Abstract
Kallikrein-related peptidase (KLK) family is one of the major serine proteases in tumor microenvironment, which plays a crucial role in cancer invasion and metastasis. A number of KLK family members have been found to be upregulated or downregulated in some cancers, and some KLKs may be potential biomarkers for cancers. However, little is known about the role of KLKs in endometrial carcinoma (EC). In this study, we analyzed the mRNA sequencing data of EC from The Cancer Genome Atlas (TCGA) public database and found that the higher expression of KLK family members 5-8 (KLK5-8) was associated with an aggressive clinicopathologic phenotype and worse prognosis in EC patients. High expression of KLK5-8 was also confirmed in our patients with advanced stage and high-grade EC, as well as in a highly invasive cell line. Our study also demonstrated the differences between the subcellular localization of KLK5-8 and the co-expression of different splicing variants of KLK5-8 in EC cells, suggesting that various isoforms of KLK5-8 may work synergistically to regulate invasion and migration. We found that the elevation of more KLKs in a patient’s sample indicated a higher risk of worse survival. Combination of KLK5-8 was shown to be an independent prognostic factor for overall survival by multivariate Cox regression (hazard ratio: 2.215, 95% confidence interval: 1.045-4.694, P=0.038), and may be a promising biomarker.
Keywords: Kallikrein-related peptidases, endometrial carcinoma, splicing variant, prognosis, TCGA database
Introduction
Endometrial carcinoma (EC) is a common gynecologic cancer that is most commonly found in perimenopausal and postmenopausal women. Although it is usually diagnosed at an early stage with an overall favorable prognosis, up to 20% of patients with EC have a risk of recurrence and death [1]. Classification of EC is based on the FIGO stage, pathologic grade, and histologic type. Poor prognosis is usually associated with advanced stage, higher grade, serious type, and lymphovascular invasion. However, recurrence and distant metastasis also occur in 8% to 10% of patients with early-stage ECs [2]. The diversity of molecular alterations may be the fundamental cause of heterogeneity in prognosis. Based on a combined analysis of tumor mutation burden, somatic copy number alterations, and microsatellite instability, The Cancer Genome Atlas (TCGA) has classified EC into the following 4 groups: POLE ultramutated (POLE), microsatellite instability hypermutated (MSI), copy number high (CNH), and copy number low (CNL) [3]. These four molecular subtypes are prognostically significant, but are impractical for clinical application. Thus, efforts have been made to identify more accurate molecular markers to complement the existing prognostic factors.
The kallikrein-related peptidase (KLK) family comprises 15 serine proteases with trypsin- or chymotrypsin-like activities. They are involved in a broad spectrum of physiological processes, including extracellular matrix (ECM) remodeling, skin desquamation, pro-hormone processing, and neural plasticity [4]. Deregulated expression of some of the family members has been reported in several human cancers, such as ovarian cancer, colon carcinoma, and prostate cancer [5-8]. Several KLK genes have been studied to explore their use as biomarkers for the diagnosis and prognosis in various cancer types [9]. For example, KLK3 (also known as prostate-specific antigen) has been widely used as a tumor marker in the diagnosis and screening of prostate cancer [10]. Other KLKs can also degrade ECM proteins, thereby facilitating tumor cell detachment from primary tumor sites and leading to tumor dissemination and metastasis [11]. However, little is known about the role of KLKs in EC.
TCGA has provided comprehensive, multi-dimensional genomic and clinicopathological data on 33 types of cancer for the research community, which facilitates research on genes involved in cancer development and progression, and enables the identification of promising biomarkers for cancer prognosis.
In this study, we analyzed the expression of 15 members of the KLK family in EC using data from TCGA and defined the relationships between KLKs and outcomes in EC patients to provide new insights into the molecular pathogenesis and progression of EC, as well as to identify useful novel biomarker candidates. As most of the KLKs have several alternative splicing variants, their expression patterns and functions in cancer may be different from those in normal tissues where they have physiological functions. We therefore tried to identify tumor-specific splicing variants and to determine whether they are meaningful in the pathogenesis and prognosis of EC.
Materials and methods
Data on endometrial cancer from TCGA public database
Level 3 mRNA expression data (HTSeq-FPKM) on 583 endometrial cancers, corresponding clinical information (version 11-27-2017) of 605 patients, and survival data of 592 patients (version 10-25-2017) were downloaded from the UCSC xena browser (https://xenabrowser.net/datapages/). After excluding repetitive specimens, recurrent samples, and normal solid tissue samples, 543 patients with both sequencing and clinical data were selected for this study.
We grouped the patients according to histological grade, FIGO stage, histological type, molecular subtype, lymph node invasion, peritoneal cytology, and status of recurrence and metastasis of cancer (Table 1). Information of molecular subtype classification was obtained from the cBioportal (www.cbioportal.org), in which 232 patients were divided into the following 4 groups: POLE, MSI, CNL, and CNH.
Table 1.
Characteristics of patients in TCGA database
| Variable | No. of patients | Percentage |
|---|---|---|
| FIGO Stage | ||
| I | 339 | 62.43% |
| II | 51 | 9.39% |
| III | 124 | 22.84% |
| IV | 29 | 5.34% |
| Histological grade | ||
| G1 | 98 | 18.05% |
| G2 | 120 | 22.10% |
| G3 | 325 | 59.85% |
| Histological Pathology type | ||
| Endometrioid | 407 | 74.95% |
| Mixed | 22 | 4.05% |
| Serous | 114 | 21.00% |
| Molecular subtype | ||
| POLE | 17 | 7.33% |
| MSI | 65 | 28.01% |
| CNL | 90 | 38.79% |
| CNH | 60 | 25.86% |
| Lymph node status | ||
| Positive | 84 | 18.79% |
| Negative | 363 | 81.20% |
| Peritoneal cytology | ||
| Positive | 57 | 14% |
| Negative | 350 | 86% |
| Recurrence & metastasis | ||
| Positive | 70 | 15.12% |
| Negative | 393 | 84.88% |
Statistics
GraphPad Prism Software (v.7.0, La Jolla, CA, USA) and SPSS (v.20.0, Inc., Chicago, IL, USA) were used for statistical analysis and graph plotting. The normality of distribution of variables was evaluated by the Shapiro-Wilk test. The Mann-Whitney U-test was used to compare the means between two groups. The Kruskal-Wallis test was used for comparison among three or more groups. P<0.05 was considered statistically significant.
X-tile software version 3.6.1 (Yale University School of Medicine, New Haven, CT, USA) [12] was used to determine the best cut-off points between high- and low-expression categories for survival analysis. Kaplan-Meier overall survival (OS) curves were plotted for gene expression below and above the cut-off point, and the log-rank test was used to examine the differences between two groups. OS time was calculated from the date of initial diagnosis to death or date of the last follow up. All deaths from any cause were considered as an event. Relapse-free survival (RFS) curves were also plotted and the recurrence, metastasis and deaths from any cause were considered an event.
The Cox proportional hazards regression model was used for multivariate analysis to compare the influence of expression of KLKs on survival. Clinicopathologic characteristics, including stage, grade, age, lymph node status, peritoneal cytology, and histological subtype, were selected through univariate analysis and were included in the model for comparison. Results were reported as hazard ratios (HR) with 95% confidence intervals (CI). P<0.05 were considered to be statistically significant.
Cell lines and tissue samples
Human EC cell lines (HEC-1A, HEC-1B, Ishikawa, AN3CA, RL95-2, and KLE) were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in our laboratory. Ishikawa, RL-95-2, and KLE cells were cultured in DMEM/F12 (Gibco-BRL, Gaithersburg, MD, USA). AN3CA and HEC-1B cells were cultured in EMEM (Gibco-BRL), and HEC-1A cells was cultured in McCoy’s SA (Gibco-BRL). All of the cell lines were maintained in a culture medium containing 10% fetal bovine serum (Hyclone, Logan, UT, USA) under 5% CO2 at 37°C.
EC tissues were obtained from 40 patients treated at the Department of Obstetrics and Gynecology of Peking People’s Hospital (Beijing, China). The histological grade and FIGO stage of endometrial cancer were confirmed by the pathologists in our hospital. The study was approved by the Ethics Committee of the People’s Hospital, Peking University.
RNA extraction and reverse transcription
Total RNA was isolated from 6 endometrial carcinoma cell lines and 40 cancerous tissues using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then 1.5 µg total RNA was reverse transcribed using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix II kit (Transgene, Beijing, China). Both oligo-dT and random primers were included in cDNA synthesis. The cDNAs were used for quantitative PCR (qPCR) and variant-specific PCR amplification.
qPCR and variant-specific PCR
The qPCR reaction was conducted on Opticon2 (BioRad, Hercules, CA, USA) using the SYBR Green Real-Time PCR Master Mix kit (Toyobo, Osaka, Japan) and primers listed in Table 2 to evaluate the expression levels of KLKs. Relative expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase as the internal reference.
Table 2.
Primers for qPCR and variant-specific PCR
| Gene | Sequence | Position |
|---|---|---|
| Primers for qPCR | ||
| KLK5F | GGATGCTTACCCGAGACAGAT | exon 6 |
| KLK5R | CAGGGAGCCATTGCAGACC | exon 7 |
| KLK6F | CCAGCCAAACTCTCTGAACT | exon 5 |
| KLK6R | CTGGATGGTGTCAGGGAAATC | exon 5 |
| KLK7F | ACGAGCCCAGATGTGACCTT | exon 4-5 |
| KLK7R | CCTTGTAAACCTTCGTGCAGTC | exon 5 |
| KLK8F | CAGATCACAGATGGCATGGTCT | exon 5 |
| KLK8R | GGAGTGCACCATCACACACC | exon 6 |
| Primers for variant-specific PCR | ||
| KLK6 AD F | GACAAAGCCCGATTGTTCCT | exon 1 |
| KLK6 AD R | AACAGCATGATGTCCTGGTC | exon 5 |
| KLK6 BCE F | ACAGAACCAGCCTCTTCCAG | exon 1 |
| KLK6 BCE R | ACAACAGAACTCTGCTCCTG | exon 4 |
| KLK7 1 F | CCAGCAGAGGGATGAAGATTT | exon 1 |
| KLK7 1 R | TGTCGCCCAGCGTATCA | exon 4 |
| KLK7 2 F | TCCAAGCCTGACTCTGCTCT | exon 1 |
| KLK7 2 R | TGTCGCCCAGCGTATCA | exon 4 |
| KLK8 1 F | ACGTGGATGTTCCTGCTCTT | exon 2 |
| KLK8 1 R | TCCTAGAATCAGCCCTTGCT | exon 6 |
Note: KLK6 AD F & AD R: variant-specific primer for variants A and D in KLK6; KLK6 BCE F & BCE R: variant-specific primer for variants B, C, and E in KLK6; KLK7 1 F & 1 R: variant-specific primer for variants 1, 3, and 4 in KLK7; KLK7 2 F & 2 R: variant-specific primer for variant 2 in KLK7; KLK8 1 F & 1 R: variant-specific primer for variants 1, 2 , 3 , 4, and 5 in KLK8.
Variant-specific PCR was conducted to examine the splicing variants of KLK6, KLK7, and KLK8 expressed in EC using specially designed primers (Table 2) based on the sequences of known splicing variants from the NCBI database (https://www.ncbi.nlm.nih.gov/). PCR products were electrophoresed on 3.5% NuSieve agarose gels (FMC Bio Products, Rockland, ME, USA). Unexpected DNA bands were recovered and sequenced.
Western blotting
The Tricine-SDS-Page system was used for Western blotting to detect low molecular weight protein isoforms. Equal amounts of 20 μg total protein were loaded onto 15.5% acrylamide Tricine-SDS gels (Solarbio, Beijing, China) and subjected to electrophoresis. Then the proteins were transferred onto polyvinylidene difluoride membranes. After blocking in 5% skim milk, the membranes were incubated with primary antibodies (the same antibodies used in immunohistochemistry [IHC]) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody (ZSGB-BIO, Beijing, China) at room temperature for 1 h. The bands were visualized using SuperSignal™ West Pico Chemiluminescent Substrate from Thermo Fisher Scientific (Waltham, MA, USA).
Immunohistochemistry
All of the tissue sections underwent an antigen retrieval step (water bath at 98°C for 20 min in EDTA buffer, pH=9.0) before immunohistochemical staining. Anti-KLK6 goat antibodies (Cat: ab223230) and anti-KLK8 (Cat: ab150395) rabbit antibodies were purchased from Abcam (Cambridge, UK). Anti-KLK7 rabbit antibodies (Cat: TA324188) were purchased from OriGene (Rockville, MD, USA). Tissue slides were incubated overnight with primary antibody at 4°C. Following a TBS wash, the slides were incubated with secondary antibody for 30 min using PV-9000 (containing horseradish peroxidase [HRP]-conjugated anti-rabbit antibody) and PV9003 (containing HRP-conjugated anti-goat antibody) kits purchased from ZSGB-BIO (Beijing, China). Then the slides were stained with DAB substrate and counterstained with hematoxylin.
Results
TCGA data analysis
Expression of KLK family members and association with clinicopathologic features of EC patients
We analyzed the expression of KLK1-15 in patients with different clinicopathologic phenotypes of EC and found that the expression levels of KLK5-KLK8 were significantly higher (Figure 1) while KLK12 and KLK14 were significantly lower in advanced stage, serious type, higher grade, and CNH subtype, and in samples with positive lymph nodes. As downregulation of KLK12 and KLK14 in aggressive EC samples cannot be explained by known biological effects and upregulation of KLK12 and KLK14 has been detected in other cancer types [13,14], we focused our study on KLK5, KLK6, KLK7, and KLK8.
Figure 1.
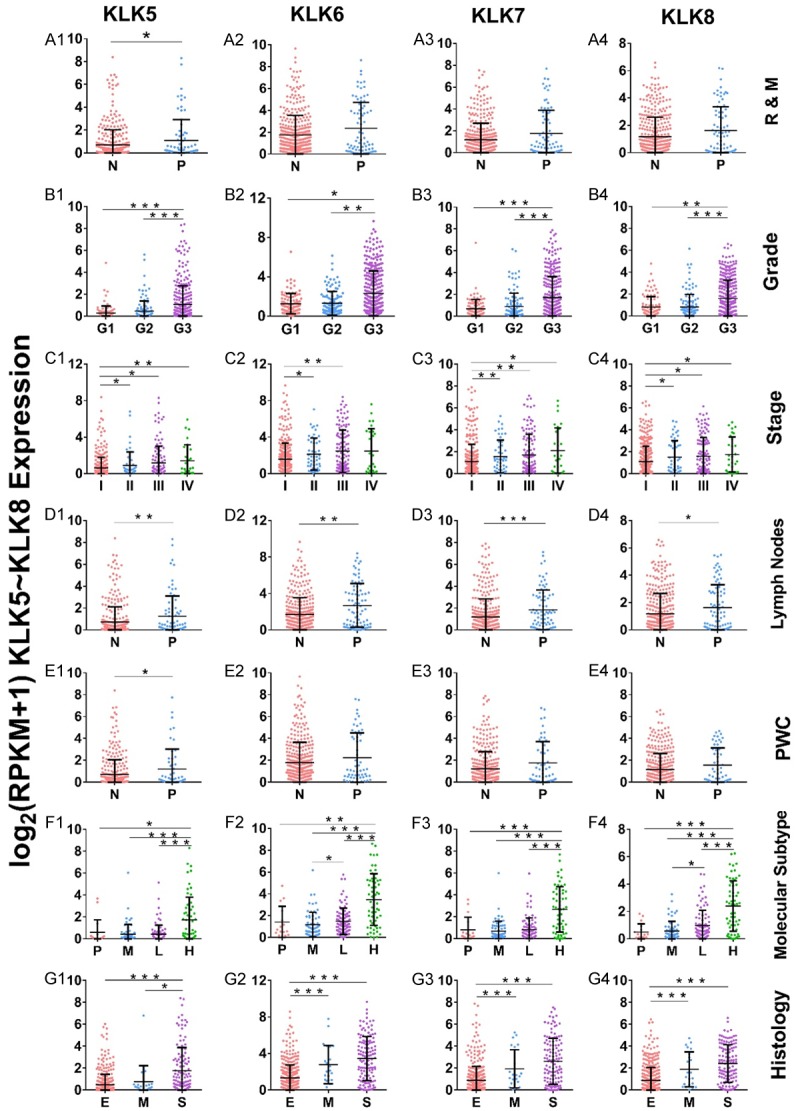
Differential expression of KLK5, KLK6, KLK7, and KLK8 in different clinicopathologic phenotypes of EC. Expression level and characteristics of KLK5-8 were shown in different recurrence and metastasis status (A1-A4), grades (B1-B4), stages (C1-C4), lymph nodes status (D1-D4), peritoneal washing cytology status (E1-E4), molecular subtypes (F1-F4), and histological types (G1-G4). N: negative; P: positive; R&M: recurrence and metastasis; PWC: peritoneal washing cytology; P: POLE; M: MSI; L: CNL; H: CNH; E: endometrioid EC; M: mixed EC; S: serous. *P≤0.05; **P≤0.01; ***P≤0.001.
As shown in Figure 1, KLK5-8 were also up-regulated in samples with positive peritoneal lavage cytology and local recurrence or distant metastasis, although the result was not statistically significant for KLK6, 7, and 8 (possibly because of limited cases available for peritoneal washes, and recurrence or distant metastasis status).
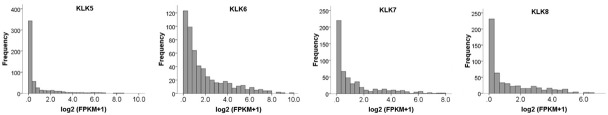
The expression of KLK5-8 in EC showed the following characteristics: there was little or no expression of KLK5-8 in most of the EC specimens. Only a small amount of specimens showed high gene expression with dispersal distribution (Figure 2). The samples showing high expression were mainly found in advanced stage, serous type, higher grade, and CNH molecular subtype EC.
Figure 2.
Frequency distributions of KLK5-KLK8 expression. There was little or no expression of KLK5-8 in most of the EC specimens. Only a small amount of specimens showed high gene expression with dispersal distribution.
As KLK5-KLK8 showed similar expression features, we performed a pairwise correlation analysis. The results showed that KLK5-KLK8 were positively correlated with each other (KLK5/KLK6 r=0.658, KLK5/KLK7 r=0.709, KLK5/KLK8 r=0.538, KLK6/KLK7 r=0.799, KLK6/KLK8 r=0.693, KLK7/KLK8 r=0.819) with statistical significance on analysis by the Spearman test (P<0.001).
Survival analysis based on KLK5-8 expression
In this study, we did not use a median cut-off point approach in survival analysis because of the skewed distribution of KLKs expressed in patients’ samples. Better results were achieved in our Kaplan-Meier survival analysis by using the powerful X-tile tool, which uses the maximum statistic to define the best split of patients.
As shown in Kaplan-Meier plots (Figure 3), higher expression levels of KLK5, 6, 7, and 8 were associated with a worse prognosis of EC patients (log-rank test, P<0.05). Univariate Cox regression verified their association with OS (Table 3A). However, multivariate Cox regression did not show independent prognostic significance of any of these genes on an individual basis.
Figure 3.

Kaplan-Meier analysis of OS based on KLK5, KLK6, KLK7, and KLK8 expression. A-D: OS analysis of KLK5-8 based on low- and high-expression levels using X-tile determined cut-off point; E: Integrated analysis of OS of KLK5-8 based on the number of high-expressed KLK5-8 genes; F: Integrated analysis of RFS of KLK5-8 based on the number of high-expressed KLK5-8 genes. Group 1: all KLK5-8 genes below the cut-off point; Group 2: 1-3 KLK5-8 genes above the cut-off point; Group 3: all KLK5-8 genes above the cut-off point.
Table 3A.
Univariate Cox analysis Regression analysis of prognostic factors for overall survival
| Clinicopathologic variable | HR (95% CI) | p-value |
|---|---|---|
| Lymph Node Status (positive vs. negative) | 3.797 (2.324-6.204) | <0.001 |
| Peritoneal Cytology (positive vs. negative) | 4.622 (2.798-7.632) | <0.001 |
| Histology (mixed or serous vs. endometrioid) | 3.026 (1.991-4.599) | <0.001 |
| Stage (III or IV vs. I or II) | 3.806 (2.502-5.792) | <0.001 |
| Grade (3 vs. 1 or 2) | 3.336 (1.941-5.734) | <0.001 |
| Age (continuous) | 1.035 (1.015-1.056) | <0.001 |
| KLK5 (categorical, above or below the cut-off point) | 2.057 (1.353-3.127) | <0.001 |
| KLK6 (categorical, above or below the cut-off point) | 2.917 (1.834-4.639) | <0.001 |
| KLK7 (categorical, above or below the cut-off point) | 2.326 (1.524-3.550) | <0.001 |
| KLK8 (categorical, above or below the cut-off point) | 2.246 (1.453-3.473) | <0.001 |
| KLK5-8 combination (at least 1 gene > the cut-off point) | 2.901 (1.766-4.764) | <0.001 |
Note: HR: Hazard ratio; CI: confidence interval.
As KLK5-8 were co-expressed, they may work synergistically to promote cancer invasion. Therefore, we performed an integrated analysis of KLK5, 6, 7, and 8. In this integrated analysis, patients were grouped into KLK-high and KLK-low groups, and KLK-high was defined when the expression level of at least one KLK gene was above the cut-off point determined by the X-tile method. The four-gene combination was analyzed by multivariate Cox regression, and showed independent prognostic significance compared with other clinicopathologic characteristics (significantly associated with outcomes by univariate analysis), with a HR of 2.215 (95% CI: 1.045-4.694) (Table 3B).
Table 3B.
Multivariate Cox Regression analysis of prognostic factors for overall survival (backward method)
| Clinicopathologic variable | HR (95% CI) | p-value |
|---|---|---|
| Stage (III or IV vs. I or II) | 2.819 (1.505-5.280) | 0.001 |
| Peritoneal Cytology (Positive vs. Negative) | 1.990 (1.044-3.794) | 0.037 |
| Histology (Mixed or Serous vs. Endometrioid) | 1.888 (1.020-3.494) | 0.043 |
| KLK5-8 combination (High vs. Low) | 2.215 (1.045-4.694) | 0.038 |
Note: HR: Hazard ratio; CI: confidence interval.
Although the expression levels of KLK5, 6, 7, and 8 were positively correlated, they were not up-regulated simultaneously in the same sample. Therefore, we stratified the patients into 3 groups by the number of high-expression KLK genes in their samples. We found that patients with low-expressed KLK5-8 showed the best OS and RFS on Kaplan-Meier analysis, with a 5-year survival rate of 86.1%. Patients with four highly expressed KLK genes showed the worst OS and RFS, with a 5-year survival rate of 61.9%; and those with 1-3 highly expressed KLK genes showed intermediate OS and RFS (Figure 3). There were more samples showing grade 3, advanced stage, serous type, CNH, and recurrence and metastasis in groups that have more highly expressed KLK5-8 genes (Table 4).
Table 4.
Proportion of aggressive clinicopathologic phenotypes in the three groups classified based on the number of high-expression KLKs
| Group 1 | Group 2 | Group 3 | |
|---|---|---|---|
| Mixed & Serous | 8.80% | 25.60% | 56% |
| Grade 3 | 48.20% | 60.30% | 91.30% |
| Recurrence & metastasis | 13.10% | 13.10% | 28.30% |
| Stage III & IV | 22.40% | 25.20% | 52.50% |
Note: Group 1: All KLK5-8 genes below the cut-off point; Group 2: 1-3 KLK5-8 genes above the cut-off point; Group 3: All KLK5-8 genes above the cut-off point.
Expression of KLK5-KLK8 in EC cell lines and tissues samples
The expression levels of KLK5-8 were evaluated by qPCR in six EC cell lines and EC samples from 40 Chinese patients. Most of the tissue samples showed very low expression, and high expression levels were found in only a small number of patients with higher grade, advanced stage, and serous type (Figure 4). These findings were similar to those found in TCGA cohort. A similar phenomenon was also observed in EC cell lines. The expression levels of KLK5-KLK8 were very low in Ishikawa, HEC-1B, and AN3CA cell lines, but much higher expression was detected in KLE and RL95-2 cell lines (Figure 4). It has been reported that KLE cells have stronger proliferation and invasion abilities than Ishikawa and HEC-1B cells [15], which supports the effects of KLK5-8 on promotion of progression.
Figure 4.
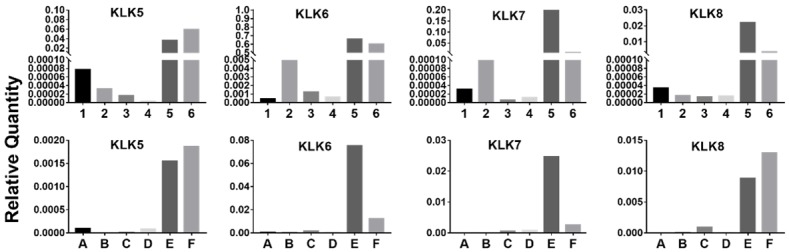
KLK5-8 expression in EC cell lines and representative tissue samples detected by qPCR. 1-6 were different EC cell lines. 1: AN3CA; 2: HEC-1A; 3: HEC-1B; 4: ISHIKAWA; 5: KLE; 6: RL95-2. A-F were different patients. A: Grade 1, stage I, endometrioid; B: Grade 1, stage I, endometrioid; C: Grade 2, stage I, endometrioid; D: Grade 2, stage II, endometrioid; E: Grade 3, stage III, serous; F: Grade 3, stage III, endometrioid.
Transcript variants detected in EC cell lines and tissues
The presence of several alternative splicing variants of the same gene is common among kallikreins. Therefore, we performed variant-specific PCR and DNA electrophoresis to study the splicing variants of KLK genes using representative samples selected on the basis of qPCR results. Considering the very low levels of KLK5 in our samples, this gene was excluded from variant-specific PCR.
Five protein-coding transcripts of the KLK6 gene have been annotated to date, and they are designated as KLK6 variants A, B, C, D, and E (accession numbers: NM_002774.3, NM_001012964.2, NM_001012965.2, NM_001319948.1, and NM_001319949.1, respectively). We used two sets of primer pairs (Table 2) to detect variants A and D and variants B, C, and E, respectively. We found that the main PCR products of KLK6 were variants A and B. (Figure 5A, 5B).
Figure 5.

Transcript Variants expressed in EC cell lines and representative tissues. DNA electrophoresis of variant-specific PCR products amplified with primers for KLK6 variants A and D (A); KLK6 variants B, C, and E (B); KLK7 variant 2 (C); KLK7 variants 1, 3, and 4 (D); KLK8 variants 1-5 (E). Marker: 100 bp DNA ladder. Lane 1-8 are different EC tissue samples or cell lines. 1: Grade 3, stage III, endometrioid; 2: Grade 3, stage IV, endometrioid; 3: Grade 1, stage I, endometrioid; 4: Grade 1, stage I, endometrioid; 5: normal endometrium; 6: ISHIKAWA cell line; 7: KLE cell line; 8: RL95-2 cell line.
KLK7 has four splicing variants V1-V4 (accession numbers: NM_005046.3, NM_139277.2, NM_001207053.1, and NM_001243126.1, respectively). We used two sets of primer pairs (Table 2) to detect variant 2 and variants 1, 3, and 4, respectively. The main expressed KLK7 variants were variants 1 and 2. Another new splice variant (216 bp, with exon 2 deletion) of KLK7 was detected and verified through sequencing (Figure 5C, 5D).
KLK8 has five splicing variants T1-T5 (accession numbers: NM_007196.3, NM_144505.2, NM_144506.2, NM_144507.2, and NM_001281431.1, respectively). We used one primer pair (Table 2) to detect all of the variants. The main expressed KLK8 variants were variants T1, T3, T4 and T5 (Figure 5E).
As we used multiplex PCR to amplify the splicing variants, the multiplex products had common sequences that easily formed a hybrid. There were several unexpected bands in agarose gels, which were proven to be hybrids through sequencing.
Protein isoforms of KLK7-8 detected in EC cell lines and tissues
The Western blot results are shown in Figure 6. Relative higher abundance of protein expression in KLK7 and KLK8 was observed in aggressive clinicopathologic phenotypes of EC.
Figure 6.
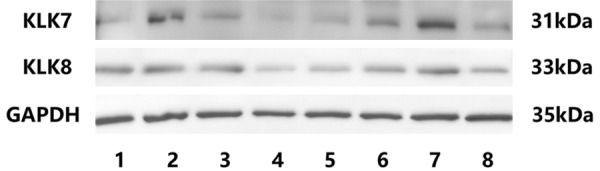
Western blotting results of KLK7 and KLK8 in EC cell lines and tissues. Lane 1-8 are different EC tissue samples or cell lines. 1: Grade 3, stage III, endometrioid; 2: Grade 3, stage IV, endometrioid; 3: Grade 1, stage I, endometrioid; 4: Grade 1, stage I, endometrioid; 5: normal endometrium; 6: ISHIKAWA cell line; 7: KLE cell line; 8: RL95-2 cell line.
A band of approximate 31 kDa was detected by anti-KLK7 antibody. This band represents protein isoform 1, which is the product of transcript variant 1 and variant 2. Other protein isoforms were not detected.
Anti-KLK8 antibody detected a band of approximate 33 kDa. This band represents protein isoform 1, which is the product of variant T1. KLK8 isoforms 3, 4 and 5 were not detected by Western blotting, although the high abundance of KLK8 transcript variant T3 and T4 was observed in variant-specific PCR. This might be due to the advantageous amplification of the small DNA fragments in PCR, and the difficulties to detect the extreme low molecular weight and low abundance proteins in Western blotting. The anti-KLK6 antibody we used is not recommended for western blotting. Even though, we conducted the experiment many times, but didn’t find a band of 30 KD, except some higher molecular non-specific bands.
Results of IHC
The characteristics of expression and localization in KLK6, KLK7, and KLK8 were observed by IHC in representative samples and cells selected on the basis of qPCR results. KLK6 and KLK7 showed homogeneous staining of the cytoplasm and KLK8 showed granular staining in the cytoplasm. Samples showing high expression levels detected by qPCR showed stronger staining. Considering the very low levels of KLK5 in our samples, this gene was excluded from IHC examination. Representative IHC images are shown in Figure 7.
Figure 7.
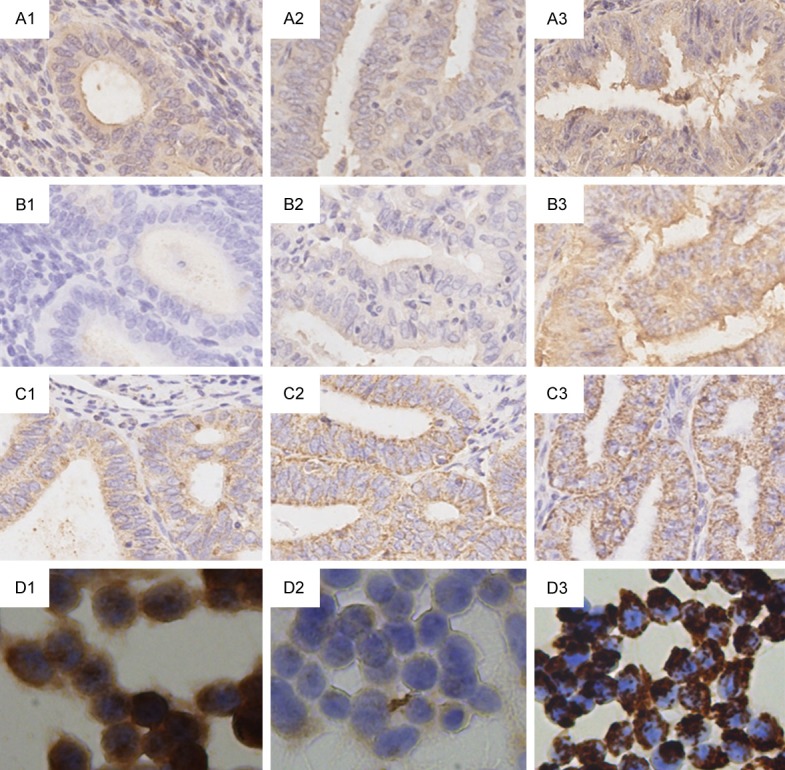
Representative pictures of KLK6, KLK7, and KLK8 expression detected by IHC. A1-A3. Immunoperoxidase (DAB) labelling of KLK6 in EC tissue with Grades 1-3. B1-B3. Immunoperoxidase labelling of KLK7 in EC tissue with Grades 1-3. C1-C3. Immunoperoxidase labelling of KLK8 in EC tissue with Grades 1-3. D1-D3. Immunoperoxidase labelling of KLK6, 7, 8 in RL 95-2 cell line. Magnification was 200× for all pictures.
Discussion
In the study, we analyzed the expression of KLK family members in EC using TCGA public data. The results showed that higher expression levels of KLK5-8 was associated with aggressive clinicopathologic phenotypes, including advanced stage, serious type, higher grade, and positive lymph nodes. High expression of KLK5-8 was also confirmed in our patients with advanced stage and high-grade EC, as well as in a high invasive cell line. Kaplan-Meier analysis showed that the higher expression levels of KLK5-8 were associated with the worse prognosis of EC patients. However, multivariate Cox regression did not show independent prognostic significance of any of these genes on an individual basis, possibly because of relatively fewer cases in the high expression group divided by the X-tile determined cut-off point. When these four genes were combined for multivariate Cox regression analysis, they showed independent prognostic significance.
KLK5-8 genes are located in a tandem cluster on chromosome 19, and they share a certain degree of sequence similarity [16]. Although they all have chymotrypsin-like activity and are often co-expressed, their locations in the cell compartment are different, which indicates that they are not tautological. A proteolytic cascade has been described for the skin desquamation process, in which KLK7 is activated by activated KLK5 [17]. Although the exact mechanism of interaction among KLK members 5-8 is not clear, our finding that a higher number of elevated KLKs in a patient’s sample indicates a higher risk of worse survival provides implications of their biologically synergistic effect in cancer.
The chymotrypsin-like activity and ability to degrade ECM proteins are the main mechanisms underlying the facilitation of tumor invasion and metastasis by KLKs. The three residues, histidine, aspartate, and serine, located on exons 2, 3 and 5, form the serine-type catalytic triad of KLKs [18]. Most of the splicing variants detected in EC contain exons 2, 3, and 5, except for KLK8 T3, T4, and T5. Variants without the catalytic triad are non-proteolytic, and their roles in cancer progression require further study. KLK8 T3 and KLK8 T4 have also been reported to be the most abundant forms in lung cancer [19]; thus, reconfirming their cancer-related function. Although we failed to find a tumor-specific splicing variant, our study showed that co-expression of different splicing variants of KLK5-8 existed in EC cells, which suggests that they may work synergistically.
The upregulation of KLK5-KLK8 and their association with patients’ prognosis have been reported in many cancers, including ovarian, prostate, renal cell, and colon cancers [5,6,20]. No systematic study of KLK5-KLK8 in EC has been published. One report showed significantly higher KLK6 expression levels in serous type (n=13) compared with endometrioid EC (n=13) by qPCR [21]. Another study showed that KLK8 expression was significantly higher in early-stage and low-grade EC samples than in advanced-stage and higher-grade EC samples [22]. Although this finding is contradictory to our results, it is not convincing because only a few cases of advanced stage and higher-grade samples were included in their studies. The expression of KLK5 and KLK7 and their association with the outcome of EC have not been reported. In addition, there is a great deficiency of studies concerning splicing variants in tumors. Further intensive studies are required to better understand their roles in tumor progression and their clinical significance.
Prognostic outcome prediction in EC is usually based on FIGO stage, pathologic grade, and histologic type. However, interobserver disagreement in grade and histotype assignment is common [23], and some patients with early-stage EC also develop recurrence and metastasis [1]. Molecular subtypes can better stratify patients with different prognosis, which is more convincing as the molecular changes are the basis of different phenotypes. However, bioinformatics analysis of second-generation sequencing data is required to determine a sample’s molecular subtype. Samples showing high expression of KLK5-8 were mainly found in the CNH molecular subtype, which is characterized by high copy number, high grade, more serous type, and poor prognosis. Furthermore, samples showing very low expression could be negative if detected by IHC and they can be easily distinguished from samples showing high expression, which are practical for clinical application. Thus, the prognosis analysis based on the number of high-expression KLK5-8 could be a promising molecular marker.
In summary, higher expression of KLK5-8 was associated with an aggressive clinicopathologic phenotype and worse prognosis of EC patients. Various isoforms of KLK5, 6, 7, and 8 may work synergistically to regulate invasion and migration. The combination of KLK5-8 was shown to be an independent prognostic factor for OS in EC patients, and it could be complementary to the present clinicopathological prognostic criteria. To the best of our knowledge, this is the first study to demonstrate the prognostic and clinical value of KLK5, KLK6, KLK7, and KLK8 in EC.
Acknowledgements
Supported by Special Projects for Strengthening Basic Research of Peking University (BMU2018JC005).
Disclosure of conflict of interest
None.
References
- 1.Suhaimi SS, Ab Mutalib NS, Jamal R. Understanding molecular landscape of endometrial cancer through next generation sequencing: what we have learned so far? Front Pharmacol. 2016;7:409. doi: 10.3389/fphar.2016.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdulfatah E, Ahmed Q, Alosh B, Bandyopadhyay S, Bluth MH, Ali-Fehmi R. Gynecologic cancers: molecular updates 2018. Clin Lab Med. 2018;38:421–438. doi: 10.1016/j.cll.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yousef GM, Diamandis EP. An overview of the kallikrein gene families in humans and other species: emerging candidate tumour markers. Clin Biochem. 2003;36:443–52. doi: 10.1016/s0009-9120(03)00055-9. [DOI] [PubMed] [Google Scholar]
- 5.Vakrakou A, Devetzi M, Papachristopoulou G, Malachias A, Scorilas A, Xynopoulos D, Talieri M. Kallikrein-related peptidase 6 (KLK6) expression in the progression of colon adenoma to carcinoma. Biol Chem. 2014;395:1105–1117. doi: 10.1515/hsz-2014-0166. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Kaushal A, Brattsand M, Nicklin J, Clements JA. Differential splicing of KLK5 and KLK7 in epithelial ovarian cancer produces novel variants with potential as cancer biomarkers. Clin Cancer Res. 2003;9:1710–1720. [PubMed] [Google Scholar]
- 7.Yousef GM, Diamandis EP. The human kallikrein gene family: new biomarkers for ovarian cancer. Cancer Treat Res. 2009;149:165–187. doi: 10.1007/978-0-387-98094-2_8. [DOI] [PubMed] [Google Scholar]
- 8.Avgeris M, Scorilas A. Kallikrein-related peptidases (KLKs) as emerging therapeutic targets: focus on prostate cancer and skin pathologies. Expert Opin Ther Targets. 2016;20:801–818. doi: 10.1517/14728222.2016.1147560. [DOI] [PubMed] [Google Scholar]
- 9.Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP. Kallikrein gene downregulation in breast cancer. Br J Cancer. 2004;90:167–172. doi: 10.1038/sj.bjc.6601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberts AR, Schoots IG, Roobol MJ. Prostate-specific antigen-based prostate cancer screening: Past and future. Int J Urol. 2015;22:524–532. doi: 10.1111/iju.12750. [DOI] [PubMed] [Google Scholar]
- 11.Kim JT, Song EY, Chung KS, Kang MA, Kim JW, Kim SJ, Yeom YI, Kim JH, Kim KH, Lee HG. Up-regulation and clinical significance of serine protease kallikrein 6 in colon cancer. Cancer. 2011;117:2608–2619. doi: 10.1002/cncr.25841. [DOI] [PubMed] [Google Scholar]
- 12.Camp RL, Dolled-Filhart M, Rimm DL. X-tile a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 13.Zhao EH, Shen ZY, Liu H, Jin X, Cao H. Clinical significance of human kallikrein 12 gene expression in gastric cancer. World J Gastroenterol. 2012;18:6597–6604. doi: 10.3748/wjg.v18.i45.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kontos CK, Adamopoulos PG, Papageorgiou SG, Pappa V, Scorilas A. mRNA overexpression of kallikrein-related peptidase 14 (KLK14) is an independent predictor of poor overall survival in chronic lymphocytic leukemia patients. Clin Chem Lab Med. 2016;54:315–324. doi: 10.1515/cclm-2015-0456. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Wang M, Fang S, Wang Q, Fang R, Chen J. Fibulin-4 is associated with prognosis of endometrial cancer patients and inhibits cancer cell invasion and metastasis via Wnt_β-catenin signaling pathway. Oncotarget. 2017;8:18991–19012. doi: 10.18632/oncotarget.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clements J, Hooper J, Dong Y, Harvey T. The expanded human kallikrein (KLK) gene family: genomic organisation, tissue-specific expression and potential functions. Biol Chem. 2001;382:5–14. doi: 10.1515/BC.2001.002. [DOI] [PubMed] [Google Scholar]
- 17.Stefanini AC, da Cunha BR, Henrique T, Tajara EH. Involvement of kallikrein-related peptidases in normal and pathologic processes. Dis Markers. 2015;2015:946572. doi: 10.1155/2015/946572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kryza T, Silva ML, Loessner D, Heuze-Vourc’h N, Clements JA. The kallikrein-related peptidase family: dysregulation and functions during cancer progression. Biochimie. 2016;122:283–299. doi: 10.1016/j.biochi.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Planque C, Choi YH, Guyetant S, Heuze-Vourc’h N, Briollais L, Courty Y. Alternative splicing variant of kallikrein-related peptidase 8 as an independent predictor of unfavorable prognosis in lung cancer. Clin Chem. 2010;56:987–997. doi: 10.1373/clinchem.2009.138917. [DOI] [PubMed] [Google Scholar]
- 20.Gabril M, White NM, Moussa M, Chow TF, Metias SM, Fatoohi E, Yousef GM. Immunohistochemical analysis of kallikrein-related peptidases in the normal kidney and renal tumors: potential clinical implications. Biol Chem. 2010;391:403–409. doi: 10.1515/BC.2010.025. [DOI] [PubMed] [Google Scholar]
- 21.Santin AD, Diamandis EP, Bellone S, Soosaipillai A, Cane S, Palmieri M, Burnett A, Roman JJ, Pecorelli S. Human kallikrein 6: a new potential serum biomarker for uterine serous papillary cancer. Clin Cancer Res. 2005;11:3320–3325. doi: 10.1158/1078-0432.CCR-04-2528. [DOI] [PubMed] [Google Scholar]
- 22.Michaelidou K, Ardavanis A, Scorilas A. Clinical relevance of the deregulated kallikrein-related peptidase 8 mRNA expression in breast cancer: a novel independent indicator of disease-free survival. Breast Cancer Res Treat. 2015;152:323–336. doi: 10.1007/s10549-015-3470-8. [DOI] [PubMed] [Google Scholar]
- 23.Hoang LN, Kinloch MA, Leo JM, Grondin K, Lee CH, Ewanowich C, Köbel M, Cheng A, Talhouk A, McConechy M, Huntsman DG, McAlpine JN, Soslow RA, Gilks CB. Interobserver agreement in endometrial carcinoma histotype diagnosis varies depending on the cancer genome atlas (TCGA)-based molecular subgroup. Am J Surg Pathol. 2017;41:245–252. doi: 10.1097/PAS.0000000000000764. [DOI] [PubMed] [Google Scholar]