. (I and J) qRT-PCR (I) and Western blot (J) analysis of N-Cadherin and E-Cadherin in NC and miR-223-3p over-expressed RKO cells. **P < 0.01 and ***P < 0.001 compared with NC group. (K and L) qRT-PCR (K) and Western blot (L) analysis of N-Cadherin and E-Cadherin in NC and miR-223-3p silenced HCT 116 cells. **P < 0.01 compared with NC group.
PRDM1 is a direct target for miR-223-3p
Above results shown that miR-223-3p was critical for the growth and invasion of human colon cancer cells. Then, RKO cells infected with lentivirus expressing control or miR-223-3p were subjected to microarray assay for the global differential gene expression. We identified 588 differentially expressed genes, including 330 up-regulated genes and 258 down-regulated genes (Figure 4A). To explore the miR-223-3p targeted downstream genes, we queried the TargetScan, microRNA.ORG and miRDB databases. Hundreds of genes were potentially regulated by miR-223-3p (Figure 4B). Since miR-223-3p served as an oncogenic microRNA, we focused on tumor suppressors among the overlap 99 predicted downstream targets. We found a putative binding sites of miR-223-3p in the 3’UTR of PRDM1 (Figure 4C). Western blot result demonstrated that PRDM1 was negatively regulated by miR-223-3p (Figure 4D). Co-transfection with miR-223-3p and wild-type (WT) PRDM1 significantly suppressed the luciferase activity, while miR-223-3p inhibitors and mutant (MUT) PRDM1 co-transfection had marginal effect on luciferase activity (Figure 4E), suggesting that miR-223-3p directly targeted PRDM1. TCGA database shown that PRDM1 was down-regulated in colon adenocarcinoma tissues (Figure 4F). qRT-PCR analysis confirmed that PRDM1 expression was also reduced in colon cancer tissues comparing with the adjacent normal tissues (Figure 4G). These results indicated that PRDM1 was a direct target for miR-223-3p in colon cancer.
Figure 4.
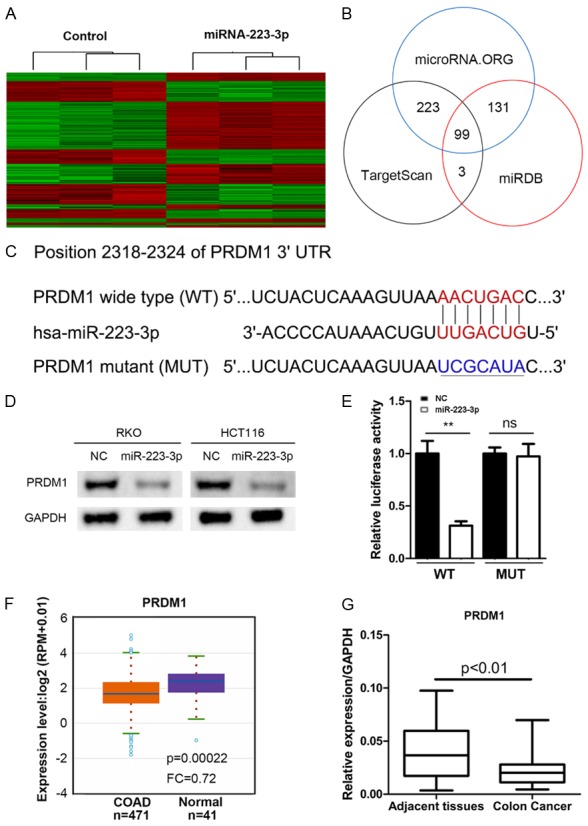
miR-223-3p directly binds to the 3’UTR region of PRDM1. A. Heatmap showed the differentially expressed genes in RKO cells with and without miR-223-3p overexpression. (Criteria: P < 0.05, absolute fold change > 1.5). B. The diagram of miR-223-3p predicted targets based on the microRNA. ORG, TargetScan and miRDB algorithms. C. The putative binding sites for miR-223-3p in the 3’UTR of PRDM1 mRNA. D. Western blot result of PRDM1 in NC and miR-223-3p over-expressed RKO and HCT 116 cells. E. Relative luciferase activity in HCT-116 cells co-transfected with negative control (NC) or miR-223-3p mimic and psiCheck2 vector/psiCheck2-PRDM1-WT/MUT. **P < 0.01 and ns (no significance) compared with NC group. F. TCGA analysis of PRDM1 mRNA expression in colon adenocarcinoma (COAD) and normal tissues. P = 0.00022 compared with normal group. G. qRT-PCR analysis of PRDM1 expression in colon cancer and adjacent normal tissues. P < 0.01 compared with adjacent tissues.
Discussion
For the past decades, the prognostic value and precise function of miRNAs have been widely investigated in colon cancer progression, either by acting as oncogenes or tumor suppressors [16-20]. In this study, we observed that miR-223-3p was elevated in colon cancer tissues and cells. miR-223-3p ectopic expression promoted the proliferation, growth, invasion and migration and EMT, while inverse results were observed in miR-223-3p knockdown HCT 116 cells. Cell apoptosis was negatively regulated by miR-223-3p in colon cancer cells.
Increasing of studies have reported the up-regulation of miR-223 in cancer tissues compared with their normal tissues, including gastric cancer [21], breast cancer [22], lung cancer [23], and pancreatic cancer [24]. Likewise, miR-223-3p expression is increased in ovarian cancer tissues and promotes the proliferation, migration, and invasion of ovarian cancer cells [14]. However, miR-223-3p was also found down-regulated and negatively regulated cell proliferation and migration in osteosarcoma and glioblastoma [15,25]. Nevertheless, the involvement of miR-223-3p in colon cancer remains unknown. In the present study, we shown that miR-223-3p was significantly up-regulated in cancer tissues as compared with the adjacent normal tissues in 30 pairs of the patients. TCGA database was consistent with the results. Comparing with the normal colon epithelial cells, miR-223-3p abundance was increased in colon cancer cell lines RKO, HCT 116 and SW620. Based on lentivirus-mediated over-expression and knockdown, we demonstrated that miR-223-3p activated the proliferation, growth and EMT, but suppressed apoptosis of colon cancer cells. Therefore, we suggested that miR-223-3p served as an oncogenic microRNA in colon cancer.
PRDM1, also known as “B lymphocyte-induced maturation protein” (Blimp-1), is a zinc finger protein that regulates the transcription of downstream genes [26]. Recently, PRDM1 has been identified as a tumor suppressor in various cancers [27]. Inactivation of PRDM1 is involved in the poor prognosis of the patients with activated B-cell-like diffuse large B-cell lymphoma [28]. In colon cancer patients, PRDM1 low expression predicts poor survival. Ectopic expression of PRDM1 prevents the formation and growth of colon cancer organoids [29]. In addition, genkwadaphnin inhibits the cell cycle progression and proliferation of colon cancer cells through inducing PRDM1 expression [30]. However, the correlation between miR-223-3p and PRDM1 I colon cancer is unclear. Here, based on microarray assay, we shown that numerous genes were dysregulated after miR-223-3p, among which included PRDM1. Western blot shown that miR-223-3p over-expression suppressed the expression of PRDM1, while its knockdown promoted PRDM1 expression. Luciferase results indicated that miR-223-3p directly binds the wide-type 3’UTR region of PRDM1, but not the mutant PRDM1. Furthermore, TCGA and our clinical data showed that PRDM1 mRNA level was reduced in colon cancer tissues as compared with the normal tissues. These result indicated that miR-223-3p promoted colon cancer development at least partly through repressing the expression of PRDM1.
In summary, we provided for the first evidence that miR-223-3p functioned as an oncogenic microRNA in colon cancer. miR-223-3p promotes the proliferation, growth, migration, invasion and EMT. Inverse results were observed in miR-223-3p silencing colon cancer cells. Furthermore, our findings showed that miR-223-3p promoted colon cancer development through negatively regulating PRDM1.
Acknowledgements
This work was sponsored by grants from the Shanxi Provincial International Collaborative Program (2015081038) and the Shanxi Provincial Youth Fund (201701D221176).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Alwan A. World Health Organization. Disaster Med Public Health Prep. 2007;1:7–8. doi: 10.1097/DMP.0b013e3180676d32. [DOI] [PubMed] [Google Scholar]
- 3.Chau I, Cunningham D. Adjuvant therapy in colon cancer--what, when and how? Ann Oncol. 2006;17:1347–1359. doi: 10.1093/annonc/mdl029. [DOI] [PubMed] [Google Scholar]
- 4.Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai Q, Zeng C, Schmit SL, Shin A, Matsuo K, Jee SH, Kim DH, Kim J, Wen W, Shi J, Guo X, Li B, Wang N, Zhang B, Li X, Shin MH, Li HL, Ren Z, Oh JH, Oze I, Ahn YO, Jung KJ, Conti DV, Schumacher FR, Rennert G, Jenkins MA, Campbell PT, Hoffmeister M, Casey G, Gruber SB, Gao J, Gao YT, Pan ZZ, Kamatani Y, Zeng YX, Shu XO, Long J, Matsuda K, Zheng W. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156:1455–1466. doi: 10.1053/j.gastro.2018.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakayama M, Oshima M. Mutant p53 in colon cancer. J Mol Cell Biol. 2019;11:267–276. doi: 10.1093/jmcb/mjy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandellini P, Rancati T, Valdagni R, Zaffaroni N. miRNAs in tumor radiation response: bystanders or participants? Trends Mol Med. 2014;20:529–539. doi: 10.1016/j.molmed.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q, Yang Z, Shi Y, Fan D. MiRNAs in human cancers: the diagnostic and therapeutic implications. Curr Pharm Des. 2014;20:5336–5347. doi: 10.2174/1381612820666140128204914. [DOI] [PubMed] [Google Scholar]
- 11.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Chen Y, Liu F, Yin M. Overexpression of miRNA-143 inhibits colon cancer cell proliferation by inhibiting glucose uptake. Arch Med Res. 2018;49:497–503. doi: 10.1016/j.arcmed.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Sun Z, Su L, Wang F, Jiang Y, Yu D, Zhang F, Sun Z, Liang W. miRNA-185 serves as a prognostic factor and suppresses migration and invasion through Wnt1 in colon cancer. Eur J Pharmacol. 2018;825:75–84. doi: 10.1016/j.ejphar.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Fang G, Liu J, Wang Q, Huang X, Yang R, Pang Y, Yang M. MicroRNA-223-3p regulates ovarian cancer cell proliferation and invasion by targeting SOX11 expression. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18061208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ, Wang Y. miR-223-3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther. 2018;26:1299–1312. doi: 10.1016/j.ymthe.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, Liu F, Dong L, Yang H, He X, Li L, Zhao L, Jin S, Li G. miR-1273g silences MAGEA3/6 to inhibit human colorectal cancer cell growth via activation of AMPK signaling. Cancer Lett. 2018;435:1–9. doi: 10.1016/j.canlet.2018.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Smith AR, Marquez RT, Li J, Li K, Lan L, Wu X, Zhao L, Ren F, Wang Y, Wang Y, Jia B, Xu L, Chang Z. MicroRNA-383 acts as a tumor suppressor in colorectal cancer by modulating CREPT/RPRD1B expression. Mol Carcinog. 2018;57:1408–1420. doi: 10.1002/mc.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao JP, Ma A. The ratio of miR-21/miR-24 as a promising diagnostic and poor prognosis biomarker in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:8649–8656. doi: 10.26355/eurrev_201812_16629. [DOI] [PubMed] [Google Scholar]
- 19.Shirafkan N, Shomali N, Kazemi T, Shanehbandi D, Ghasabi M, Baghbani E, Ganji M, Khaze V, Mansoori B, Baradaran B. microRNA-193a-5p inhibits migration of human HT-29 colon cancer cells via suppression of metastasis pathway. J Cell Biochem. 2018 doi: 10.1002/jcb.28164. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, Hassan EA, Fathy W, El-Deek HEM. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1alpha-VEGF signaling pathway. Mol Cell Biochem. 2019;454:177–189. doi: 10.1007/s11010-018-3462-1. [DOI] [PubMed] [Google Scholar]
- 21.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, Xiao B. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, Liu C, Xu L, Wang J, Shen H, Zhang J, Zen K, Zhang CY. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 24.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 25.Ding Q, Shen L, Nie X, Lu B, Pan X, Su Z, Yan A, Yan R, Zhou Y, Li L, Xu J. MiR-223-3p overexpression inhibits cell proliferation and migration by regulating inflammation-associated cytokines in glioblastomas. Pathol Res Pract. 2018;214:1330–1339. doi: 10.1016/j.prp.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 27.Mzoughi S, Tan YX, Low D, Guccione E. The role of PRDMs in cancer: one family, two sides. Curr Opin Genet Dev. 2016;36:83–91. doi: 10.1016/j.gde.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Xu-Monette ZY, Tzankov A, Li X, Manyam GC, Murty V, Bhagat G, Zhang S, Pasqualucci L, Visco C, Dybkaer K, Chiu A, Orazi A, Zu Y, Richards KL, Hsi ED, Choi WW, van Krieken JH, Huh J, Ponzoni M, Ferreri AJ, Moller MB, Parsons BM, Winter JN, Piris MA, Westin J, Fowler N, Miranda RN, Ok CY, Li Y, Li J, Medeiros LJ, Young KH. Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of activated B-cell-like diffuse large B-cell lymphoma. Leukemia. 2017;31:625–636. doi: 10.1038/leu.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Banister CE, Weige CC, Altomare D, Richardson JH, Contreras CM, Buckhaults PJ. PRDM1 silences stem cell-related genes and inhibits proliferation of human colon tumor organoids. Proc Natl Acad Sci U S A. 2018;115:E5066–E5075. doi: 10.1073/pnas.1802902115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang HB, Lee HR, Jee da J, Shin SH, Nah SS, Yoon SY, Kim JW. PRDM1, a tumor-suppressor gene, is induced by genkwadaphnin in human colon cancer SW620 cells. J Cell Biochem. 2016;117:172–179. doi: 10.1002/jcb.25262. [DOI] [PubMed] [Google Scholar]