Abstract
Background
Acetyl-CoA is an important metabolic intermediate and serves as an acetylation precursor for the biosynthesis of various value-added acetyl-chemicals. Acetyl-CoA can be produced from glucose, acetate, or fatty acids via metabolic pathways in Escherichia coli. Although glucose is an efficient carbon source for acetyl-CoA production, the pathway from acetate to acetyl-CoA is the shortest and fatty acids can produce acetyl-CoA through fatty acid oxidation along with abundant NADH and FADH2. In this study, metabolically engineered E. coli strains for efficiently supplying acetyl-CoA from glucose, acetate, and fatty acid were constructed and applied in one-step biosynthesis of N-acetylglutamate (NAG) from glutamate and acetyl-CoA.
Results
A metabolically engineered E. coli strain for NAG production was constructed by overexpressing N-acetylglutamate synthase from Kitasatospora setae in E. coli BW25113 with argB and argA knockout. The strain was further engineered to utilize glucose, acetate, and fatty acid to produce acetyl-CoA. When glucose was used as a carbon source, the combined mutants of ∆ptsG::glk, ∆galR::zglf, ∆poxB::acs, ∆ldhA, and ∆pta were more efficient for supplying acetyl-CoA. The acetyl-CoA synthetase (ACS) pathway and acetate kinase-phosphate acetyltransferase (ACK-PTA) pathway from acetate to acetyl-CoA were investigated, and the ACK-PTA pathway showed to be more efficient for supplying acetyl-CoA. When fatty acid was used as a carbon source, acetyl-CoA supply was improved by deletion of fadR and constitutive expression of fadD under the strong promoter CPA1. Comparison of acetyl-CoA supply from glucose, acetate and palmitic acid revealed that a higher conversion rate of glutamate (98.2%) and productivity (an average of 6.25 mmol/L/h) were obtained when using glucose as a carbon source. The results also demonstrated the great potential of acetate and fatty acid to supply acetyl-CoA, as the molar conversion rate of glutamate was more than 80%.
Conclusions
Metabolically engineered E. coli strains were developed for NAG production. The metabolic pathways of acetyl-CoA from glucose, acetate, or fatty acid were optimized for efficient acetyl-CoA supply to enhance NAG production. The metabolic strategies for efficient acetyl-CoA supply used in this study can be exploited for other chemicals that use acetyl-CoA as a precursor or when acetylation is involved.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1177-y) contains supplementary material, which is available to authorized users.
Keywords: N-Acetylglutamate, Acetyl-CoA, Glucose, Acetate, Fatty acid
Background
Acetyl-CoA, a metabolite in primary carbon metabolism, is involved in various biological processes. It can act as an intermediate to transfer an acetyl group during the biosynthesis of diverse acetyl-chemicals [1]. Acetyl-CoA is also a platform chemical for producing various high-value products, such as isoprenoids (used as flavors, biofuels, pharmaceuticals, and vitamins) [2], 1-butanol [3], 3-hydroxypropionate (one of the 12 biology-based chemical products with the most potential according to the Department of Energy, USA) [4], and polyhydroxyalkanoates [5]. Metabolic engineering targeting the intracellular acetyl-CoA pool is typically conducted to enhance the acetyl-CoA supply for producing high-value added chemicals using acetyl-CoA as a precursor [6, 7].
Acetyl-CoA can be synthesized from glucose, acetate, and fatty acid in Escherichia coli (Fig. 1). Glucose is the most commonly used carbon source in E. coli, which produces acetyl-CoA via an efficient glycolysis pathway. Glucose is first converted to pyruvate through the glycolytic pathway, and then into acetyl-CoA by pyruvate dehydrogenase under aerobic conditions or by pyruvate-formate lyase under anaerobic conditions with the release of CO2. The loss of carbon in this step is associated with a decrease in the theoretical carbon recovery rate of 66.7% when using glucose as an acetyl-CoA source (Table 1). In recent studies, Mainguet et al. [8] and Bogorad et al. [9] had reported that 3 mol acetyl-CoA could be produced from 1 mol glucose by a reconstructed glyoxylate shunt or a non-oxidative glycolysis. The studies are still in the stage of proof of concept and more works are needed to make it possible for industrial application. Recently, acetate was used as an alternative carbon source and to produce chemicals such as succinate [10, 11], itaconic acid [12], isobutanol [13], and polyhydroxyalkanoates [14]. Two pathways can convert acetate to acetyl-CoA: (1) the reversible acetate kinase-phosphate acetyltransferase (ACK-PTA) pathway and (2) the irreversible ACS pathway catalyzed by acetyl-CoA synthetase (ACS). ACS is a high-affinity enzyme with a low Km of 200 μM for acetate [15], while the ACK-PTA pathway can only act at a relatively high concentration, as the Km values of both enzymes for their substrates are 7–10 mM [16]. More ATP is consumed in the ACS pathway when AMP is formed compared to the ACK-PTA pathway in which ADP is formed [17, 18]. Fatty acids can also be used to produce acetyl-CoA along with abundant NADH and FADH2 via fatty acid oxidation [19]. For example, oxidation of 1 mol palmitic acid (a type of fatty acid) can produce 7 mol NADH and 7 mol FADH2. The used of fatty acids is very efficient and can result in product yields that are significantly higher than those obtained from other carbon sources because of its high atom economy [20]. The theoretical carbon recovery rate of acetate and fatty acid as an acetyl-CoA source is 100%, which is higher than that of glucose.
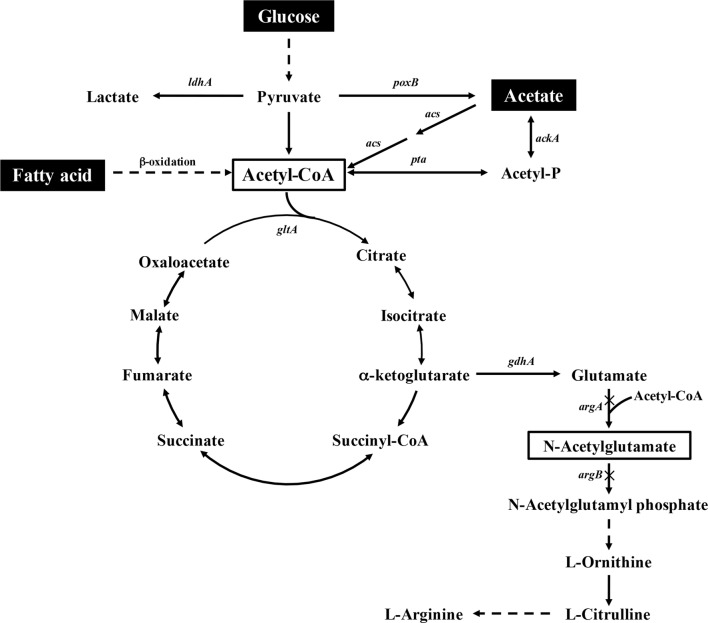
Fig. 1.
Overview of acetyl-CoA metabolism in Escherichia coli and the biosynthesis pathway of NAG
Table 1.
The equation of different carbon sources producing acetyl-CoA
| Carbon source | Equation | Carbon recovery rate |
|---|---|---|
| Glucose | C6H12O6 → 2 Acetyl-CoA + 2 CO2 + 4 NADH + 2 ATP | 2/3 |
| Acetate | C2H4O2 + ATP → Acetyl-CoA | 1 |
| Palmitic acid | C16H32O2 + 2 ATP → 8 Acetyl-CoA + 7 NADH + 7 FADH2 | 1 |
NAG is the initial precursor of arginine in the arginine biosynthetic pathway in microorganisms and plants and is essential for the biosynthesis of l-citrulline. In addition, NAG is an essential allosteric cofactor of mitochondrial carbamyl phosphate synthetase I, the first and rate-limiting enzyme of the urea cycle in ureotelic animals. Studies of NAG synthesis are important for improving the biosynthesis of l-citrulline, l-arginine, and other derivatives. Acetyl-CoA is used to provide an acetyl group in this one-step reaction of NAG synthesis from glutamate.
In this study, a metabolically engineered E. coli was developed for NAG production by introducing genes that catalyze NAG formation into the modified host strain. We focused on metabolic engineering for efficiently supplying acetyl-CoA from glucose, acetate, and fatty acid (particularly, palmitic acid). The results provide insight into methods for efficiently supplying acetyl-CoA for chemical synthesis using acetyl-CoA as a precursor or requiring acetylation. Additionally, metabolic engineering for NAG synthesis provides a foundation for industrial production of NAG and biosynthesis of arginine and its derivatives.
Results
Construction of biosynthetic pathway of NAG in E. coli
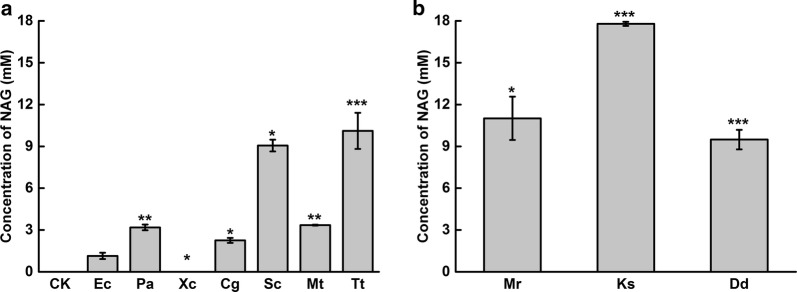
In E. coli, NAG was the precursor for arginine synthesis. N-acetylglutamate kinase encoded by argB and bi-functional N-acetylglutamate synthase encoded by argA were involved in the arginine biosynthesis pathway (Fig. 1). For NAG accumulation, argB and argA in E. coli BW25113 were knocked out resulted in strain N00 which was used as chassis for NAG production. N-Acetylglutamate synthase (NAGS) with high catalytic efficiency was then screened for efficient synthesis of NAG. The reported NAGS genes from E. coli, Pseudomonas aeruginosa, Xanthomonas campestris, Corynebacterium glutamicum, Streptomyces coelicolor, Mycobacterium tuberculosis, and Thermus thermophiles [21–27] were overexpressed in E. coli under the control of the pBAD promoter in plasmids pN01, pN02, pN03, pN05, pN06, pN08, and pN14, respectively. The plasmid was transformed into the host strain N00. The strains were cultured and used for whole-cell biocatalysis of NAG. After 8 h of whole-cell bioconversion, 10.12 mM NAG was obtained from 50 mM sodium glutamate and 50 mM glucose by the strain with NAGS from T. thermophiles which was higher than that from other NAGSs (Fig. 2a). However, the protein expression of Tt-NAGS (NAGS from T. thermophiles) was unstable. Although the strategies of codon-optimization, co-expression with chaperone, and lower temperature (data not shown) had been tried, no significant improvement on Tt-NAGS expression was observed. Therefore, we search for new NAGS in the NCBI database based on the reported amino acid sequences of the NAGS from S. coelicolor and T. thermophiles. NAGSs from Meiothermus ruber, Kitasatospora setae, and Deinococcus deserti were selected and cloned in host N00. A higher amount of NAG (17.89 mM) was obtained by the strain 0019 with Ks-NAGS. Enzyme measurements of Ec-NAGS, Tt-NAGS and Ks-NAGS from E. coli, T. thermophiles and K. setae were performed using purified recombinant proteins. The enzymatic analysis of the specific activity of Ec-NAGS, Tt-NAGS and Ks-NAGS showed that Ks-NAGS from K. setae had higher specific activity than the other two (Table 2). Thus, strain 0019 was used thereafter for metabolic engineering of acetyl-CoA supply from different carbon sources.
Fig. 2.
Effects of N-acetylglutamate synthase (NAGS) from different species on NAG production. a Effects of NAGS reported in references on NAG production. b Effects of NAGS not previously reported on NAG production. Ec = E. coli, Pa = Pseudomonas aeruginosa, Xc = Xanthomonas campestris, Cg = Corynebacterium glutamicum, Sc = Streptomyces coelicolor, Mt = Mycobacterium tuberculosis, Tt = Thermus thermophilus, Mr = Meiothermus ruber, Ks = Kitasatospora setae, Dd = Deinococcus deserti
Table 2.
The specific activity of Ec-NAGS, Tt-NAGS and Ks-NAGS from E. coli, T. thermophiles and K. setae respectively
| Enzyme | Specific activity (μmol/min/mg) |
|---|---|
| Ec-NAGS | 3.7 ± 0.1 |
| Tt-NAGS | 5.4 ± 0.4 |
| Ks-NAGS | 8.0 ± 0.1 |
Metabolic engineering for efficient supply of acetyl-CoA from glucose
When using glucose as a carbon source to produce acetyl-CoA, the glucose utilization system was first modified by replacing native ptsG and galR with glk and zglf (galactose: H+ symporter from Zymomonas mobilis) in the chromosome of host N00 based on a previous study which showed that PTS-– strains recruiting Z. mobilis glucose facilitator Glf and E. coli Glk had higher glucose utilization rates than wild-type strain [28]. The resulting host N01 (BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf) was then transformed with plasmid pN19. The recombinant strain 0119 exhibited increased NAG production to 20.95 mM after 8 h of bioconversion (Fig. 3, shown as N01).
Fig. 3.
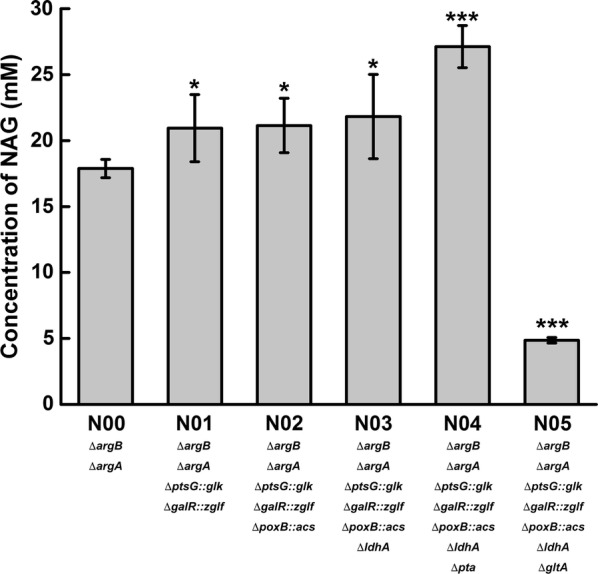
Production of NAG by engineered strain from glutamate and glucose. Engineered host strains transformed with plasmid pNAG19 were induced and suspended in a reaction mixture containing 50 mM sodium glutamate and 50 mM glucose. The bioconversion reactions were performed at 37 °C and 200 rpm for 8 h
Because acetyl-CoA was converted via pyruvate when using glucose as a carbon source, the bypass pathways of pyruvate were then blocked in host strain N01 to eliminate by-products and increase the pyruvate pool. The host strain N02 was obtained by replacing the pyruvate oxidase gene, poxB, with the acetyl-CoA synthetase gene, acs. The lactate dehydrogenase gene, ldhA, was then inactivated in N02 to produce the host N03. As shown in Fig. 3, this manipulation slightly increased NAG production.
pta and gltA, which react in the bypass pathways of acetyl-CoA, were inactivated in host N03 to produce hosts N04 and N05, respectively. When using recombinant strain 0419 as a whole-call biocatalyst, NAG production was clearly improved. In contrast, deletion of gltA greatly reduced NAG production (Fig. 3, shown as N05). Based on these results, modification of the glucose utilization system and disruption of the bypass pathways of pyruvate and acetyl-CoA increased the acetyl-CoA pool. This led to a 51.6% increase in NAG production to 27.12 mM.
Metabolic engineering of the strain for efficient supply of acetyl-CoA from acetate
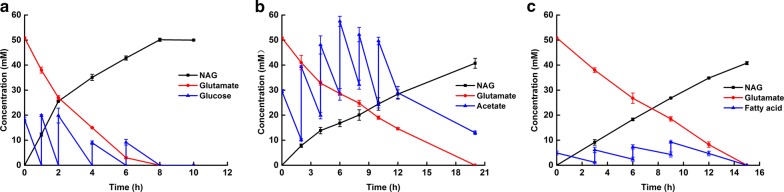
There are two pathways in E. coli that can convert acetate to acetyl-CoA: (1) ACS pathway can function at a low concentration of acetate but consumes more ATP. (2) ACK-PTA pathway requires less ATP but can only function at a relatively high concentration of acetate. These two pathways were investigated in our study by overexpressing acs or ackA-pta in E. coli. The plasmids overexpressing ACS (pAcs) or AckA-Pta (pAck) were co-transformed with pN19 into the host strain N00 to form the recombinant strains 0019Acs and 0019Ack, respectively. In the whole-cell bioconversion acetate was used as substrate for acetyl-CoA synthesis and low concentration of glucose (2 mM) was added to the system for initial supply of ATP. As shown in Fig. 4, after 20 h of bioconversion from 50 mM sodium glutamate and 100 mM sodium acetate, 6.52 mM NAG was produced by strain 0019Acs overexpressing the ACS pathway, while 17.88 mM NAG was produced by strain 0019Ack overexpressing the ACK-PTA pathway. Strain 0019Acs produced even less NAG than the control strain 0019, which produced 10.05 mM NAG after 20 h of bioconversion. Additionally, after overnight induction, the biomass of strain 0019Acs was 40% lower than those of the other two strains (data not shown). This indicated that the overexpression of acs might affect the normal metabolism in this strain.
Fig. 4.
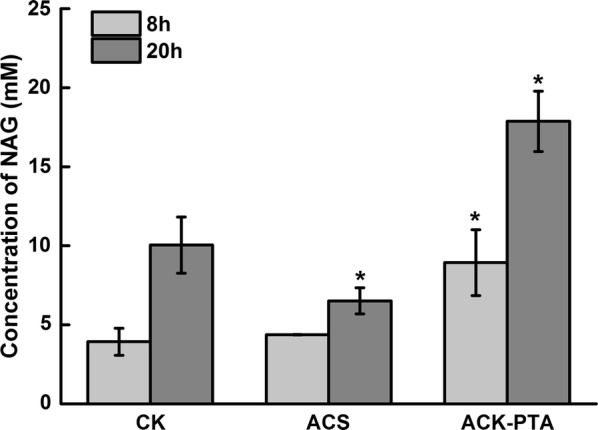
Production of NAG by engineered strain from glutamate and acetate. The bioconversion medium used here contained 50 mM sodium glutamate and 100 mM sodium acetate. The bioconversion reactions were performed at 37 °C and 200 rpm. H2SO4 was added at different reaction times to adjust the pH value
The above results demonstrated that overexpression of the ackA-pta operon in a plasmid can enhance the pathway that converts acetate to acetyl-CoA, leading to a 77.9% increase in the acetyl-CoA supply for NAG production when using acetate as a carbon source.
Metabolic engineering for efficient supply of acetyl-CoA from fatty acid
Fatty acid is catabolized through the β-oxidation pathway into acetyl-CoA. Metabolic engineering for supplying acetyl-CoA from fatty acid was manipulated by derepression of fadR (a major regulator that negatively controls the fatty acid degradative regulon) and constitutive expression of fadD (a transporter involved in the transport and activation of exogenous fatty acid).
Host strain N06 was constructed by deletion of fadR and replacement of the native promoter of fadD with a strong promoter CPA1 in host N00. Strain 0619 with plasmid pN19 in host N06 used fatty acid efficiently and produced 31.92 mM NAG after 20 h of bioconversion with 50 mM sodium glutamate and 16 mM palmitic acid as substrates (Fig. 5). These results confirmed that an increased acetyl-CoA pool can be efficiently achieved from fatty acids through a modified β-oxidation pathway which increased NAG production by 3.8-fold.
Fig. 5.
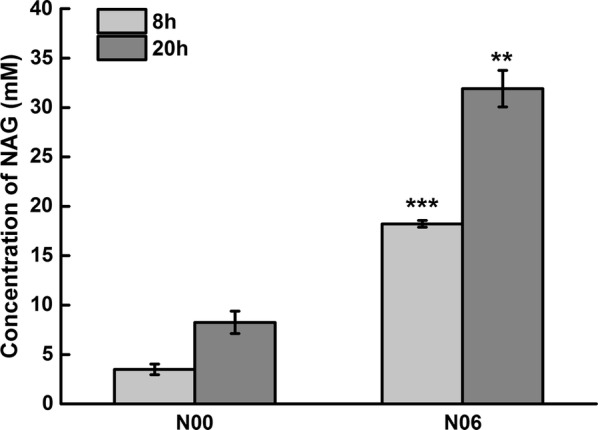
Production of NAG by engineered strain from glutamate and fatty acid. Concentrations of 50 mM sodium glutamate and 15 mM palmitic acid were used for NAG production. The bioconversion reactions were performed at 37 °C and 200 rpm
Comparison of three sources for acetyl-CoA supply
To compare the efficiency of different carbon sources for acetyl-CoA supply, 80 mM glucose, 160 mM sodium acetate, or 20 mM palmitic acid were used as substrate which produce mole equivalent of acetyl-CoA theoretically (Table 1). The carbon sources were added to the reaction system with 50 mM sodium glutamate for whole-cell bioconversion to achieve NAG production.
When glucose was used as an acetyl-CoA source, all 50 mM glutamate was transformed to NAG after 8 h of conversion (Fig. 6a) by strain 0419 with 98.2% and 62.4% molar conversion ratios of NAG from glutamate and glucose, respectively. When using acetate to produce acetyl-CoA, the reaction rate was much slower than that when using glucose. After 20 h of bioconversion, 40.73 mM NAG was obtained from strain 0019Ack and 13 mM acetate remained (Fig. 6b). After 20 h, the molar conversion ratios of NAG from glutamate and acetate were 81.4% and 27.7%, respectively. When using palmitic acid, a fatty acid, as the source of acetyl-CoA, 40.80 mM NAG was obtained by strain 0619 after 15 h of bioconversion. The titer of NAG was equal to that using acetate as a source by 0019Ack (Fig. 6c), but the productivity was higher. The molar conversion ratios of NAG from glutamate and palmitic acid were 81.6% and 204%, respectively.
Fig. 6.
Comparison of glucose, acetate, and fatty acid as source of acetyl-CoA. The reaction mixture containing 50 mM sodium glutamate and 1 × M9 salts buffer were used. a Glucose (80 mM) was supplemented at rates of 20, 20, 20, 10, and 10 mM at 0, 1, 2, 4, and 6 h of bioconversion. b Acetate (160 mM) was supplemented at rates of 30, 30, 30, 30, 20, and 20 mM at 0, 2, 4, 6, 8, and 10 h of bioconversion. Additionally, 2 mM glucose was added at 0 h of bioconversion to supply ATP for the acetylation of acetate. c Palmitic acid (20 mM) was supplemented at 5 mM at each addition at 0, 3, 6, and 9 h of bioconversion
The results showed that the strain engineered to use glucose (strain 0419) exhibited the highest conversion rate of glutamate and the highest productivity (an average of 6.25 mmol/L/h), indicating that the highest productivity of acetyl-CoA was achieved when glucose was used as a carbon source. The productivity of acetyl-CoA from acetate and fatty acid was not as efficient as that from glucose.
Bioconversion for NAG production by engineered strain 0419
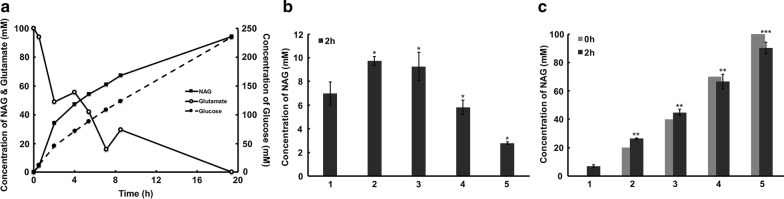
Strain 0419 produced NAG with a high yield and conversion rate using glucose as a carbon source for acetyl-CoA supply. Thus, scale-up bioconversion using strain 0419 was carried out in a 1-L fermenter containing 500 mL medium for NAG production. The substrate sodium glutamate was added at 100 mM in one portion, and the substrate glucose was added in a fed-batch manner. After 23 h of bioconversion, 94 mM NAG was obtained, while 230 mM glucose was consumed (Fig. 7a). The molar conversion rate of NAG from glucose was much lower in the fermenter than in the shaking flasks. This may be because acetyl-CoA was consumed in the tricarboxylic acid (TCA) cycle. The scale-up bioconversion process was also carried out with higher concentrations of glutamate (200 or 500 mM) added once or multiple times, but the NAG yield did not increase. The results indicated that there might be substrate inhibition or product inhibition in this reaction.
Fig. 7.
Bioconversion for NAG by engineered strain 0419. a Scale-up bioconversion of NAG production in a 1-L fermenter. The black square indicates the concentration of NAG. The hollow circle represents the concentration of glutamate. The black circle represents the concentration of glucose consumed. b Substrate inhibition. The concentrations of glutamate used were: 1: 50 mM; 2: 100 mM; 3: 200 mM; 4: 500 mM; 5: 1 M. The glucose concentration was 50 mM. The reaction was carried out for 2 h. c Product inhibition. The concentration of NAG at 0 h represented the initially added concentration. In detail, the concentrations of NAG added initially were: 1: 0 mM; 2: 20 mM; 3: 40 mM; 4: 70 mM; 5: 100 mM. The bioconversion reactions were performed at 37 °C and 200 rpm for 2 h
As shown in Fig. 7b, as glutamate levels increased, NAG production did not increase accordingly. When the concentration of glutamate was increased to 200 mM or more, the NAG yield decreased. These results suggest that glutamate inhibited NAG production. NAG production was reduced when the amount of NAG added initially was more than 40 mM. When the initial NAG concentration was more than 70 mM, NAG degradation was faster than NAG production (Fig. 7c). These results confirmed that a high concentration NAG inhibited NAG synthesis. The effects of NAG and glutamate on the Ks-NAGS activity were also analyzed in vitro, and the specific activity of Ks-NAGS reduced when the concentration of glutamate was higher than 20 mM and the concentration of NAG was higher than 0.5 mM (Additional file 1: Figure S1). The result was consistent with the in vivo experiment. The above results suggested inhibition of both substrate inhibition and product inhibition of NAGS from Kitasatospora setae. This limited the increase in NAG production during scale-up bioconversion.
Discussion
Acetyl-CoA is the precursor of many high-cost chemicals and can provide an acetyl-group precursor for producing various industrially useful acetyl-metabolites. Metabolic engineering of E. coli for efficiently supplying acetyl-CoA would enhance the production of acetyl-chemicals. In this study, the biosynthesis of NAG, which is synthesized from glutamate and acetyl-CoA, was used to develop an efficient strategy for supplying acetyl-CoA. We first constructed a NAG-producing E. coli strain 0019 by knocking out native argB and argA and overexpressing a new NAGS from K. setae. However, Ks-NAGS was inhibited by both its substrate glutamate and its product NAG. Screening of NAGS with higher enzyme activity or the evolution of NAGS from K. setae may further improve the NAG production.
The NAG producing strain 0019 was further optimized by metabolic engineering using different carbon sources, including glucose, acetate, and fatty acid. Glucose was more efficient than acetate and fatty acid for supplying acetyl-CoA with the highest NAG productivity, but the yield of NAG from glucose remained far below the theoretical conversion rate. The active TCA cycle, which can consume large amounts of acetyl-CoA, may explain this discrepancy. Further studies to balance the acetyl-CoA flux to NAG and TCA cycle are needed to increase the yield of acetyl-CoA from glucose.
Acetate is a promising alternative carbon source for supplying acetyl-CoA in complex reactions because of its short pathway. Acetate has been used as a carbon source in the production of polyhydroxyalkanoates, itaconic acid, and other chemicals. Chen et al. used acetate as a main carbon source to synthesize polyhydroxyalkanoates, and strains overexpressing the ACK-PTA pathway produced nearly fourfold levels of poly-3-hydroxybutyrate, twofold levels of poly(3-hydroxybutyrate-co-4-hydroxybutyrate), and twofold levels of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) compared to control strains [14]. By overexpressing acs and inactivating iclR, the conversion rate of acetate to itaconic acid was improved to 16.7% of the theoretical maximum yield [12]. In this study, a 27.7% conversion rate of acetate to NAG was obtained by overexpression of the ACK-PTA pathway and NAGS from K. setae. However, the reaction rate when using acetate as a carbon source was very slow, and the conversion rate remained far below the theoretical conversion rate. Further studies are needed to evaluate the assimilation of acetate, ATP supply, and TCA consumption for efficient conversion of acetate to acetyl-CoA.
Fatty acid is advantageous for synthesizing compounds such as n-butanol and 3-hydroxypropionate in processes that require a high reducing power. Liu et al. [29] developed an engineered E. coli strain that can efficiently use fatty acid as feedstock and produced 3-hydroxypropionate with a yield of 1.56 g/g FAs in a 5-L bioreactor. In this study, fatty acid was used to supply acetyl-CoA for NAG synthesis with an acceptable titer. The results indicated that fatty acid can also be used to provide an acetyl group for producing value-added acetyl-chemicals. Except for palmitic acid, other types of fatty acids can be tested in further studies, and more targets of the β-oxidation pathway should be identified to further improve the assimilation and utilization efficiency of fatty acid.
Conclusions
In this study, systemic metabolic engineering of E. coli strains for efficiently supplying acetyl-CoA from glucose, acetate, and fatty acid was carried out and applied in the biosynthesis of NAG. Deletion of the native argB and argA and introduction of an efficient NAGS from K. setae resulted in an engineered E. coli strain for NAG production. Metabolic engineering strategies were then applied in the E. coli strain using different carbon sources to supply the precursor acetyl-CoA. The recombinant strains 0419, 0019Ack, and 0619 were used to compare acetyl-CoA supplied from 80 mM glucose, 160 mM acetate, and 20 mM palmitic acid for NAG production, respectively. The molar weights of glucose, acetate, and palmitic acid used theoretically produced 160 mM acetyl-CoA. For NAG synthesis, glucose was the most efficient carbon source, but acetate and fatty acid also showed good performance. The results demonstrated the great potential of acetate and fatty acid for supplying acetyl-CoA. The strategies developed in this study may be potentially applicable for producing not only acetyl-CoA-derived metabolites, but also a wide range of useful compounds that require acetylation in the pathway. However, further engineering is required to enhance the utilization of acetate and fatty acid by E. coli strains.
Methods
Strains and plasmids
The bacteria strains and plasmids used in this study are listed in Table 3. Escherichia coli DH5α was used for molecular cloning. Escherichia coli K12 (BW25113) was used as the parental strain for genetic modification and NAG production. Gene knockout strains were obtained from the KEIO collection (National BioResource Project) [30]. The plasmids pRB1 s and pSB1a used for gene expression were derived from expression vectors previously developed in our laboratory (unpublished) and have the following features: a promoter (araBAD), multiple cloning sites, rrnB terminator, origin of replication RSF1030 and pSC101, and streptomycin and ampicillin resistance genes. The nucleotide sequence of genes encoding enzymes that can catalyze NAG formation were obtained by PCR amplification and ligated into pRB1 s at the NcoI and EcoRI sites by the Gibson assembly method [31]. The templates for NAG synthase genes from T. thermophilus, M. ruber, K. setae, and D. deserti were synthesized and codon-optimized by Generay (Shanghai, China). Because NAGS from E. coli and P. aeruginosa have both an acetyl transferase domain and amino acid kinase domain [22], only the C-terminal proteins of E. coli ArgA and P. aeruginosa ArgA were amplified and cloned into pRB1 s. The plasmids pAcs and pAck were constructed by introducing the acs and ackA-pta operons of E. coli into the NcoI and EcoRI sites of pSB1a. For chromosomal phenotype integration, P1 virus-mediated transfection was performed as described by Thomason et al. [32]. The plasmid pCP20 was used to remove the FLP recognition target-flanked resistant marker in E. coli K12 [33]. For gene and promoter replacement, the CRISPR-Cas9 system was used [34]. The primers used in this study are listed in Additional file 1: Table S1.
Table 3.
Strains and plasmids used in this study
| Strain/plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−endA1 glnV44 thi−1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZ ∆M15 ∆(lacZYA-argF)U169, hsdR17(r−K m+K), λ− | Invitrogen |
| E. coli BW25113 | lacIqrrnBT14∆lacZWJ16hsdR514∆araBADAH33∆rhaBADLD78 | Invitrogen |
| N00 | E. coli BW25113, ∆argB, ∆argA | This study |
| N01 | E. coli BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf | This study |
| N02 | E. coli BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf, ∆poxB::acs | This study |
| N03 | E. coli BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf, ∆poxB::acs, ∆ldhA | This study |
| N04 | E. coli BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf, ∆poxB::acs, ∆ldhA, ∆pta | This study |
| N05 | E. coli BW25113, ∆argB, ∆argA, ∆ptsG::glk, ∆galR::zglf, ∆poxB::acs, ∆ldhA, ∆gltA | This study |
| N06 | E. coli BW25113, ∆argB, ∆argA, ∆fadR, PfadD::PCPA1 | This study |
| Plasmids | ||
| pRB1 s | RSF1030 origin, Pbad promoter, StrR | Our lab |
| pSB1a | pSC101 origin, Pbad promoter, AmpR | Our lab |
| pN01 | argA from E. coli cloned into pRB1 s | This study |
| pN02 | argA from Pseudomonas aeruginose cloned into pRB1 s | This study |
| pN03 | argA from Xanthomonas campestris cloned into pRB1 s | This study |
| pN05 | argA from Corynebacterium glutamicum cloned into pRB1 s | This study |
| pN06 | argA from Streptomyces coelicolor cloned into pRB1 s | This study |
| pN08 | argA from Mycobacterium tuberculosis cloned into pRB1 s | This study |
| pN14 | argA from Thermus thermophilus cloned into pRB1 s | This study |
| pN18 | argA from Meiothermus ruber cloned into pRB1 s | This study |
| pN19 | argA from Kitasatospora setae cloned into pRB1 s | This study |
| pN20 | argA from Deinococcus deserti cloned into pRB1 s | This study |
| pAcs | acs from E. coli cloned into pSB1a | This study |
| pAck | ackA-pta from E. coli cloned into pSB1a | This study |
Culture conditions
Luria-Bertani (LB) medium (per liter: tryptone 10 g, yeast extract 5 g, NaCl 10 g) was used for all molecular construction experiments and strain cultures. Strains harboring the temperature-sensitive plasmid pCP20 were incubated at 30 °C and 220 rpm, while the other strains were grown at 37 °C and 220 rpm. For protein expression, the strains were pre-cultured in LB medium supplemented with appropriate antibiotics (streptomycin 50 mg/L, ampicillin 100 mg/L) at 37 °C for 4–6 h, and then transferred into auto-inducing ZYM medium (per liter: tryptone 10 g, yeast extract 5 g, glycerol 5 g, glucose 0.5 g, l-arabinose 2 g, Na2HPO4 25 mM, KH2PO4 25 mM, NH4Cl 50 mM, Na2SO4 5 mM, MgSO4 2 mM and trace elements containing 0.05 mM FeCl3, 0.02 mM CaCl2, 0.01 mM MnCl2, 0.01 mM ZnSO4, and 0.002 mM each of CoCl2, NiCl2, Na2Mo4, Na2SeO3, and H3BO3) [35] with 1% inoculum and induced at 37 °C for 12–16 h.
Whole-cell biocatalysis conditions
Whole-cell biocatalysis was used for NAG production [36]. Cells induced overnight were harvested by centrifugation at 4000 rpm for 10 min and washed with 0.85% NaCl solution once. The cells were suspended in 1 mL bioconversion medium containing 50 mM sodium glutamate and 1 × M9 salts (per liter: 12.8 g Na2HPO4.7H2O, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl) and different carbon sources to form a cell suspension from a starting biomass of OD600nm = 30. The bioconversion reaction was performed at 37 °C and 200 rpm in a flask. In experiments comparing the three sources of acetyl-CoA, a reaction mixture containing 50 mM sodium glutamate and 1 × M9 salts was used. The substrate glucose, acetate, or fatty acid was supplemented frequently in small amounts based on the experimental conditions.
Scale-up of bioconversion for NAG production using glucose as a substrate
The scale-up of bioconversion was carried out in a 1-L fermenter containing 500 mL bioconversion medium. Cells were induced first in a 5-L fermenter containing 2 L auto-inducing ZYM medium similar to that in the shake flasks, and then the cells were harvested by centrifugation. The induced cells were suspended in bioconversion medium containing 100 mM glutamate and 1 × M9 salts with an initial cell density of OD600nm = 30. The cells were cultured at 37 °C with 30% dissolved oxygen. The pH was maintained at 7.0 by feeding of sodium hydroxide. Glucose was fed at an initial rate of 25 mM/h and slowed after 2 h when it accumulated. The feeding rate was then controlled to ensure accumulation of no more than 1 g/L.
Enzymatic analysis of NAGS
NAGS activity was assayed spectrophotometrically by measuring the increase in absorbance at 412 nm due to the formation of 5-thio-2-nitrobenzoate resulting from the reaction between the sulfhydryl group of CoASH, generated by the amino acid acetylating activity, and 5,5-dithio-bis(2-nitrobenzoic acid) (DTNB) as previously reported [37]. Reaction mixtures contained 50 mM Tris–HCl (pH 8.0), 2 mM AcCoA, 20 mM sodium glutamate, 10 mM MgCl2, 0.2 mM DTNB and 0.65 μg enzyme in a total volume of 300 μL. Color production was linear with time for > 1 min, One unit of enzyme activity is defined as the amount of enzyme required to produce 1 μmol of NAG per minute at 37 °C.
Substrate and product inhibition
For substrate inhibition analysis, 5 groups of bioconversion media were used containing 50 mM glucose and different concentrations of glutamate (50, 100, 200, and 500 mM and 1 M). Cells after culture were harvested and suspended in 1 mL conversion medium, and the reaction was carried out for 2 h at 37 °C and 200 rpm.
The product inhibition experiment was carried out by adding different concentrations of NAG (0, 20, 40, 70 and 100 mM) to the conversion medium containing 50 mM glutamate and 50 mM glucose. After conducting the reaction at 37 °C and 200 rpm for 2 h, NAG in the supernatants was detected.
Analytical methods
Cell density was estimated by measuring the optical density at 600 nm. The concentrations of NAG, glucose, and organic acids in the supernatant were determined by high-performance liquid chromatography equipped with a Bio-Rad Aminex HPX-87H Ion Exclusion column (7.8 × 300 mm; Hercules, CA, USA), refractive index detector, and diode-array detector. Analysis was performed at 65 °C with a mobile phase of 5 mM H2SO4 at a flow rate of 0.5 mL/min.
Additional file
Additional file 1: Figure S1. The effects of NAG and glutamate on Ks-NAGS activity. Table S1 Primers used in this study.
Acknowledgements
This work was supported by National Natural Science Foundation of China Grant 31670051. We would like to thank Editage (http://www.editage.cn) for English language editing.
Abbreviations
- NAG
N-acetlyglutamate
- NAGS
N-acetlyglutamate synthase
- Ec
E. coli
- Pa
Pseudomonas aeruginose
- Xc
Xanthomonas campestris
- Cg
Corynebacterium glutamicum
- Sc
Streptomyces coelicolor
- Mt
Mycobacterium tuberculosis
- Tt
Thermus thermophiles
- Mr
Meiothermus ruber
- Ks
Kitasatospora setae
- Dd
Deinococcus deserti
- TCA
tricarboxylic acid
- LB
Luria–Bertani
Authors’ contributions
S-SZ carried out the main work, collected and analyzed the data, and drafted the manuscript. WY, HC and BL participated in the research. B-XL supervised the work and participated in data analysis, and revised the manuscript. YT participated in the conception and design of the study and finalized the manuscript. All authors read and approved the final manuscript.
Funding
National Natural Science Foundation of China Grant 31670051.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shasha Zhang, Email: zhss31@126.com.
Wei Yang, Email: 18518281172@163.com.
Hao Chen, Email: 1020786970@qq.com.
Bo Liu, Email: ycliubo@qq.com.
Baixue Lin, Email: linbaixue@126.com.
Yong Tao, Email: taoyong@im.ac.cn.
References
- 1.Jope RS, Jenden DJ. The utilization of choline and acetyl coenzyme A for the synthesis of acetylcholine. J Neurochem. 1980;35:318–325. doi: 10.1111/j.1471-4159.1980.tb06267.x. [DOI] [PubMed] [Google Scholar]
- 2.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 3.Lian J, Si T, Nair NU, Zhao H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Kildegaard KR, Jensen NB, Schneider K, Czarnotta E, Ozdemir E, Klein T, Maury J, Ebert BE, Christensen HB, Chen Y, et al. Engineering and systems-level analysis of Saccharomyces cerevisiae for production of 3-hydroxypropionic acid via malonyl-CoA reductase-dependent pathway. Microb Cell Fact. 2016;15:53. doi: 10.1186/s12934-016-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen G-Q, Jiang X-R. Engineering microorganisms for improving polyhydroxyalkanoate biosynthesis. Curr Opin Biotechnol. 2018;53:20–25. doi: 10.1016/j.copbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Krivoruchko A, Zhang Y, Siewers V, Chen Y, Nielsen J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab Eng. 2015;28:28–42. doi: 10.1016/j.ymben.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Zhang B, Jiang R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol Biofuels. 2017;10:41. doi: 10.1186/s13068-017-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mainguet SE, Gronenberg LS, Wong SS, Liao JC. A reverse glyoxylate shunt to build a non-native route from C4 to C2 in Escherichia coli. Metab Eng. 2013;19:116–127. doi: 10.1016/j.ymben.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Bogorad IW, Lin TS, Liao JC. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature. 2013;502:693–697. doi: 10.1038/nature12575. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Huang B, Wu H, Li Z, Ye Q, Zhang YP. Production of succinate from acetate by metabolically engineered Escherichia coli. ACS Synth Biol. 2016;5:1299–1307. doi: 10.1021/acssynbio.6b00052. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, Yang H, Fang G, Zhang X, Wu H, Li Z, Ye Q. Central pathway engineering for enhanced succinate biosynthesis from acetate in Escherichia coli. Biotechnol Bioeng. 2018;115:943–954. doi: 10.1002/bit.26528. [DOI] [PubMed] [Google Scholar]
- 12.Noh MH, Lim HG, Woo SH, Song J, Jung GY. Production of itaconic acid from acetate by engineering acid-tolerant Escherichia coli W. Biotechnol Bioeng. 2018;115:729–738. doi: 10.1002/bit.26508. [DOI] [PubMed] [Google Scholar]
- 13.Song H-S, Seo H-M, Jeon J-M, Moon Y-M, Hong JW, Hong YG, Bhatia SK, Ahn J, Lee H, Kim W, et al. Enhanced isobutanol production from acetate by combinatorial overexpression of acetyl-CoA synthetase and anaplerotic enzymes in engineered Escherichia coli. Biotechnol Bioeng. 2018;115:1971–1978. doi: 10.1002/bit.26710. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Li W, Zhang ZZ, Tan TW, Li ZJ. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb Cell Fact. 2018;17:102. doi: 10.1186/s12934-018-0949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown TDK, Jones-Mortimer MC, Kornberg HL. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. Microbiology. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 17.Enjalbert B, Millard P, Dinclaux M, Portais JC, Letisse F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci Rep. 2017;7:42135. doi: 10.1038/srep42135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valgepea K, Adamberg K, Nahku R, Lahtvee PJ, Arike L, Vilu R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst Biol. 2010;4:166. doi: 10.1186/1752-0509-4-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JJ, Battaile KP. Burning fat: the structural basis of fatty acid beta-oxidation. Curr Opin Struct Biol. 2002;12:721–728. doi: 10.1016/S0959-440X(02)00390-1. [DOI] [PubMed] [Google Scholar]
- 20.Dellomonaco C, Rivera C, Campbell P, Gonzalez R. Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl Environ Microbiol. 2010;76:5067–5078. doi: 10.1128/AEM.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldovic L, Ah Mew N, Shi D, Morizono H, Yudkoff M, Tuchman M. N-acetylglutamate synthase: structure, function and defects. Mol Genet Metab. 2010;100(Suppl 1):S13–S19. doi: 10.1016/j.ymgme.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Errey JC, Blanchard JS. Functional characterization of a novel ArgA from Mycobacterium tuberculosis. J Bacteriol. 2005;187:3039–3044. doi: 10.1128/JB.187.9.3039-3044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petri K, Walter F, Persicke M, Ruckert C, Kalinowski J. A novel type of N-acetylglutamate synthase is involved in the first step of arginine biosynthesis in Corynebacterium glutamicum. BMC Genomics. 2013;14:713. doi: 10.1186/1471-2164-14-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramon-Maiques S, Fernandez-Murga ML, Gil-Ortiz F, Vagin A, Fita I, Rubio V. Structural bases of feed-back control of arginine biosynthesis, revealed by the structures of two hexameric N-acetylglutamate kinases, from Thermotoga maritima and Pseudomonas aeruginosa. J Mol Biol. 2006;356:695–713. doi: 10.1016/j.jmb.2005.11.079. [DOI] [PubMed] [Google Scholar]
- 25.Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Mechanism of arginine regulation of acetylglutamate synthase, the first enzyme of arginine synthesis. FEBS Lett. 2009;583:202–206. doi: 10.1016/j.febslet.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Functional dissection of N-acetylglutamate synthase (ArgA) of Pseudomonas aeruginosa and restoration of its ancestral N-acetylglutamate kinase activity. J Bacteriol. 2012;194:2791–2801. doi: 10.1128/JB.00125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi D, Allewell NM, Tuchman M. The N-acetylglutamate synthase family: structures, function and mechanisms. Int J Mol Sci. 2015;16:13004–13022. doi: 10.3390/ijms160613004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J, Zhu X, Lu J, Liu P, Xu H, Tan Z, Zhang X. Recruiting alternative glucose utilization pathways for improving succinate production. Appl Microbiol Biotechnol. 2013;97:2513–2520. doi: 10.1007/s00253-012-4344-1. [DOI] [PubMed] [Google Scholar]
- 29.Liu B, Xiang S, Zhao G, Wang B, Ma Y, Liu W, Tao Y. Efficient production of 3-hydroxypropionate from fatty acids feedstock in Escherichia coli. Metab Eng. 2018;51:121–130. doi: 10.1016/j.ymben.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2006(2):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 32.Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007;79(1):1.17.1–1.17.8. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Lin B, Tao Y. Whole-cell biocatalysts by design. Microb Cell Fact. 2017;16:106. doi: 10.1186/s12934-017-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Site-directed mutagenesis studies of acetylglutamate synthase delineate the site for the arginine inhibitor. FEBS Lett. 2008;582:1081–1086. doi: 10.1016/j.febslet.2008.02.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The effects of NAG and glutamate on Ks-NAGS activity. Table S1 Primers used in this study.
Data Availability Statement
Not applicable.