Abstract
Much uncertainty exists in science and herbal products referencing turmeric (T), turmeric extract (TE), curcuminoid-enriched turmeric extract (CTE), further processed curcuminoid-enriched materials (CEM), or curcumin as a single-chemical entity. To facilitate the rational chemical and biological assessment of turmeric-derived NPs, we introduced the DESIGNER approach of Depleting and Enriching Select Ingredients to Generate Normalized Extract Resources to Curcuma longa preparations. Countercurrent separation of a commercial CTE yielded four key materials: lipophilic metabolites; purified curcumin (“purcumin”); a mixture of curcumin, demethoxycurcumin, and bisdemethoxycurcumin (“purcuminoids”); and hydrophilic metabolites, and enabled production of a curcuminoid-free TE (“nocumin”). Their characterization utilized TLC, 1H (q)NMR spectroscopy, and HPLC.
Graphical Abstract
Turmeric spice is produced from Curcuma longa L. (Zingiberaceae) rhizomes and represents the authentic crude source of the single phytochemical compound, curcumin (1 in Scheme 1). Dried C. longa rhizomes contain 3–15% of curcumin and its congeneric metabolites, collectively referred to as curcuminoids.1 The volume of the scientific literature that describes the chemical and biological properties of “curcumin” is astounding. The Curcumin Resource Database, created in 2015 boasts >9,000 peer-reviewed publications.2 However, the material referred to as “curcumin” in much of the literature is very rarely the single-chemical entity 1 in pure, or even highly purified, form. The materials that have received the moniker “curcumin” may be divided into five categories: (i) turmeric (T); the raw C. longa rhizome; (ii) turmeric extract (TE), a solvent extract of dried or fresh C. longa rhizomes; (iii) curcuminoids-enriched turmeric extract (CTE), typically the deep-yellow precipitate collected from the solvent extract upon concentration under reduced temperature;3 (iv) curcuminoids-enriched material (CEM), obtained via further, typically large-scale chromatographic purification of CTE; and (v) curcumin as single-chemical entity, in a purity that is commensurate with that of a validated and/or metrological reference material. The deficiency in the adequate chemical characterization of the immense diversity of crude turmeric (T), extracts (CE), enriched materials (CTE, CEM), and even materials considered “pure 1“, augmented by the fact that the materials often share the same name (“curcumin”), casts doubt upon the usefulness of the biological data acquired from the plethora of these preparations.4 Furthermore, attributing biological activity of a complex mixture solely to the major component, however prominent, it unjustifiably naïve.5
Scheme 1.
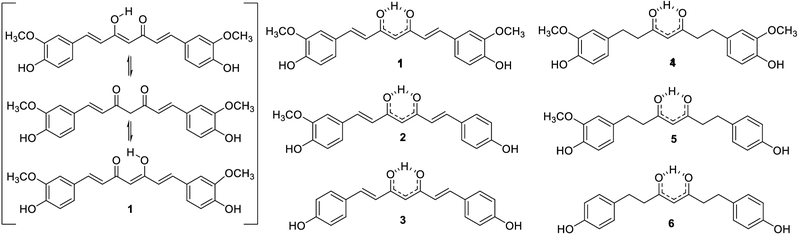
The three major curcuminoids found in turmeric and its extracts: curcumin (1), demethoxycurcumin (2), and bisdemethoxycurcumin (3), along with their tetrahydro congeners tetrahydrocurcumin (4), tetrahydrodemethoxycurcumin (5), and tetrahydrobisdemethoxycurcumin (6); shown as tautomers for 1 (left) and corresponding resonance hybrids (right).
Ascertaining the chemical composition of a sample being assessed for biological activity is a fundamental requirement for studying the potential medicinal value of natural products (NPs), ranging from crude extracts to “pure” compounds.5 Simply reporting the content (often termed “purity”) of the major component ignores potential contributions of other constituents (“impurities”) to the observed biological activity. The term “residual complexity” (RC; go.uic.edu/residualcomplexity) offers a more holistic view of the metabolomic profiles of impure NPs in reference to the complex nature of the source material, the parameters of the separation methods employed, and the dynamic transformations of the chemical constituents themselves. The RC of any given NP material may be the static remnants of metabolites accompanying the major compound that are determined, largely, by the purification protocol. A second source of RC is the dynamic chemical transformations of the constituents subjected to chemical and physical stressors during extraction, purification, and handling. The bioassay creates a third source of RC as the biological system can affect the chemistry of the components. As demonstrated recently, RC can affect drug discovery profoundly, even at very low and seemingly inconsequential levels (0.24% w/w).6
Customary solvent extraction of C. longa rhizomes is widely considered to exclude compounds insoluble in organic solvents. Normally, C. longa rhizomes are dried before processing. The dried rhizomes are typically “defatted” with an appropriate non-polar solvent. This removes the oleoresins, which consist primarily of congeneric sesquiterpenes, including aromatic turmerone, α-santalene, β-sesquiphellandrene, α-zingerberene, β-curcumene, and β-bisabolene.7 A number of monoterpenes are also removed: eucalyptol, terpinen-4-ol, α-terpineol, p-cymene, and p-cymen-8-ol.1,7 Next, the rhizomes are extracted with a medium-polarity organic solvent, such as acetone or ethanol, to solubilize the curcuminoids. Subsequent precipitation of the extract at low temperature further enriches the material in curcuminoids due to intermolecular interactions between these aromatic compounds, such as π-stacking.
However, an appreciable diversity of lipophilic and hydrophilic components persist during both the extraction and the enrichment processes, conceivably due to non-specific interactions with curcuminoids and with each other. Importantly, the presence of inherent natural deep eutectic solvents (NADES) in the plant material contributes to the solubility of a larger-than-expected diversity of metabolites in solvents, in which they are “normally” considered insoluble. Most notably, NADES moderate the polarities of both lipophilic and hydrophilic components. Interestingly, little has been done to characterize the hydrophilic metabolites in C. longa rhizomes, other than noting that turmeric contains a considerable amount of starch.8 During “curcumin” purification, significant RC arises from the diarylheptanoid congeners (curcuminoids) that likely persist even in highly purified 1. Although there are numerous diarylheptanoids that fall under the general description of “curcuminoids,” the three major curcuminoids are curcumin (1), demethoxycurcumin (2), and bisdemethoxycurcumin (3; also often called bisdesmethoxycurcumin, didesmethoxycurcumin, or didemethoxycurcumin) (Scheme 1). Collectively, the occurrence of both much more lipophilic and hydrophilic constituents in less refined materials such as CTEs and CMEs, and even in “pure“ curcuminoids is inevitable, but frequently overlooked.
In addition to the extraction-related considerations that lead to static RC, degradation of curcuminoids is another well-known contributor to the dynamic RC complexity of “curcumin” preparations. Schneider et al. reported extensively on the degradation of 1 under conditions encountered in most biological assays.9,10 The primary degradation pathways involve double bond hydrogenation, auto-oxidation, and conjugation by nucleophilic addition. The myriad of known degradation products undoubtedly influences observed in vivo and in vitro activities, but are largely underrecognized in the literature.
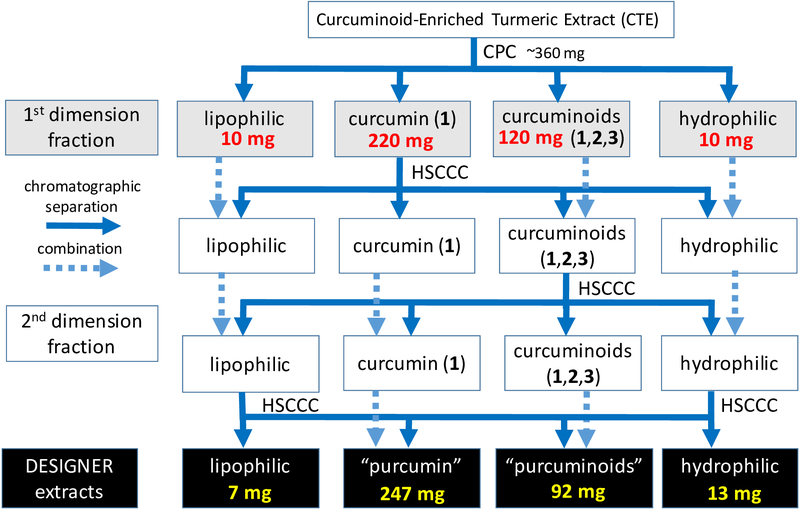
The countercurrent chromatographic approach of Depleting and Enriching Select Ingredients to Generate Normalized Extract Resources (DESIGNER) provides a rational and systematic means of separating components of biologically active source materials. Due to the high-resolution capabilities of countercurrent separation (CCS), DESIGNER methodology permits purposeful investigation of RC and enables the assessment of the therapeutic value of NPs, particularly extracts and other chemically complex mixtures.11–13 The challenge of DESIGNER formulations, in contrast to phytochemical-driven chromatographic fractionation, is that the entirety of the original sample must be accounted for in the subsequent fractions. A carefully designed fractionation protocol should not be directed at purification of target compounds, but also strive to minimize compound overlap between fractions. This is in contrast to the common practice of chromatographic “heart cuts” that yield higher purity compounds (RC still applies) at the expense of overall recovery. Minimizing compound overlap between fractions is especially challenging when processing curcumin-enriched turmeric extracts (CTEs) where curcuminoids as major compounds “bleed” into many fractions. The rational distribution of fractions for the CTE targeted in this study were: (i) lipophilic metabolites, which have been described as being part of the turmeric oleoresins; (ii) purified 1 (“purcumin”); (iii) a mixture of exclusively 1, 2, and 3 (“purcuminoids”); and (iv) hydrophilic metabolites.
This study employed countercurrent separation (CCS), which includes both the hydrostatic (CPC) and hydrodynamic (HSCCC) instruments.14 CCS is superior to solid phase-based column chromatography for the preparation of DESIGNER materials, because it avoids the absorption of constituents onto solid phases and the chemical changes that these phases can induce.14 In addition, CCS combines preparative loading capacity with high resolution of target compounds. Tailing of major metabolites is minimized with the resulting “clean cuts” between fractions.15 CCS is well suited for recovery of lipophilic and hydrophilic compounds at each polarity extreme of the chromatographic run and focuses on the high-efficiency separation of individual curcuminoids at the center.
RESULTS AND DISCUSSION
A commercially available curcuminoid-enriched turmeric extract (CTE) was used for the proof of principle in this study. A quantitative 1H NMR (qHNMR) spectroscopy analysis revealed 90.0% total curcuminoids with mass percentages of 73.0, 15.0, and 2.0 % of 1, 2, and 3, respectively (Figure S1, Supporting Information). Subsequent NMR spectroscopy studies showed a high degree of similarity between this preparation and other commercial curcuminoid-enriched turmeric extracts commonly referred to as “95% curcuminoids”. Nonetheless, variations are likely to exist in the total curcuminoid content and residual complexity (RC) signatures of products from different manufacturers, due to differences in extraction solvents and protocols, as well as natural variability of the starting botanicals.
Solvent system (SS) selection in CCS involves finding a SS with satisfactory resolution of the target analytes. Extensive SS investigations have already been documented for the three major curcuminoids.16 An n-hexane/CHCl3/MeOH/water biphasic system was determined to be optimal. Subsequently, during this work, this SS was modified slightly in order to optimize separation parameters such as stationary phase volume retention.17 Recently, a centrifugal partition centrifugation separation of 1-3 has been described with a heptane/CHCl3/MeOH/water SS.18 The only other publication dealing with the separation of curcuminoids with CCS used pH zone refining, which introduced pH changes that foster the degradation of curcuminoids and other metabolites.19 CCS resolution may be enhanced by (i) fine-tuning the SS; (ii) optimizing operating parameters that increase the stationary phase retention; (iii) changing the mobile phase; and (iv) decreasing column loading. All four of these avenues were explored to obtain an optimal resolution of 1-3, between themselves, as well as from other constituents.
In addition, CH2Cl2 was studied as a CHCl3 substitute for safety and environmental reasons.20 This involved measurement of the respective partition coefficients (K values) of a series of n-hexane/CH2Cl2/MeOH/water SSs. Increasing the proportion of n-hexane in this 2:7:7:3, 3:7:7:3, and 4:7:7:3 SS series increased the K values for all three major curcuminoids. Increasing the percentage of n-hexane in this SS series had the effect of repelling medium polarity compounds into the predominantly aqueous methanolic upper phase. The SS composition was investigated by GC-FID. In these studies, n-hexane was found almost exclusively in the lower phase, while water was largely predominant in the upper layer. The proportions of both MeOH and CH2Cl2 in the lower layer decreased as the amount of n-hexane increased due to the increase of the total volume of the lower phase, whereas the proportions of n-hexane, CH2Cl2, MeOH, and water in the upper phase changed very little (Figure S3, Supporting Information).
An important consideration in CCS is the choice of stationary phase. For SSs containing CHCl3 or CH2Cl2, the lower phase is organic, while the upper phase is aqueous. If the lower phase is mobile, normal phase elution order is observed. An effort was made to optimize both SS loading capacity and resolution by using the n-hexane/CH2Cl2/MeOH/water SS in reversed-phase (upper phase as mobile phase) elution mode. A reversed-phase separation with a 2:7:7:3 ratio was performed on a hydrodynamic CCS instrument. While a reversed-phase elution order was observed, neither the stationary phase retention nor the resolution were improved over the n-hexane/CH2Cl2/MeOH/water 3:7:7:3 normal phase mode. Another reversed-phase separation with the same solvents (1:7:7:3) in ascending mode was performed on a hydrostatic CCS instrument. In this case, reversed-phase elution was not observed, and all three major curcuminoids (1-3) eluted together, with appreciable tailing of 1.
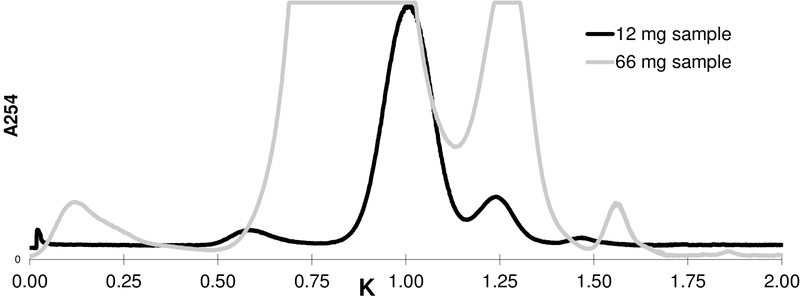
Unfortunately, the CTE displayed insufficient solubility in the SS that gave an optimal CCS resolution of 1-3. In the formation of the n-hexane/CH2Cl2/MeOH/water SS, MeOH and CH2Cl2 entrained 1-3 into the upper and lower phases, respectively, while hexane and water pushed 1-3 into the upper and lower phases, respectively. This combination of pulling and pushing created a biphasic system where 1 was nearly equally distributed in the upper and lower phases at the expense of overall solubility. The most suitable sample load for the n-hexane/CH2Cl2/MeOH/water 2:7:7:3 SS was 15 mg/mL in a 1:1 biphasic mixture. The same solvents at a 3:7:7:3 ratio had a sample load of 14 mg/mL, whereas a saturated mixture of the same solvents (4:7:7:3) allowed loading of merely 8.7 mg/mL. While increased sample load had a positive effect on chromatographic throughput, it came at the expense of increasing overlap of 1-3. Additionally, high percentage of 1 caused it to disperse into neighboring fractions (Figure 1). Addition of DMSO significantly enhanced the overall solubility of CTE in either the biphasic n-hexane/CH2Cl2/MeOH/water 3:7:7:3 or the lower phase alone. At the CPC processing stage, adding 1.0 mL DMSO to 8.0 mL of lower phase increased the solubility of CTE to 40 mg/mL. Thus, CPC served as a high-capacity first step towards the production of four DESIGNER materials from CTE (Figure S2, Supporting Information). The DMSO eluted with several curcuminoid-containing fractions and was removed by co-evaporation with MeOH and water.
Figure 1.
Hydrodynamic (HSCCC) countercurrent separation (CCS) of 12 and 66 mg of curcuminoid-enriched turmeric extract (CTE), dissolved in 15 mL each (280 mL column volume, 2 mL/min flow rate, 900 rpm, Sf = 0.27). The chromatogram is represented as a K-value based Reciprocal Symmetry (ReS) plot to facilitate instrument and operator independent reproducibility.30
Sample recovery was monitored carefully throughout the separation protocol as shown in Figure 2. While it is often emphasized that, by default, CCS has high sample recovery and lacks indiscriminant absorption, the actual recoveries are seldom documented; the present study provides experimental evidence for this assertion. Sample loss may occur, through (the mechanics of) sample handling, from vial to sample loop and fraction collection (e.g. switch valves in high-flow fraction collectors). Other liquid handling sample loss may take place during fraction consolidation procedures. In addition, the combined masses of multiple test tubes to a single sample loading mass is influenced by the propagation of weighing errors caused by the relatively high tare weight of vials. Importantly, the losses experienced in CCS practice are inconsequential for subsequent chemical and biological applications, as they are non-specific and typically do not translate into changes of the chemical profiles of the DESIGNER materials.
Figure 2.
Flowchart for the preparation of four DESIGNER extracts from curcuminoid-enriched turmeric extract (CTE). The final products targeted in the present study are the lipophilic, “purcumin”, “purcuminoids”, and hydrophilic fractions. When applied to other starting materials, their relative amounts are expected to vary substantially.
Based on the results of the CCS method development studies, a two-step preparative purification procedure was adopted for the production of DESIGNER materials from CTE. Initial fractionation used 266 mL CPC with a ~360 mg sample load, yielding a total of 80 fractions using an elution-extrusion procedure (EECCC). The resulting fractions were combined into four primary fractions and re-chromatographed on an HSCCC instrument of similar column volume (280 mL), but employing lower loading capacity. The limited solubility of the primary fraction enriched in 1 required the sample resulting from step one to be divided into five runs in step two. The resulting secondary fractions were recombined into four congruent subfractions following the scheme in Figure 2, and the resulting re-combined secondary fractions were analyzed by RP HPLC and qHNMR spectroscopy.
Performing fraction monitoring on TLC plates impregnated with a fluorescent indicator, the curcuminoids gave visibly yellow bands and were also revealed as dark and fluorescent spots under 254 and 366 nm UV light, respectively. In addition, a complex pattern of yellow bands was observed before the curcuminoid-containing fractions. A colorless compound, tetrahydrocurcumin (4), was revealed under UV light with a higher Rf value than 1, overlapping with the beginning of the test tubes containing 1. The hydrophilic compounds eluting after 3 were less highly colored on the TLC plates, and only faintly visible under 254 nm UV light.
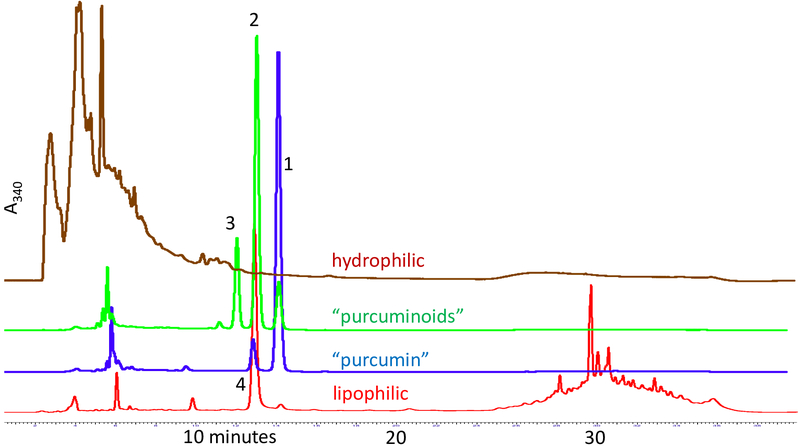
RP HPLC analysis was performed according to the method of Inoue et al.16 The order of elution for the three major curcuminoids was 3, 2, and 1 (Figure 3). The retention time for 4 was very similar to that of 2. Both the lipophilic and hydrophilic fractions revealed the large diversity of compounds in those mixtures. This provided an excellent illustration of the ability of CCS to target the purification of specific compounds and at the same time recover compounds both more lipophilic and more hydrophilic than the target(s). This separation pattern reminded the authors of what in NMR terminology is considered an “AMNX signal pattern”. In this figurative analogy, “A” and “X” represent the extreme polarities of the CCS chromatogram, whereas the “MN” part is equivalent to a pair of targeted curcuminoids. Interestingly, in our experience, CCS outcomes frequently represent similar “ANMX” separation patterns. Furthermore, it should be noted that the CTE starting material used in this study was already highly enriched in curcuminoids (reported as 95%). Therefore, it is significant that residual complexity at both polarity extremes is present in the sample. Identification of the individual components in these “nocumin” fractions will require extensive investigation. The composition of these factions will depend on the degree of refinement of the starting material and may offer an insight into the physical and chemical provenance of the initial sample.
Figure 3.
The RP-HPLC chromatograms (UV 340 nm) of the four DESIGNER materials derived from the CTE illustrates two important aspects: the high selectivity of the preparative two-step CCS scheme; and the substantial residual complexity (RC) of the CTE starting material, which in the literature is customarily designated as “curcumin”. CCS not only provides access to purified curcumin (1; “purcumin”) and its congeners (2–6; 5 and 6 detected by NMR and not labeled here) including a truly refined mixture of the three main curcuminoids (1–3; “purcuminoids”), but also separates and recovers the lipophilic and hydrophilic C. longa metabolites into the “A” and “X” fractions of the “AMNX” like separation pattern that is typical for CCS.
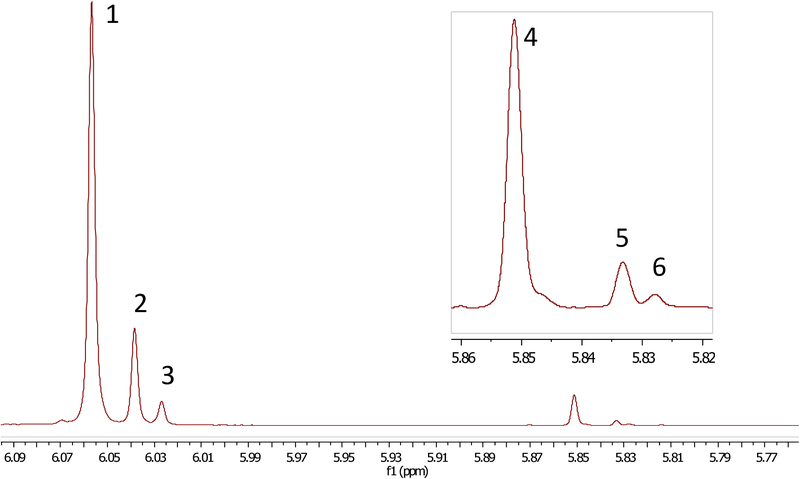
The 1H NMR spectroscopic analysis of the initial CTE revealed an array of signals in the 5.25 to 8.00 ppm region, denoting curcuminoids. Signals between 5.25 and 6.25 ppm indicated the presence of the hydrogen in the 1,3 diketone enol form. Quantitation of 1-3 was achieved by either direct integration or by integration after deconvolution of the signals at 6.057, 6.038, and 6.023 ppm, respectively (Figure 4). The 1H NMR spectroscopic analysis of individual test tubes revealed the presence of a significant amount of 4 eluting immediately before and overlapping with the test tubes containing 1. The tetrahydro congeners of 1-3 could be detected by their corresponding enol hydrogens, leading to the identification of tetrahydrocurcumin (4), tetrahydrodemethoxycurcumin (5), and tetrahydrobisdemethoxycurcumin (6) at 5.851, 5.833, and 5.828 ppm, respectively (Figure 4). Tetrahydrocurcumin has been reported in turmeric extracts on many occasions and has been tested alongside “curcumin” for biological activity.21,22 The other reduced curcuminoids, 5 and 6, while less likely to be associated with C. longa extracts, but are more abundant in “white turmeric” (Curcuma zedoaria (Christm.) Roscoe).23
Figure 4.
Interpretation of the 5.77 to 6.09 ppm interval of the 1H NMR spectrum of the CTE reveals that, along with the widely recognized curcuminoids, 1-3, their corresponding tetrahydro derivatives, 4-6, are also present.
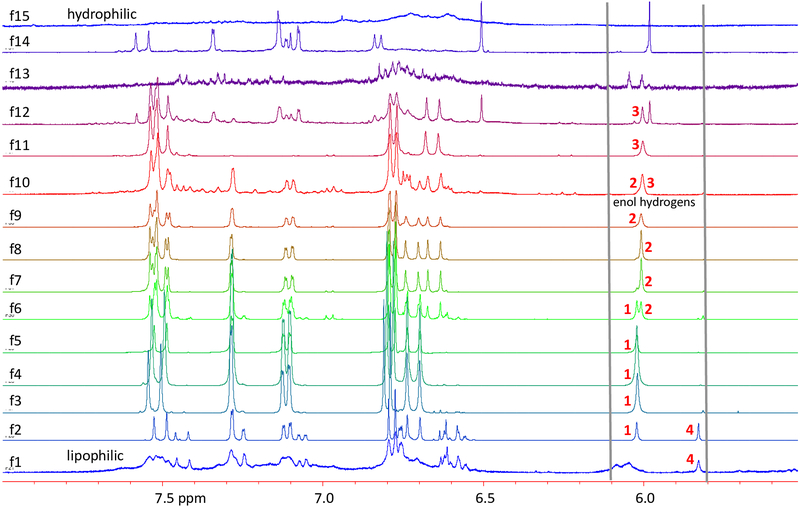
The stacked 1H NMR spectra of the 5.5 to 8.0 ppm region of the HSCCC fractions of the CTE (Figure 5) showed the separation pattern of 1-4 as well as the presence of low concentration compounds with related structures. Indeed, a wide array of compounds have been described in turmeric extracts, including at least 22 curcuminoid diarylheptanoids.1 Comparative 1H NMR spectroscopic analysis of HSCCC fractions provided a qualitative and quantitative complement to TLC and HPLC.
Figure 5.
The 5.5 to 8.0 ppm 1H NMR regions of the CCS fractions acquired from the normal phase separation of curcuminoid-enriched turmeric extract (CTE) by HSCCC. Fractions 4, 8, and 11 served as reference standards for the generation of HiFSA profiles of 1, 2, and 3, respectively.
The qHNMR spectroscopy purity analysis of DESIGNER “purcumin” via the relative 100% method24 indicated that the sample contained 95.56 w/w % 1 and 3.19% 4 along with residues arising from chromatographic solvents (accounting for the residual 1.25%). The purity analysis of DESIGNER “purcuminoids” determined that the sample contained 91.75% of the three curcuminoids, 1–3, and 2.54% of the three corresponding tetrahydrocurcuminoids, 4–6. The remaining 5.71% was attributed to solvents. Further peak-fitting guided qHNMR spectroscopic analysis (PF-qHNMR spectroscopy, via global spectral deconvolution [GSD] in MNova) of the enol hydrogen signals between 5.25 – 6.25 ppm provided further refinement of the previous values of 1, 2, and 3 to 37.96%, 43.88%, and 9.49% (updated sum: 91.33%), respectively, and the tetrahydrocurcuminoids to 2.97%. Quantum-mechanics based qHNMR (QM-qHNMR) spectroscopy of the same sample using 1H iterative full-spin analysis (HiFSA)25 indicated that the mass ratios of 1/2/3 were 44.14/45.82/10.01 (Table S4, Supporting Information), which is in good agreement with the GSD results (41.57/48.04/10.39). This also confirmed that the underlying spin systems are essentially first order and, thus, amenable to PF-qHNMR spectroscopy.
Apart from looking specifically at known curcuminoids as marker compounds, the qNMR spectroscopic analysis of the four DESIGNER fractions was extended to seven significant regions that afforded insight into the RC of these fractions. This was accomplished by choosing seven δ regions (shown in Figure S5, Supporting Information) that are significant in the analysis of curcuminoids and may be integrated separately: (i) the 0.10–2.48 ppm region represents aliphatic hydrogens; (ii) the DMSO-d5 resonance is found in the 2.48–2.52 ppm region; (iii) 2.52–3.70 ppm is a region where some alkoxy hydrogen signals are found, as well as those of the methylene hydrogens of tetrahydrocurcuminoids; (iv) the curcuminoid methoxy hydrogens resonate in the 3.70–4.00 ppm region; (v) the 4.00–5.25 ppm region was rather barren in these studies; (vi) the 5.25–6.25 ppm region reflects the enol hydrogens associated with curcuminoids; and (vii) 6.25–8.00 ppm region contains the aromatic hydrogen resonances associated with curcuminoids but also other metabolites. The % total integration values were calculated without the DMSO signal. The “purcumin” and “purcuminoids” DESIGNER fractions showed a similar relative distribution of regions iv, vi, and vii of 36, 4, and 57% and 27, 4, 65%, respectively. The lipophilic fraction had a distribution that favored regions i, iii, and vii at 54, 20, and 14%, indicative of the presence of aliphatic compounds with a significant amount of aromatic moieties. The hydrophilic fraction had a total % integration distributed between regions i, iii, iv, and vii at 54, 7, 9, and 27%, which indicates a significant amount of aliphatic and aromatic hydrogens with likely more oxygenation present. All four of the DESIGNER extracts exhibited signals in the 5.25–6.25 ppm region, which indicates that congeneric curcumin derivatives may populate even the lipophilic and hydrophilic fractions. For example, lipophilic terpecurcuminoids that could partially account for the RC of the lipophilic fraction have been reported recently.26 Considering the solvent extraction process and impact of NADES (see introduction), the presence of constituents with vastly different polarity in enriched or even “pure” materials is plausible. Accordingly, the presence of lipophilic Curcuma oleoresin terpenoid chemistry in curcuminoid fractions is nearly inevitable, in quantities that obviously depend largely on the process that generated the material. To support future investigations of the phytoconstituents in the residually complex lipophilic and hydrophilic fractions, the raw NMR data of the present study are made available to the reader (see Supporting Information).
In conclusion, application of the DESIGNER approach to a CTE, via a series of CCS steps with a single SS afforded the four chemically highly distinct and well-defined fractions of turmeric extract: (i) the lipophilic (residual oleoresin) metabolites; (ii) purified 1 (“purcumin”); (iii) a mixture of 1-3 that is largely free of other compounds (“purcuminoids”); and (iv) the hydrophilic metabolites. The starting material of this study, CTE, represents an enriched but specialized form of crude turmeric, notably still not identical with turmeric nor with curcumin. Accordingly, fractionation of raw turmeric or crude CE under identical conditions would have yielded many more constituents populating the fractions, particularly metabolites that are more and/or less lipophilic than the curcuminoids, 1–3. The same CCS protocol may be employed reproducibly with any turmeric extract to produce similar DESIGNER fractions. Even synthetic curcumin may be fractionated by this approach to reveal a very different residual complexity landscape. It is important to note that, depending on the starting material, the lipophilic and hydrophilic DESIGNER fractions (“A” and “X” fractions; see above) can be expected to vary substantially in composition and weight: both “A” and “X” will increase significantly in starting materials such as T and TE that are cruder and/or more chemically complex, relative to the CTE used in this study (see also Figure 3).
A combination of TLC, HPLC, 1H-NMR and qHNMR methodologies were used to analyze the DESIGNER materials, with qHNMR being the principal method for quantitation. The “purcumin” DESIGNER material was >95% 1 with the remainder being residual solvents. Exogenous solvents are inevitably introduced into the sample during extraction, chromatography, and sample workup. Most analytical methods including routine LC will not detect and quantify residual solvents to the extent readily possible through NMR spectroscopy. As can be expected, even a highly selective two-step separation scheme will produce target compounds, such as 1 in the “purcumin” DESIGNER fraction, with an inevitable level of residual complexity (RC). This may be addressed by careful evaluation of the RC via LC and qNMR. The “purcuminoids” DESIGNER fraction was primarily a combination of 1-3 (over 91.5%) along with other curcuminoids (nearly 3%). The remainder was again identified as residual solvents. Depending on the desired use of the DESIGNER materials, further fractionation with orthogonal methods such as a third CCS step or preparative HPLC may be undertaken. Potential considerations for future studies include a full metabolomic characterization of the DESIGNER materials and development of custom CSS purification protocols for 2 and 3.
In addition to testing the four DESIGNER fractions separately, Figure 6 shows other potential DESIGNER materials that may be produced for biological testing by formulating a series of enrichment, depletion, and/or knockout steps. Employed individually or in combination, these C. longa DESIGNER materials can be viewed as unique biological tools that can help fill the gap between chemical composition and biological endpoints of turmeric-derived NP bioassays. Of particular interest will be the role 1 plays, both as a single-chemical entity (SCE; tested as e.g. “purcumin”) and as a RC component, in the biological activities that have been attributed to it without direct testing of either purified 1, or a turmeric extract depleted in 1 (“curcumin knockdown”), or an extract without any curcuminoids (“nocumin”; Figure 6). This will likely shed new light on the many studies that have attributed the biological activity of turmeric (T) and turmeric extracts (TE) to 1 simply because it is the most prominent constituent of the formulation.4,27
Figure 6.
Deplete and Enrich Select Ingredients to Generate Normalized Extract Resources (DESIGNER) scheme for extracts that can be produced from turmeric by selective removal of, e.g., pure 1 (“purcumin”) and/or a highly refined mixture of the curcuminoids 1–6 (“purcuminoids”). This enables the generation of unique pharmacological tools, such as a curcuminoid-free C. longa knockout extract (“nocumin”; bottom).
Considering the impact of compound instability (see above), and combined with systematic studies of the chemical variability of crude turmeric, this can contribute to new insights regarding the truly bioactive species and their mechanisms of action.28 The biological activity associated with an extract, such as TE, is not conclusive until each constituent has been tested individually along with every possible combination of constituents that may reveal additive, subtractive, synergistic, or antagonistic properties. This is a fairly unrealistic goal, however, given the complexity of plant extracts. Meanwhile, the DESIGNER approach allows these types of studies to be conducted with more feasibility and avoids the reductionist approach of evaluating single-chemical entities as representative of the substantial herbal metabolomic complexity. The present outcomes enable such studies by providing a two-step preparative procedure to the production of unique C. longa DESIGNER materials, along with their comprehensive qHNMR-based characterization.
EXPERIMENTAL SECTION
Materials and Reagents.
CTE was obtained from Sigma (St. Louis, MO, USA), as article C1386, with a declared purity ≥65% by HPLC. Other sources of curcuminoid preparations were: Jarrow Formulas, Inc. (Los Angeles, CA, USA) Curcumin 95 (reported 95% of 1) and Alfa Aesar (Tewksbury, MA, USA) B21573 (reported 95% of 1).
Samples were stored at −20 °C until analyzed. DMSO-d6 (99.9% D) was obtained from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). The other chemicals were reagent grade from Sigma-Aldrich Inc. (St. Louis, MO, USA). All organic solvents were analytical grade, purchased from Pharmco-AAPER (Crookfield, CT, USA), and distilled before use.
CCS Instruments.
CPC employed a SCPC-250 (Armen Instrument, Saint Ave, France) chromatograph equipped with a 266 mL rotor containing 16 stacked disks with a total of 864 twin-cells. The total active volume was 210 mL and the volume of the interconnecting ducts was 56 mL. The rotation speed could be adjusted from 500 to 3000 rpm, thus producing a g-force in the cells up to 700 g. Samples were injected through a 10 mL sample loop. The solvents were pumped through a semi-preparative 4-way binary high-pressure pump (50 mL/min maximum flow rate, 150 bar). The detection was performed by a UV-Vis DAD detector at 210 and 254 nm. Fractions were collected with an Armen Fraction Collector LS-5600. Chromatographic data were acquired by using the Armen Glider CPC Control Software V2.9.2.9 and then transferred to an Excel worksheet for further processing.
For hydrodynamic countercurrent chromatography (HSCCC), a TBE-300A (Shanghai Tauto Biotech Co., Shanghai, People’s Republic of China) instrument was used, consisting of three multilayer coil separation columns connected in series (1.9 mm tubing i.d.) to give a 280 mL total column volume, and equipped with a 20-mL sample loop. The revolution radius or the distance between the holder axis and the central axis of the centrifuge (R) was 5 cm, and the β values of the multilayer coil varied from 0.5 at the internal terminal to 0.8 at the external terminal (β = r/R where r is the distance from the edge of the coil to the holder shaft). The rotational speed of the apparatus could be regulated with a speed controller with the range 0–1000 rpm. A Neslab RTE7 constant temperature-circulating bath (Thermo Fisher Scientific, Newington, NH, USA) was used to control the separation temperature within the range of 5–35 °C. The CCC system was equipped with a ChromTech (Apple Valley, MN, USA) Series I digital single-piston solvent pump, a JMST Systems (Colorado Springs, CO, USA) VUV-14D fixed wavelength UV–vis detector (254 nm filter) with preparative flow cell, and an Advantec (Dublin, CA, USA) CHF122SC fraction collector. Data were recorded on a PEAK-ABC Chromatography Data Handling System and then transferred to an Excel worksheet for further processing.
General CCS Methods.
Unless otherwise indicated, all experiments were performed using the upper phase as the stationary phase. The system was first filled with the stationary phase (20 mL/min CPC and 3 mL/min HSCCC). The flow rate was then adjusted to the experimental value and rotation initiated. The mobile phase was then pumped in the descending or head-to-tail direction through the column (10 mL/min CPC and 2 mL/min HSCCC). The sample was injected either before or after column equilibration had been achieved. The stationary phase volume retention ratio (Sf value) was measured either by the void volume (injection before equilibration) or graduated cylinder (injection after equilibration) method. Sweep elution followed by extrusion was initiated by pumping the upper phase. Fractions were collected in 15-mL (CPC) or 10-mL (HSCCC) volumes.
Sample Preparation.
CCS samples were dissolved in equal volumes of upper and lower phase to a total volume of 10 to 15 mL. Samples were sonicated and filtered (0.45 μm Millipore glass fiber syringe filter) before introduction into the sample loop. CPC samples were dissolved in 1 mL DMSO, sonicated, and diluted to 9 mL with n-hexane/CH2Cl2/MeOH/water 3:7:7:3 lower phase before filling the sample loop.
For (q)NMR analysis, samples were dissolved in 200 μL DMSO-d6, measured with a VICI Pressure-Lok precision analytical syringe (Valco Instruments, Baton Rouge, LA, USA) in 3 mm Norell S-3_HT-7 NMR tubes.
Thin-Layer Chromatography.
Collected fractions were dried and analyzed by TLC (mobile phase: lower phase of n-hexane/CH2Cl2/MeOH/water 3:7:7:3). The chromatograms were visualized under visible light, and at 254 nm and 360 nm UV light without chemical derivatization. TLC plates were Macherey-Nagel Alugram SIL G/UV254 10 × 20 cm, with 0.2 mm silica gel layer thickness (Easton, PA, USA).
Gas Chromatography.
A Perkin Elmer (Shelton, CT, USA) Clarus 600 instrument with a Restek (Bellefonte, PA, USA) Stabilwax-DA 30 m column (0.53 mm i.d. and 1.0 μm df) was used for GC analysis. Sample injection volume was 1.0 μL. The GC instrument was interfaced with a computer running Totalchrome Navigator Workstation v. 6.8 software.
Solvent System Composition Studies.
Solvent system (SS) composition samples were prepared by mixing a total volume of 100 mL of biphasic SS, separating the two layers. Subsequently, 50 μL of each phase was mixed with 1.5 mL acetone (performed in triplicate). Standard curves were created for n-hexane, CH2Cl2, CHCl3, and MeOH to determine their respective response factors. The oven program was held at 30 °C for 4 min, increased to 50 °C at 2 °C/min, held for one min, and then increased to 200 °C at 10 °C/min. The flow rate was 80.0 mL min with a 19:1 split ratio. A flame ionization detector (FID) was held at 325 °C with a H2 (45.0 mL/min) and air (450.0 mL/min) feed.
HPLC.
A Perkin Elmer series 200 HPLC with a double headed pump, autosampler, UV/Vis detector, solvent degasser, and dot LINK data handling system was employed. The instruments were interfaced with a computer running Totalchrome Navigator Workstation v. 6.8 software. A HICHROM Apollo 5 2μm C18 250 × 4.6 mm column was used. The solvent feed was acetonitrile and 0.1% aqueous formic acid in a 55:45 ratio at a 0.8 mL/min flow rate. The injection volume was 10 μL, with UV detection at 340 nm. The solvent gradient method started with acetonitrile/0.1% aqueous formic acid held at 55:45 for 20 min. This was followed by a five-min linear gradient to acetonitrile/0.1% aqueous formic acid 100:0. This ratio was held for five min. Afterward, the column was re-conditioned by a five-min linear gradient to acetonitrile/0.1% aqueous formic acid 55:45, which was held for five more min.
Partition Coefficient Measurements.
Sigma C1386 CTE (9 – 11 mg) was weighed into a 20 mL vial. The partition coefficient was measured as the ratio of the area under the peak for a particular compound in the upper and lower phases. Next, 5 mL of each upper and lower phases were added, the mixture was sonicated for 10 min, and the upper and lower phases allowed to separate. A 100 μL aliquot of each phase was diluted to 0.5 mL with MeOH for HPLC analysis. The partition coefficient was measured as the ratio of the peak areas for a particular compound in the upper and lower phases.
Sample Loading Studies.
Sigma C1386 CTE (50 – 60 mg) was weighed, to the nearest mg, into a 20 mL vial. To that vial, 2 mL of each of the upper and lower phases were added, the mixture sonicated for 10 min, and the solution filtered through filter paper into a pre-weighed side-arm test tube. The solvents were evaporated under forced air and the tube weighed to the nearest mg. Each experiment was done in triplicate.
Quantitative 1H NMR (qHNMR) Spectroscopic Analysis.
The NMR spectra were acquired on a JEOL (Jeol Resonance Inc., Peabody, MA, USA) ECZ 400 MHz spectrometer. A total of 64 scans (NS) were acquired, collecting 64 k of time domain (TD) data, and using a 45 degree excitation pulse as well as a relaxation delay (D1) of 30 sec. Post-acquisition processing used NUTS (Acorn NMR Inc. Livermore, CA) and MestReNova-10.0.1–14719 (Mestrelab Research, Santiago de Compostela, Spain) software. Zero-filling was applied to at least 512 k prior to Fourier transformation of the FID. Line shape and resolution were improved with Gaussian-Lorentzian window functions (LB −0.3 and GF 0.05). Phase correction was implemented manually, and baseline correction used a 5th order polynomial function. Integrals were used as default quantitative measures. Peak-fitting (PF) via global spectral deconvolution (GSD), and quantum-mechanics (QM)-based methods was conducted as described previously and as given below.29
1H NMR Full Spin Analysis (HiFSA).
PERCH NMR software (v.2010.1, PERCH Solutions Ltd., Kuopio, Finland) was used for full spin analysis. The resolution-enhanced 1H NMR spectra were imported into PERCH as JCAMP-DX files and subjected to baseline correction, peak picking, and integration. The three-dimensional models of 1-3 were built using the molecular modeling system (MMS) module. After geometry optimization and molecular dynamics simulations, 1H NMR parameters in DMSO-d6 were predicted. After a manual examination of the 1H NMR assignments, the calculated 1H NMR chemical shifts, signal line widths, and J-couplings were refined using both the integral-transform (D) and total-line-fitting (T) modes, until an agreement between the observed and simulated spectra was reached (iteration convergence) to yield the HiFSA fingerprints of 1–3.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NCCIH and ODS of the NIH through grant U41 AT008706. The authors are grateful to the greater CENAPT team for valuable discussions and general help with laboratory procedures. The authors also wish to acknowledge the excellent CPC support from Gregoire Audo, of Armen Instruments, Saint Ave, France and Gilson Inc., Middleton (WI); the extensive technical and CCS knowledge provided by Samuel Pro of Wrightwood Technologies Inc., Chicago (IL); the kind technical support by Tommy Li Deng of Tauto Biotech, Shanghai, People’s Republic of China; the expert NMR support from Ashok Krishnaswami, of JEOL Resonance Inc., Peabody (MA); and the advice regarding quantum mechanical NMR analysis from Matthias Niemitz of NMR Solutions, Kuopio, Finland. Finally, we are grateful to Jonathan Bisson, Anton Bzhelyansky, Jayme Dahlin, James Graham, Kathryn Nelson, Charlotte Simmler, and Michael Walters for fruitful and inspiring discussions of the extended “curcumin” topic.
Footnotes
Supporting Information
The Supporting Information is available free of charge at the ACS Publications website at DOI: pending NMR spectra in DMSO-d6.
The original NMR data (FIDs) of the spectra in the figures, and the DESIGNER lipophilic, “purcumin,” “purcuminoids,” and hydrophilic extracts are made available at DOI: http://dx.doi.org/10.7910/DVN/ERQ7AV.
The authors declare no competing financial interests.
REFERENCES
- (1).Li S; Yuan W; Deng G; Wang P; Ynag P; Aggarwal BB Pharm. Crops 2011, 2, 28–54. [Google Scholar]
- (2).Kumar A; Chetia H; Sharma S; Kabiraj D; Talukdar NC; Bora U Database 2015, 2015, bav070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Antony B U.S. Patent US8859020B2. 2014.
- (4).Nelson KM; Dahlin JL; Bisson J; Graham J; Pauli GF; Walters MA J. Med. Chem 2017, 60, 1620–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pauli GF; Chen SN; Friesen JB; McAlpine JB; Jaki BU J. Nat. Prod 2012, 75, 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Choules MP; Klein LL; Lankin DC; McAlpine JB; Cho SH; Cheng J; Lee H; Suh JW; Jaki BU; Franzblau SG; Pauli GF J. Org. Chem 2018, 83, 6664–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Singh G; Kapoor IP; Singh P; de Heluani CS; de Lampasona MP; Catalan CA Food Chem. Toxicol 2010, 48, 1026–1031. [DOI] [PubMed] [Google Scholar]
- (8).Kuttigounder D; Lingamallu JR; Bhattacharya SJ Food. Sci 2011, 76, C1284–C1291. [DOI] [PubMed] [Google Scholar]
- (9).Gordon ON; Luis PB; Sintim HO; Schneider CJ Biol. Chem 2015, 290, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schneider C; Gordon ON; Edwards RL; Luis PB J. Agric. Food Chem 2015, 63, 7606–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ramos Alvarenga RF; Friesen JB; Nikolic D; Simmler C; Napolitano JG; van Breemen R; Lankin DC; McAlpine JB; Pauli GF; Chen SN J. Nat. Prod 2014, 77, 2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chen SN; Turner A; Jaki BU; Nikolic D; van Breemen RB; Friesen JB; Pauli GF J. Pharm. Biomed. Anal 2008, 46, 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dietz BM; Chen SN; Alvarenga RFR; Dong H; Nikolic D; Biendl M; van Breemen RB; Bolton JL; Pauli GF J. Nat. Prod 2017, 80, 2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kamto EL; Carvalho TS; Mbing JN; Matene MC; Pegnyemb DE; Leitao GG J. Chromatogr. A 2017, 1480, 50–61. [DOI] [PubMed] [Google Scholar]
- (15).Chadwick LR; Fong HHS; Farnsworth NR; Pauli GF J. Liq. Chromatog. Relat. Technol 2005, 28, 1959–1969. [Google Scholar]
- (16).Inoue K; Nomura C; Ito S; Nagatsu A; Hino T; Oka HJ Agric. Food Chem 2008, 56, 9328–9336. [DOI] [PubMed] [Google Scholar]
- (17).Liu Y; Friesen JB; Grzelak EM; Fan Q; Tang T; Duric K; Jaki BU; McAlpine JB; Franzblau SG; Chen SN; Pauli GF J. Chromatogr. A 2017, 1504, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kukula-Koch W; Grabarska A; Luszczki J; Czernicka L; Nowosadzka E; Gumbarewicz E; Jarzab A; Audo G; Upadhyay S; Glowniak K; Stepulak A Phytother. Res 2018, 32, 933–942. [DOI] [PubMed] [Google Scholar]
- (19).Patel K; Krishna G; Sokoloski E; Ito YJ Liq. Chromatog. Relat. Technol 2000, 23, 2209–2218. [Google Scholar]
- (20).Alfonsi K; Colberg J; Dunn PJ; Fevig T; Jennings S; Johnson TA; Kleine HP; Knight C; Nagy MA; Perry DA; Stefaniak M Green Chem. 2008, 10, 31–36. [Google Scholar]
- (21).Huang MT; Ma W; Lu YP; Chang RL; Fisher C; Manchand PS; Newmark HL; Conney AH Carcinogenesis 1995, 16, 2493–2497. [DOI] [PubMed] [Google Scholar]
- (22).Pari L; Murugan P Pharmacol. Res 2004, 49, 481–486. [DOI] [PubMed] [Google Scholar]
- (23).Lobo R; Prabhu KS; Shirwaikar A; Shirwaikar AJ Pharm. Pharmacol 2009, 61, 13–21. [DOI] [PubMed] [Google Scholar]
- (24).Pauli GF; Chen SN; Simmler C; Lankin DC; Godecke T; Jaki BU; Friesen JB; McAlpine JB; Napolitano JG J. Med. Chem 2014, 57, 9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Napolitano JG; Godecke T; Rodriguez-Brasco MF; Jaki BU; Chen SN; Lankin DC; Pauli GF J. Nat. Prod 2012, 75, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lin X; Ji S; Li R; Dong Y; Qiao X; Hu H; Yang W; Guo D; Tu P; Ye MJ Nat. Prod 2012, 75, 2121–2131. [DOI] [PubMed] [Google Scholar]
- (27).Nelson KM; Dahlin JL; Bisson J; Graham J; Pauli GF; Walters MA ACS Med. Chem. Lett 2017, 8, 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Booker A; Frommenwiler D; Johnston D; Umealajekwu C; Reich E; Heinrich MJ Ethnopharmacol. 2014, 152, 292–301. [DOI] [PubMed] [Google Scholar]
- (29).Phansalkar RS; Simmler C; Bisson J; Chen SN; Lankin DC; McAlpine JB; Niemitz M; Pauli GF J. Nat. Prod 2017, 80, 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Friesen JB; Pauli GF Anal. Chem 2007, 79, 2320–2324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.