Abstract
DNA double strand breaks (DSBs) are one of the most deleterious DNA lesions that promote cell death, genomic instability and carcinogenesis. The two major cellular mechanisms that repair DSBs are Nonhomologous End-Joining (NHEJ) and Homologous Recombination Repair (HRR). NHEJ is the predominant pathway, in which XLF (also called Cernunnos) is a key player. Patients with XLF mutation exhibit microcephaly, lymphopenia, and growth retardation, and are immunodeficient and radiosensitive. During NHEJ, XLF interacts with XRCC4-Ligase IV, stimulates its ligase activity, and forms DNA-binding filaments of alternating XLF and XRCC4 dimers that may serve to align broken DNA and promote ligation of noncomplementary ends. Despite its central role in NHEJ, the effects of XLF deficiency are surprisingly variable in different biological contexts, and different individual cell lines. This review summarizes the role of XLF in NHEJ, and the unexpected complexity of its interplay with other repair factors in supporting radiosurvival and V(D)J recombination.
Keywords: Nonhomologous End-Joining (NHEJ), XLF/Cernunnos, XRCC4, V(D)J Recombination
Introduction:
Mammalian cells are constantly subjected to a wide variety of attacks both endogenously via normal cellular processes and exogenously via environmental agents. These assaults give rise to a multitude of DNA lesions, the most cytotoxic of which are DSBs and interstrand crosslinks. DSBs can arise from diverse sources including reactive oxygen species produced by normal cellular metabolism, collision of replication forks with single strand breaks, V(D)J and class switch recombination processes for T and B-cell development, as well as radiation and radiomimetic or topoisomerase-targeted anti-cancer drugs [1–4]. DSBs that remain unrepaired or which undergo defective repair could lead to senescence, aging, chromosomal aberrations, and ultimately cancer. As a result, eukaryotic cells have evolved mechanisms namely Homologous Recombination Repair (HRR) and Nonhomologous End-Joining (NHEJ) that can counter these lesions. NHEJ is a complex process that involves crosstalk between a large number of proteins. Unlike HRR, NHEJ does not require a homologous template and can mediate the direct religation of broken DNA ends, allowing it to function in all phases of the cell cycle, while at the same time rendering it highly error-prone. The basic steps in NHEJ mainly include; 1) Recognition of the damaged ends and assembly of the NHEJ complex; 2) Stabilization of the NHEJ complex at the DSB ends; 3) DNA end-processing and 4) Ligation of the DSB broken ends and clearing of the NHEJ complex [5–7]. These steps involve interaction of a variety of proteins the most essential of which are Ku heterodimer, DNA-PKcs, Artemis, XRCC4, XLF, and DNA Ligase IV (LIG4).
XLF (XRCC4-like factor), also called Cernunnos, was originally identified as the protein defective in a subset of RS-SCID patients who, in addition to immunodeficiency and radiosensitivity, displayed growth retardation, microcephaly, and lymphopenia. The radiosensitivity of cells derived from these patients could be rescued by complementing with a wild type XLF cDNA [8]. Independently, XLF was identified as a protein that interacts with the XRCC4-DNA Ligase IV complex (X4L4) during NHEJ [9]. XLF is a homologue of Nej1, one of the NHEJ factors identified in yeast [10,11]. Data from multiple studies suggest that although XLF may not be strictly required for NHEJ, the loss of XLF does affect NHEJ and interestingly, symptoms of XLF patients (radiosensitivity and impaired DSB repair) bear resemblance to those of the more severe LIG4 syndrome patients [12]. Thus, XLF inactivation manifests a condition similar to partial loss of LIG4. During NHEJ, the interactions of XLF with X4L4 are important for successful NHEJ. This review will specifically focus on the functions of the XLF protein and discuss its interplay with major players involved in NHEJ and V(D)J recombination.
XLF Structure:
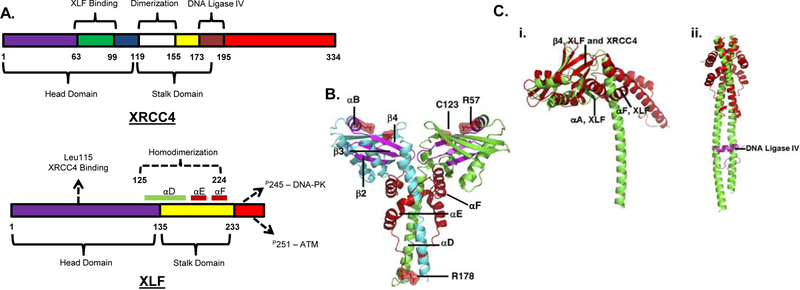
NHEJ is strongly dependent on 5 core proteins: Ku, DNA-PKcs, XRCC4, LIG4 and XLF/Cernunnos with lesser contributions from a host of additional factors. Although XLF has only limited sequence homology to XRCC4, the two proteins are structurally quite similar [13]. Both mainly exist as dimers and can bind DNA [14]. XRCC4 has a globular head domain (amino acids 1–119), an elongated α-helical stalk domain (also called the coiled coil domain, amino acids 120–180) and a C-terminal domain whose function is not yet determined [15,16] (Fig. 1A). Similarly, XLF has a globular head domain (amino acids 1–135), a stalk domain (amino acids 136–233) and a C-terminal domain whose structure was not determined [17] (Fig. 1A, B). Both proteins form homodimers through interaction of regions mostly within the stalk (amino acids 119–155 in XRCC4 [15,18] and amino acids 125–224 in XLF [17,19]). However, X-ray crystallography shows that XLF differs from XRCC4 in that the angle formed between the globular head and coiled coil domain is larger than in XRCC4 [19]. Furthermore, whereas the stalk domain of XRCC4 is a single helix that extends away from the globular head domain with a slight conformational change at the LIG4 binding site, the stalk region of XLF is split into three helices and folds back on itself, precluding the binding of LIG4 to XLF [19] (Fig 1C).
Figure 1.
XLF structure and comparison with XRCC4. A. Domain structures of NHEJ proteins showing select binding sites. XRCC4: The head domain comprises amino acids 1–119 and the stalk domain 120–180 [16]. XLF: The head domain comprises amino acids 1–135 and the stalk domain 136–233. α-Helices D, E and F constituting the stalk are highlighted. The Leu115 residue is important for XRCC4 binding. DNA-PK phosphorylates Ser245 and ATM phosphorylates Ser251. Adapted from [13]. B. Structure of the XLF dimer as determined from crystallography of the 1–233 fragment. Cyan and green α-helices distinguish the two monomers. Segments that would be deleted as a result of a disease-associated nonsense mutation at R178 are shown in red. R57 and C123 are sites of disease-associated point mutations. The region spanning A25 to R57 is shown in magenta. (Image reproduced with permission from [17]). C. (i). Comparison of XLF (red) and XRCC4 (green) structures. α-Helices and antiparallel β-sheets are nearly superimposable in the head domains, but the stalk domains diverge. The angle between the head and stalk is larger in XLF than in XRCC4 because of the insertion of helices αF and αA. (ii). The coiled-coil in XLF is much shorter than that in XRCC4, and does not contain an equivalent region to the LIG4 binding site of XRCC4. LIG4 fragment bound to XRCC4 is shown in magenta. (Image reproduced with permission from [17].)
DNA Binding
The 68-aa C-terminal region of XLF confers its DNA-binding abilities, and the binding of XLF to DNA is length-dependent. As determined by electrophoretic mobility shift assays, the minimum size of DNA required for XLF binding is ~83bp [20] as compared to ~200bp for XRCC4 [21]. However, when both Ku and XLF were added to a shorter 65-bp duplex, a distinct, discrete band corresponding to the putative ternary complex of DNA, Ku and XLF could readily be detected [22], in contrast to the diffused and misshapen bands typically seen in gel shifts by XLF alone. Thus, Ku recruits XLF to DNA and presumably targets it to DNA ends.
Interaction with XRCC4
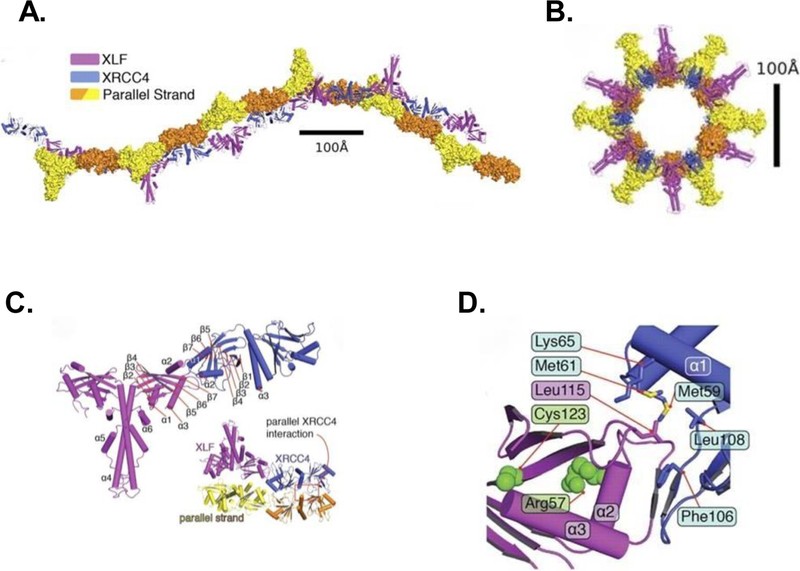
Many proteins interact with XRCC4 and among them, XLF interacts with amino acids 63–99 in the head domain [19], while LIG4 interacts with amino acids 173–195 in the stalk (Fig. 1Cii) [23]. XRCC4/XLF binding occurs via Leu115 of XLF occupying a hydrophobic pocket on XRCC4 [17,19]. This interaction creates a helical filament of alternating XRCC4 and XLF dimers (Fig 2A–D). Structural analysis of the XRCC4/XLF filament indicates that it forms an extended grooved channel that is positively charged and binds DNA [24]. It is likely that the filament thereby stabilizes DNA end synapsis or aids in end alignment but the precise geometry by which it does so is not clear.
Figure 2.
Structure of the XLF-XRCC4 filament as determined by crystallography. A. Two orthogonal views of the biological unit (one helical turn) of the filament formed by XLF(1–224) (magenta) and XRCC4(1–140) (blue), as seen in the crystal lattice. Parallel strands (yellow/orange) of a second filament are in surface representation. B. View of the biological unit rotated by 90°. C. Overall architecture of the XLF(1–224)-XRCC4(1–140) complex, colored magenta and blue, respectively. Helices α4-α6 correspond to helices αD-αF in Figure 1. Inset shows interaction between XRCC4 dimers in parallel filaments. D. Interface between head domains of XLF(1–224) (magenta) and XRCC4(1–140) (blue). Leu115 of XLF occupies a hydrophobic pocket formed by XRCC4 residues Leu108, Phe106, Met59, Met61, and Lys65. (Image reproduced with permission from [24].)
HA immunoprecipitates from 293T cells transfected with HA-tagged XLF showed co-immunoprecipitation of XRCC4 and LIG4, verifying interactions of the two proteins in cells [9]. Furthermore, purified GST-XLF protein efficiently co-precipitated XRCC4 and LIG4 from HeLa nuclear extracts. XRCC4 is needed for the stability of LIG4 against proteolysis [25], and in XRCC4-knockdown cells, XLF does not compensate for the XRCC4 deficiency. XLF does not contribute to LIG4 stability and no known complex of XLF and LIG4 without XRCC4 is detected in cells. However, in the absence of LIG4, the interaction of XLF with XRCC4 was reduced, suggesting that LIG4 is needed for optimal formation of an XRCC4/XLF/LIG4 complex in cells [26,27].
XLF and NHEJ
NHEJ is a dynamic process that involves a systematic recognition of the DNA DSBs followed by recruitment of repair proteins and subsequent processing and ultimate religation of the DNA ends. NHEJ is initiated by the binding of the Ku heterodimer, which stabilizes broken DNA ends and protects them from nucleolytic degradation [28]. Ku then recruits the DNA-PK catalytic subunit (DNA-PKcs), forming the DNA-PK holoenzyme [29,30]. The DNA-PK holoenzyme facilitates the recruitment of the X4L4 complex [26,31] which, although it can bind DNA on its own, is ineffective in specifically recognizing DSB ends unless recruited to those sites by Ku and DNA-PK [31–33]. Once bound to DNA, DNA-PK undergoes autophosphorylation to recruit end-processing enzymes such as Artemis to process modified DNA ends [34]. Fibroblasts that lack detectable XLF, for example the 2BN cell line from an RS-SCID patient, show a profound DSB repair defect as indicated by persistent γ-H2AX foci and broken DNA as assessed by pulsed field gel electrophoresis. On complementation of these cells with wild-type XLF, the repair-proficient phenotype was restored [8,9]. Similarly, an XLF−/− derivative of HCT116 colorectal cancer cells shows a severe deficiency in end joining of transfected plasmid substrates and sensitivity to etoposide, which induces topoisomerase II-associated DSBs [35].
Functionally, XLF enhances the ligation activity of the X4L4 complex. Using purified proteins and quantitative PCR, it was shown that XLF also facilitates the joining of noncohesive ends by X4L4 in the presence of Ku [36]. For example, it promotes the joining of two noncomplementary 4-base 3′-overhangs and, to a lesser extent, noncomplementary 5′-overhangs, as well as the joining of either a 3′ or a 5′ overhang to a blunt end (without ligation of the recessed complementary strand). While the absolute extent of end joining is less for noncomplementary than for cohesive ends, the degree of stimulation by XLF is much greater, with joining of several non-cohesive substrates being stimulated by ~100-fold in the presence of XLF. While addition of DNA-PKcs increased ligation efficiency, it decreased the degree of stimulation by XLF. A similar study using 32P-radiolabeled substrates showed that X4L4 is able to ligate noncomplementary overhangs, that the extent of ligation is strongly dependent on the exact sequence of the overhangs, and that, in general, the least favorable substrates for X4L4-mediated ligation showed the strongest stimulation by XLF. Ligation of single strands (i.e. dT30) by X4L4 was similarly enhanced by XLF. For some substrates but not others, the effect of XLF could be mimicked by higher-than-physiologic Mg2+, suggesting a possible charge neutralization function for XLF. Overall, it was concluded that X4L4 can optimally align some substrates on its own, while other substrates required XLF for optimum alignment [37].
Using blunt-ended DSB substrates harboring the oxidatively modified nonplanar base thymine glycol (Tg) at the first, second, third or fifth positions from one 3′ terminus (Tg1, Tg2, Tg3 and Tg5 respectively), NHEJ of damaged DSB ends was examined in human whole-cell extracts [38]. In XLF-deficient extracts, end joining of all substrates, including unmodified blunt ends, was completely dependent on addition of recombinant XLF. On the other hand, in ligation reactions using only purified Ku, X4L4 and XLF, ligation of both Tg3 and Tg5 (as well as unmodified blunt ends) was efficient and only partly XLF-dependent. However, Tg1 and Tg2 ligation was completely XLF-dependent. These results suggest a possible role for XLF in alignment of DSB ends containing structurally disruptive lesions such as Tg, that otherwise would be poor ligation substrates. Such clustered lesions are induced at relatively high frequency by ionizing radiation. The strict requirement for XLF for ligation of all substrates in extracts suggests that there are additional proteins present that block X4L4-mediated ligation and that XLF addition somehow relieves that block.
Polymerases µ and λ are required to fill gaps during NHEJ [39]. The immunodepletion of Pol λ and not Pol µ affects NHEJ in cell extracts [40]. Pol µ interacts with Ku and XRCC4 and is recruited to DNA damage foci [41]. In XLF-deficient Bustel cell extracts, addition of recombinant XLF was required for Pol µ- and λ-mediated gap filling in aligned DSB ends [42] indicating a significant role of XLF in NHEJ beyond merely enhancing X4L4 ligation activity.
All these results would be consistent with a role for XLF in promoting alignment of DNA ends, leading to speculation that such alignment is a function of the XRCC4/XLF filament (Fig. 2). The extended, positively charged channel created by the filament suggests that bound DNA would likely be collinear with the filament [24]. Thus, a filament spanning a DSB could position two DSB ends in juxtaposition (Fig. 3A). However in this model, it is unclear how DNA-PKcs, LIG4 or the processing enzymes like PNKP gain access to the DNA ends. It could be that XRCC4-XLF filaments form only after the autophosphorylation and subsequent release of DNA-PKcs. Alternatively, the presence of LIG4, recruited to DNA ends by interactions with Ku [43], could hinder the XRCC4-XLF filament formation. Therefore, it is likely that XRCC4 homodimers involved in end joining could be complexed with LIG4 while the XRCC4 units within the XRCC4-XLF filaments involved in DNA end alignment would be free of LIG4. Roy et al [44] proposed that phosphorylation events could also influence filament remodeling.
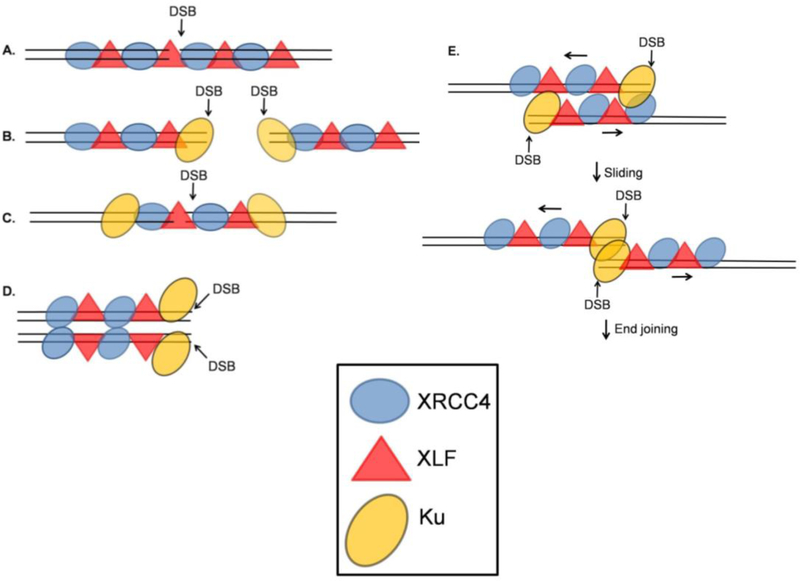
Figure 3.
Different possible arrangements of XRCC4/XLF filaments with respect to DSBs. A. An XRCC4/XLF filament spans a DSB, aligning the DNA ends on either side of the break. This model does not clearly explain how DNA-PKcs and other processing enzymes gain access to the DNA DSB. B. XRCC4/XLF filaments form adjacent to terminally bound Ku. C. An XRCC4/XLF filament spans a DSB after translocation of Ku away from the DSB end. D. XRCC4/XLF filaments aligned side-by-side position two DNA molecules such that processing and ligation can occur at the exposed DSB ends. E. XRCC4/XLF filaments, each binding a DNA segment near a DSB, align side-by-side, then slide to bring the DSB ends into juxtaposition. Adapted from [45, 46, 47].
Super-resolution fluorescence microscopy [45] clearly demonstrates in cells the formation of XRCC4/XLF filaments, often terminated by Ku or extending from Ku in both directions. These images would be consistent with a model wherein filaments form along DNA on both sides of a DSB, interior to Ku (Fig. 3B), prior to synapsis of the two ends. This model allows for DNA-PKcs to access the DNA ends, and its subsequent autophosphorylation and removal would allow other DNA end processing enzymes like Artemis, PNKP and LIG4 to be recruited to the DNA ends. In addition, it is compatible with the existence of two populations of XRCC4, one in filamentous form and one complexed with LIG4. However, as pointed out by Mahaney et al. [46], this raises somewhat of a conundrum because Ku, bound to and completely encircling the DNA ends, would disrupt the continuity of the filament, and so its end-tethering function would presumably be lost. Although Ku could in principle migrate into the interior of the DNA molecule allowing the filament to span the break (Fig. 3C), that arrangement still does not easily explain how DNA-PKcs and associated end-processing enzymes (Artemis, pol µ/λ) gain access without disrupting the tethering.
The crystal lattice of XRCC4/XLF filaments suggests that interactions between stalk domains can stabilize side-by-side alignment of filaments (Fig. 2) [46,47], raising the possibility that two parallel filaments could tether two DNA molecules and bring the ends into proximity yet leave them accessible to processing (Fig. 3D). However, in this case a major structural rearrangement would seem to be required to reposition the ends for ligation. Based on single-molecule FRET studies, an alternative model has been proposed wherein two XRCC4/XLF filaments, each binding a segment of DNA near a DSB end, initially line up side-by-side, then slide until the two Ku-bound ends meet, at which point end joining presumably proceeds [45] (Fig. 3E). Unlike the models described above (Fig. 3A–D), this model has the advantage that it utilizes XRCC4/XLF filaments for alignment, yet allows for access to DNA ends by processing enzymes.
A more recent study using a combination of optical tweezers and fluorescence microscopy [48] showed that XLF and XRCC4 form heteromeric complexes capable of binding to and diffusing along the DNA. These XRCC4-XLF complexes form sliding sleeve-like structures that can act as a tether between two separate DNA molecules. However, these images show the XRCC4/XLF-bound DNA molecules maintaining contact with each other only at a single point along their length, rather than adopting an extended side-by-side alignment [48].A separate single-molecule FRET study in Xenopus laevis egg extracts showed that synapsis occurs in two stages [49]; a long range complex involving Ku70/80 and DNA-PKcs without DNA-PK’s catalytic activity and a short range complex that requires DNA-PK’s catalytic activity, XLF and X4L4 but not ligase catalytic activity. Thus, while all these studies suggest that XLF, in conjunction with XRCC4, can promote synapsis, tethering and alignment of DNA ends, different analytical techniques have tended to suggest diverse structural geometries rather than a single coherent model.
The X4L4-mediated ligation reaction that is stimulated by XLF begins with a charging step that generates a LIG4-adenylate complex. AMP is then transferred from LIG4 to the 5′ phosphate of DNA, forming a DNA-adenylate conjugate and releasing uncharged LIG4 that must be re-adenylated for subsequent ligation reactions. Re-adenylation is a slow process and is rate-limiting under in vitro conditions [50,51]. Experiments with recombinant X4L4 that had been de-adenylated by exposure to pyrophosphate showed that XLF stimulates ligase adenylation in the X4L4 complex, thereby enhancing its ATP-mediated ligation activity [52].
The Leu115 residue in XLF is important for XRCC4 interaction [19,53]. Biochemical studies with an L115A mutant showed that its interaction with XRCC4 as well as its DNA end-bridging functions were disrupted, while its stimulation of LIG4 activity was retained [54]. An L115D mutation disrupted both functions. The sensitivity of several XLF-deficient cell lines to the radiomimetic agent Zeocin was corrected by expression of the L115A but not the L115D mutant, suggesting that LIG4 stimulation by XLF, but not filament formation, is essential for radioresistance and therefore critical for DSB repair. Curiously, neither allele reversed the Zeocin sensitivity of XLF−/− HCT116 cells at all (even though L115A restored end joining of transfected plasmid substrates [35]), suggesting that both LIG4 stimulation and filament formation were important for DSB repair in these cells. Conversely, both mutant alleles almost fully reversed cell sensitivity to hydroxyurea, suggesting a third functional role for XLF, the molecular basis of which remains to be determined, in protection from replicative stress [54]. Overall, these studies show that XLF plays an important role in DNA bridging and stimulation of LIG4 activity, with as yet unexplained cell line-specific differences in the importance of these functions for DSB repair.
After a DNA DSB is formed, the DNA damage response is triggered to signal recruitment and/or activation of diverse repair factors for proper repair to occur. During NHEJ, it was shown in cells treated with calicheamicin that XLF is co-recruited with other NHEJ factors. However, XLF is not required for the localization of other NHEJ factors to DSB-damaged chromatin [55]. XLF recruitment to DSBs in vivo requires the 10 amino acids in the C-terminus, and the C-terminal deletions in XLF [XLF (1–270) and XLF (1–289)] also abolished its interaction with Ku [56]. This result contrasts with an earlier finding wherein XLF with a 68-amino acid C-terminal truncation fully restored V(D)J recombination and bleomycin resistance in XLF-deficient human fibroblasts [57]. These conflicting results are difficult to reconcile, but it is possible that a reduced level of recruitment of the truncated XLF was sufficient to promote NHEJ even though it was below the threshold of detection by fluorescence microscopy.
XLF and Post-Translational Modifications
Like XRCC4, Artemis and DNA-PKcs itself, XLF is phosphorylated by DNA-PKcs after its recruitment to DSB sites and this phosphorylation is abolished in DNA-PKcs-defective cell lines like Fus9 and HeLa-shKcs. Two major sites, Ser245 and Ser251, for in vitro phosphorylation by DNA-PK were identified, but whereas Ser245 is phosphorylated by DNA-PK in vivo, Ser251 is phosphorylated by ATM [58]. Neither of these phosphorylations appears to have any significant effect on DNA binding, recruitment to DSB sites, DSB repair or radiosensitivity in cells, nor are they needed for XLF’s end joining activity in cell extracts [42].
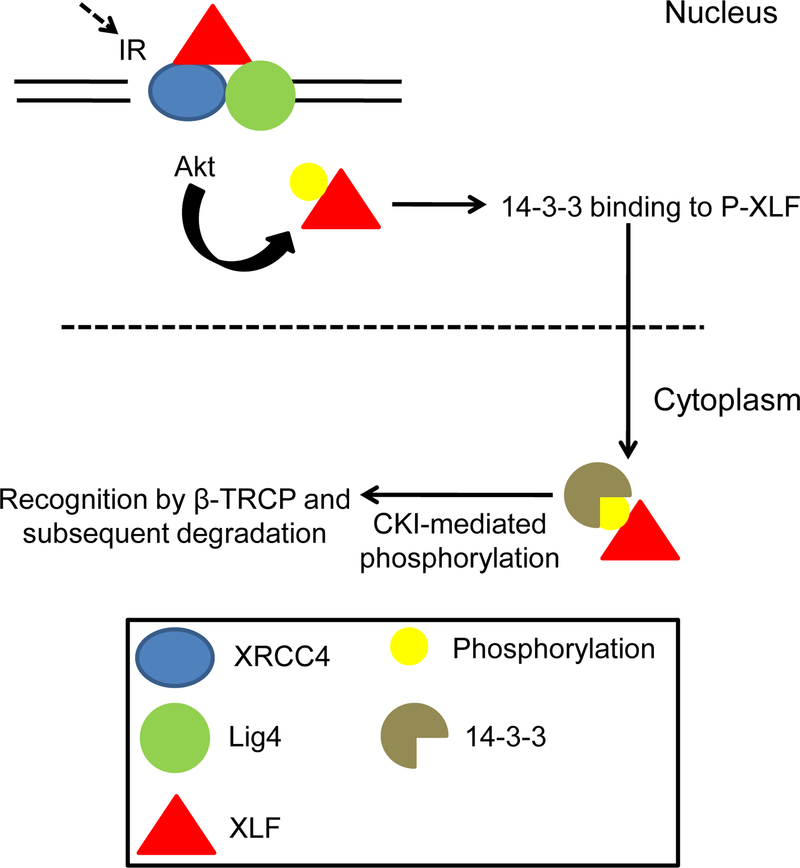
XLF is also phosphorylated by Akt at Thr181, resulting in the dissociation of phosphorylated XLF from X4L4, export to the cytoplasm by 14–3-3 protein, casein kinase I-mediated phosphorylation at Ser170 and Thr173, and finally recognition by β-TRCP for ubiquitination and degradation [59] (Fig. 4). Expression of a phosphomimetic T181E mutant XLF in XLF-deficient 2BN cells conferred defective and delayed DNA repair, as compared to cells expressing wild-type XLF, suggesting a potent inhibitory effect of this phosphorylation on NHEJ.
Figure 4.
Akt-mediated phosphorylation and degradation of XLF. Phosphorylation at Thr181 leads to dissociation from the XRCC4-DNA Ligase IV complex. Phosphorylated XLF is then recognized by 14–3-3 protein and translocated to the cytoplasm where it could be further phosphorylated by CKI. It is then recognized by β-TRCP for ubiquitination and subsequent degradation. Adapted from [59].
XLF and V(D)J Recombination
V(D)J recombination or antigen receptor gene rearrangement is an essential process in developing lymphocytes, that assembles the immunoglobulin variable domains and T-cell receptor genes. It occurs via the introduction of DNA DSBs between the V, (D), and J segments and the flanking recombination signal sequences (RSS). Subsequently, the RSS are joined precisely, while the coding regions are modified via nucleotide loss and addition. These joining steps are carried out by NHEJ proteins and the error-prone nature of NHEJ is an important factor in the generation of antibody diversity. As a result, mutations in or absence of NHEJ proteins can have drastic V(D)J-related consequences including immunodeficiencies and cancers, especially lymphoma and leukemia [60,61]. For example, DNA-PKcs-deficient scid mice are radiosensitive, and show defective lymphocyte development resulting from deficiency in formation of coding joints, while signal joints are not affected [62–64]. Similarly, using CHO cells (XR-1), Roy et al [44] showed that XRCC4 mutants unable to interact with XLF can restore signal joints but not coding joints to XRCC4-deficient cells, suggesting a specific role for XLF in repair of DSBs that require end processing before religation.
During V(D)J recombination, several processes contribute to sequence diversity in coding joints, including asymmetric opening of the hairpins to yield palindromic (P) nucleotides, deletion of terminal nucleotides by exonucleases and nontemplated (N) nucleotides inserted by terminal deoxynucleotidyl transferase (TdT). Patients with mutant XLF alleles showed reduced N-nucleotide addition, ultimately resulting in defective T and B-cell development and immunodeficiency [65]. Although in XRCC4-deficient and LIG4-deficient patients, IgH rearrangements also showed fewer N-nucleotides, the deficiency was not as great as that seen in the XLF-deficient patients [65]. Thus, the ligation complex is important for N-nucleotide addition, with XLF playing a dominant role. Human patients harboring a mutation in XLF also show impaired class switch recombination (CSR) and low lymphocyte numbers [66].
On the other hand, XLF-deficient mice have lymphocyte numbers that are just slightly reduced as compared to wild type. Although XLF-deficient murine embryonic fibroblasts show a severe deficiency in V(D)J recombination, murine pro B-cells from the same mice can carry out V(D)J recombination at almost wild-type levels, indicating that the developing lymphocytes have factors that could compensate for XLF deficiency [66]. ATM appears to be one such factor, as a combined XlfΔ/ΔAtm−/−genotype (but not either mutation alone) abolishes V(D)J recombination, disrupts B- and T-cell development, and impairs c-NHEJ-mediated CSR, without affecting the residual CSR mediated by Alt-NHEJ [67]. Further, using XlfΔ/Δ mice that were also homozygous for a LoxP-flanked H2ax allele, XLF was shown to have overlapping V(D)J functions with H2AX, a substrate of ATM phosphorylation following DNA damage [67]. Thus, in the absence of XLF, its functions may be compensated by ATM and certain ATM substrates.
V(D)J recombination occurs during the G1 phase of the cell cycle and is mainly mediated by the recombination activating gene products, RAG1 and RAG2 that together constitute the RAG endonuclease. These enzymes induce DNA DSBs between the V, (D), and J coding segments and the flanking RSS [68]. RAG-induced DSBs are repaired by NHEJ and knockout of XLF revealed a cryptic function of the RAGs in promoting rejoining. Thus, although XLF is not needed for V(D)J recombination, Rag2c/cXlf−/− mice (harboring a C-terminal RAG2 deletion) show blocked lymphocyte development at the progenitor stage, and v-Abl-transformed pro-B cells from these mice show chromosomal breaks and translocations reflecting a severe impairment in repairing RAG DSBs [69]. Furthermore, Rag2c/cXlf−/−p53−/− mice showed a high incidence of pro B-cell lymphomas harboring translocations and gene amplifications. Taken together, these results suggest that in B cells RAG2 and XLF function redundantly to enable end joining for V(D)J recombination.
Studies by two separate groups [70,71] also have shown a functional redundancy between XLF and 53BP1 wherein a loss of both XLF and 53BP1 had drastic effects on lymphocyte development characterized by reduced splenic B cells and developmental impairment at the CD43+B220+ pro–B-cell stage. Southern blotting of the recombination products of chromosomally integrated V(D)J reporter constructs in v-Abl-transformed pro-B cell lines indicated a defect in NHEJ in cells from XlfΔ/Δ53bp1−/− mice [70]. Further it was shown that in XLF-deficient cells, the Tudor domains of 53BP1 play a key role in both end joining and end protection during V(D)J recombination [71]. A similar functional redundancy was reported between XLF and DNA-PKcs wherein XlfΔ/ΔDna-PKcs−/− mice showed abnormal growth and viability. Furthermore, in XLF-deficient pro-B cells, the kinase activity of DNA-PKcs was required not only for coding joint formation, but also for signal joint formation during V(D)J recombination. XLF and DNA-PKcs also have redundant activities during CSR and in maintaining genomic stability [72].
In summary, XLF deficiency in mice is associated with a V(D)J phenotype that is milder than in human, but that when combined with deficiency in any of several NHEJ proteins, confers a severe defect in V(D)J coding joint formation (Table I). In many cases either the double-mutant cells are also radiosensitive, or the corresponding mice are inviable, suggesting a general NHEJ defect.
Table I.
XLF Redundancies
| Protein | System | Function | References |
|---|---|---|---|
| ATM | Mice (XlfΔ/ΔAtm−/−) | V(D)J recombination, B- and T-cell development, C-NHEJ-mediated CSR | [67] |
| H2AX | Mice (XlfΔ/ΔH2ax−/−, XlfΔ/ΔH2axF/F) | V(D)J recombination, End-protection | [67] |
| PAXX | G1 phase enriched murine v-Abl cells | Sensitivity to ionizing radiation-induced DSBs | [75–78] |
| PAXX | Mice | Viability | [76] |
| 53BP1 | v-Abl transformed pro-B murine cell lines generated from WT, XlfΔ/Δ, 53bp1−/−, and XlfΔ/Δ53pb1−/− mice | Lymphocyte development, V(D)J recombination | [70,71] |
| DNA-PKcs | Mice (XlfΔ/ΔDna-PKcs−/−) | Growth/viability; V(D)J recombination (signal joints) and NHEJ; CSR | [72] |
| RAG | Mice (Rag2c/cXlf−/−p53−/−) | Lymphocyte development at the progenitor stage | [69] |
XLF and PAXX
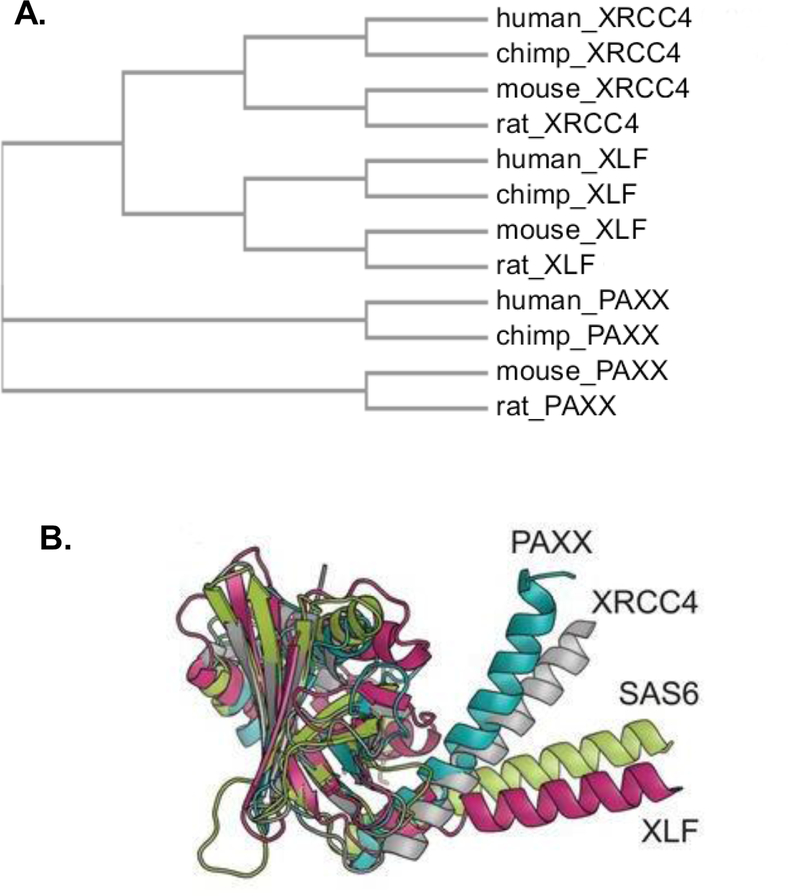
Ochi et al. [73] and Xing et al. [74] identified a paralog of XRCC4 and XLF, PAXX (PAralog of XRCC4 and XLF), that has structural similarities to XRCC4, interacts with Ku and is recruited to damage sites [73], suggesting a role in DSB repair. In sequence homology, XLF and XRCC4 are more similar to each other than to PAXX (Fig. 5A). Crystallographic studies showed that PAXX residues 1–113 constitute a globular head similar in structure to the head domains of XLF, XRCC4 and SAS6 [73,74] (Fig 5B). In some ways, PAXX is more similar to XRCC4 than to XLF, but more similar to XLF in other ways. The angle formed between the head domain and the helical stalk in both XRCC4 and PAXX is ~45° while in XLF it is ~90°. On the other hand, PAXX, like XLF, has a coiled-coil stalk that is much shorter than that of XRCC4. Furthermore, both PAXX and XLF lack any structure corresponding the LIG4 binding motif of XRCC4, and conversely, both PAXX and XLF contain a Ku-binding region at the extreme C-terminus that is essential for efficient recruitment to DSB ends [56,74] and is not found in XRCC4.
Figure 5.
Comparison of XRCC4 superfamily members. A. Phylogenetic tree of the three proteins, constructed using Clustal Omega. B. Superposition of the crystal structures of PAXX (cyan), XRCC4 (silver), XLF (magenta), and SAS6, a spindle assembly protein (green). (Image reproduced with permission from [73]).
Reflecting these structural similarities, PAXX can serve in several contexts as a substitute for XLF. PAXX promotes X4L4-mediated ligation of DSB ends in the absence of XLF in vitro, as well as assembly of core NHEJ proteins on damaged chromatin [73]. PAXX is not required for the repair of DSBs arising from V(D)J recombination in G1-arrested murine v-Abl-transformed cells, for IgH CSR in cycling CH12 cells or for radioresistance [75]. However, although Paxx−/− mice, like Xlf−/− mice, are viable and only mildly radiosensitive, Paxx−/−Xlf−/− double knockouts exhibit embryonic lethality, defective lymphogenesis, and nervous system-associated cell death, suggesting synthetic lethality of the combined deficiency [76]. Nevertheless, using CRISPR it is possible to generate viable Paxx−/−Xlf−/−v-Abl-transformed pro-B cells, and unlike either of the single mutants, such cells are profoundly radiosensitive and show a severe defect in rejoining of coding ends for V(D)J recombination, resulting in a high level of chromosome instability [75,77,78]. Furthermore, epistasis studies in DT40 chicken cells suggest that PAXX and XLF function redundantly in repair of etoposide-induced DSBs, which bear covalently linked topoisomerase II at their 5′ termini [74].
In other situations, however, PAXX and XLF are not functionally equivalent. For example, in the same DT40 cell model, PAXX shows no redundancy with XLF in repair of radiation damage [74]. Similarly, human retinal RPE-1 cells with a PAXX−/− mutation are radiosensitive, but knocking down XLF does not further increase sensitivity [73]. The two paralogues also differ in their interplay with ATM. As mentioned earlier, XLF is redundant with ATM and with the RAG complex in repairing broken DNA ends. In contrast, in experiments with v-Abl-expressing mouse pro-B cells harboring the artificial V(D)J substrate pMX-INV, PAXX/ATM double-deficient cells still showed substantial RAG-induced V(D)J recombination, similar to cells with ATM deficiency alone. Further, whereas ATMi treatment of Xlf−/−pro-B cells almost completely blocked V(D)J rearrangement, similar treatment in Paxx−/− pro-B cells did not affect the rearrangement [77]. Similarly, in the same experimental system, Paxx−/−Rag2c/c pro-B cells, unlike Xlf−/−Rag2c/c cells, retained V(D)J recombination proficiency, even in the presence of ATMi. Thus, in contrast to XLF, PAXX does not function redundantly with either ATM or the RAG complex.
XLF regulation
Although the role of XLF in DNA repair has been studied extensively, relatively little work has been done on its transcription, translation or regulation. However, Liu et al [79], showed that the Werner Syndrome protein WRN positively regulates the transcription of XLF. WRN binding to the XLF promoter was detected via ChIP [79,80] and WRN-deficient fibroblasts showed a decrease in XLF protein levels. It was suggested that compromised NHEJ, due to reduced XLF expression, could contribute to premature cellular senescence in WRN-deficient cells. Also, the WRN protein was shown to interact with the C-terminal domain of p53, a region highly involved in the functionality of p53. WRN overexpression showed an increase in p53 transcriptional activity and subsequently increased p21 protein expression, suggesting a role for WRN-p53 interaction in maintaining genomic stability [81]. In a related study, the levels of XLF and mutant p53 were shown to have an inverse correlation in head and neck cancers harboring mutant p53 wherein it was concluded that mutant p53 might affect the activity of WRN, in the process significantly decreasing XLF expression, leading to genomic instability and cellular senescence [80].
Conclusions
XLF performs at least three functions in NHEJ: stimulating LIG4 activity, promoting LIG4 adenylation, and aiding in DSB end alignment through formation of XRCC4/XLF filaments. For repair of DSBs that require significant processing prior to ligation, XLF appears to be essential in some contexts but dispensable in others. Perhaps the simplest explanation of these enigmatic data is that, when end alignment by XRCC4/XLF filaments fails, the remaining NHEJ machinery is able to bring two DNA ends into juxtaposition in most cases. When that machinery becomes compromised as well, then the alignment function of XLF may be critical. This variability in the degree to which XLF is essential might actually improve prospects for XLF as a therapeutic target, since tumor cells often have multiple cryptic defects in repair pathways, including NHEJ. Thus, an XLF inhibitor might in principle be selectively toxic to a limited class of tumor cells with specific NHEJ defects. While scaffolding proteins with large interaction surfaces are typically poor candidates for pharmacological intervention, the leucine-binding pocket of XRCC4 could potentially be targeted by a small-molecule inhibitor that would block XLF binding, prevent filament formation, and thereby chemo/radiosensitize certain cells with cryptic NHEJ defects.
Acknowledgments
We thank the authors of refs. 17, 24 and 73 for permission to reproduce images of crystal structures. Preparation of this review was supported by Grant CA166264 from the National Cancer Institute.
References
- [1].Povirk LF DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes, Mutat. Res 355 (1996) 71–89. [DOI] [PubMed] [Google Scholar]
- [2].Hutchinson F Chemical changes induced in DNA by ionizing radiation, Prog. Nucleic Acid Res. Mol. Biol 32 (1985) 115–154. [DOI] [PubMed] [Google Scholar]
- [3].Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D Oxidatively generated complex DNA damage: Tandem and clustered lesions, Cancer Lett 327 (2012) 5–15. [DOI] [PubMed] [Google Scholar]
- [4].Valerie K, Povirk LF Regulation and mechanisms of mammalian double-strand break repair, Oncogene 22 (2003) 5792–5812. [DOI] [PubMed] [Google Scholar]
- [5].Walker JR, Corpina RA, Goldberg J Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair, Nature 412 (2001) 607–614. [DOI] [PubMed] [Google Scholar]
- [6].Davis AJ, Chen DJ DNA double strand break repair via non-homologous end-joining, Transl. Cancer. Res 2 (2013) 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lieber MR The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway, Annu. Rev. Biochem 79 (2010) 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly, Cell 124 (2006) 287–299. [DOI] [PubMed] [Google Scholar]
- [9].Ahnesorg P, Smith P, Jackson SP XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining, Cell 124 (2006) 301–313. [DOI] [PubMed] [Google Scholar]
- [10].Kegel A, Sjostrand JO, Astrom SU Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast, Curr. Biol 11 (2001) 1611–1617. [DOI] [PubMed] [Google Scholar]
- [11].Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP Cernunnos interacts with the XRCC4 x DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1, J. Biol. Chem 281 (2006) 13857–13860. [DOI] [PubMed] [Google Scholar]
- [12].Girard PM, Kysela B, Harer CJ, Doherty AJ, Jeggo PA Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: The impact of two linked polymorphisms, Hum. Mol. Genet 13 (2004) 2369–2376. [DOI] [PubMed] [Google Scholar]
- [13].Mahaney BL, Meek K, Lees-Miller SP Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining, Biochem. J 417 (2009) 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos, J. Biol. Chem 281 (2006) 37517–37526. [DOI] [PubMed] [Google Scholar]
- [15].Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W Crystal structure of the Xrcc4 DNA repair protein and implications for end joining, EMBO J 19 (2000) 5962–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA XLF regulates filament architecture of the XRCC4.ligase IV complex, Structure 18 (2010) 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ, EMBO J 27 (2008) 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leber R, Wise TW, Mizuta R, Meek K The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase, J. Biol. Chem 273 (1998) 1794–1801. [DOI] [PubMed] [Google Scholar]
- [19].Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS Crystal structure of human XLF: A twist in nonhomologous DNA end-joining, Mol. Cell 28 (2007) 1093–1101. [DOI] [PubMed] [Google Scholar]
- [20].Lu H, Pannicke U, Schwarz K, Lieber MR Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity, J. Biol. Chem 282 (2007) 11155–11162. [DOI] [PubMed] [Google Scholar]
- [21].Modesti M, Hesse JE, Gellert M DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity, EMBO J 18 (1999) 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ Ku recruits XLF to DNA double-strand breaks, EMBO Rep 9 (2008) 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L Crystal structure of an Xrcc4-DNA ligase IV complex, Nat. Struct. Biol 8 (2001) 1015–1019. [DOI] [PubMed] [Google Scholar]
- [24].Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair, J. Biol. Chem 286 (2011) 32638–32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bryans M, Valenzano MC, Stamato TD Absence of DNA ligase IV protein in XR-1 cells: Evidence for stabilization by XRCC4, Mutat. Res 433 (1999) 53–58. [DOI] [PubMed] [Google Scholar]
- [26].Calsou P, Delteil C, Frit P, Drouet J, Salles B Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment, J. Mol. Biol 326 (2003) 93–103. [DOI] [PubMed] [Google Scholar]
- [27].Jayaram S, Ketner G, Adachi N, Hanakahi LA Loss of DNA ligase IV prevents recognition of DNA by double-strand break repair proteins XRCC4 and XLF, Nucleic Acids Res 36 (2008) 5773–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 18597–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Weterings E, van Gent DC The mechanism of non-homologous end-joining: A synopsis of synapsis, DNA Repair (Amst) 3 (2004) 1425–1435. [DOI] [PubMed] [Google Scholar]
- [30].Walker JR, Corpina RA, Goldberg J Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair, Nature 412 (2001) 607–614. [DOI] [PubMed] [Google Scholar]
- [31].Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA Ku recruits the XRCC4-ligase IV complex to DNA ends, Mol. Cell. Biol 20 (2000) 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Teo SH, Jackson SP Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks, Curr. Biol 10 (2000) 165–168. [DOI] [PubMed] [Google Scholar]
- [33].Smith GC, Jackson SP The DNA-dependent protein kinase, Genes Dev 13 (1999) 916–934. [DOI] [PubMed] [Google Scholar]
- [34].Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP DNA-PK autophosphorylation facilitates artemis endonuclease activity, EMBO J 25 (2006) 3880–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fattah FJ, Kweon J, Wang Y, Lee EH, Kan Y, Lichter N, Weisensel N, Hendrickson EA A role for XLF in DNA repair and recombination in human somatic cells, DNA Repair (Amst) 15 (2014) 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tsai CJ, Kim SA, Chu G Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 7851–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: Influence of terminal DNA sequence, Nucleic Acids Res 35 (2007) 5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Almohaini M, Chalasani SL, Bafail D, Akopiants K, Zhou T, Yannone SM, Ramsden DA, Hartman MC, Povirk LF Nonhomologous end joining of complex DNA double-strand breaks with proximal thymine glycol and interplay with base excision repair, DNA Repair (Amst) 41 (2016) 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Daley JM, Laan RL, Suresh A, Wilson TE DNA joint dependence of pol X family polymerase action in nonhomologous end joining, J. Biol. Chem 280 (2005) 29030–29037. [DOI] [PubMed] [Google Scholar]
- [40].Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts, J. Biol. Chem 279 (2004) 805–811. [DOI] [PubMed] [Google Scholar]
- [41].Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA Association of DNA polymerase mu (pol mu) with Ku and ligase IV: Role for pol mu in end-joining double-strand break repair, Mol. Cell. Biol 22 (2002) 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Akopiants K, Zhou RZ, Mohapatra S, Valerie K, Lees-Miller SP, Lee KJ, Chen DJ, Revy P, de Villartay JP, Povirk LF Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts, Nucleic Acids Res 37 (2009) 4055–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hsu HL, Yannone SM, Chen DJ Defining interactions between DNA-PK and ligase IV/XRCC4, DNA Repair (Amst) 1 (2002) 225–235. [DOI] [PubMed] [Google Scholar]
- [44].Roy S, Andres SN, Vergnes A, Neal JA, Xu Y, Yu Y, Lees-Miller SP, Junop M, Modesti M, Meek K XRCC4’s interaction with XLF is required for coding (but not signal) end joining, Nucleic Acids Res 40 (2012) 1684–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, Oksuz BA, Fenyo D, Lieber MR, Ramsden DA, Rothenberg E Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair, Proc. Natl. Acad. Sci. U. S. A 112 (2015) E2575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair, Biochem. Cell Biol 91 (2013) 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL Non-homologous end-joining partners in a helical dance: Structural studies of XLF-XRCC4 interactions, Biochem. Soc. Trans 39 (2011) 1387–92, suppl 2 p following 1392. [DOI] [PubMed] [Google Scholar]
- [48].Brouwer I, Sitters G, Candelli A, Heerema SJ, Heller I, Melo de AJ, Zhang H, Normanno D, Modesti M, Peterman EJ, Wuite GJ Corrigendum: Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA, Nature 543 (2017) 742. [DOI] [PubMed] [Google Scholar]
- [49].Graham TG, Walter JC, Loparo JJ Two-stage synapsis of DNA ends during non-homologous end joining, Mol. Cell 61 (2016) 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang Y, Lamarche BJ, Tsai MD Human DNA ligase IV and the ligase IV/XRCC4 complex: Analysis of nick ligation fidelity, Biochemistry 46 (2007) 4962–4976. [DOI] [PubMed] [Google Scholar]
- [51].Riballo E, Doherty AJ, Dai Y, Stiff T, Oettinger MA, Jeggo PA, Kysela B Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity, J. Biol. Chem 276 (2001) 31124–31132. [DOI] [PubMed] [Google Scholar]
- [52].Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA XLF-cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation, Nucleic Acids Res 37 (2009) 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, Callebaut I, de Villartay JP Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF, J. Biol. Chem 285 (2010) 26475–26483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Roy S, de Melo AJ, Xu Y, Tadi SK, Negrel A, Hendrickson E, Modesti M, Meek K XRCC4/XLF interaction is variably required for DNA repair and is not required for ligase IV stimulation, Mol. Cell. Biol 35 (2015) 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wu PY, Frit P, Malivert L, Revy P, Biard D, Salles B, Calsou P Interplay between cernunnos-XLF and nonhomologous end-joining proteins at DNA ends in the cell, J. Biol. Chem 282 (2007) 31937–31943. [DOI] [PubMed] [Google Scholar]
- [56].Yano K, Morotomi-Yano K, Lee KJ, Chen DJ Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF, FEBS Lett 585 (2011) 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Malivert L, Callebaut I, Rivera-Munoz P, Fischer A, Mornon JP, Revy P, de Villartay JP The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo, Mol. Cell. Biol 29 (2009) 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks, DNA Repair (Amst) 7 (2008) 1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu P, Gan W, Guo C, Xie A, Gao D, Guo J, Zhang J, Willis N, Su A, Asara JM, Scully R, Wei W Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair, Mol. Cell 57 (2015) 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Revy P, Buck D, le Deist F, de Villartay JP The repair of DNA damages/modifications during the maturation of the immune system: Lessons from human primary immunodeficiency disorders and animal models, Adv. Immunol 87 (2005) 237–295. [DOI] [PubMed] [Google Scholar]
- [61].Nussenzweig A, Nussenzweig MC Origin of chromosomal translocations in lymphoid cancer, Cell 141 (2010) 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bosma GC, Custer RP, Bosma MJ A severe combined immunodeficiency mutation in the mouse, Nature 301 (1983) 527–530. [DOI] [PubMed] [Google Scholar]
- [63].Lieber MR, Hesse JE, Lewis S, Bosma GC, Rosenberg N, Mizuuchi K, Bosma MJ, Gellert M The defect in murine severe combined immune deficiency: Joining of signal sequences but not coding segments in V(D)J recombination, Cell 55 (1988) 7–16. [DOI] [PubMed] [Google Scholar]
- [64].Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak Rothstein A, Jeggo PA, Herrera VL Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity, Immunity 9 (1998) 355–366. [DOI] [PubMed] [Google Scholar]
- [65].IJspeert H, Rozmus J, Schwarz K, Warren RL, van Zessen D, Holt RA, Pico-Knijnenburg I, Simons E, Jerchel I, Wawer A, Lorenz M, Patiroglu T, Akar HH, Leite R, Verkaik NS, Stubbs AP, van Gent DC, van Dongen JJ, van der Burg M XLF deficiency results in reduced N-nucleotide addition during V(D)J recombination, Blood 128 (2016) 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination, Mol. Cell 31 (2008) 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks, Nature 469 (2011) 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schatz DG, Swanson PC V(D)J recombination: Mechanisms of initiation, Annu. Rev. Genet 45 (2011) 167–202. [DOI] [PubMed] [Google Scholar]
- [69].Lescale C, Abramowski V, Bedora-Faure M, Murigneux V, Vera G, Roth DB, Revy P, de Villartay JP, Deriano L RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair, Nat. Commun 7 (2016) 10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Oksenych V, Kumar V, Liu X, Guo C, Schwer B, Zha S, Alt FW Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, Blundell TL, Jackson SP DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with ku to promote DNA double-strand break repair, Science 347 (2015) 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, Yan Z, Li W, Wang Q, Hou M, Dong C, Guo R, Gao G, Ji J, Zha S, Lan L, Liang H, Xu D Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway, Nat. Commun 6 (2015) 6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kumar V, Alt FW, Frock RL PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line, Proc. Natl. Acad. Sci. U. S. A 113 (2016) 10619–10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Balmus G, Barros AC, Wijnhoven PW, Lescale C, Hasse HL, Boroviak K, le Sage C, Doe B, Speak AO, Galli A, Jacobsen M, Deriano L, Adams DJ, Blackford AN, Jackson SP Synthetic lethality between PAXX and XLF in mammalian development, Genes Dev 30 (2016) 2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lescale C, Lenden Hasse H, Blackford AN, Balmus G, Bianchi JJ, Yu W, Bacoccina L, Jarade A, Clouin C, Sivapalan R, Reina-San-Martin B, Jackson SP, Deriano L Specific roles of XRCC4 paralogs PAXX and XLF during V(D)J recombination, Cell. Rep 16 (2016) 2967–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hung PJ, Chen BR, George R, Liberman C, Morales AJ, Colon-Ortiz P, Tyler JK, Sleckman BP, Bredemeyer AL Deficiency of XLF and PAXX prevents DNA double-strand break repair by non-homologous end joining in lymphocytes, Cell. Cycle (2016) 0. [DOI] [PMC free article] [PubMed]
- [79].Liu D, Deng X, Yuan C, Chen L, Cong Y, Xu X Werner syndrome protein positively regulates XRCC4-like factor transcription, Mol. Med. Rep 9 (2014) 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Feng S, Rabii R, Liang G, Song C, Chen W, Guo M, Wei X, Messadi D, Hu S The expression levels of XLF and mutant P53 are inversely correlated in head and neck cancer cells, J. Cancer 7 (2016) 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Blander G, Kipnis J, Leal JF, Yu CE, Schellenberg GD, Oren M Physical and functional interaction between p53 and the werner’s syndrome protein, J. Biol. Chem 274 (1999) 29463–29469. [DOI] [PubMed] [Google Scholar]