Abstract
Breast cancer is one of the most common cancers with the highest morbidity and mortality among women despite the treatment approaches have advanced including surgery, endocrine therapy and targeted therapy. Novel biomarkers are warranted to be discovered for the early detection, treatment and prognosis for breast cancer. CircRNA is a class of covalently closed single-stranded circular RNA molecules without free 5’ or 3’ end which makes them well expressed and more stable than their linear counterparts. In this review, we mainly discuss the oncogenic or anti-oncogenic roles of circRNAs can be utilized in the treatment and prognosis of breast cancer. A large number of circRNAs have shown great potential to function in carcinogenesis, metastasis or chemoresistance of breast cancer through transcriptional regulation of RNAs including miRNA and mRNA, in addition to their promise as stable biomarkers that can be used for monitoring breast cancer progression. However, the translation phenomenon of circRNAs in breast cancer and the diagnostic value of circRNAs in breast cancer requires further investigation for which the detection of circRNAs in plasma exosomes could be worthy of a try. Above all, engineered exosomes preloaded with engineered anti-oncogenic circRNAs are likely to provide a novel direction in the personal medicine of breast cancer.
Keywords: CircRNA, breast cancer, metastasis, chemoresistance, exosome, fusion-circRNA
Background
Breast cancer (BC) is a globally common cancer which is the most frequently diagnosed cancer in the vast majority of the countries and the leading cause of cancer death among females in over 100 countries [1]. According to the newly data from Global Cancer Statistics 2018, over 2 million females are newly diagnosed with BC (24.2% of the total cases) and more than 60 thousand deaths are caused by BC (15.0%). In addition, incidence rates for BC have been rising in the last decade and have far exceeded those for other cancers in both developed and developing countries. Despite the surgery, endocrine therapy and targeted therapy, the morbidity and mortality of BC remain highest in female patients. Novel biomarkers are warranted to be discovered for the early detection, treatment and prognosis for BC.
Circular RNAs (circRNAs) were discovered in plants and shown to encode subviral agents [2]. It is a class of covalently closed single-stranded circular RNA molecules formed by back-splicing. CircRNA can escape exonucleic acid degrading enzymes due to lack of a free 5’ or 3’ end which makes them well expressed and more stable than their linear counterparts. Alternative circularization coupled with alternative splicing can produce a variety of additional circRNAs from one gene, making such molecules have enriched sequences in conserved nucleotides [3]. There are basically four types of circRNAs including exon circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), exon-intron circRNAs (EIciRNAs), intergenic circRNAs or fusion circRNAs (f-circRNAs), which means they derive often but not always from coding exons [4-7].
A vast number of circRNAs have been discovered in a variety of cancers and they are activated in either inhibiting tumor progression or promoting tumorigenesis. On one hand, circ-PTK2 inhibits TGF-β-induced epithelial mesenchymal transition (EMT) and metastasis by controlling TIF1γ in non-small cell lung cancer (NSCLC) [8]. Circ-MTO1 inhibits Hepatocellular Carcinoma (HCC) progression [9]. Circ-LARP4 suppress cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression [10]. Circ-FBXW7 in represses glioma tumorigenesis and its expression positively associated with glioblastoma patient overall survival [11]. Circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression [12]. On the other hand, circRNA PRMT5 promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30 c to induce epithelial - mesenchymal transition [13]. Circ-CCDC66 promotes colon cancer growth and metastasis [14]. In addition to solid tumor, circRNAs are also found in hematologic malignancies like acute myeloid leukemia [15]. Moreover, discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA further indicate the prominent and extensive potential of circRNAs in malignancies [16].
Main functions of circRNA
CircRNAs can exert various functions according to their parental genes [17], herein we mainly discuss the functions of circRNAs as miRNA sponges, in regulation of parental genes and translation.
miRNA sponges
MicroRNAs (miRNAs) are important post-transcriptional regulators of gene expression that acts by direct base pairing to target sites within untranslated regions of messenger RNAs. In the recent decade, miRNA activity has been proved to be affected by miRNA sponge transcripts, as known as competing endogenous RNA (ceRNA). circRNA contains selectively conserved miRNA target sites that can strongly suppresses miRNA activity, resulting in suppressed effects of miRNAs [18]. A circRNA can serves as a sponge for multiple miRNAs. For instance, circHIPK3 can sponge 9 miRNAs which all have been reported to serve as tumor-suppressive miRNAs and thus regulate cancer cell growth [19]. So far, a few circRNAs have been found in BC to serve as miRNA sponges and the detailed information is listed in Table 1.
Table 1.
Functional circRNA-miRNA-mRNA axis in breast cancer
| circRNA | Parental gene | Sponged miRNA | Targeted mRNA | Specimen type | Pairs of specimens | Reference |
|---|---|---|---|---|---|---|
| Oncogenic | ||||||
| hsa_circ_0000479 | EPSTI1 | miR-4753/6809 | BCL11A | Tissue | 37 | [37] |
| hsa_circ_0001783 | EBLN3 | miR-200c-3p | ZEB1/2, ETS1 | Tissue | 18 | [35] |
| hsa_circ_0001846 | UBAP2 | miR-661 | MTA1 | Tissue | 78 | [38] |
| hsa_circ_0001982 | RNF111 | miR-143 | N/A | Tissue | 29 | [33] |
| hsa_circ_0005230 | DNM3OS | miR-618 | CBX8 | Tissue | 76 | [43] |
| hsa_circ_0005239 | GFRA1 | miR-34a | GFRA1 | Tissue | 51 | [20] |
| hsa_circ_0005505 | IRAK3 | miR-3607 | FOXC1 | Tissue | 35 | [25] |
| hsa_circ_0006528 | PRELID2 | miR-7-5p | Raf1 | Tissue | 20 | [45] |
| hsa_circ_0007294 | ANKS1B | miR-148a-3p | USF1 | Tissue | 20 | [32] |
| miR-152-3p | ||||||
| hsa_circ_0007534 | DDX42 | miR-593 | MUC19 | Tissue | 40 | [40] |
| hsa_circ_0008039 | PRKAR1B | miR-432-5p | E2F3 | Tissue | 38 | [36] |
| hsa_circ_0008717 | ABCB10 | miR-1271 | N/A | Tissue | 32 | [34] |
| hsa_circ_0011946 | SCMH1 | N/A | RFC3 | Tissue | 12 | [39] |
| hsa_circ_0052112 | ZNF83 | miR-125a-5p | ZNF83 | N/A | N/A | [23] |
| hsa_circ_0058514 | AGFG1 | miR-195-5p | CCNE1 | Tissue | 40 | [44] |
| hsa_circ_0072995 | ARHGEF28 | miR-30c-2-3p | ARHGEF28 | N/A | N/A | [24] |
| Anti-oncogenic | ||||||
| N/A | ITCH | miR-214/miR-17 | ITCH | Tissue | 91 | [21] |
| hsa_circ_0000911 | IFNGR2 | miR-449a | Notch1 | Tissue | 35 | [51] |
| hsa_circ_0001098 | BARD1 | miR-3942 | BARD1 | Tissue | 15 | [22] |
| hsa_circ_0007874 | MTO1 | N/A | TRAF4/Eg5 | N/A | N/A | [46] |
| hsa_circ_0087378 | NTRK2 | miR-1260b | SFRP1 | Tissue | 23 | [52] |
Regulation of parental genes
CircRNAs can regulate their parental genes directly or indirectly. Indirectly, some circRNAs regulate parental genes through the ceRNA mechanism. For instance, circGFRA1 might function as miR-34a sponge to regulate parental mRNA GFRA1 expression [20]. Circ-ITCH regulates ITCH by sponging miR-214 and miR-17 [21], has-circ-0001098 regulates BARD1 by sponging miR-3942 [22], hsa_circ_0052112 regulates ZNF83 by sponging miR-125a-5p [23], hsa_circ_0072995 regulates ARHGEF28 by sponging miR-30c-2-3p [24] and thus impact the expression of their parental mRNAs in BC. Intriguingly, some circRNA like circ_IRAK3 regulates IRAK3 through a positive-feedback loop circ_IRAK3/miR-3607/FOXC1/IRAK3 loop in which the target mRNA FOXC1 binds to the promotor of parental gene IRAK3 [25]. Directly, circRNAs can regulate their parental genes in cis. On one hand, circRNAs like circ_CNOT2 which include the start-codon could potentially influence the expression of the linear gene because the linear transcript from which the circRNA was spliced is now forced to use another start codon for its translation, which makes some circRNAs negatively correlated with their parental mRNAs [26]. On the other hand, EIciRNAs, a kind of circRNAs circularized with introns ‘retained’ between exons, have been found predominantly localized in nucleus and can enhance the expression of their parental genes in cis by interacting with Pol II, U1 snRNP and parental gene promoters [6].
Translation
CircRNAs have been predominantly regarded as a type of non-coding RNA since the advent of high-throughput RNA sequencing (RNA-seq). Most circular RNA like circ-CCDC66 promotes cancer growth and metastasis through noncoding mechanism [14]. However, a subset of circRNAs contains an open reading frame and is translated. Pamudurti et al. verified in Drosophila, mouse and rat and showed that a subset of circRNAs like circ-Mb1 generally share the start codon with the hosting RNA, encode proteins with specific protein domains, and are translated in a cap-independent manner [27]. In another study, circ-ZNF609 can also be translated into a protein in a splicing dependent/cap-independent manner. Driven by internal ribosome entry site, circ-FBXW7 encodes a novel protein termed FBXW7-185aa and thus inhibited proliferation and cell cycle acceleration [11]. Moreover, translation of circRNAs is likely to be regulated by multiple mechanisms [28]. For instance, translation of circMb1 may be regulated by starvation and FOXO and that of circ-ZNF609 can be modulated by stress conditions [27,28]. Whether circRNAs founded in breast cancer can be translated remains unknown.
Regulation of circRNAs biogenesis
CircRNAs were commonly recognized as the production of back-splicing of pre-mRNA transcripts and can resistance against RNase R. To the best of our knowledge, biogenesis of circRNA can be regulated in several mechanisms. (1) RNA binding protein (RBP): Simon J. Conn observed that the level of RNA binding protein Quaking (QKI) is regulated in the EMT and the QKI can regulate circRNAs in EMT [29]. During the process, the QKI acts by binding to recognition elements in the introns that near the circRNA-forming splice sites. Besides, the insertion of QKI motifs can also induce circRNA formation; (2) Competitive synthesis mediated by the general splicing factors: The splicing factor muscleblind (MBL) can promote circRNA biogenesis, which is based on the abundant MBL binding sites in introns flanking sequence and that its protein can directly interact with circ-Mbl and balance its own mRNA and circ-Mbl production [30]. Moreover, it was found that once the canonical pre-mRNA processing event was disturbed it will shift toward circRNA production [31]; (3) Positive circRNA/miRNA/mRNA loop: Circ_ANKS1B (hsa-circ-0007294) abundantly sponges miR-148a-3p and miR-152-3p to increase the expression of transcription factor USF1 while circ-ANKS1B biogenesis in BC was promoted by splicing factor ESRP1 whose expression was also regulated by USF1 [32]. Similarly, circ-IRAK3 expression can be enhanced by its targeting mRNA FOXC1 binding to its parental gene IRAK3 promoter [25]. These regulation mechanisms of circRNAs may help us to better understand and utilize the functions of different circRNAs and we believe that there could be more mechanistic regulation of circRNA which remain to be discovered.
CircRNAs validated in BC
Recently, an increasing number of circRNAs have been discovered in BC through next-generation sequencing and circRNA microarray and have been proven to show potential in diagnosis, treatment and prognosis. These circRNAs are activated in BC progression including carcinogenesis, metastasis and chemoresistance (Figure 1).
Figure 1.
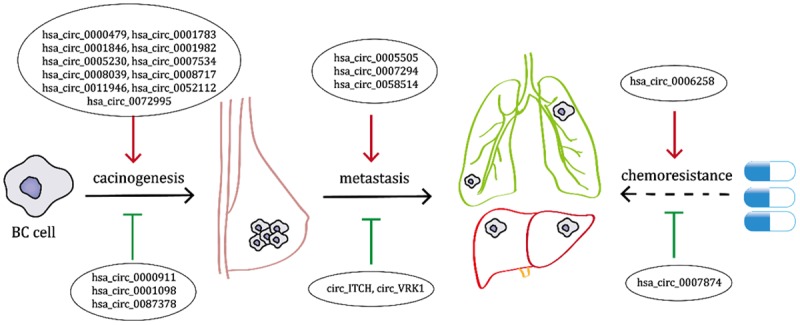
CircRNAs that are found activated in BC progression including carcinogenesis, metastasis and chemoresistance.
Carcinogenesis associated circRNAs
Among all the validated aberrantly expressed circRNAs in BC, the upregulated circRNAs are more often found to serve as oncogenic circRNAs or vice versa. For instance, hsa_circ_0001982 is upregulated in BC and promotes BC cell carcinogenesis through decreasing miR-143 (microarray, in vitro) [33]. Similarly, circ_ABCB10 (hsa_circ_0008717) promotes BC proliferation and progression through sponging miR-1271 [34]. More convincingly, hsa_circ_001783 regulates BC progression (in vitro) via sponging miR-200c-3p to regulate ZEB1/2 and ETS1 and correlated with poor clinical outcomes in BC patients [35]. Hsa_circ_0008039 depletion significantly suppressed the proliferation (in vivo), arrested cell-cycle progression and reduced migration mechanistically as ceRNA of miR-432-5p which functionally regulated E2F3 [36]. In TNBC, Circ_EPSTI1 (hsa_circ_000479)-miR-4753/6809-BCL11A axis was proven to affect the proliferation and apoptosis of TNBC (in vivo) also through the mechanism of ceRNA. In addition, circ_EPSTI1 serve as an independent prognostic marker for survival in patients with TNBC [37]. Another in vivo study showed that upregulation of circ_UBAP2 (hsa_circ_0001846) promotes TNBC progression through the miR-661/MTA1 pathway (in vivo) and also predicts poor prognosis [38]. Biomathematical prediction and in vitro experiments manifested downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3 (RNA-seq) [39]. Downregulation of hsa_circ_0007534, having been proven up-regulated in CRC and glioma tissue, also suppresses BC cell proliferation and invasion by targeting miR-593/MUC19 axis which is different from that in glioma [40-42]. More circRNA regulation axis have been and are being validated in vitro, including hsa_circ_0052112/miR-125a-5p/ZNF83 [23], hsa_circ_0072995/miR-30c-2-3p/ARHGEF28 (in vitro) [24], hsa_circ_0005230/miR-618/CBX8 [43] and so on.
Metastasis associated circRNAs
Not only do circRNAs participate in the carcinogenesis of BC, but also play pivotal roles in metastasis of BC. Circ_IRAK3 (hsa_circRNA_0005505) was greatly upregulated in metastatic BC cells, including brain metastatic cells, bone metastatic cells and lung metastatic cells, and is correlated with a high risk of recurrence in distant organs and the poor prognosis of BC patients [25]. Consistently, the knockdown of circ_IRAK3 can obviously repress BC lung metastasis in vivo, and FOXC1 expression was simultaneously decreased in the BC lung metastatic lesions. Circ_ANKS1B (hsa_circ_0007294) abundantly sponges miR-148a-3p and miR-152-3p to increase the expression of transcription factor USF1 which can transcriptionally up-regulate TGF-β1 expression, subsequently activating TGF-β1/Smad signaling to promote EMT [32]. Circ_AGFG1 (hsa_circ_0058514) acts as a sponge of miR-195-5p to promote TNBC proliferation, mobility and invasion as well as tumorigenesis and metastasis through regulating CCNE1 expression in vivo [44]. Therefore, circRNA including circ_IRAK3, circ_ANKS1B and circ_AGFG1 might act as potential predictors and therapeutic targets for metastatic BC.
Chemoresistance associated circRNAs
A few studies which analyzed the expression of circRNAs in chemoresistant breast cancer revealed that circRNAs may play a role in either promoting or reversing BC chemoresistance. Hsa_circ_0006528 was identified as upregulated in Adriamycin (ADM) resistant MCF-7 and MDA-MB-231 cells and validated in ADM-resistant tissues. Bioinformatic and correlation analysis showed that circ_0006528/miR-7-5p/Raf1 might be an active axis participating in chemotherapeutic resistance in BC, which requires further verification and functional analysis [45]. Circ_MTO1 (hsa_circ_0007874), downregulated in monastrol resistant cell lines (MCF-7R and MDA-MB-231R), can suppress BC cell viability and promote monastrol-induced cytotoxicity through regulating the TRAF4/Eg5 axis by targeting Eg5 protein and sequestering TRAF4 from binding to Eg5 gene [46]. Still, our knowledge of the role of circRNAs in chemotherapeutic resistance is far limited due to the lack of deep mechanistic investigations and in vivo studies. Whether circRNAs could be a future direction to overcome BC chemoresistance requires further exploration.
Anti-oncogenic circRNAs
In previous studies, most circRNAs proven downregulated in BC play anti-oncogenic roles in BC progression via ceRNA mechanisms. Circ-ITCH is a renowned anti-oncogenic circRNA involved in cancer progression and it is markedly down-regulated in esophageal squamous cell carcinoma [47], colorectal cancer [48], lung cancer [49], glioma [50] and bladder cancer [12]. Similarly, Circ-ITCH can regulate TNBC progression suppresses TNBC growth and metastasis in vivo by up-regulating the expression of its parental gene ITCH through sponging miR-214 and miR-17, and thereby activating Wnt/β-catenin pathway [21]. Hsa_circ_000911 exerts its anti-oncogenic function by sponging miR-449a and thereby elevating the expression of Notch1 and promoting the function of the NF-κB signaling pathway [51]. Hsa_circ_0087378 was also validated to be downregulated in ER-positive BC. Bioinformatic and correlation analysis showed that hsa_circ_0087378/miR1260b/SFRP1 axis was proposed to be a key regulatory pathway [52]. Zhao et al. found an up-regulated anti-oncogenic circRNA Circ_BARD1 (hsa_circ_0001098) which is induced by TCDD treatment in BC cells. Also, circ_BARD1 can inhibit cell proliferation, block cell cycle, enhance apoptosis and restrain tumor growth in vivo via miR-3942/BARD1 axis [22]. Some circRNAs suppress BC progression through breast cancer stem cells (BCSCs). BCSCs have capacities including self-renewal and pluripotency, which are considered as the cause of treatment failure and are liable for metastatic dissemination. Yan et al. discovered a list of aberrantly expressed circRNAs in BCSCs and matched non-BCSCs and validated that loss of circ_VRK1 can enhance BCSCs’ expansion capacities [53]. Together, anti-oncogenic circRNAs may serve as a promising therapeutic target for patients with BC and provide a future direction in the development of a novel treatment strategy for BC.
All these circRNAs involved in BC progression were corroborated to act as miRNA sponge to regulate their target gene. However, rarely has research further investigate the deeper mechanisms in the regulation of these circRNAs. Although some circRNAs are common in several cancers such as circ_ITCH [12,21,47,49], hsa_circ_0007534 [40,42], hsa_circRNA_0002908 hsa_circRNA_0000950 and so on [54], few of these circRNAs has been proven specific in BC instead of other malignancies. Moreover, a clinical trial on the diagnostic value of circRNA in cardiac disease is ongoing (https://clinicaltrials.gov/ct2/show/NCT03170830?term=circRNA&rank=1), which indicates whether circRNA could serve as diagnostic biomarker for BC should also be taken in to consideration. In addition, circRNAs involving complex background of BC like hypoxic environment are rarely discovered except for HIF1α-associated circ_DENND4C which promotes proliferation of BC cells [55]. Together, more researches are warranted to find more BC-associated and BC-specific circRNAs and further investigate the mechanisms behind the regulation of circRNA.
Future exploration
Mystery of F-circRNA
It has been well-established that cancer-associated chromosomal translocations give rise to fusion proteins [56,57]. Similarly, fusion-circRNAs (f-circRNAs) are produced from transcribed exons of distinct genes affected by aberrant chromosomal translocations. Some f-circRNAs, like f-circM9 (MLL/AF9) and f-circEA-2a (EML4/ALK) have been proven to be oncogenic circRNAs that not only contribute to cellular transformation and promote cell viability but also confer resistance to therapy and have tumor-promoting properties in models of leukemia and lung cancer respectively [5,58]. Apparently, our knowledge of f-circRNAs is limited. Do f-circRNAs all play oncogenic roles in cancer? How about the expression and functions of f-circRNAs in breast cancer? Whether engineered f-circRNAs with pivotal anti-oncogenic nucleic acid fragments can be synthesized and serve as innovative treatment method? More evidences are warranted to further elucidate the role of f-circRNAs.
Detection of circRNA in BC plasma exosome
Most circRNAs are detected in cell of tissue RNA sample in previous studies on BC, but few is done in plasma (Table 1). Detection of circRNA in BC plasma exosome is worthy of trying. Exosomes are 30-100 nm sized microvesicles secreted by most cells, containing a variety of functional proteins and RNAs and have been found in both the blood of healthy individuals and patients with malignancies. Exosomes can be isolated from the blood through various methods, including chemical binding, immunoaffinity capture and differential ultracentrifugation and serve as biomarkers for diagnosis and the monitoring of tumour progression [59]. Over 58 330 circRNAs, 15 501 lncRNAs and 18 333 mRNAs have been identified in exosomes with expression level and possible original tissue [60]. More than 1,000 circRNAs were identified in human serum exosomes, including circ-KLDHC10 identified in serum of CRC patient, which means circRNAs originated from cancer could enter the circulation and be readily measured in the serum [61] Circ-PDE8A was first identified in liver-metastatic pancreatic cells and furtherly analyzed in exosome. Exosome circRNA secreted from adipocytes like circ-deubiquitination promotes the growth of HCC [62]. Intriguingly, exosomal circRNA derived from tumor like ciRS-133 can promote white adipose browning. However, there are 12,316 lncRNAs and 17,102 mRNAs but no circRNAs yet having been identified in exosomes in BC (www.exorbase.org). Whether exosome circRNA also exist in serum of BC patients? Do adipocytes secrete circRNA and thus regulate the progress of BC? More relevant investigations are warranted to elucidate these questions to further confirm whether the innovation of cancer exosomal circRNA-based diagnostic routines can be applied in the clinic for BC. Since engineered exosomes can be therapeutic options for BC [59], whether engineered exosomes preloaded with engineered anti-oncogenic circRNAs would potentiate a route towards the development of personalized medicine for BC is worthy of exploration (Figure 2).
Figure 2.
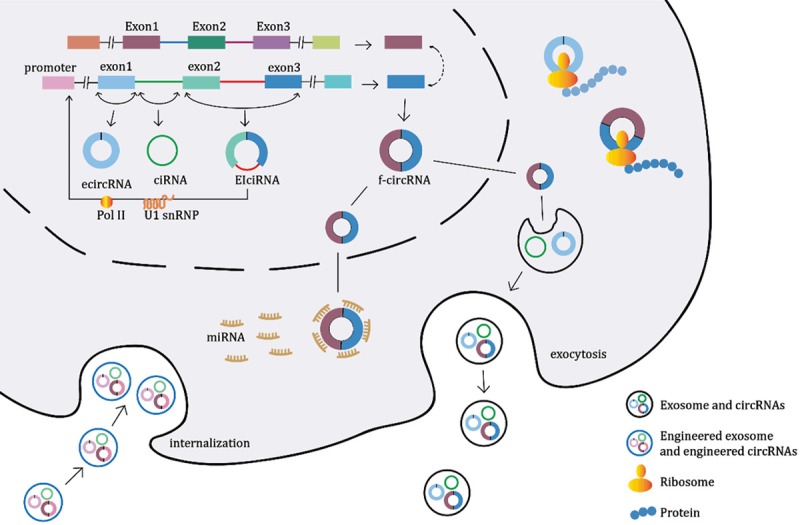
Engineered exosomes preloaded with engineered circRNAs may serve as a therapeutic option for BC. The formation and secretion of circRNAs.
Conclusions
In summary, a large number of circRNAs, including circ_ANKS1B, circ_AGFG1, circ_IRAK3, circ_GFRA1 and circ_EPSTI1, show great potential to function in carcinogenesis, metastasis or chemoresistance of breast cancer via transcriptional regulation of RNAs including miRNA and mRNA, in addition to their promise as stable biomarkers that can be used for monitoring breast cancer progression. The oncogenic or anti-oncogenic roles of circRNAs can be utilized in the treatment and prognosis of breast cancer. However, the translation phenomenon of circRNAs in breast cancer and the diagnostic value of circRNAs in breast cancer requires further investigation for which the detection of circRNAs in plasma exosomes could be worthy of trying. Moreover, engineered exosomes preloaded with engineered anti-oncogenic circRNAs may are likely to provide a novel direction in the personal medicine of breast cancer.
Acknowledgements
This work was supported by an individualized support project for original scientific research of Fudan University (XM03180313), a Grant (SHDC12014207) from the Shen Kang Hospital Development Center Foundation, and a Grant (14-E-34) from the Funded Project of Baoshan District Science and Technology Commission, Shanghai.
Disclosure of conflict of interest
None.
Abbreviations
- BC
breast cancer
- BCSC
breast cancer stem cell
- CRC
colorectal cancer
- ecircRNAs
exon circRNAs
- ciRNAs
circular intronic RNAs
- EIciRNAs
exon-intron circRNAs
- NSCLC
non-small cell lung cancer
- TCDD
tetrachlorodibenzo-p-dioxin
- TNBC
triple negative breast cancer
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly basepaired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 7.Han B, Chao J, Yao HH. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, Su Z, Zhang C, Liu X, Xu G, Zhang HT. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2019;111:435. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, Xiao KH, Liu ZW, Luo JH, Zhou FJ, Xie D. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch S, Blatte TJ, Grasedieck S, Cocciardi S, Rouhi A, Jongen-Lavrencic M, Paschka P, Kronke J, Gaidzik VI, Dohner H, Schlenk RF, Kuchenbauer F, Dohner K, Dolnik A, Bullinger L. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica. 2017;102:2039–2047. doi: 10.3324/haematol.2017.172866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquina AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R, Ziegelbauer JM. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A. 2018;115:12805–12810. doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He RF, Liu P, Xie XM, Zhou YJ, Liao QJ, Xiong W, Li XL, Li GY, Zeng ZY, Tang HL. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, Wang R, Wei Q, Kang YH, Meng R, Feng XH. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. 2019;66:232–239. doi: 10.4149/neo_2018_180710N460. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Zou H, Han C, Ma J, Zhao J, Tang J. Circlular RNA BARD1 (Hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle. 2018;17:2731–2744. doi: 10.1080/15384101.2018.1556058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhou SY, Zhu LP, Li J, Wang DD, Sun DW, Ji ZL, Tang JH. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342–1353. doi: 10.1016/j.biopha.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP, Wang DD, Yang SJ, He YJ, Mao CF, Yang Y, Wang JY, Zhang Q, Xu HZ, Yu DD, Zhao JH, Tang JH, Ji ZL. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10:1229–1242. doi: 10.2217/epi-2018-0002. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Jiang ZR, Chen C, Hu QS, Fu ZY, Chen JJ, Wang ZD, Wang Q, Li AP, Marks JR, Guo CY, Chen Y, Zhou JW, Yang LQ, Lin CR, Wang SY. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 26.Smid M, Wilting S, Uhr K, Rodriguez-Gonzalez G, de Weerd V, Prager-Van der Smissen W, van der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A, Martin S, Davies H, Staaf J, van de Vijver M, Richardson A, MacGrogan G, Salgado R, van den Eynden G, Purdie C, Thompson A, Caldas C, Span P, Sweep F, Simpson P, Lakhani S, van Laere S, Desmedt C, Paradiso A, Eyfjord J, Broeks A, Vincent-Solomon A, Futreal A, Knappskog S, King T, Viari A, Borresen-Dale AL, Stunnenberg H, Stratton M, Foekens J, Sieuwerts A, Martens J. The circular RNome of primary breast cancer. Genome Res. 2019;29:356–366. doi: 10.1101/gr.238121.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Mol Cell. 2017;66:9–21. e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, Cherry S, Wilusz JE. The output of protein-coding genes shifts to circular RNAs When the Pre-mRNA Processing Machinery Is limiting. Mol Cell. 2017;68:940–954. e943. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, Liu X, Sun H, Pan Y, Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17:160. doi: 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 34.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E, Gong C. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YH, Lu CT, Zhou YZ, Zhang ZH, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502:358–363. doi: 10.1016/j.bbrc.2018.05.166. [DOI] [PubMed] [Google Scholar]
- 37.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y, Xue X, Meng R, Wei Q, Feng X. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505:996–1002. doi: 10.1016/j.bbrc.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Zhang WW, Peng F, Sun JY, He ZY, Wu SG. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. doi: 10.2147/CMAR.S155923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song LL, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503:2603–2610. doi: 10.1016/j.bbrc.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499:765–771. doi: 10.1016/j.bbrc.2018.03.219. [DOI] [PubMed] [Google Scholar]
- 42.Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:118–126. doi: 10.26355/eurrev_201801_14108. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, Cui Y. Increased expression of circular RNA circ_ 0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell Physiol Biochem. 2018;51:1710–1722. doi: 10.1159/000495675. [DOI] [PubMed] [Google Scholar]
- 44.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Gao DF, Zhang XF, Liu BB, Meng D, Fang K, Guo ZJ, Li LH. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. doi: 10.2217/epi-2017-0055. [DOI] [PubMed] [Google Scholar]
- 46.Liu YX, Dong YY, Zhao LP, Su LH, Luo J. Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752–1762. doi: 10.3892/ijo.2018.4485. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Ma K, Sun M, Shi S. Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am J Transl Res. 2018;10:1373–1386. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Wang HL, Xiao Y, Wu L, Ma DC. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan C, Zhou L, Zhang L, Yin K, Peng J, Sha R, Zhang S, Xu Y, Sheng X, Wang Y, Lin Y, Xu S, Yin W, Lu J. Identification and integrated analysis of key differentially expressed circular RNAs in ER-positive subtype breast cancer. Epigenomics. 2019;11:297–321. doi: 10.2217/epi-2018-0147. [DOI] [PubMed] [Google Scholar]
- 53.Yan NN, Xu HY, Zhang JN, Xu L, Zhang YY, Zhang L, Xu YC, Zhang FC. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704–95718. doi: 10.18632/oncotarget.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi PY, Sung J, He BY, Song H, Li ZQ, Kong WM, Wang JP, Wang JM, Xue HC. Profiles of differentially expressed circRNAs in esophageal and breast cancer. Cancer Manag Res. 2018;10:2207–2221. doi: 10.2147/CMAR.S167863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1alpha-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37:4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, Satoh Y, Okumura S, Nakagawa K, Mano H, Ishikawa Y. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 57.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19:4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, Li J, Wei YQ, Liu L, Zhao Y, Peng Y. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17:138. doi: 10.1186/s12943-018-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Qiu Y, Lu W, Jiang Y, Wang J. Immunotherapeutic interventions of Triple Negative Breast Cancer. J Transl Med. 2018;16:147. doi: 10.1186/s12967-018-1514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, Zheng Q, Li Y, Wang P, He X, Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Zheng QP, Bao CY, Li SY, Guo WJ, Zhao J, Chen D, Gu JR, He XH, Huang SL. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


