Abstract
Bacterial biofilms are complex, multicellular communities made up of bacteria enmeshed in a self-produced extracellular matrix (ECM) that protects against environmental stress. The ECM often comprises insoluble components, which complicates the study of biofilm composition, structure, and function. Wrinkled, agar-grown Escherichia coli biofilms require 2 insoluble macromolecules: curli amyloid fibers and cellulosic polymers. We quantified these components with solid-state nuclear magnetic resonance (NMR) and determined that curli contributed 85% of the isolated uropathogenic E coli ECM dry mass. The remaining 15% was cellulosic, but, surprisingly, was not ordinary cellulose. We tracked the identity of the unanticipated peak in the 13C NMR spectrum of the cellulosic component and discovered that E coli secrete phosphoethanolamine (pEtN)-modified cellulose. Cellulose is the most abundant biopolymer on the planet, and this marked the first identification of a naturally, chemically modified cellulose. To investigate potential roles of pEtN cellulose, we customized a newly designed live-cell monolayer rheometer and demonstrated that pEtN cellulose facilitated E coli attachment to bladder epithelial cells and acted as a glue, keeping curli cell associated. The discovery of pEtN cellulose opens questions regarding its biological function(s) and provides opportunities in materials science to explore this newly discovered biopolymer.
Keywords: biofilms, phosphoethanolamine cellulose, E coli, curli, bacterial adhesion
Comment on: Hollenbeck EC, Antonoplis A, Chai C, Thongsomboon W, Fuller GG, Cegelski L. Phosphoethanolamine cellulose enhances curli-mediated adhesion of uropathogenic Escherichia coli to bladder epithelial cells. Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10106-10111. doi: 10.1073/pnas.1801564115. Epub 2018 Sep 19. PubMed PMID: 30232265; PubMed Central PMCID: PMC6176564.
Biofilms: Integral Components to Life As We Know It
Bacterial biofilms are surface-associated communities or aggregates of bacteria encapsulated in a self-produced extracellular matrix (ECM). Biofilms exist at solid-air, liquid-air, and solid-liquid interfaces. They are ubiquitous in nature and confer protection against environmental stresses such as UV radiation, desiccation, predation, antimicrobials, and starvation. Biofilms play crucial ecological roles. For example, rhizobia biofilms assemble on legume plant roots to fix nitrogen for their host. Aquatic bacterial biofilms contribute to the carbon biogeochemistry of rivers and streams by affecting the dissolved organic carbon profile.1 Humans harness biofilm metabolism to clean wastewater (bioremediation) and produce various fermented food products: yogurt, kimchi, and kombucha, to name a few. While many biofilms are beneficial, there are also harmful biofilms. Biofouling and biocorrosion of water pipes reduce water quality and system efficiency. Plant-associated biofilms can ruin crop yields. Biofilm-contaminated food and food-processing equipment cause disease outbreaks. Multiple other serious infections are biofilm-related including cystic fibrosis airway infections, endocarditis, periodontitis, urinary tract infections (UTIs), catheter-associated infections, and infections of other indwelling devices.2 As such, understanding biofilm structure, function, metabolism, and pathogenicity remains at the forefront of multidisciplinary fields including microbiology, biochemistry, and biophysics. Studying biofilm structure and assembly is quite challenging as the ECM materials are often insoluble, complex mixtures and the exact composition may be species specific. To fully connect biofilm assembly with function and to gain further insight into the fascinating macromolecular assembly phenomena that take place beyond the cell surface, we must identify the building materials and how they interact with each other to ultimately understand the construction of the biofilm ECM.
Quantitative Determination of Biofilm ECM Composition
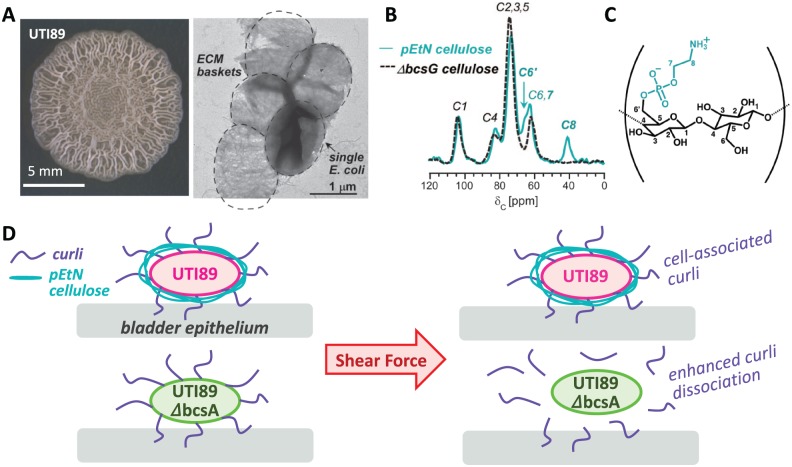
Biofilms are heterogeneous structures consisting of cells, proteins, and polysaccharides with pores and channels running through them for nutrient and waste transport. Molecular biology and biochemistry techniques provide a “parts list” of biofilm components. For example, a large family of Escherichia coli biofilms are composed primarily of two macromolecules: curli, an amyloid fiber, and a cellulosic polysaccharide. The curli-specific genes (csg) and bacterial cellulose synthesis (bcs) genes are required for ECM production. Due in part to the insolubility of ECM components, a quantitative accounting of the ECM had remained elusive. We overcame this challenge by designing solid-state nuclear magnetic resonance (NMR) measurements to access composition and architecture of the insoluble extracted ECM. We first employed a standard biochemical approach to semi-quantitatively consider the amount of curli through protein gel analysis and optimized assays for formic acid–treated depolymerized curli. We ascribed the remainder of the ECM to cellulose and other potential components. However, as with previous biofilm matrix analyses of other systems, methods that rely on dissolution, acid hydrolysis, or enzymatic hydrolysis of the isolated ECM suffer from the potential to miss components that are not easily digested or are not able to be quantified (as for insoluble aggregates). Solid-state NMR, however, can be performed on insoluble and heterogeneous macromolecular aggregates and even intact bacterial and mammalian cells.3 We have implemented solid-state NMR approaches to evaluate composition in biofilms formed by E coli as well as Vibrio cholerae and the fungus Aspergillus fumigatus.4–7 For E coli, we spectroscopically examined the ECM of uropathogenic E coli (UPEC) biofilms in a bottom-up approach, using the wild-type ECM material, pure curli from a strain that only produces curli (MC4100), and the purified cellulosic material from a curli mutant (UTI89ΔcsgBA). Homogenization of the biofilm separates the matrix components from bacteria without cell lysis. Based on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis and 13C CPMAS NMR on the extracted material, we determined that curli is the major proteinaceous ECM component.8 Flagella were observed but removed with SDS, and they were not integral to the observed architectures. In a sum-of-the-parts approach, we discovered that a spectral sum of the curli and cellulose 13C NMR spectra perfectly matched that of the 13C NMR spectrum of the extracted ECM. Thus, the UTI89 ECM appeared simpler than we anticipated with only two major parts by mass, curli and cellulose, in a 6:1 ratio.8 In other E coli strains we have examined that exhibit the hallmark wrinkled morphologies, this mass ratio for the extracted ECM typically varies between approximately 6:1 (with ~15% pEtN cellulose) and 3:1 (with ~25% pEtN cellulose). The UTI89 ECM study provided the first determination for the composition of the intact bacterial biofilm ECM with no inferences based on amyloid disassembly and matrix digestibility.
E coli Produce Phosphoethanolamine Modified Cellulose
The bottom-up sum-of-the-parts NMR analysis of the E coli ECM described above also yielded a surprise that led to the identification of the pEtN modification on cellulose. 13C CPMAS NMR spectra of the cellulosic material always contained an additional peak at 41 ppm not ascribed to cellulose, which as a polymer of glucose contains six distinct carbons with chemical shifts between 60 and 110 ppm. A combination of rotational-echo double-resonance (REDOR) NMR experiments to examine couplings between 13C, 15N, and 31P nuclei revealed that E coli produce pEtN cellulose (Figure 1B), with about half of the glucose units modified.9 Solution-state NMR of acid-digested material corroborated the C6 carbon as the site of modification and mass spectrometry of acid hydrolyzed material yielded detection of a species with a mass corresponding to a pEtN glucose unit as well as other hydrolysis products (glucose, glucose 6-phosphate, and ethanolamine). Previous work with isolated and digested cellulosic material, from what we now know are often pEtN cellulose producers, reported the detection of glucose and did not appear to look for products such as ethanolamine. This contributes to how this structure escaped detection until our close inspection of the intact polymer by solid-state NMR. This is the first discovery of a chemically modified cellulose produced in nature.
Figure 1.
(A) Two-day-old UTI89 Escherichia coli macrocolony biofilm on YESCA agar and transmission electron micrograph of extracted UTI89 ECM.6 (B) 13C CPMAS solid-state nuclear magnetic resonance spectra of pEtN cellulose and of the cellulosic component isolated from UTI89 ΔbcsG.9 The peaks at 41 and 63 ppm correspond to the C-8 and C-7 in the phosphoethanolamine functional group, respectively. The C-6 is shifted from 62 to 66 ppm in the pEtN cellulose. (C) Plausible representation of pEtN cellulose with alternating glucose and pEtN glucose, although modification patterning still must be determined. (D) Model depicting the role of pEtN cellulose in facilitating E coli attachment to bladder epithelium.12 pEtN cellulose acts as a type of glue to enhance curli association at the bacterial surface. ECM indicates extracellular matrix.
In a quest to understand how E coli modifies its cellulose, we identified the genetic and molecular basis for installation of the pEtN modification. We targeted our initial search to just several genes, including a few genes in the cellulose synthesis operons that are present in some organisms but had not been ascribed specific roles in cellulose synthesis. Through solid-state NMR analysis of the resulting cellulosic materials, we identified BcsG as a phosphoethanolamine transferase. In collaboration with Hengge and coworkers, bacterial two-hybrid experiments to evaluate protein-protein interactions elucidated that BcsG, BcsE, and BcsF participate in a novel transmembrane c-di-GMP signaling pathway to efficiently produce pEtN cellulose. Other microbes with the bcsEFG operon also produce pEtN cellulose, eg, Salmonella enterica, and we demonstrated that S enterica also produces pEtN cellulose. Colony morphology and electron microscopy images further demonstrated that pEtN-modified cellulose is important for E coli biofilm structure, enabling basket-like structures to fully enmesh bacteria and connect community members.8,9,10 The pEtN modification likely participates in direct interactions with the curli amyloid fibers in the ECM and enhances the physical properties of biofilms by increasing their strength and/or elasticity. Future work needs to elucidate the potential roles of pEtN cellulose in promoting biofilm cohesion, adhesion, and protection against predators and antimicrobials. The determination of atomic-level curli-pEtN cellulose intermolecular contacts will reveal fundamental molecular design principles for these fascinating matrix architectures.
Why Do Some Microbes Produce pEtN Cellulose?
Some bacterial cellulose producers, such as Komagataeibacter xylinus, lack bcsG and produce unmodified cellulose. Yet, other bacteria have evolved to produce pEtN cellulose. This raises the natural questions of why this modified cellulose is produced and how it affects bacterial biofilms. One hypothesis is that pEtN cellulose promotes bacterial adhesion. For one test of this, we investigated the role of pEtN cellulose in UPEC adhesion to bladder epithelial cells. UPEC are the major causative agent of UTIs and catheter-associated biofilm infections, and about half of all women will experience at least one UTI in their lifetime.11 While some UTIs are uncomplicated, others result in serious health outcomes including renal damage in youth, premature delivery, and sepsis.11,12
To test the role of curli and pEtN cellulose in UPEC adhesion, we developed a new live-cell monolayer rheometer–based approach to quantitatively measure attachment strength and dynamics of bacteria brought into contact with a monolayer of bladder epithelial cells, all housed within a microscope for concomitant visual detection. We reported the remarkable ability of purified curli to directly mediate adhesion to bladder cells. Curliated bacteria coproducing cellulose also exhibited strong adhesion, but curli mutants exhibited very little adhesion. Somewhat surprisingly, E coli that produce curli but lack pEtN cellulose were also deficient in bladder cell adhesion, with reduced relaxation moduli compared with wild-type cells.13 These results suggested a model wherein pEtN cellulose acts as a “glue” keeping the curli associated with the E coli cells to permit strong attachment to bladder epithelial cells. We performed a biochemical assay to corroborate this model. We vortexed suspensions of UTI89 and UTI89ΔbcsA to apply shear force, pelleted the cells with low-speed centrifugation, and then examined whether curli remained cell-associated or were removed from the cell surface and observed in the supernatant. Indeed, Western blot analysis and electron microscopy revealed that without pEtN cellulose, curli fibers dissociated from E coli under shear stress and were detected in the supernatant, whereas curli from pEtN cellulose producers remained cell-associated and was detected only in the cell pellet. These results support a model in which pEtN cellulose acts as a “glue” to keep curli cell-associated and enhances curli-mediated adhesion (Figure 1D). Future work with UTI89ΔbcsG will examine whether the pEtN modification specifically is required for this phenomenon.
This study elucidates just one role of pEtN cellulose in UPEC. We also consider, as supported by previous in vivo studies, that UPEC production of the modified cellulose and its interactions with curli help bacteria to evade immune recognition by masking curli, which are antigenic.14 Future work is needed to survey the prevalence of pEtN cellulose production among E coli and other organisms and to evaluate how else the pEtN modification might influence UPEC pathogenesis in vivo. Interestingly, most K12 strains do not produce cellulose, including MG1655, MC4100, and W3110, because the bcs gene cluster is not fully functional. These strains contain a point mutation introducing a stop codon in the bcsQ open reading frame, abrogating expression of bcsQ and downstream bcsA. This has been repaired in W3110 to generate the strain AR3110 that does produce cellulose and exhibits the hallmark curli-cellulose biofilm phenotypes.10 Still, UTI pathogenesis experiments have only scratched the surface in evaluating the influence of curli and cellulose or pEtN cellulose. Most UTI infection model studies focus on E coli strains that produce type 1 pili, a well-studied adhesive fiber mediating adhesion to and invasion into bladder epithelial cells, yet exhibit little or no production of curli and cellulose under the conditions that promote type 1 pili production at 37°C. Many UPEC, however, do produce curli and cellulose at 37°C, and studies have detected or strongly supported their production in vivo.14–19 Thus, functional studies of curli and cellulose will help to more fully understand the roles of the various ECM components in E coli pathogenesis. Finally, many other bacterial species contain the cellulose modification machinery and research is needed to understand what benefits might be conferred to these bacteria that have evolved to produce a modified cellulose.
Future Applications of pEtN Cellulose
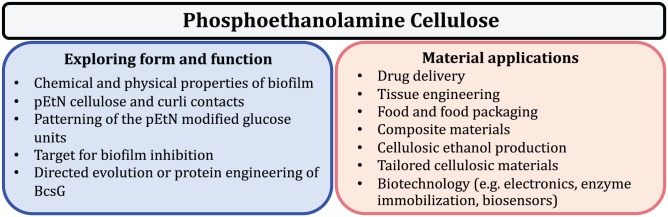
Cellulose is the most abundant biopolymer on earth. This simple macromolecule consists of a chain of repeating glucose units connected by β-1,4-glycosidic bonds. Individual cellulose polymers can join together to form fibrils that exhibit high tensile strength due to the extensive network of inter- and intramolecular hydrogen bonds within its structure. pEtN cellulose is a newly discovered, naturally produced polymer whose material properties are not yet characterized. Although we know that approximately half of the glucose residues are modified in pEtN cellulose samples, the patterning of the modified glucose units along the polymer is not yet defined. Also, the average length of the pEtN cellulose polymer and its ability to form fibrils have not been determined.
Furthermore, cellulose is found in a broad spectrum of consumer products, including in food packaging, food thickeners, wound dressings, paper products, eye drops, and hydrogels.20,21 The pEtN modification may exhibit superior properties for multiple applications (Figure 2). The terminal amine group of the pEtN is ripe for further modification, with applications in drug delivery, sensor devices, or enzyme immobilization. Hydrolysis of the organophosphate bond connecting the modification to the cellulose backbone could additionally provide a mechanism for controlled release of cargo attached to the amine.
Figure 2.
Fundamental, open questions and potential applications of pEtN cellulose.
Cellulose is frequently combined with other polysaccharides and with proteins and nanoparticles to form composites with applications as tissue engineering scaffolds, packaging materials, biosensors, electronics, and low-calorie foods.20,21 It is an attractive scaffold as cellulose is a renewable resource with high water holding capacity that is generally biodegradable and biocompatible. pEtN cellulose could similarly be combined with other polymers and/or proteins to create new composite materials. The E coli biofilm ECM is, indeed, a composite material itself, composed of pEtN cellulose and curli. A polysaccharide-protein fiber combination is not unique to E coli and is also found in biofilms formed by other pathogens such as Pseudomonas aeruginosa and the soil-dwelling organism Bacillus subtilis. Rigorous examination of the chemical and mechanical properties of the individual components and matrix composites can guide the design of novel composite materials and products and may contribute to understanding the evolutionary advantages of combining polysaccharides and protein fibers in the ECM. Moreover, rational design or directed evolution approaches could be employed to engineer the pEtN modification machinery to biosynthetically add a functional group of choice to cellulose and perhaps, to other polysaccharides. In this way, bacterial biofilms could be exploited as highly specific, polymer-producing factories.
To conclude, the discovery of pEtN cellulose ignites a multitude of investigations spanning biology, chemistry, materials science, and chemical engineering. Future studies promise to further reveal the biological significance of this first-identified naturally, chemically modified cellulose. Finally, the discovery of pEtN cellulose in commonly studied E coli biofilms makes us wonder whether other chemically modified celluloses are lurking out there in nature.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: L.C. acknowledges support from the National Science Foundation CAREER/PECASE Award 1453247. J. J. acknowledges support through the National Human Genome Research Institute of the National Institutes of Health under Award Number T32HG000044. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation or National Institutes of Health.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JJ and LC composed the manuscript. All authors reviewed and edited the manuscript and approve of its content.
ORCID iD: Lynette Cegelski  https://orcid.org/0000-0002-0978-1814
https://orcid.org/0000-0002-0978-1814
References
- 1. Fischer H, Sachse A, Steinberg CEW, Pusch M. Differential retention of dissolved organic carbon (DOC) by a bacterial community in river sediments. Limnol Oceanogr. 2002;47:1702–1711. [Google Scholar]
- 2. Del Pozo JL. Biofilm-related disease. Expert Rev Anti-Infect Ther. 2018;16:51–65. [DOI] [PubMed] [Google Scholar]
- 3. Werby SH, Cegelski L. Spectral comparisons of mammalian cells and intact organelles by solid-state NMR. J Struct Biol. 2018;206:49–54. doi: 10.1016/j.jsb.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reichhardt C, Ferreira JA, Joubert LM, Clemons KV, Stevens DA, Cegelski L. Analysis of the Aspergillus fumigatus biofilm extracellular matrix by solid-state nuclear magnetic resonance spectroscopy. Eukaryot Cell. 2015;14:1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reichhardt C, Fong JC, Yildiz F, Cegelski L. Characterization of the Vibrio cholerae extracellular matrix: a top-down solid-state NMR approach. Biochim Biophys Acta. 2015;1848:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cegelski L. Bottom-up and top-down solid-state NMR approaches for bacterial biofilm matrix composition. J Magn Reson. 2015;253:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reichhardt C, Cegelski L. Solid-state NMR for bacterial biofilms. Mol Phys. 2014;112:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCrate OA, Zhou X, Reichhardt C, Cegelski L. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J Mol Biol. 2013;425:4286–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thongsomboon W, Serra DO, Possling A, Hadjineophytou C, Hengge R, Cegelski L. Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science. 2018;359:334–338. [DOI] [PubMed] [Google Scholar]
- 10. Serra DO, Richter AM, Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195:5540–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113:5–13. [DOI] [PubMed] [Google Scholar]
- 12. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hollenbeck EC, Antonoplis A, Chai C, Thongsomboon W, Fuller GG, Cegelski L. Phosphoethanolamine cellulose enhances curli-mediated adhesion of uropathogenic Escherichia coli to bladder epithelial cells. Proc Natl Acad Sci U S A. 2018;115:10106–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kai-Larsen Y, Luthje P, Chromek M, et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. Plos Pathog. 2010;6:e1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. [DOI] [PubMed] [Google Scholar]
- 16. Lim JY, Pinkner JS, Cegelski L. Community behavior and amyloid-associated phenotypes among a panel of uropathogenic E. coli. Biochem Biophys Res Commun. 2013;443:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Antypas H, Choong FX, Libberton B, Brauner A, Richter-Dahlfors A. Rapid diagnostic assay for detection of cellulose in urine as biomarker for biofilm-related urinary tract infections. NPJ Biofilms Microbiomes. 2018;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. Escherichia coli global gene expression in urine from women with urinary tract infection. Plos Pathog. 2010;6:e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nhu NTK, Phan MD, Peters KM, et al. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. MBio. 2018;9:e01462–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalia S, Dufresne A, Cherian BM, et al. Cellulose-based bio- and nanocomposites: a review. Int J Polym Sci. 2011;2011:837875. [Google Scholar]
- 21. Sharma A, Thakur M, Bhattacharya M, Mandal T, Goswami S. Commercial application of cellulose nano-composites—a review. Biotechnol Rep (Amst). 2019;21:e00316. [DOI] [PMC free article] [PubMed] [Google Scholar]