Abstract
In this issue of Immunity, Iwata et al. (2017) report that the transcription factor T-bet acts as a selective repressor of the type I interferon (IFN) transcriptional program in response to IFN-γ signaling.
Differentiation of CD4+ T helper (Th) cells into distinct effector subsets is critical for the generation of an effective immune response against the diverse world of pathogenic microorganisms that we encounter in our lifetime. Each CD4+ Th cell subset is characterized by a specialized gene-expression program that is under the control of a master transcription factor, which ensures proper lineage specification by activating one genetic program while simultaneously silencing the gene-expression program of the alter-native CD4+ Th fates. Currently, four CD4+ Th effector subsets have been identified: Th1, Th2, Th17, and T follicular helper (Tfh) cells, which express distinct sets of cytokines specifically tailored to fight particular classes of pathogens. Th1 cells primarily produce interferon-γ (IFN-γ), which provides effective protection against intracellular bacteria; conversely, Th2 cells, which secrete interleukin-4 (IL-4), IL-5, and IL-13, coordinate anti-hel-minth responses. Th17 cells have evolved to provide protection against extracellular bacteria and fungi, whereas Tfh cells support the generation of antiviral humoral immunity. Integration of multiple signals contributes to the generation of CD4+ Th cell subsets with distinct effector functions. The ability of CD4+ T cells to sense, interpret, and respond to extracellular stimuli by modifying gene expression through the actions of signal-dependent transcription factors and lineage-defining master transcription factors will deter-mine the ultimate phenotype of an individual differentiating CD4+ Th progenitor cell (Weinmann, 2014).
Despite their commitment to a specific Th cell lineage, CD4+ T cells retain a certain degree of developmental flexibility and, upon changes in the cytokine environment, can acquire gene-expression profiles associated with other CD4+ Th cell lineages. This important feature of CD4+ Th cells allows them to adapt to changes in the environment and appropriately match their response to a specific microbial challenge. In this context, CD4+ Th1-cell-lineage-specifying transcription factors are critically important not only for controlling fate determination but also for fine-tuning the plasticity that allows the emergence of the required Th cell phenotype (Weinmann, 2014). The dual functionality is best exemplified by the CD4+ Th1-cell-lineage-specifying transcription factor T-bet (T-box expressed in T cells). T-bet is rapidly induced in response to T cell receptor (TCR), IFN-γ, and IL-12 signaling. T-bet in turn activates IFN-γ expression and establishes a self-amplifying feed-forward loop between T-bet and IFN-γ to ensure continuous T-bet expression. In addition to depending on feed-forward loops, the reinforcement of the Th1 cell differentiation program is dependent on concomitant suppression of alternative lineage transcriptional programs. T-bet regulates this either by blocking the expression of other lineage-specifying transcription factors or by interfering with their transcriptional activity. For example, T-bet inhibits Th17 cell lineage commitment by blocking Runx1-mediated induction of the Th17- cell-specific master transcription factor RORγt, whereas T-bet suppresses the Th2 cell transcriptional program by interfering with the transcriptional activity of the Th2-cell-specific master regulator Gata3 (Hwang et al., 2005; Lazarevic et al., 2011). Interestingly, after expo-sure to Th1-cell-polarizing cytokines, committed Th2 and Th17 cells can upregulate T-bet expression and become more like Th1 cells (Hegazy et al., 2010; Wang et al., 2014). Acquisition of T-bet and its differentiation program produces a population of CD4+ effector Th cells with increased functional potential, and if not properly controlled, these can lead to the development of autoimmune and inflammatory diseases. Transcriptional regulatory networks that direct the transition of Th2 and Th17 cells to a Th1-like state in response to Th1-cell-polarizing cytokines have been well characterized (Hegazy et al., 2010; Wang et al., 2014); however, very little is known about how Th1 cells manage to avoid an excessive Th1-cell-polarizing environment induced by elevated IFN-γ levels that could lead to autoimmune sequelae.
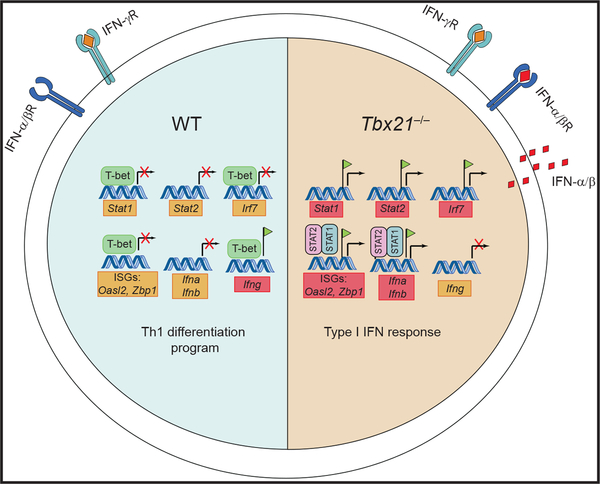
The study by Iwata et al. (2017) in this issue of Immunity investigates the role of the transcription factor T-bet in regulating Th1 cell adaptation to the IFN-γ-rich environment that is generated during type 1 immune responses. By performing genome-wide transcriptional profiling on T-bet-deficient and wild-type (WT) CD4+ T cells, the authors identified a fingerprint of 529 genes that were significantly altered by T-bet deficiency in response to IFN-γ signaling. Chromatin immune-precipitation sequencing revealed that T-bet bound to the regulatory regions of 35%–42% of these differentially ex-pressed genes. The most striking feature of the adaptation response to IFN-γ was the suppression of canonical type I IFN response genes. Type I IFNs (IFN-α and IFN-β) are produced by innate immune cells and virally infected cells. Although they share similar names, the transcriptomes induced by IFN-α, IFN-β, and IFN-γ differ significantly. Interestingly, the gene-expression profile of IFN-γ-treated T-bet-deficient CD4+ T cells resembled that of IFN-α- and IFN-β-treated WT CD4+ T cells, indicating that in the absence of T-bet, CD4+ T cells were unable to integrate and respond appropriately to IFN-γ signaling. Instead, IFN-γ-treated T-bet-deficient CD4+ T cells were now aberrantly ex-pressing the type I IFN transcriptional program. The authors demonstrated that T-bet-deficient CD4+ T cells increased IFN-α and IFN-β production and that anti-IFNα receptor antibody treatment reduced the expression of IFN-stimulated genes (ISGs), indicating that T-bet sup-presses auto-amplifying type I IFN circuitry in CD4+ T cells in an IFN-γ-rich environment. Mechanistically, T-bet inhibited the expression of type-I-IFN-specific transcription factors–signal transducer and activator of transcription 1 (STAT1), STAT2, and IFN regulatory factor 7 (IRF7). Additionally, T-bet interfered with STAT2 transcriptional activity by competing with STAT2 for binding to canonical ISG regulatory elements of approximately 26% ISGs (Figure 1).
Figure 1. T-bet Is a Negative Regulator of Type I IFN Response in CD4+ Th1 Cells.
In the presence of IFN-γ, T-bet blocks the type I IFN response in CD4+ T cells by inhibiting the expression of type I IFN transcription factors (STAT1, STAT2, and IRF7) and approximately 26% of ISGs. In contrast, in CD4+ T cells deficient in T-bet (Tbx21−/−), IFN-γ treatment causes an upregulation of STAT1, STAT2, and IRF7 expression and enhancement of STAT2 binding at ISG regulatory regions, leading to elevated production of IFN-α and IFN-β. The IFN-α and IFN-β produced form a self-amplifying feed-forward loop that reinforces the type I IFN response.
The study by Itawa et al. raises an intriguing question about the importance of silencing type I IFN responses in CD4+ T cells during Th1 cell differentiation. Studies have shown that the involvement of type I IFNs in the regulation of CD4+ Th1 cell responses is rather complex. Type I IFNs can promote CD4+ Th1 cell differentiation by inducing IFN-γ via STAT4 activation (Nguyen et al., 2002). Additionally, type I IFNs have been shown to increase T cell survival and clonal expansion during viral infections (Havenar-Daughton et al., 2006), indicating that type I IFNs can promote T cell fitness during an inflammatory response. Thus, enhanced type I IFN responses could be potentially beneficial in the context of T-bet deficiency. Paradoxically, however, IFN-β is the most widely accepted treatment for multiple sclerosis (MS). In the experimental autoimmune encephalomyelitis (EAE) model of MS, IFN-β was exceptionally effective in suppressing EAE symptoms induced by Th1 cells via an IL-10-dependent mechanism (Axtell et al., 2010), suggesting that type I IFNs could have a negative regulatory role on Th1 cells. Because of the pleiotropic functions of T-bet in the immune system, T-bet-deficient mice show increased susceptibility to viral and bacterial infections; thus, it is challenging to investigate bio-logical consequences of misdirected, aberrant type I IFN responses in T-bet-deficient backgrounds in vivo. To evaluate whether T-bet selectively inhibits type I IFN circuitry in vivo, Iwata et al. reconstituted Rag2−/− mice with a mix of both WT and T-bet-deficient naive CD4+ T cells before infection with Toxoplasma gondii (a strong IFN-γ inducer) or lymphocytic choriomeningitis virus (LCMV, a strong IFN-α and IFN-β inducer). At the transcriptional level, the upregulation of type-I-IFN-specific transcription factors was evident only in T-bet-deficient CD4+ T cells isolated from the peritoneal exudate during T. gondii infection and not during LCMV infection, demonstrating a T-cell-intrinsic role of T-bet in suppressing type I IFN circuitry in vivo during a strong Th1 cell response. Although not investigated here, it would be interesting to know whether, in this mixed co-transfer model, autocrine type I IFN signaling increases the survival of T-bet-deficient Th1 cells or whether the type-I-IFN-rich environment generated by T-bet deficiency boosts the functional fitness of WT Th1 cells and improves T. gondii clearance. It remains to be determined whether an enhanced type I transcriptional program is a common phenotype of T-bet-deficient CD4+ Th cells in other inflammatory settings. For example, T-bet is expressed in Th17 cells exposed to IL-12 or IL-23, and its expression is required for the pathogenicity of Th17 cells in the context of neuroinflammation (Wang et al., 2014; Yang et al., 2009). Because Th17 cells lacking T-bet cannot cause disease in a passive EAE model, it would be interesting to determine whether the non-pathogenicity of T-bet-deficient Th17 cells could be explained by the aberrant production of type I IFNs, which is known to be protective in MS and its animal model, EAE. Future studies will reveal whether targeting T-bet-mediated regulation of type I IFN circuitry will be useful for ameliorating CD4+ Th-cell-induced autoimmune pathologies.
REFERENCES
- Axtell RC, de Jong BA, Boniface K, van der Voort LF, Bhat R, De Sarno P, Naves R, Han M, Zhong F, Castellanos JG, et al. (2010). Nat. Med 16, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Kolumam GA, and Murali-Krishna K (2006). J. Immunol 176, 3315–3319. [DOI] [PubMed] [Google Scholar]
- Hegazy AN, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, and Löhning M (2010). Immu- nity 32, 116–128. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, and Glimcher LH (2005). Science 307, 430–433. [DOI] [PubMed] [Google Scholar]
- Iwata S, Mikami Y, Sun H-W, Brooks SR, Jankovic D, Hirahara K, Onodera A, Shih H-Y, Kawabe T, Jiang K, et al. (2017). Immunity 46, this issue, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, and Glimcher LH (2011). Nat. Immunol 12, 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, and Biron CA (2002). Science 297, 2063–2066. [DOI] [PubMed] [Google Scholar]
- Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, and Lazarevic V (2014). Immunity 40, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS (2014). Adv. Immunol 124, 171–206. [DOI] [PubMed] [Google Scholar]
- Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, Peng H, Cravens PD, Racke MK, and Lovett-Racke AE (2009). J. Exp. Med 206, 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]